Abstract
The main extracellular matrix binding component of the dystrophin-glycoprotein complex, α-dystroglycan (α-DG), which was originally isolated from rabbit skeletal muscle, is an extensively O-glycosylated protein. Previous studies have shown α-DG to be modified by both O-GalNAc- and O-mannose-initiated glycan structures. O-Mannosylation, which accounts for up to 30% of the reported O-linked structures in certain tissues, has been rarely observed on mammalian proteins. Mutations in multiple genes encoding defined or putative glycosyltransferases involved in O-mannosylation are causal for various forms of congenital muscular dystrophy. Here, we explore the glycosylation of purified rabbit skeletal muscle α-DG in detail. Using tandem mass spectrometry approaches, we identify 4 O-mannose-initiated and 17 O-GalNAc-initiated structures on α-DG isolated from rabbit skeletal muscle. Additionally, we demonstrate the use of tandem mass spectrometry-based workflows to directly analyze glycopeptides generated from the purified protein. By combining glycomics and tandem mass spectrometry analysis of 91 glycopeptides from α-DG, we were able to assign 21 different residues as being modified by O-glycosylation with differing degrees of microheterogeneity; 9 sites of O-mannosylation and 14 sites of O-GalNAcylation were observed with only two sites definitively exhibiting occupancy by either type of glycan. The distribution of identified sites of O-mannosylation suggests a limited role for local primary sequence in dictating sites of attachment.
Keywords: Carbohydrate Structure, Glycoconjugate, Glycoprotein, Glycosylation, Mass Spectrometry (MS), Muscular Dystrophy, Dystroglycan, Glycomics, Glycoproteomics, Site-mapping
Introduction
Defects in protein glycosylation related to human disease were first reported in the 1980s, and since then, about 40 various types of congenital disorders of glycosylation have been reported (1). The term congenital disorders of glycosylation was first used to describe alterations of the N-glycosylation pathway and was later expanded to include the O-glycosylation pathways (1–3). The importance and complexity of O-linked glycosylation have only recently begun to be appreciated (1, 3, 4). In particular, mutations in genes encoding (putative) glycosyltransferases, which catalyze the addition and extension of O-linked mannose-initiated glycans, have garnered increased attention in the last decade given that they are causative for several forms of congenital muscular dystrophy (5, 6).
The most common forms of O-glycosylation on secretory proteins are the mucin-like O-GalNAc structures that are initiated by polypeptide N-α-acetylgalactosaminyltransferases in the endoplasmic reticulum-Golgi intermediate compartment and/or early cis-Golgi (7). Additionally, other O-linked structures are initiated with alternative monosaccharides, such as O-mannose, O-glucose, O-fucose, O-xylose, and O-GlcNAc on Ser/Thr residues and the O-galactose modification of hydroxylysine residues in collagen domains (4). The diversity of O-mannosylated proteins in mammals, although quite abundant in some tissues (∼30% of O-glycans released from mouse brains (8)), has not been well characterized. The only clearly identified mammalian protein modified by O-mannosylation is α-dystroglycan (α-DG)2 (9).
α-DG is a subunit of dystroglycan and was originally isolated from rabbit skeletal muscle as the extracellular matrix-binding component of the dystrophin-glycoprotein complex (10). The binding to extracellular components such as laminin is dependent upon the addition of O-linked oligosaccharides. Numerous studies have shown that proper post-translational processing of α-DG through the addition of O-mannose structures is crucial for proper muscle and brain development (5, 6, 9). Of particular interest, several distinct forms of congenital muscular dystrophy have been linked to defects in glycosyltransferases involved in the O-mannosylation of α-DG (5, 6, 9). Defects in glycosyltransferases involved in O-mannose attachment and extension, including POMT1/2 and POMGnT1, as well as the putative glycosyltransferase LARGE, are present in various forms of congenital muscular dystrophy including Walker-Warburg syndrome and Muscle-Eye-Brain disease (5, 6, 9). Furthermore, ablation of these gene products in mouse model systems recapitulates much of the pathophysiology of the corresponding human diseases (5, 6, 9).
Given the importance of O-mannosylation to the function of α-DG, we undertook glycomics and glycoproteomic site mapping of α-DG isolated from rabbit skeletal muscle. Because α-DG contains both O-Man- and O-GalNAc-initiated structures, the use of tagging strategies following β-elimination (such as BEMAD (11)) cannot distinguish glycan type at individual sites. Therefore, we developed and employed methodology for the direct assignment of glycopeptides when coupled with glycomic analysis. O-Glycan analysis was performed on released permethylated glycans using MSn tandem mass spectrometry to define the structural diversity of O-Man- and O-GalNAc-initiated glycans present on the purified α-DG. We then performed direct analysis of the peptides/glycopeptides of α-DG, following tryptic digestion with and without glycosidase treatment, via tandem mass spectrometry. Taking advantage of the ability of an ion trap instrument to perform pseudo-neutral loss-triggered MS3 analysis, we were able to assign specific O-glycan structures to peptides and in many cases to the exact sites of addition on α-DG. This study, which is the first to map endogenously added O-mannose sites from purified functional α-DG, facilitates our understanding of O-mannosylation in general. With respect to α-DG, the study highlights the interplay between the O-Man and O-GalNAc classes of O-glycosylation, which will further the development of future studies designed to unravel structure/function relationships for this important glycoprotein as it relates to the pathophysiology of congenital muscular dystrophy.
EXPERIMENTAL PROCEDURES
Protein Purification
α-DG was extracted and purified exactly as described previously (12).
Silver Staining and Western Blotting of Gels
SDS-PAGE was performed on a 4–20% Tris-HCl precast gel purchased from Bio-Rad. Silver staining was conducted using an adapted protocol from Shevchenko et al. (13). Western blots were performed on semi-dry transferred polyvinylidene difluoride membranes using the VIA4 and IIH6 monoclonal antibodies followed by ECL detection as described previously (12).
Glycosidase Treatment
The enzyme-treated α-DG sample was prepared by combining 1.5 μg of α-DG in 15 μl of water, 4 μl of 5× incubation buffer (Prozyme), and 1.5 μl of 100 mm dithiothreitol and 6 μl of enzyme mixture (GlycoProTM deglycosylation kit (Prozyme) combined with the prO-LINK extender kit (Prozyme) containing the following enzymes: peptide:N-glycosidase, sialidase A (Arthrobacter ureafaciens), O-glycosidase, β(1–4)-galactosidase, chondroitinase, and β-N-acetylglucosaminidase. The mock-digested α-DG was prepared similarly but lacked enzyme. Both the mock-digested and glycosidase-treated fractions of α-DG were incubated overnight at 37 °C.
Immobilization of Laminin-1 on the Sensor Surface and Surface Plasmon Resonance
Murine laminin-1 was not stable at acidic pH values for coupling to the CM5 chips using the standard amine coupling chemistry. In preparation for binding to the streptavidin chip (SA), laminin-1 was first treated with 4-(2-aminoethyl)-benzenesulfonyl fluoride as described previously by Colognato et al. (14). Briefly, a 100 mm solution of the serine protease inhibitor 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride containing laminin-1 was incubated overnight on ice. Free 4-(2-aminoethyl)-benzenesulfonyl fluoride was removed using a Microcon-10 (Amicon) microconcentrator. 4-(2-Aminoethyl)-benzenesulfonyl fluoride-treated laminin-1 was then biotinylated at a molar ratio of 40:1 with NHSLC-biotin (Pierce) according to the manufacturer's instructions. The reaction product was dialyzed free of unreacted NHSLC-biotin and the reaction product confirmed by 4′-hydroxyazobenzene-2-carboxylic acid assay (data not shown). Approximately 609 resonance units of biotinylated laminin-1 was bound on the SA chip during a 70-μl injection (5 μl/min) of 100 μg/ml biotinylated laminin-1 in phosphate-buffered saline. Samples of glycosidase-treated and -untreated α-DG were tested at a flow rate of 10 μl/min over the immobilized laminin-1 for 1 min, followed by a 2-min delayed wash to allow the dissociation phase to be recorded. Binding analysis of α-DG to laminin-1 was performed using a Biacore 3000 (Pharmacia Biosensor AB, Uppsala, Sweden). Binding causes a change in the surface plasmon resonance, which was detected optically and measured in resonance units. Sensograms were collected as the difference in binding to the laminin-1 versus a blank reference channel. The sensor chip surface was regenerated using 20 μl of glycine HCl solution (pH 2.5) after each round of binding. The BIAevaluation software 3.0 (BIAcore) was used to analyze binding data.
Release of O-Linked Glycans
Purified α-DG, ∼22 μg, was transferred to a glass tube and stored at −80 °C prior to drying on a lyophilizer. To remove any residual detergent that might have been present from the purification process, the dried protein powder was resuspended in acetone and centrifuged. The acetone supernatant was decanted from the protein pellet, and the pellet and any remaining acetone were removed under a stream of nitrogen gas with mild warming (45 °C). The dried sample was resuspended in 440 μl of Milli-Q water, and a 200-μl aliquot was taken for release of O-linked glycans. The aliquot was re-lyophilized and subjected to reductive β-elimination (1 m NaBH4 in 50 mm NaOH, 18 h at 45 °C). The reaction was neutralized by adding 10% acetic acid dropwise while vortexing. The completely neutralized sample was desalted by loading onto a small column of AG50-X8 (1-ml bed volume). Released oligosaccharides were eluted from the column with 3 volumes of 5% of acetic acid, collected, and evaporated to dryness using a SpeedVac. Borate was removed as an azeotrope with methanol and acetic acid by resuspending the dried sample in 9:1 methanol/acetic acid and then drying under a nitrogen stream at 37 °C four times.
Permethylation and Analysis of Released O-Linked Glycans
To aid in analysis of O-linked glycan structures, the released oligosaccharide mixture was permethylated according to the method of Ciucanu and Kerek (15). Permethylated glycans were analyzed as described previously (16). Briefly, following permethylation, glycans were dissolved in 1 mm NaOH in 50% methanol. Using a nanoelectrospray source, the O-glycan mixture was directly infused into a linear ion trap mass spectrometer (LTQ, Thermo Fisher) at a flow rate of 0.4 μl/min. As described previously, total ion mapping was used to detect and quantify the prevalence of individual glycans (17). The suggested nomenclature from The Consortium for Functional Glycomics was used for all representations of glycan structures in the figures and tables with the undefined hexose or HexNAc species displayed in gray.
Protein Digestion
α-DG purified from rabbit skeletal muscle was digested using either sequence grade trypsin (Promega) alone or in combination with endoproteinases Lys-C (Sigma). The samples were diluted to 40 mm ammonium bicarbonate and reduced with 100 mm dithiothreitol for 1 h at 56 °C, carboxyamidomethylated with 55 mm iodoacetamide in the dark for 45 min, and then protease-digested overnight at 37 °C. In the case of Lys-C, the digest was carried out in 6 m urea, and the reduction temperature was held at 37 °C followed by dilution to 1 m urea with 40 mm ammonium bicarbonate and then overnight trypsin digest. After digestion, the reaction was quenched with 1% trifluoroacetic acid making the final concentration ∼0.1% trifluoroacetic acid. The resulting peptides were dried down using a SpeedVac and stored at −20 °C until ready to analyze.
β-Elimination Followed by Michael Addition of Dithiothreitol (BEMAD)
The application of BEMAD to tryptic peptides was as described previously (11).
Nano-LC-MS3
α-DG glycopeptides were analyzed on a linear ion trap mass spectrometer (LTQ; ThermoFisher) using an MS3 data-dependent neutral loss method. The glycopeptides were resuspended in 0.5 μl of solvent B (0.1% formic acid, 80% acetonitrile) and 19.5 μl of solvent A (0.1% formic acid), filtered using a 0.2-μm spin filter at 12,000 rpm, and loaded on a 75 μm × 8.5-cm C18 reverse phase column/emitter (packed in-house, YMC GEL ODS-AQ120ÅS-5) using a nitrogen pressure bomb. Peptides were eluted over a 160-min linear gradient increasing from 5 to 100% solvent B over 90 min at a flow rate of 250 nl/min. Each full MS scan from 300 to 2000 m/z yielded five MS/MS scans of the top five most intense peaks with a dynamic exclusion of two for 30 s. Data-dependent MS3 scans were triggered if a neutral loss was observed equal to the singly or doubly charged mass of hexose, HexNAc, fucose, or Neu5Ac (sialic acid) within the top three peaks from the MS/MS scan.
Data Analysis
The acquired data were searched against a nonredundant rabbit data base (generated March 26, 2004) obtained from the National Center for Biotechnology Information (NCBI) using the TurboSequest algorithm (Bio-Works, Thermo Fisher). To aid in identification of glycopeptides, we allowed for a mass increase of 162.1, 203.1, and 365.2 daltons on both threonines and serines looking for the addition of Man, GalNAc, and Hex-HexNAc, respectively. Additionally, peptides that were subjected to BEMAD were searched looking for a mass increase of 136.2 daltons on both serines and threonines as described previously. Output files that failed to a yield a final score (Sf) and probability score (P) above 0.45 and 30, respectively, were not considered further. All remaining spectra were manually evaluated for the presence of glycopeptides and sites of modification and were further validated by TurboSequest searches against the rabbit α-DG FASTA sequence combined with the TurboSequest common contaminants data base.
RESULTS
Characterization of Purified α-DG from Rabbit Skeletal Muscle
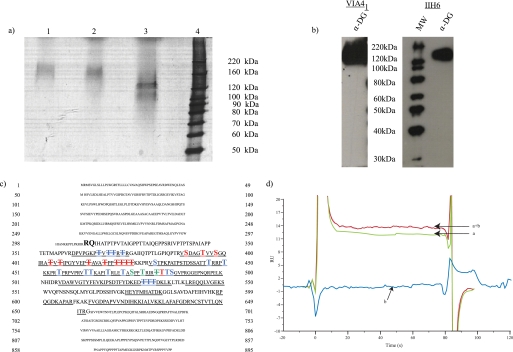
α-DG was purified from rabbit skeletal muscle as described previously and validated for purity via silver staining (Fig. 1a) and for functional glycosylation using the IIH6 and VIA41 antibodies (Fig. 1b) that have previously been demonstrated to bind functional and glycosylated α-DG, respectively (12). Purity of the sample was also determined via trypsin digestion followed by LC-MS/MS. Based on the full-length sequence of α-DG, coverage at 1% false-discovery rate was only 13%. Decorin and calsequestrin were also identified in the sample but contributed less than 5% of the total spectral counts assigned to proteins and thus represent minor co-purifying/contaminating proteins. To increase coverage, α-DG was subjected to glycosidase treatment (with N-glycosidase, sialidase A (A. ureafaciens), O-glycosidase, β(1–4)galactosidase, and β-N-acetylglucosaminidase) that increased the mobility of the protein upon electrophoresis (Fig. 1a). Furthermore, mature α-DG is known to be processed by proteases that cleave off the N terminus (18), and we were unable to detect any peptides corresponding to this region (Fig. 1c, attempts to determine the N terminus by automated Edman degradation sequencing were unsuccessful suggesting that the N terminus is blocked; data not shown). LC-MS/MS analysis following glycosidase and trypsin/endoproteinase-LysC treatment increased overall coverage to 65% (Fig. 1c) when one takes into account the proposed cleavage site for the mature protein by Kanagawa et al. (18) and the glycopeptides we observed (supplemental Table 1). Furthermore, surface plasmon resonance experiments were used to confirm that the purified α-DG could bind to laminin in agreement with the method of purification (laminin affinity column) and antibody binding (Fig. 1d). As observed previously, using different methodologies (12), treatment of α-DG with sialidase and galactosidase did not have a detrimental effect on laminin binding. Thus, this characterization of the starting material made us confident in moving forward with further analysis of functionally active, glycosylated α-DG.
FIGURE 1.
Purified, functionally glycosylated α-DG from rabbit skeletal muscle. a, silver staining following SDS-PAGE of purified α-DG (lane 1). Lanes 2 and 3 represent mock or glycoside (N-glycosidase F, sialidase, endo-O-glycosidase, β(1–4)-galactosidase, and β-N-acetylglucosaminidase)-treated α-DG. b, Western blot analysis following SDS-PAGE of purified α-DG with the glycan-dependent anti-α-DG monoclonal VIA41 and IIH6, which specifically recognizes fully glycosylated, functionally active α-DG. c, protein sequence derived from the dystroglycan gene with the capitalized boldface sequence representing the predicted mature α-DG protein. Peptides assigned by tandem mass spectrometry are underlined. Sites detected to be modified by GalNAc are highlighted in blue; sites of O-mannosylation are highlighted with red; residues observed to be modified by both GalNAc and mannose are green. Sites of potential modification are highlighted similarly and distinguished by striking through the modified residue. d, untreated α-DG (a), α-DG treated with β-galactosidase and sialidase (a + b), or glycosidases alone (b, enzymes without α-DG present) binding to immobilized laminin-1 as measured by surface plasmon resonance.
O-Glycans Released from α-DG
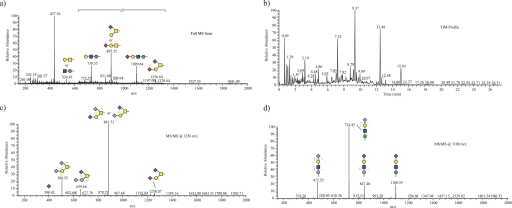
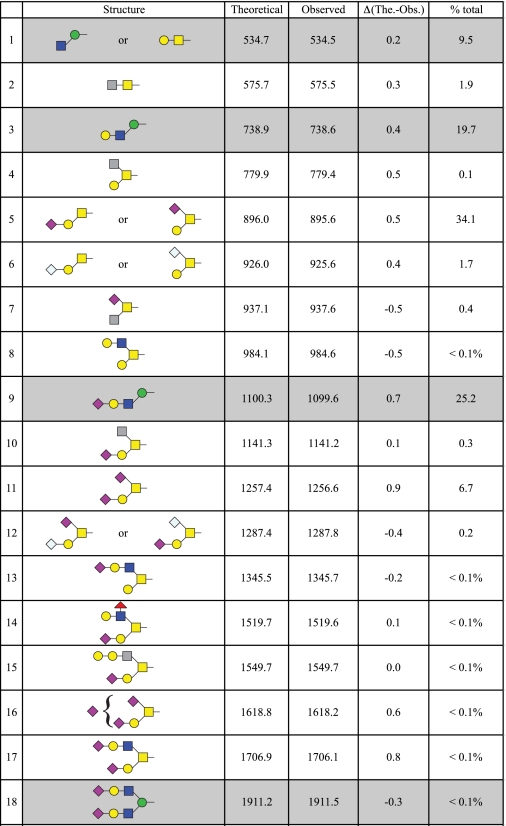
O-Linked glycans were released from α-DG purified from rabbit skeletal muscle by β-elimination, permethylated, and analyzed by nanospray ionization-MS/MS. The generated full scans allowed for detection of released O-linked glycans (Fig. 2a), and structure of the O-glycans observed in the full MS was assigned based on MS/MS fragmentation. To detect glycans in an unbiased manner, the sample was subjected to total ion mapping as described previously (Fig. 2b) (17). Total ion mapping generates MS/MS fragmentation profiles in small overlapping m/z ranges, allowing the detection of fragments that predict the presence of glycans across the full range of detected m/z values. Detected glycans were further confirmed by MSn fragmentation as needed to define the structure (data not shown). In Fig. 2, c and d, we present two such MS/MS profiles (from a total of over 700) to display the identification of an O-GalNAc- (disialylated T antigen) and an O-Man (the classical O-Man tetrasaccharide, Siaα2–3Galβ1–4GlcNAcβ1–2Man)-initiated structure. Table 1 includes a list of all of the glycan structures observed from rabbit skeletal muscle α-DG. Although there are more total O-GalNAc-initiated structures observed, O-Man-initiated structures represent ∼50% of the structures by prevalence.
FIGURE 2.
O-Man- and O-GalNAc-initiated glycans of α-DG. a, full MS scan of the released and permethylated O-linked glycan structures on α-DG purified from rabbit skeletal muscle. b, total ion mapping (TIM) profile of the released O-linked glycans allowing for quantifying prevalence of each structure as well as confirmation of structure by fragmentation. c, MS/MS fragmentation at m/z 1256 demonstrates the presence of an O-GalNAc initiated structure SA-Gal-(SA)-GalNAc. d, fragmentation of m/z 1100 demonstrates the presence of an O-Man-initiated structure SA-Gal-GlcNAc-Man.
TABLE 1.
Quantification of O-glycans released from α-DG purified from rabbit skeletal muscle
Assignment of Glycopeptides and Sites of Attachment
Having established the range of structures observed on α-DG, we set out to assign these structures to the polypeptide backbone. Purified α-DG was digested using sequence grade trypsin alone or in combination with the endoproteinase Lys-C to increase protein coverage and/or glycosidase treatment to improve digestion, yielding a mixture of peptides and glycopeptides. The resulting mixtures were then analyzed via LC-MS3 using a linear ion trap mass spectrometer. By taking advantage of the capabilities of the linear ion trap mass spectrometer, we were able to apply MS3 fragmentation to glycopeptides that generated neutral losses of glycans in MS/MS. To identify the glycopeptides, a full MS scan was acquired from 300 to 2000 m/z (Figs. 3a and 4a). From the acquired full scan, MS/MS fragmentation spectra were generated for the top five peaks (Figs. 3b and 4b). Upon fragmentation, if a predetermined neutral loss corresponding to a glycan was observed, a data-dependent MS3 scan was triggered on the neutral loss peptide that yielded further fragmentation data for the glycopeptide (Figs. 3, b and c, and 4, b and c, and supplemental figures).
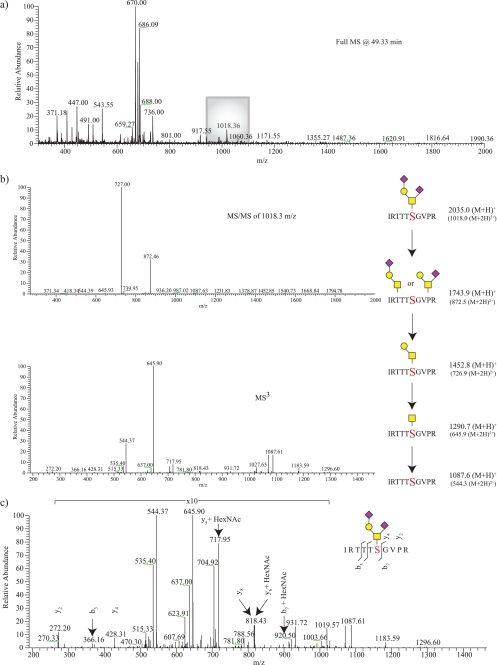
FIGURE 3.
Assignment of an O-GalNAc α-DG glycopeptide. From the full scan acquired at 49.33 min (a), a peak at 1018.3 m/z was selected for fragmentation (b). The resulting MS/MS of 1018.3 m/z yielded the neutral loss of two terminal SA residues, which was then followed by MS3 fragmentation indicating the loss of a Gal residue followed by a reducing end GalNAc. The combined glycan structure was determined to belong to a peptide with 1087.6 m/z. From examining the MS/MS spectra (not shown) and the neutral loss-triggered MS3 spectra (c), the site of post-translational modification to the serine within the peptide IRTTTSVGPR is assigned.
FIGURE 4.
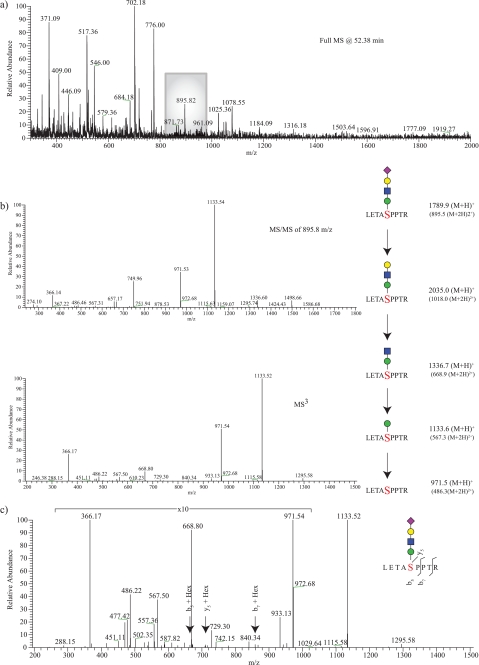
Assignment of an O-Man α-DG glycopeptide. From the full scan acquired at 52.38 min (a), a peak at 895.8 m/z was selected for fragmentation. b, resulting MS/MS of 895.8 m/z yielded the neutral loss of terminal SA residue, which was then followed by MS3 fragmentation indicating the neutral loss of a Gal-GalNAc residue followed by a reducing end Man. The combined glycan structure was determined to belong to a peptide with 971.5 m/z. From examining the MS/MS spectra (data not shown) and the neutral loss-triggered MS3 spectra (c), the site of post-translational modification to the serine within the peptide LETASPPTR is assigned.
Through application of this pseudo-neutral loss-triggered MS3 method, we were able to observe, in many cases, sequential monosaccharide losses, defining the glycan structure from its distal end to its glycosidic attachment to Ser/Thr. The observed losses of glycan (hexose, HexNAc, and Neu5Ac) species were then fitted to the existing confirmed structures on α-DG that had been determined through reductive β-elimination, permethylation, and MSn analysis (Table 1). The modified peptide was able to be determined upon calculating the neutral loss of glycans and the generation of b and y ions in MS/MS and/or MS3. The peptide sequence was able to be determined by comparing a list of the generated peptide (M + H)+ values against a theoretical list of generated peptides for the α-DG protein sequence using the MS digest application from the Prospector website created by the University of California, San Francisco. We also used the BEMAD method to aid in mapping sites modified by O-linked glycans (11). Although this method proved to be beneficial by indicating the modified residue in a limited set of cases (supplemental Table 1), it is not capable of distinguishing between O-GalNAc- or O-mannose-initiated glycan structures. Thus, to make more confident assignments of the glycan structure responsible for modification at a particular Ser/Thr residue, we examined the b and y ions that were generated from MS/MS and MS3 fragmentation of the peptide backbone. By comparing the theoretical b and y ions of O-GalNAc- or O-Man-containing fragments with those that were observed in the two spectra, we were able to determine in many cases the exact residue modified. For example, Figs. 3c and 4c show glycopeptides from α-DG that were modified by the addition of an O-GalNAc-initiated glycan structure, disialylated T antigen, and an O-mannose-initiated glycan structure, Siaα2–3Galβ1-4GlcNAcβ1–2Man, at Ser-475 and Ser-485, respectively. Upon fragmentation, b and y ions still modified by hexose or HexNAc allow unequivocal assignment of the structures to specific residues. Similar strategies were applied for all O-Man- and O-GalNAc-initiated structures, and the results are summarized in Table 2 and supplemental Table 1.
TABLE 2.
Summary of sites of modification and modifying residue(s)
DISCUSSION
O-Linked glycans containing mannose were first isolated nearly 30 years ago from an enriched mixture of brain chondroitin sulfate proteoglycans, with a core structure suggested to be Galβ1–4GlcNAcβ-1–2Man-Ser/Thr (19). However, sites of modification have not previously been mapped from native sources for the most well characterized O-mannosylated protein, α-DG. Given the importance of O-glycosylation for proper function of α-DG, we sought here to map defined glycan structures to sites of attachment on the polypeptide from endogenously glycosylated α-DG isolated from rabbit skeletal muscle.
α-DG was purified from rabbit skeletal muscle as described previously and shown to be highly enriched and functionally glycosylated (Fig. 1) (12). To map glycan structures to specific sites, we first released and permethylated the glycans from the glycoprotein so that we could get detailed fragmentation defining the full set of glycans present on α-DG (Fig. 2). This allowed us to determine that there were at least 21 different O-linked glycans present on α-DG purified from rabbit skeletal muscle. Four of these structures were initiated by O-Man with the classical, previously described (20), Siaα2–3Galβ1-4GlcNAcβ1–2Man tetrasaccharide structure being the most prevalent (Table 1). Only one branched O-Man structure was observed in rabbit skeletal muscle-derived α-DG at less than 0.1% prevalence (Table 1), consistent with the proposal that the brain-specific enzyme, GnT-Vb (GnT-IX), is responsible for O-Man branching (21).
With the glycans on α-DG defined, direct glycopeptide analysis following enzymatic digestion of the protein was performed via pseudo-neutral loss-triggered MS3 analysis (Figs. 3 and 4). This procedure relies on the neutral loss of a glycan mass to trigger further fragmentation of the glycopeptide. Given the lability of the glycosidic linkage, most glycopeptides generate dominant neutral loss peaks associated with glycan fragmentation upon collision-induced dissociation (Figs. 3 and 4). Further fragmentation of the species that has undergone a neutral loss provides further glycan losses, as well as peptide b and y ions to assist in the assignment of the peptide and the site(s) of glycosylation. To facilitate improved digestion and better coverage for the resulting peptides, endoproteinase Lys-C under denaturing conditions followed by trypsin digestion was used. Furthermore, we found that partial deglycosylation greatly facilitated digestion and glycopeptide assignments. Although incomplete exoglycosidase treatment allowed discrimination between sites of O-Man and O-GalNAc initiation, it limited mapping of intact glycan structures at many sites (Table 2 and supplemental Table 1). In several cases, there was not sufficient fragmentation information (i.e. fragments containing glycans) to map the exact site of attachment, but we could assign the glycan to a particular peptide or subset of residues in the peptide (Table 2 and supplemental Table 1). Of note, stretches of Thr residues can be particularly problematic for mapping sites of GalNAc attachment because the molecular weight of two adjacent Thr residues is almost identical to the weight of a HexNAc.
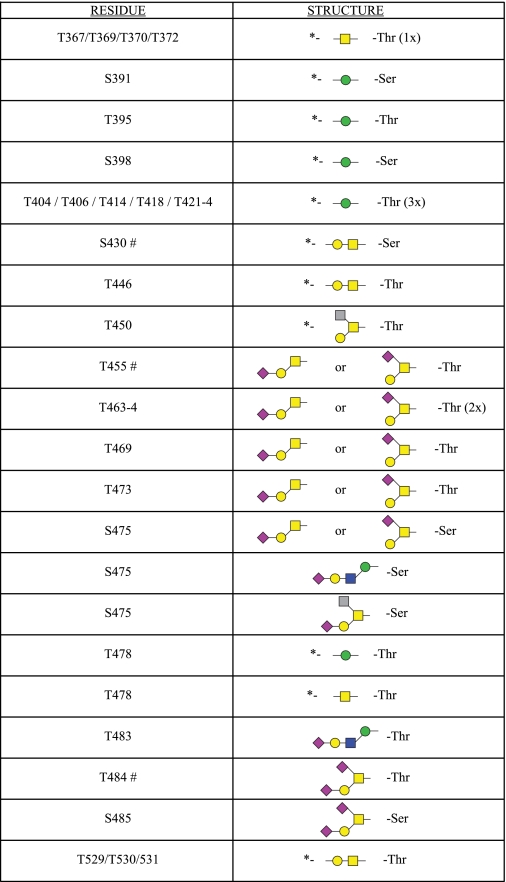
In total, we observed 91 glycopeptides in our analyses that allowed us to assign 16 specific O-glycosylated residues within α-DG (Fig. 1c and supplemental figures). In addition, another 16 sites of modification were restricted to a small subset of possible residues (Fig. 1c). As expected, for many of the sites of modification, we saw microheterogeneity; glycopeptides with different glycan structures on the identical residues were observed (supplemental Table 1). We also observed that two sites of glycosylation (Ser-475 and Thr-478) could accept O-Man- and O-GalNAc-initiated glycan structures. This suggests that O-mannosylation, at least on these sites, is substoichiometric because the enzymes for O-Man attachment are localized in the endoplasmic reticulum and likely precede the O-GalNAc machinery that is localized to the cis-Golgi and/or endoplasmic reticulum-Golgi intermediate compartment (7, 9). In total, we observed 24 sites of glycosylation on rabbit skeletal muscle α-DG, including 9 sites of O-Mannosylation.
Recently, an in vitro study by Manya et al. (22), using recombinant POMT1/2 enzymes and synthetic peptides derived from α-DG, concluded that mammalian O-mannosylation is dependent upon a consensus sequence (IXPT(P/X)TXPXXXXPTX(T/X)XX). When we compared the nine sites we identified as O-mannosylated with the proposed consensus site, six of our defined sites do not fit this model. However, in our studies, there are three other unresolved sites of O-Man within a single peptide, containing eight potential sites of modification, that are consistent with their model. The sites reported in the in vitro study, Thr-404, Thr-406, and Thr-414, potentially overlap with the three sites of O-Man modification localized between residues 404 and 424 on endogenously glycosylated α-DG. In 2008, Breloy et al. (23), relying on data generated via overexpression of fragments of human α-DG in epithelial cells, argued that O-mannosylation was regulated in a much more complex manner than a simple local primary sequence. Alignment of our O-Man sites generated from endogenous rabbit skeletal muscle α-DG provided no obvious local consensus site for attachment (supplemental Table 2), and thus our findings are in agreement with Breloy et al. (23). Therefore, we conclude that O-mannosylation of particular residues is not regulated solely by a local consensus sequence. Further work is needed to determine the mechanism by which residues for O-Man addition are selected and to elucidate the effect of O-Man modification on the further modification of the glycoprotein by O-GalNAc-initiated structures.
Campbell and co-workers (24) recently demonstrated that O-mannosyl phosphorylation was present on α-DG and was required for laminin binding. Furthermore, that study placed phosphomannose at Thr-379 on a human α-DG construct isolated from cell lines with minimal LARGE activity. Of particular interest to this study, we were unable to observe the analogous rabbit peptide (residues 374–389) by our methods. Presumably, this is because of an unknown LARGE-dependent modification of the phosphorylated O-Man structure. Without knowledge of the complete chemical nature of the LARGE-dependent modification, peptides bearing this structure would be missed. For completeness, it should also be noted that Thr-381 and Thr-388 of this peptide were also observed to be O-Man modified in the human overexpression study (24), and they are in a stretch of sequence similar, but not identical, to the proposed consensus sequence of Endo and co-workers (22).
Interestingly, beyond this missed peptide that contains the phosphomannose-containing trisaccharide extended with an unknown LARGE-dependent modification (24), we did not detect four other predicted tryptic peptides of greater than four amino acids in length. Two of these peptides are contiguous and represent the extreme N terminus of the fully processed polypeptide. Given that the N terminus of the mature protein was apparently blocked based on results of automated Edman sequencing, it is not surprising that the extreme N-terminal peptide was not observed as the nature of the moiety blocking the N terminus is unknown. This leaves us with three peptides that we failed to detect, all of which contain Ser/Thr residues. Two of these peptides (residues 338–359 and 550–572) are quite large and contain six and five potential sites of glycosylation, respectively. Thus, it is likely that if multiple sites were utilized for glycosylation, these peptides would exceed the upper mass limit of the instrument (2000 m/z). The remaining unexplained absent peptide is residues 583–597. This peptide only contains one potential site of O-linked glycosylation (Ser-586). Given the modest size of this peptide, its lack of multiple sites of glycosylation, and the fact that both peptides flanking this sequence were assigned, failure to detect this peptide is difficult to explain unless, like peptide 374–389, this peptide also contains the LARGE-modified phosphomannose trisaccharide at Ser-586. Given that we chose to map sites on fully functional α-DG, it is not surprising that we were unable to map the phosphomannose-containing trisaccharide peptide(s) that we had previously assigned in a LARGE-deficient cell line (24). However, based on the absence of detection in this study, we speculate that other peptides, including 583–597 and possibly 338–359 and 550–572, may indeed be modified in a LARGE-dependent manner as well.
In conclusion, we have developed and implemented a work flow that enabled us to assign defined O-glycan structures to specific residues on the polypeptide by utilizing both glycomics and glycopeptidomics. The resulting site map describing both O-Man and O-GalNAc initiated glycosylation on α-DG isolated from rabbit skeletal muscle provides a framework for elucidating structure/function relationships for this complex glycoprotein. It should also facilitate a greater understanding of the interplay between O-GalNAcylation and O-mannosylation, two glycosylation pathways that theoretically are competing for the same sites of modification. Given that O-mannosylation is defective in multiple forms of congenital muscular dystrophy, is required for α-DG function, and is likely found on other yet-to-be-identified mammalian proteins, the work presented here lays an essential groundwork for future functional studies.
Supplementary Material
Acknowledgments
We thank Ariana Combs for purifying the α-DG used in this study and all of the Wells, Bergmann, and Tiemeyer laboratory members for helpful discussions.
Note Added in Proof
Recently, Nilsson et al. (25) reported using MS methods similar to the ones described here for the analysis of α-DG glycopeptides isolated from human skeletal muscle. From their analysis, they identified 25 glycopeptides, which were observed to be modified with either O-GlcNAc or O-mannose initiated glycan structures.
This work was supported, in whole or in part, by National Institutes of Health Grant P41 RR018502 from NCRR (to L. W. and M. T.). This work was also supported by the Muscular Dystrophy Association (to J. M. E. and L. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and figures.
- α-DG
- α-dystroglycan
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MS
- mass spectrometry
- SA
- streptavidin
- BEMAD
- β-elimination followed by Michael addition of dithiothreitol.
REFERENCES
- 1.Jaeken J., Hennet T., Freeze H. H., Matthijs G. (2008) J. Inherited Metab. Dis. 31, 669–672 [DOI] [PubMed] [Google Scholar]
- 2.Aebi M., Helenius A., Schenk B., Barone R., Fiumara A., Berger E. G., Hennet T., Imbach T., Stutz A., Bjursell C., Uller A., Wahlström J. G., Briones P., Cardo E., Clayton P., Winchester B., Cormier-Dalre V., de Lonlay P., Cuer M., Dupré T., Seta N., de Koning T., Dorland L., de Loos F., Kupers L. (1999) Glycoconj. J. 16, 669–671 [DOI] [PubMed] [Google Scholar]
- 3.Cohn R. D. (2005) Neuromuscul. Disord. 15, 207–217 [DOI] [PubMed] [Google Scholar]
- 4.Haltiwanger R. S., Lowe J. B. (2004) Annu. Rev. Biochem. 73, 491–537 [DOI] [PubMed] [Google Scholar]
- 5.Barresi R., Campbell K. P. (2006) J. Cell Sci. 119, 199–207 [DOI] [PubMed] [Google Scholar]
- 6.Martin P. T. (2007) Curr. Mol. Med. 7, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ten Hagen K. G., Fritz T. A., Tabak L. A. (2003) Glycobiology 13, 1R–16R [DOI] [PubMed] [Google Scholar]
- 8.Chai W., Yuen C. T., Kogelberg H., Carruthers R. A., Margolis R. U., Feizi T., Lawson A. M. (1999) Eur. J. Biochem. 263, 879–888 [DOI] [PubMed] [Google Scholar]
- 9.Endo T., Manya H. (2006) Methods Mol. Biol. 347, 43–56 [DOI] [PubMed] [Google Scholar]
- 10.Ervasti J. M., Campbell K. P. (1993) J. Cell Biol. 122, 809–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., Hart G. W. (2002) Mol. Cell. Proteomics 1, 791–804 [DOI] [PubMed] [Google Scholar]
- 12.Combs A. C., Ervasti J. M. (2005) Biochem. J. 390, 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 14.Colognato H., Winkelmann D. A., Yurchenco P. D. (1999) J. Cell Biol. 145, 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciucanu I., Kerek F. (1984) Carbohydr. Res. 131, 209–217 [Google Scholar]
- 16.Aoki K., Porterfield M., Lee S. S., Dong B., Nguyen K., McGlamry K. H., Tiemeyer M. (2008) J. Biol. Chem. 283, 30385–30400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoki K., Perlman M., Lim J. M., Cantu R., Wells L., Tiemeyer M. (2007) J. Biol. Chem. 282, 9127–9142 [DOI] [PubMed] [Google Scholar]
- 18.Kanagawa M., Saito F., Kunz S., Yoshida-Moriguchi T., Barresi R., Kobayashi Y. M., Muschler J., Dumanski J. P., Michele D. E., Oldstone M. B., Campbell K. P. (2004) Cell 117, 953–964 [DOI] [PubMed] [Google Scholar]
- 19.Finne J., Krusius T., Margolis R. K., Margolis R. U. (1979) J. Biol. Chem. 254, 10295–10300 [PubMed] [Google Scholar]
- 20.Smalheiser N. R., Haslam S. M., Sutton-Smith M., Morris H. R., Dell A. (1998) J. Biol. Chem. 273, 23698–23703 [DOI] [PubMed] [Google Scholar]
- 21.Alvarez-Manilla G., Troupe K., Fleming M., Martinez-Uribe E., Pierce M. (2010) Glycobiology 20, 166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manya H., Suzuki T., Akasaka-Manya K., Ishida H. K., Mizuno M., Suzuki Y., Inazu T., Dohmae N., Endo T. (2007) J. Biol. Chem. 282, 20200–20206 [DOI] [PubMed] [Google Scholar]
- 23.Breloy I., Schwientek T., Gries B., Razawi H., Macht M., Albers C., Hanisch F. G. (2008) J. Biol. Chem. 283, 18832–18840 [DOI] [PubMed] [Google Scholar]
- 24.Yoshida-Moriguchi T., Yu L., Stalnaker S. H., Davis S., Kunz S., Madson M., Oldstone M. B., Schachter H., Wells L., Campbell K. P. (2010) Science 327, 88–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson J., Nilsson J., Larson G., Grahn A. (2010) Glycobiology DOI 10.1093/glycob/cwg082 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.