Abstract
Asukamycin, a member of the manumycin family metabolites, is an antimicrobial and potential antitumor agent isolated from Streptomyces nodosus subsp. asukaensis. The entire asukamycin biosynthetic gene cluster was cloned, assembled, and expressed heterologously in Streptomyces lividans. Bioinformatic analysis and mutagenesis studies elucidated the biosynthetic pathway at the genetic and biochemical level. Four gene sets, asuA–D, govern the formation and assembly of the asukamycin building blocks: a 3-amino-4-hydroxybenzoic acid core component, a cyclohexane ring, two triene polyketide chains, and a 2-amino-3-hydroxycyclopent-2-enone moiety to form the intermediate protoasukamycin. AsuE1 and AsuE2 catalyze the conversion of protoasukamycin to 4-hydroxyprotoasukamycin, which is epoxidized at C5–C6 by AsuE3 to the final product, asukamycin. Branched acyl CoA starter units, derived from Val, Leu, and Ile, can be incorporated by the actions of the polyketide synthase III (KSIII) AsuC3/C4 as well as the cellular fatty acid synthase FabH to produce the asukamycin congeners A2–A7. In addition, the type II thioesterase AsuC15 limits the cellular level of ω-cyclohexyl fatty acids and likely maintains homeostasis of the cellular membrane.
Keywords: Antibiotics, Anticancer Drug, Bacterial Metabolism, Epoxygenase Pathway, Fatty Acid Synthase, Biosynthetic Pathway, Natural Product, Polyketide, Streptomyces, Thioesterase
Introduction
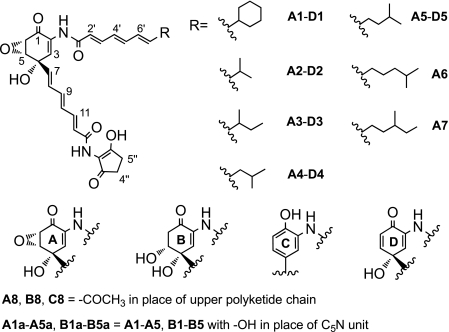
The metabolites of the manumycin family are known to have a broad range of antibiotic functions including antibacterial, anticoccidial, and antifungal activities (1). Manumycin compounds also display a strong activity against farnesyltransferase, IκB kinase β, interleukin-1β-converting enzymes, and acetylcholinesterase and are considered as drug candidates to treat cancers, inflammation, and Alzheimer disease (1–4). Asukamycin A1, a manumycin-type metabolite, contains a unique 2-amino-4-hydroxy-5,6-epoxycyclohex-2-enone (mC7N)3 core and two trans-triene polyketide chains, in which the upper one starts with a cyclohexane ring, and the lower one terminates in the five-membered ring of a 2-amino-3-hydroxycyclopent-2-enone (C5N) moiety (Fig. 1) (5). Asukamycin, together with a series of congeners A2–A7 carrying branched alkyl groups in place of the cyclohexane moiety, were isolated from Streptomyces nodosus subsp. asukaensis (6–8). Like in a majority of the manumycin-type metabolites, the C5,C6 epoxy group can be reduced to a 5-hydroxyethylene structure enzymatically or nonenzymatically to form the type II asukamycins, B1–B5 (Fig. 1) (8). The epoxide group of the manumycins is considered crucial for many biological functions, as the type II products show weaker activities (1).
FIGURE 1.
Structures of asukamycin and related metabolites. Compounds are grouped according to the main A, B, C, and D core units.
Based on precursor feeding experiments, the asukamycin mC7N core unit and the cyclohexane moiety originate from 3-amino-4-hydroxybenzoic acid (3,4-AHBA) and shikimic acid, respectively (9–11). The identical lower and upper triene polyketide chains are built up from 3,4-AHBA and cyclohexylcarbonyl CoA (CHC-CoA), respectively, by three steps of classical polyketide condensations. A gene set involved in the CHC-CoA biosynthesis of ansatrienin has been well characterized in Streptomyces collinus (12), but the genes involved in 3,4-AHBA and polyketide chain assembly of asukamycin have not been identified. The C5N ring moiety, found in several natural products including reductiomycin, moenomycin A, and ECO-02301, was suggested to be derived from a 5-aminolevulinate (5-ALA) intermediate (13–16). The only reported gene product of asuD2 (hemA-asuA), a member of the 2-oxoamine synthases, was found to be involved in asukamycin biosynthesis and to catalyze the condensation of succinyl-CoA and glycine (15). Protoasukamycin C1, the polyketide assembly product, is further oxygenated to give the final product asukamycin (1, 10, 17). The upper polyketide chain assembly of the congeners A2–A7 recruits the branched acyl CoA starters, which are predominantly used for fatty acid biosynthesis in S. nodosus subsp. asukaensis (7). Alternatively, the CHC-CoA starter of asukamycin polyketide chain assembly can also be used to form ω-cyclohexyl fatty acids as a part of cellular membrane components. As the membrane fluidity and permeability are strongly determined by the fatty acid composition, the biosynthetic regulation of each fatty acid component is crucial for membrane homeostasis in bacteria (18, 19). The content of ω-cyclohexyl fatty acids, a byproduct generated during the peak period of asukamycin biosynthesis, is maintained to be as low as 3% (7). This implies that the organism has a control mechanism to balance the metabolic flux between the production of membrane fatty acids and asukamycin. Herein, we report the cloning, mutagenic analysis, and characterization of the biosynthetic genes, which provides insight into the biosynthetic assembly of the asukamycins in S. nodosus subsp. asukaensis. Possible cross-talk between the biosynthesis of the fatty acids and the asukamycins is also discussed.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Chemicals
S. nodosus subsp. asukaensis ATCC 29757 was obtained from the American Type Culture Collection. Streptomyces lividans K4-114 was kindly provided by Dr. C. Khosla. Escherichia coli strains were obtained from the John Innes Center (Norwich, UK). The media and chemicals were purchased from Difco, Sigma-Aldrich, and EMD Chemicals.
Genomic Library and Mutant Construction
See supplemental text.
In Vivo Recombination and Heterologous Expression of the Cloned asu Gene Cluster
The cosmids 2B9 and 10D6 were linearized by SpeI and EcoRI, respectively, equally mixed, and transformed into E. coli BW25113/pIJ790 competent cells by electroporation (20). The cells were then added to 1 ml of SOC medium (20), recovered at 37 °C for 3 h, and plated out onto LB agar with 100 μg/ml apramycin. For the negative control, the linearized 2B9 and 10D6 were transformed separately by the same procedure. The recombinant clone was confirmed by matching the anticipated EcoRI digestion pattern and named pART1361. Intergeneric conjugation of the recombinant cosmids from E. coli to S. lividans K4-114 followed the established protocol (21).
Construction of pART1391, pART1361E3, and pALS4-S83T
To construct pART1391, a 1.5-kb DNA fragment from pIJ778 was PCR-amplified using primers AsuRED-4 and -6 (supplemental Table S3) to carry a spectinomycin resistance gene cassette plus two 50-bp nucleotides, designed to be identical to the boundaries of the targeted region on pART1361 (supplemental Fig. S5) (20). To obtain the recombinant strain, the gel-purified PCR fragment was introduced into E. coli BW25113/pART1361/pIJ790 by electroporation, and the transformants were screened for spectinomycin resistance. The deletion in the resulting clone was confirmed by matching up to the expected EcoRI digestion pattern and verified by PCR. The pART1361E3 construction followed the same strategy as described above but using primers AsuRED-E3F and -E3R (supplemental Table S3).
To insert the point mutation that modifies the TCC (Ser) codon into ACC (Thr) in the asuD2 gene, the internal fragment containing the BamHI-KpnI sites was PCR-amplified using primers S83TF and S83TR (supplemental Table S3). The BamHI-KpnI-cleaved fragment was then used to replace the corresponding region in the pALS3 plasmid. The further steps were identical with the pALS4 construction (15).
HPLC and LC-MS Analysis of the Asukamycin Metabolites
S. nodosus subsp. asukaensis, S. lividans K4-114, and the derived strains were cultivated as described (10, 21). Feeding experiments with 3,4-AHBA (100 μg/ml) and 4-hydroxyprotoasukamycin (20 μg/ml) were conducted by adding these compounds to 1-day-old cultures and harvesting 1 day later.
Fermented cultures were mixed thoroughly with an equal volume of ethyl acetate. The mixture was separated by centrifugation, and the ethyl acetate layer was collected and evaporated to near dryness. The crude extract was dissolved in an equal volume of methanol and analyzed using a P680 HPLC system (Dionex, Sunnyvale, CA) and an XTerra RP18 column (4.6 × 250 mm; Waters) at a flow rate of 0.5 ml/min at 25 °C. Twenty μl of crude extract were injected, equilibrated with 65% solvent B (acetonitrile with 0.1% formic acid) in solvent A (water with 0.1% formic acid) for 5 min, developed with a gradient of 65–90% solvent B in solvent A for 20 min, and washed with 90% solvent B in solvent A for 10 min. To analyze the asuD1–D3 mutants, a gradient of 50–70% solvent B in solvent A was applied instead. The extraction and HPLC analysis of 3,4-AHBA followed the described protocol (22).
High resolution MS and LC-MS analyses were carried out on a 6210-TOF LC-MS system (Agilent Technologies Inc., Santa Clara, CA) in which the LC was performed using a XDB C18 (4.6 × 50 mm, 1.8-micron particle size) high throughput column (Agilent Technologies, Inc.) with the same elution program as the HPLC analysis mentioned above. The exact masses of the identified compounds are listed in supplemental Table S1.
Fatty Acid Analysis
See supplemental text.
Purification and NMR Analysis of D1
A reliable yield of the novel intermediate 4-hydroxyprotoasukamycin D1 was obtained in S. lividans K4-114 carrying pART1361E3, which was generated from pART1361 with the λ-Red PCR strategy by replacing asuE3 with a spectinomycin resistance gene. S. lividans K4-114/pART1361E3 was inoculated into 500 ml of MS medium in 2-liter flasks with a coil and fermented on a rotary shaker at 250 rpm and 28 °C for 4–5 days. One liter of harvested culture was extracted three times with an equal volume of ethyl acetate, and the combined ethyl acetate extract was dried, concentrated, and purified on a silica gel column (100–200 mesh), eluting with dichloromethane/methanol (100:3). The compound D1 fraction was collected and further purified by semi-preparative C18 reverse phase HPLC (Dionex), eluting with 75–85% solvent C (methanol with 0.1% formic acid) in solvent A, to give 20 mg of pure compound D1. Its 1H-NMR, 13C-NMR, HSQC, and HMBC spectra were recorded in Me2SO-d6 on a Varian System 700 spectrometer (supplemental Table S2).
Purification and NMR Analysis of B8
The asuC2 mutant was inoculated into 50 ml of YMG medium in 250-ml flasks with coils and fermented at 250 rpm and 28 °C for 3 days. One liter of harvested culture was extracted three times with an equal volume of ethyl acetate. The combined ethyl acetate extract was dried, concentrated, and purified on a silica gel column (100–200 mesh), eluting with dichloromethane/methanol (90:10). The compound B8 fraction was further purified by semi-preparative C18 reverse phase HPLC (Dionex), and eluting with 35–50% solvent C in solvent A, and 2 mg of pure compound B8 was obtained. 1H NMR (700 MHz, Me2SO-d6) δH 8.91 (s, 1H, NH-14), 8.88 (s, 1H, NH-0′), 7.27 (d, 1H, J = 7.8 Hz, H-3), 7.12 (dd, 1H, J = 15.0, 11.5 Hz, H-11), 6.67 (dd, 1H, J = 15.1, 10.9 Hz, H-9), 6.47 (dd, 1H, J = 15.2, 11.0 Hz, H-10), 6.43 (dd, 1H, J = 14.9, 11.5 Hz, H-8), 6.38 (d, 1H, J = 15.0 Hz, H-12), 6.12 (d, 1H, J = 15.2 Hz, H-7), 3.85 (m, 1H, H-5), 2.71 (dd, 1H, J = 16.8, 3.2 Hz, H-6ax), 2.59 (dd, 1H, J = 16.5, 6.07 Hz, H-6eq), 1.90 (s, 3H, H-2′). 13C-NMR (700 MHz, Me2SO-d6) δC 192.6 (C-1), 169.5 (C-1′), 165.0 (C-13), 140.3 (C-11), 138.8 (C-9), 138.4 (C-7), 131.5 (C-8), 130.8 (C-2), 129.6 (C-3), 129.5 (C-10), 124.5 (C-12), 73.0 (C-4), 71.3 (C-5), 40.1 (C-6), 23.9 (C-2′).
RESULTS
Cloning of the asu Cluster
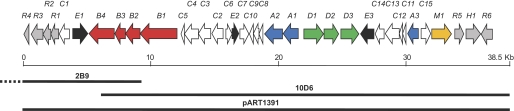
To clone the asukamycin biosynthetic gene cluster, a cosmid genomic library of S. nodosus subsp. asukaensis was constructed and screened with two 32P-labeled DNA probes. ChcA, encoding the 1-cyclohexenylcarbonyl CoA reductase of ansatrienin biosynthesis in S. collinus (12), identified two cosmids, 2B9 and 10D6, which revealed a 1.8-kb overlapping region of the cloned DNA inserts. AsuD2, encoding a 2-oxoamine synthase, hybridized only with cosmid 10D6 (15). Cosmids 2B9 and 10D6 were mapped and sequenced to disclose a total combined 63,922 bp of DNA inserts (GenBankTM accession number GQ926890). DNA sequence analysis revealed 36 potential open reading frames, tentatively assigned as the asu genes, encoding gene products potentially involved in asukamycin biosynthesis and regulatory activities (Fig. 2 and Table 1).
FIGURE 2.
Open reading frames identified in the asu cluster. The putative genes were divided into eight groups based on the functional characterization. Group A, 3,4-AHBA biosynthesis and adenylation; Group B, CHC-CoA biosynthesis; Group C, polyketide chain assembly; Group D, C5N moiety biosynthesis; Group E, oxygenation; Group R, transcriptional regulation. AsuM1, efflux protein. The relative genomic regions and overlapping inserts of three cosmid clones, 2B9, 10D6, and pART1391, are indicated with bold lines.
TABLE 1.
Deduced functions of open reading frames in the asukamycin biosynthetic gene cluster
| Protein | Amino acid | Homolog (accession no.) | Identified or proposed function |
|---|---|---|---|
| AsuA1 | 372 | GriH (YP_001825760.1) | 3,4-AHBA synthase |
| AsuA2 | 471 | ACMS I (AAD30111.1) | 3,4-AHBA carboxyl group adenylation |
| AsuA3 | 278 | GriI (BAF36651.1) | Condensation of l-aspartate-4-semialdehyde and dihydroxyacetone phosphate |
| AsuB1 | 983 | PlmJK (AAQ84158.1) | 5-Enolpyruvylshikimate-3-phosphate synthase/CHC-CoA ligase |
| AsuB2 | 394 | PlmL (AAQ84159.1) | Acyl-CoA dehydrogenase |
| AsuB3 | 277 | ChcA (AAC44655.1) | 1-Cyclohexenylcarbonyl CoA reductase |
| AsuB4 | 667 | PlmM (AAQ84161.1) | 2,4-DienoylCoA reductase |
| AsuC1 | 219 | Sfp (PDB 1QR0) | Phosphopantetheinyl transferase |
| AsuC2 | 269 | GdmF (AAO06919.1) | Arylamine N-acyltransferase |
| AsuC3 | 338 | ZhuH (PDB 1MZJ) | Asukamycin ketosynthase III |
| AsuC4 | 361 | ZhuH (PDB 1MZJ) | Asukamycin ketosynthase III |
| AsuC5 | 84 | ZhuG (AAG30194.1) | Asukamycin KSIII associated ACP |
| AsuC6 | 151 | PaaI (P76084) | Thioesterase |
| AsuC7 | 234 | Sco1815 (PDB 2NM0) | Ketoreductase |
| AsuC8 | 152 | HadC (NP_215151.1) | Acyl dehydratase |
| AsuC9 | 150 | HadB (YP_001281933.1) | Acyl dehydratase |
| AsuC10 | 94 | Putative open reading frame | Unknown |
| AsuC11 | 91 | ZhuN (AAG30201.1) | KSI/II associated ACP |
| AsuC12 | 87 | AcmACP (AAD30112.1) | 3,4-AHBA carrier protein |
| AsuC13 | 397 | FabF (PDB 2GQD) | Asukamycin ketosynthase I/II |
| AsuC14 | 240 | FabF (PDB 2GQD) | Asukamycin ketosynthase I/II |
| AsuC15 | 260 | RifR (PDB 3FLB) | Type II thioesterase |
| AsuR1 | 242 | GerE (PDB 1FSE) | Transcriptional regulation |
| AsuR2 | 191 | TetR family protein | Transcriptional regulation |
| AsuR3 | 307 | Membrane protein | Transcriptional regulation |
| AsuR4 | 141 | DNA-binding protein | Transcriptional regulation |
| AsuR5 | 254 | FarR3 (BAG74713.1) | Transcriptional regulation |
| AsuH1 | 384 | Putative open reading frame | Unknown |
| AsuR6 | 280 | GerE (PDB 1FSE) | Transcriptional regulation |
| AsuE1 | 407 | PHBH (PDB 1YKJ) | Protoasukamycin 4-hydroxylase |
| AsuE2 | 185 | PheA2 (PDB 1RZ0) | Flavin reductase |
| AsuE3 | 371 | LimB (Q9EUT9.1) | 4-Hydroxyprotoasukamycin epoxidase |
| AsuD1 | 531 | NovL (AAF67505.1) | Amide synthase |
| AsuD2 | 409 | MoeC4 (ABJ90148.1) | 2-Oxoamine synthase |
| AsuD3 | 468 | MoeA4 (ABJ90146.1) | 5-Aminolevulinate CoA ligase |
| AsuM1 | 512 | EmrB (YP_001507963.1) | Asukamycin exporter |
Heterologous Expression of the asu Cluster
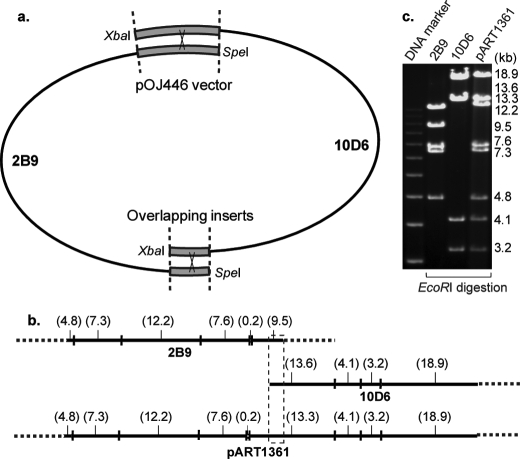
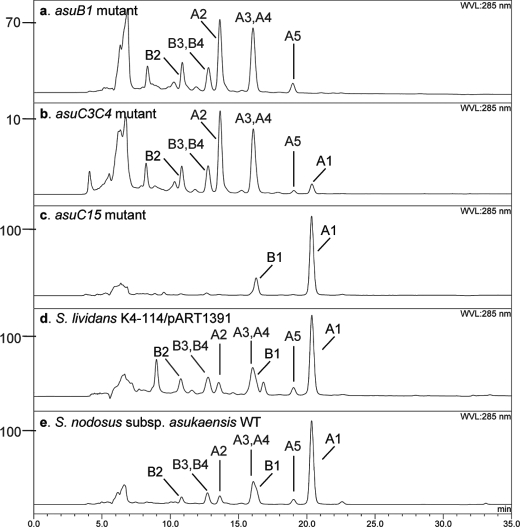
To confirm that the cloned region contains all of the genes in control of asukamycin biosynthesis, regulation, and self-immunity (Table 1), a series of heterologous expression experiments was conducted in the asukamycin nonproducing S. lividans K4-114. Because neither 2B9 nor 10D6 carries a full set of the predicted biosynthetic genes, we modified the conventional λ-Red recombination strategy (20, 23–25) and reassembled the cosmids 2B9 and 10D6 into a single clone (Fig. 3). The resulting cosmid pART1361 was introduced into S. lividans by conjugation from E. coli. Brightly yellowish metabolites were observed in the culture and were confirmed to be asukamycins A1–A7 by high resolution LC-MS. Cosmid pART1391 was further constructed from pART1361 to eliminate a 25.4-kb left-hand side insert region, in which the identified open reading frames were mainly related to primary metabolic functions in Streptomycetes. LC-MS analysis confirmed that S. lividans K4-114/pART1391 is fully capable of producing asukamycins A1–A7 (Fig. 4d). These expression results indicate that the cloned 36 asu genes in pART1391 are sufficient for asukamycin production.
FIGURE 3.
Assembly of two cosmids with overlapping inserts. a, cosmids 2B9 and 10D6 were linearized by SpeI and XbaI, respectively. Homologous recombination occurred at two overlapping insert and vector regions, indicated by heavy lines. b, the restriction map of 2B9, 10D6, and pART1361. The inserts are shown with solid lines. The EcoRI fragment sizes are indicated (kb). c, comparison of the EcoRI digestion patterns by DNA gel electrophoresis.
FIGURE 4.
HPLC analysis of asukamycin metabolites. a, 20 μl of crude culture extract of the asuB1 mutant. b, the asuC3C4 double mutant. c, the asuC15 mutant. d, S. lividans K4-114/pART1391. e, S. nodosus subsp. asukaensis wild type strain. The related peaks of asukamycins A1–A5 and B1–B4 are indicated. The y axis indicates the absorbance abundance. Panel b was calculated based on a 10× injection amount.
3,4-AHBA and Lower Polyketide Chain Formation
Two genes in the cloned asu cluster, asuA1 and asuA3, are closely related to the characterized griH and griI, respectively, which are involved in the assembly of l-aspartate-4-semialdehyde and dihydroxyacetone phosphate into the grixazone 3,4-AHBA moiety in Streptomyces griseus (22). Mutants disrupted in asuA1 and asuA3, respectively, were constructed to confirm their roles in 3,4-AHBA formation. Neither 3,4-AHBA nor asukamycin were detected in the cultures of these mutants, but the addition of 3,4-AHBA restored asukamycin production in both mutants (supplemental Fig. S1a). However, the new minor accumulated products E1 and E3/E4, which possess a C5N moiety amide-linked with the triene upper chains corresponding to A1 and A3/A4, respectively, were detected by LC-MS analysis (supplemental Fig. S6).
Presumably, 3,4-AHBA is activated by adenylation and then tethered to a specific aroyl carrier protein for the downstream chain extensions as in the aromatic polyketide and polypeptide biosynthetic systems employing an aryl carboxylate starter (26–28). AsuA2 is homologous to the actinomycin synthetase I, which initiates actinomycin formation in Streptomyces chrysomallus by activating 4-methyl-3-hydroxyanthranilate (4-MHA) in the presence of ATP (26). The asuA2 mutant failed to produce any asukamycins but accumulated 3,4-AHBA, as confirmed by HPLC comparison with standard 3,4-AHBA as well as by restoring asukamycin production upon co-culturing with the 3,4-AHBA assembly-deficient asuA1 mutant (supplemental Fig. S1b). AsuC11 and AsuC12 both belong to the acyl carrier protein (ACP) family. AsuC12 is distinctly related to AcmACP, a specific aroyl carrier protein of the actinomycin pathway, which is charged by the priming molecule 4-MHA-AMP and forwards it to the peptide synthase AcmII for chain initiation (26). The mutant in which both asuC11 and asuC12 were truncated showed a similar phenotype as the asuA2 mutant, i.e. 3,4-AHBA accumulation and no asukamycin production (supplemental Fig. S1b).
AsuC13 has strong similarity to a group of type II iterative fatty acid synthase (FAS)/polyketide synthase (29, 30). AsuC13 was confirmed to be a KSI/II polyketide synthase (31, 32) involved in the lower polyketide chain assembly, as the asuC13 mutant failed to produce any asukamycins and accumulated 3,4-AHBA, similar to the asuA2 and asuC11C12 mutants (supplemental Fig. S1b). Notably, trace amounts of E1 and E3/E4 were also found in this mutant culture as observed in the asuA1 and asuA3 mutants. The asuC14 gene, immediately downstream of asuC13, encodes a protein homologous to AsuC13, except for a truncated N terminus and lack of the characteristic Cys-His-His catalytic triad. This unusual asuC13-asuC14 pair in which an intact polyketide synthase gene accompanies a partially truncated partner is also found in several uncharacterized gene clusters from the genomic sequencing projects of Streptomyces hygroscopicus (ZP_05513707-8) and Salinispora arenicola (YP_001535964-5 and YP_001537504-5). Mutation studies are in progress, but the AsuC14 function remains unknown for the moment. Two additional sequential reactions, a β-keto reduction and a dehydration, are expected for the lower chain assembly. AsuC7 displays high homology to several polyketide ketoreductases (33, 34). Downstream of asuC7, two putative dehydratase genes, asuC8 and asuC9, were found next to each other. AsuC9 is closely related to HadB, which forms a heterodimer with either HadA or HadC to function in mycolic acid biosynthesis in Mycobacterium tuberculosis H37Rv (35). Coincidently, AsuC8 is highly related to both HadA and HadC. As asuC7 and asuC8,C9 are the only genes in the cluster that could be involved in β-keto reduction and dehydration, we suggest that they operate in the assembly of both the lower and upper trans-triene chains.
Upper Polyketide Chain Formation
Unlike other manumycins, formation of the asukamycin upper polyketide chain starts with cyclohexylcarbonyl-CoA (11). Four clustered genes, asuB1–B4, are homologous to two gene sets, ansJKLM/chcA from the ansatrienin producer S. collinus and plmJKLM/chcA from the phoslactomycin producer Streptomyces sp. HK-803, which are involved in CHC-CoA biosynthesis (12, 36). The asuB1 mutant failed to produce asukamycin, but levels of the previously identified asukamycin congeners A2–A7 were increased 3-fold compared with the wild type strain (Fig. 4a). LC-MS analysis further identified three new compounds, A8, B8, and C8 (Fig. 1). The NMR analysis of B8 revealed a B1-like structure except that the upper polyketide chain was replaced by an acetyl group. Based on the molecular weight, compounds C8 and A8 are likely the proto-form and type I form of B8, respectively. These products probably result from the absence of CHC-CoA plus an oversupply of the 3,4-AHBA-lower chain-ACP intermediate.
To initiate fatty acid or polyketide biosynthesis, acyl-CoA starters are recruited and condensed with a (methyl)-malonyl-ACP by the action of fatty acid synthase FabH or KSIII. This is distinct from the action of KSI/II, joining two ACP-bound acyl substrates in the subsequent chain extensions in many microorganisms (29, 30). The adjacent asuC3 and asuC4 are homologous to numerous genes encoding bacterial FabHs and KSIIIs including the zhuH of Streptomyces. sp. R1128 (37). Both AsuC3 and AsuC4 feature a putative CoA-binding site and a conserved Cys-His-Asn catalytic triad. As AsuC3 and AsuC4 are highly similar to each other, we assume that they might act together as a heterodimer to condense CHC-CoA and malonyl-ACP to give a 3-cyclohexyl-3-oxopropanoyl-ACP intermediate for the downstream polyketide chain assembly. Interestingly, a double mutation of asuC3 and asuC4 did not completely eliminate asukamycin production. The asuC3C4 mutant retained 0.9% asukamycin A1 and 30.0% congeners A2–A7 relative to the wild type strain yield (Fig. 4b), possibly because of the functional complementation by the cellular FabH of fatty acid biosynthesis. AsuC5, presumably organized in the same transcription unit with asuC3,C4, is a zhuG homolog (38). As ZhuG acts together with ZhuH to initiate the assembly of the antitumor polyketide R1128 (38), AsuC5 may play a crucial role in the beginning round of asukamycin upper chain assembly. So far, the KSI/II gene(s) required for the second and third round extensions of the upper chain remain to be identified.
Asukamycin C5N Moiety Formation
Three adjacent genes, asuD1, asuD2, and asuD3, appear to share one transcription unit. AsuD2, previously identified as HemA-AsuA, belongs to a group of pyridoxal phosphate-dependent 2-oxoamine synthases, including 5-aminolevulinate synthase (ALAS), FUM8, l-serine palmitoyltransferase, and 8-amino-7-oxononanoate synthase, which catalyze the decarboxylative condensation between an amino acid and an acyl-CoA (15, 39–42). Despite high overall sequence similarity, AsuD2 possesses a Ser83 instead of a strictly invariant Thr83 of the ALAS subgroup. AsuD2 functions differently from the reported ALAS forming 5-ALA in many organisms, as no asukamycins were detected in the asuD2 mutant even in the presence of 5-ALA, but the asukamycin production was restored by reintroducing the plasmid pALS4 carrying the asuD2 gene (15). The pALS4-S83T with a S83T replacement in asuD2 was constructed but failed to restore asukamycin production in the asuD2 truncated mutant (supplemental Fig. S2). Notably, pALS4-S83T complemented a 5-ALA-deficient gtr mutant of Streptomyces coelicolor A3(2), which can only grow with 5-ALA supplementation. AsuD3, an acyl-CoA ligase, is proposed to act in the C5N ring formation, since disruption of the homologous moeA4 in Streptomyces ghanaensis led to accumulation of a moenomycin A analog lacking the C5N moiety (14). AsuD1 is closely related to a group of amide synthases, including SimL for simocyclinone biosynthesis (43, 44). All of the asuD1, D2, and D3 mutants failed to produce asukamycin but accumulated a new set of metabolites, A1a–A5a, which carry a free carboxyl group in place of the amide-linked C5N moiety of A1–A5, respectively (supplemental Fig. S1c). The corresponding type II compounds, B1a–B5a, were also present in all three mutants. Furthermore, the asuD1–D3 operon was expressed in S. lividans, which resulted in attachment of the C5N moiety to added ferulic acid to form feruloyl-N-(3-hydroxycyclopent-2-enolone)acrylamide, whereas no C5N moiety was detected in the absence of asuD1, D2, or D3 (supplemental text and Fig. S2). This confirmed that asuD1, D2, and D3 are required and sufficient for C5N moiety formation.
Attachment of the Upper Polyketide Chain
Asukamycin contains two amide bonds: the linkage between the upper chain and the mC7N core and the bridge tethering the C5N moiety to the lower polyketide chain. AsuC2 belongs to the N-acyltransferase family (45). An asuC2 disrupted mutant was constructed to confirm its involvement in the attachment of the upper chain to the 3-amino group of the 3,4-AHBA primed polyketide intermediate. The production of asukamycins A1–A7 was completely blocked in the asuC2 mutant. Instead, the N-acetylated compounds A8, B8, and C8 were accumulated as in the asuB1 mutant. In view of the functional deficiency of asuC2, these shunt products presumably resulted from the action of a cellular arylamine N-acetyltransferase, commonly found to detoxify arylamine or arylhydroxylamine metabolites (46).
Oxygenation of Protoasukamycin
S. parvulus Tü 64 fermentation under an 18O2 atmosphere has demonstrated that the hydroxyl and the epoxide oxygens at C4 and C5–C6 of manumycin A originate from molecular oxygen, by either a two-step process or a dioxygenase mechanism (10, 17). Three distantly located genes, asuE1, asuE2, and asuE3, are possibly involved in the oxygenation reaction(s). AsuE1 is homologous to p-hydroxybenzoate hydroxylase, which converts p-hydroxybenzoate into 3,4-dihydroxybenzoate in Pseudomonas aeruginosa (47). Instead of producing asukamycin, the asuE1 mutant accumulated protoasukamycin C1 and its congeners C2–C5 (supplemental Fig. S1d). AsuE2 is a homolog of the flavin reductase PheA2, which recycles oxidized FADox and provides FADred to its hydroxylase partner PheA1 in the phenol hydroxylation in Bacillus thermoglucosidasius A7 (48). The asuE2 mutant produced asukamycin A1 in considerably lower yield than the wild type strain but accumulated major amounts of the protoasukamycins C1–C5 (supplemental Fig. S1d). This implies that AsuE2 is necessary for the full catalytic function of AsuE1 as a two-component flavoprotein hydroxylase.
AsuE3 is highly related to LimB, a limonene 1,2-monooxygenase that generates limonene-1,2-epoxide in Rhodococcus erythropolis (49). Instead of the recognized asukamycins, a group of bright yellowish compounds were accumulated by the asuE3 disrupted mutant (supplemental Fig. S1d). LC-MS analysis of the major peak suggested that one oxygen atom was missing compared with asukamycin A1. 1H, 13C, HMBC, and HSQC NMR analysis of the purified compound showed that a double bond at C5–C6 replaces the epoxide group of asukamycin A1 (supplemental Table S2). This new compound was therefore named as 4-hydroxyprotoasukamycin D1. Its congeners D2–D5 were also detected. D1 was converted to asukamycin A1 when fed to the asuA3 mutant culture, indicating that D1 is an intermediate that is further epoxidized by AsuE3 in the final step of asukamycin biosynthesis (supplemental Fig. S1a). Since D1 was absent in the asuE2 mutant, the epoxidation may not require AsuE2.
Thioesterase Affects the Primed CHC
AsuC15 is closely related to a group of type II thioesterases found in polyketide and nonribosomal peptide biosynthetic gene clusters, including RifR of rifamycin biosynthesis (50). As the yields of the polyketide and polypeptide products were often considerably impaired in the knock-out mutants, type II thioesterases were proposed to restore the acyl or peptidyl carrier protein function by releasing undesirable intermediates, resulting from a priming or processing error (51–54). The asuC15 mutant produced a normal amount of asukamycin A1, yet the yield of the congeners A2–A7 was significantly reduced (Fig. 4c). Since CHC-CoA is not only a building block of asukamycin but can also form ω-cyclohexyl fatty acids as membrane components in S. nodosus subsp. asukaensis (7), we analyzed the fatty acid compositions of the asuC15 mutant and the wild type strain by GC-MS (supplemental Fig. S3). Notably, 16.5% of ω-cyclohexyl-C17 fatty acid was detected in the asuC15 mutant, at least 5-fold higher than the 3.1% detected in the wild type strain. Moreover, 1.7% of ω-cyclohexyl-C19 fatty acid, undetectable in the wild type strain, was also found.
Other asu Gene Functions
AsuC6, a hot dog fold thioesterase, presumably functions to hydrolyze the thioester bond and remove the acyl intermediate from the polyketide synthase (55, 56). It could be responsible for A1a–A5a accumulation in the asuD1, D2, and D3 mutants, which are supposed to be tightly bound to the ACP AsuC11. The asuC6 mutant has not yet been constructed, and the precise AsuC6 function remains to be clarified. AsuC1, a phosphopantetheinyl transferase, presumably primes the carrier proteins AsuC5, C11, and C12, which are involved in the lower and upper chain biosynthesis. AsuM1, a homolog of numerous antibiotic efflux exporters, could release the synthesized asukamycin products from the cells. AsuR1–R6, discretely detected at the two border regions of the cluster, show homology to numerous putative transcriptional regulator genes found in Gram-positive bacteria. AsuR1 and AsuR6 are closely related to the LuxR-type transcriptional regulators (57). AsuR2 is likely a TetR-like transcriptional regulator (58). AsuR3 is a putative integral membrane sensor protein, which could work together with the DNA-binding protein AsuR4 (59). AsuR5 belongs to the Streptomyces antibiotic regulatory proteins (60). As many antibiotic biosynthesis gene clusters contain one or multiple regulatory genes, AsuR1–R6 could have regulatory functions to control the asukamycin biosynthetic gene activities. AsuH1 is a putative open reading frame with an unknown function.
DISCUSSION
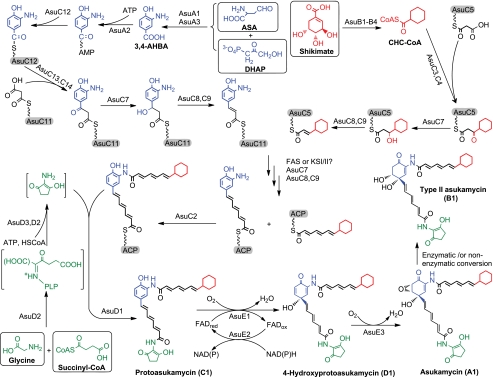
The structure of asukamycin features two triene polyketide chains connected to a 3,4-AHBA-derived mC7N core. The upper chain starts with a cyclohexane head group, and the lower one ends with a C5N structural moiety. In this study, four major groups of asu genes were identified to be involved in the structural assembly: AsuA1 and A3 for 3,4-AHBA biosynthesis, AsuB1–B4 for the conversion of shikimate into CHC-CoA, AsuD1–D34 for C5N unit formation, and the AsuC group for the lower and upper polyketide chain assembly (Fig. 5).
FIGURE 5.
Proposed asukamycin biosynthetic pathway. The primary metabolite substrates are shown in squares. The ACP and aroyl carrier protein are in capsules.
Construction of the lower polyketide chain likely requires a set of type II polyketide synthetic enzymes including the KSI/II AsuC13,C14, the polyketide ketoreductase AsuC7, and the DH AsuC8,C9, based on the sequencing and mutagenic analysis. The results indicate that 3,4-AHBA is activated by AsuA2 to form an acyl-AMP and subsequently loaded onto the aroyl carrier protein AsuC12 to initiate the lower polyketide assembly. AsuC13 and AsuC14 are most likely responsible for the initiation and two further rounds of polyketide chain extensions, as the lower chain biosynthesis was abolished in the asuC13 mutant. The presence of asuC14 may not be coincidental. AsuC13 and AsuC14 could function together to constrain the growing triene structures and control the polyketide chain length. Further studies on other manumycin biosynthetic genes should provide more clues on the function of asuC14. Since asuC11 is potentially organized in the same transcription unit with asuC12–C14, the AsuC11 ACP could be involved in the second and the third rounds of lower chain polyketide extensions.
For the upper chain, AsuC3,C4 likely initiate the condensation between CHC-CoA and malonyl-ACP, resembling the bacterial FabH and other KSIIIs, which are directly primed with an acyl-CoA. In the asuC3,C4 mutant most asukamycin A1 production was abolished, and levels of the congeners A2–A7 were reduced compared with the wild type. This reflects a possible involvement of the cellular fatty acid synthase FabH, which displays a substrate preference distinct from AsuC3,C4, priming preferentially with branched chain acyl-CoAs rather than CHC-CoA, to give a higher yield of A2–A7 relative to A1 (7). The involvement of AsuC3,C4 in the second and third chain elongations is less likely, as FabH catalyzes only the first condensation step and passes the resulting 3-ketoacyl-ACP directly to another enzyme complex for further reductions and chain elongations. Given that E1 and E3/E4 were found in the asuC13 mutant, the only recognized KSI/II gene pair, asuC13,C14, may not play a role in the upper chain assembly, although both the upper and lower polyketide chains of asukamycin share an identical triene structure. It seems that an unidentified polyketide KSI/II is required to act together with the polyketide ketoreductase AsuC7 and DH AsuC8,C9 to carry out the upper triene polyketide extension. We also cannot rule out the involvement of a cellular FabB/F-like fatty acid synthase, as the successful heterologous expression in S. lividans implies that a common KSI/II or FASI/II can fulfill the functional role of this unidentified β-keto condensation enzyme.
The formation of A1–A4 only differs at the priming step of the upper polyketide chain. A2–A4 employ the branched chain amino acid-derived starters 2-methylpropionyl-CoA and 3- and 2-methylbutanoyl-CoA, respectively, which are predominantly used for fatty acid synthesis in Streptomyces (7). In the asuB1 mutant, the absence of CHC-CoA abolished the entire A1 production and redirected the biosynthetic flux to A2–A4. Since A2–A4 formation was 10-fold higher in this mutant than in the asuC3,C4 mutant, the majority of A2–A4 upper polyketide initiation is likely performed by the KSIII AsuC3,C4, and only a minor fraction of A2–A4 production in the asuB1 mutant as well as the wild type strain is contributed by the FabH activity. The relatively small amount of A5–A7 production could result from an action of the primary FAS complex to give 4-methylpentanoyl- and 5- and 4-methylhexanoyl-ACP intermediates. These ACP-tethered thioesters could be directly recruited by an unidentified KSI/II or FASI/II to perform three additional elongations and introduce the characteristic triene in analogy with the A1–A4 upper chain biosynthesis. These observations are in agreement with the FabH involvement and suggest physiological cross-talk between polyketide and fatty acid biosynthesis.
CHC-CoA is not only the substrate for the polyketide synthase AsuC3,C4 in A1 upper chain assembly but is also recruited by the FAS complex and assembled into ω-cyclohexyl fatty acids, which account for 3.1% of total cellular fatty acids in S. nodosus subsp. asukaensis (7). The CHC-CoA utilization to form asukamycin A1 and ω-cyclohexyl fatty acids does not occur in a fixed ratio, as A1 was increased to more than 80% of total asukamycins, whereas ω-cyclohexyl fatty acids were kept under 25% of total fatty acids upon feeding excess cyclohexanecarboxylic acid (7). Besides the distinct substrate specificities of FabH and AsuC3,C4, this could also be the result of type II thioesterase AsuC15 action to release the tethered cyclohexylpropanoate from the FAS-ACP and allow the recruitment of more branched chain acyl-CoA starters, as the percentage of ω-cyclohexyl fatty acids was increased 5-fold in the asuC15 mutant. The cellular membrane fluidity is probably affected by variations in the content of ω-cyclohexyl fatty acids, changing the phase transition temperature, as observed for several thermo-acidophilic bacteria, which may contain up to 90% of ω-cyclohexyl fatty acids (18, 19, 61). Thus AsuC15 could suppress excess ω-cyclohexyl fatty acid formation and help to maintain membrane homeostasis and structural integrity, particularly when the cellular level of CHC-CoA is rising. Evidently, AsuC15 can also discharge the CHC-acyl intermediate from AsuC5-ACP to enhance the production of congeners A2–A4, as the asuC15 mutant predominantly accumulates A1 rather than an equal amount of A1 and A2–A4 observed in the wild type.
AsuD1, D2, and D3 are necessary and sufficient for the assembly and attachment of the C5N moiety to form protoasukamycin. AsuD2 bears a Ser83 in the position of the conserved Thr83 of ALAS. The distinct role of AsuD2-Ser83 in C5N moiety formation was confirmed by expression of AsuD2-S83T, which failed to complement the asuD2 mutant but sustained the ability to form 5-ALA in the gtr mutant of S. coelicolor. The fact that 5-ALA is a precursor of the C5N unit but does not complement the asuD2 mutation shows that asuD2 has a second function in C5N formation (13, 15, 16). The nascent pyridoxal phosphate-bound 5-ALA or possibly a predecarboxylation intermediate 2-amino-3-oxoadipate could be retained on AsuD2 and carboxyl-activated by the action of the CoA ligase AsuD3, followed by cyclization catalyzed by AsuD2 (Fig. 5). The dual role of AsuD2 is also supported by the results of the heterologous expression of asuD1–D3 to produce the C5N unit in the absence of any other candidate cyclase gene in S. lividans. The amide synthase AsuD1 is responsible for the attachment of the C5N unit to the C terminus of the lower chain. As all the assumed intermediates in this process are relatively unstable (10), AsuD1, D2, and D3 might work together to secure the formation of protoasukamycin.
Both one-step dioxygenase and two-step monooxygenase mechanisms have been proposed for epoxyquinol formation (1, 10, 11). The present study resolves this issue as two monooxygenations, first 4-hydroxylation of protoasukamycin C1 by AsuE1 and AsuE2, followed by epoxidation of the resulting quinol D1 by AsuE3 to form asukamycin A1 (Fig. 5). As AsuE3 specifically acts on the quinol moiety but not a phenolic structure, the protoasukamycin 4-hydroxylation is a prerequisite for epoxidation. Notably, replacement of the upper chain by an acetyl group or the absence of the C5N moiety has no impact on epoxyquinol formation as evidenced by A8 and A1a, respectively.
It seems that asukamycin biosynthesis has evolved through multiple horizontal gene transfer events and possibly an extensive genomic rearrangement in S. nodosus subsp. asukaensis. The biosynthetic genes involved in C5N moiety and CHC-CoA assembly are closely positioned and appear to be co-transcribed, but many other functionally related asu genes are not organized in any apparent order. Recently, the λ-Red recombination system has been extended to heterologous expression (23–25). Taking advantage of the shared vector and overlapping inserts, we simplified this system and assembled two cosmids in a single step by skipping the PCR cloning and multiple “stitching” processes (Fig. 3). Since the linearized cosmids are unable to replicate, the circular recombinant pART1361 could propagate and form colonies under antibiotic selection. This straightforward recombination approach is particularly useful to examine cloned genes and provides a convenient platform for further gene manipulations. For example, a reliable yield of 4-hydroxyprotoasukamycin D1 was obtained in S. lividans carrying the pART1361E3 by replacing asuE3 with a spectinomycin resistance gene.
Supplementary Material
Acknowledgments
We are grateful to C. Khosla (S. lividans K4-114) and John Innes Center (λ-Red recombination kit) for the gifts of strains and plasmids. We thank Drs. J. Chen, G. Henderson, and T. K. Weldeghiorghis for assistance with mass spectral and NMR analyses and M. Saboori for the asuC2 analysis. We thank the anonymous reviewers for constructive comments.
This work was supported, in whole or in part, by National Institutes of Health Grant CA76461 (to T.-W. Y.). This work was also supported by Czech Ministry of Education Grant 2B06154, Institutional Scientific Program AV0Z50200510 (to M. P.), a Louisiana State University Faculty Startup Fellowship (to T.-W. Y.), and a Louisiana State Economic Development Assistantship (to T.-W. Y. and Z. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1–S3, and Figs. S1–S6.
During the peer review process of this manuscript, the Walsh group reported a study that supports the proposed AsuD1–D3 function in the C5N moiety formation (62).
- mC7N
- 2-amino-4-hydroxy-5,6-epoxycyclohex-2-enone
- 3,4-AHBA
- 3-amino-4-hydroxybenzoic acid
- C5N
- 2-amino-3-hydroxycyclopent-2-enone
- 5-ALA
- 5-aminolevulinate
- ACP
- acyl carrier protein
- FAS
- fatty acid synthase
- ALAS
- 5-aminolevulinate synthase
- CHC
- cyclohexylcarbonyl
- HPLC
- high pressure liquid chromatography
- LC-MS
- liquid chromatography-mass spectrometry
- KS
- polyketide synthase
- PDB
- Protein Data Bank.
REFERENCES
- 1.Sattler I., Thiericke R., Zeeck A. (1998) Nat. Prod. Rep. 15, 221–240 [DOI] [PubMed] [Google Scholar]
- 2.Shipley P. R., Donnelly C. C., Le C. H., Bernauer A. D., Klegeris A. (2009) Int. J. Mol. Med. 24, 711–715 [DOI] [PubMed] [Google Scholar]
- 3.Bernier M., Kwon Y. K., Pandey S. K., Zhu T. N., Zhao R. J., Maciuk A., He H. J., Decabo R., Kole S. (2006) J. Biol. Chem. 281, 2551–2561 [DOI] [PubMed] [Google Scholar]
- 4.Zheng Z. H., Dong Y. S., Zhang H., Lu X. H., Ren X., Zhao G., He J. G., Si S. Y. (2007) J. Enzyme Inhib. Med. Chem. 22, 43–49 [DOI] [PubMed] [Google Scholar]
- 5.Kakinuma K., Ikekawa N., Nakagawa A., Omura S. (1979) J. Am. Chem. Soc. 101, 3402–3404 [Google Scholar]
- 6.Omura S., Kitao C., Tanaka H., Oiwa R., Takahashi Y., Nakagawa A., Shimada M., Iwai Y. (1976) J. Antibiot. 29, 876–881 [DOI] [PubMed] [Google Scholar]
- 7.Hu Y. D., Floss H. G. (2006) Heterocycles 69, 133–149 [Google Scholar]
- 8.Hu Y., Floss H. G. (2001) J. Antibiot. 54, 340–348 [DOI] [PubMed] [Google Scholar]
- 9.Hu Y. D., Melville C. R., Gould S. J., Floss H. G. (1997) J. Am. Chem. Soc. 119, 4301–4302 [Google Scholar]
- 10.Hu Y. D., Floss H. G. (2004) J. Am. Chem. Soc. 126, 3837–3844 [DOI] [PubMed] [Google Scholar]
- 11.Thiericke R., Zeeck A., Nakagawa A., Omura S., Herrold R. E., Wu S. T. S., Beale J. M., Floss H. G. (1990) J. Am. Chem. Soc. 112, 3979–3987 [Google Scholar]
- 12.Cropp T. A., Wilson D. J., Reynolds K. A. (2000) Nat. Biotechnol. 18, 980–983 [DOI] [PubMed] [Google Scholar]
- 13.Beale J. M., Lee J. P., Nakagawa A., Omura S., Floss H. G. (1986) J. Am. Chem. Soc. 108, 331–332 [Google Scholar]
- 14.Ostash B., Saghatelian A., Walker S. (2007) Chem. Biol. 14, 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrícek M., Petrícková K., Havlícek L., Felsberg J. (2006) J. Bacteriol. 188, 5113–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagawa A., Wu T. S., Keller P. J., Lee J. P., Omura S., Floss H. G. (1985) J. Chem. Soc. Chem. Comm. 519–521 [Google Scholar]
- 17.Thiericke R., Zeeck A., Robinson J. A., Beale J. M., Floss H. G. (1989) J. Chem. Soc. Chem. Comm. 402–403 [Google Scholar]
- 18.Kaneda T. (1991) Microbiol. Rev. 55, 288–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y. M., Rock C. O. (2008) Nat. Rev. Microbiol. 6, 222–233 [DOI] [PubMed] [Google Scholar]
- 20.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, The John Innes Foundation, Norwich, UK [Google Scholar]
- 22.Suzuki H., Ohnishi Y., Furusho Y., Sakuda S., Horinouchi S. (2006) J. Biol. Chem. 281, 36944–36951 [DOI] [PubMed] [Google Scholar]
- 23.Binz T. M., Wenzel S. C., Schnell H. J., Bechthold A., Müller R. (2008) ChemBioChem 9, 447–454 [DOI] [PubMed] [Google Scholar]
- 24.Wolpert M., Heide L., Kammerer B., Gust B. (2008) ChemBioChem 9, 603–612 [DOI] [PubMed] [Google Scholar]
- 25.Hu Y., Phelan V. V., Farnet C. M., Zazopoulos E., Bachmann B. O. (2008) ChemBioChem 9, 1603–1608 [DOI] [PubMed] [Google Scholar]
- 26.Pfennig F., Schauwecker F., Keller U. (1999) J. Biol. Chem. 274, 12508–12516 [DOI] [PubMed] [Google Scholar]
- 27.Zhou Z., Lai J. R., Walsh C. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreras J. A., Stirrett K. L., Lu X., Ryu J. S., Soll C. E., Tan D. S., Quadri L. E. (2008) Chem. Biol. 15, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hertweck C. (2009) Angew. Chem. Int. Ed. Engl. 48, 4688–4716 [DOI] [PubMed] [Google Scholar]
- 30.Sattely E. S., Fischbach M. A., Walsh C. T. (2008) Nat. Prod. Rep. 25, 757–793 [DOI] [PubMed] [Google Scholar]
- 31.Huang W., Jia J., Edwards P., Dehesh K., Schneider G., Lindqvist Y. (1998) EMBO J. 17, 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen J. G., Kadziola A., von Wettstein-Knowles P., Siggaard-Andersen M., Lindquist Y., Larsen S. (1999) FEBS Lett. 460, 46–52 [DOI] [PubMed] [Google Scholar]
- 33.Tang Y., Lee H. Y., Tang Y., Kim C. Y., Mathews I., Khosla C. (2006) Biochemistry 45, 14085–14093 [DOI] [PubMed] [Google Scholar]
- 34.Cohen-Gonsaud M., Ducasse S., Hoh F., Zerbib D., Labesse G., Quemard A. (2002) J. Mol. Biol. 320, 249–261 [DOI] [PubMed] [Google Scholar]
- 35.Sacco E., Covarrubias A. S., O'Hare H. M., Carroll P., Eynard N., Jones T. A., Parish T., Daffé M., Bäckbro K., Quémard A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palaniappan N., Kim B. S., Sekiyama Y., Osada H., Reynolds K. A. (2003) J. Biol. Chem. 278, 35552–35557 [DOI] [PubMed] [Google Scholar]
- 37.Pan H., Tsai S., Meadows E. S., Miercke L. J., Keatinge-Clay A. T., O'Connell J., Khosla C., Stroud R. M. (2002) Structure 10, 1559–1568 [DOI] [PubMed] [Google Scholar]
- 38.Tang Y., Lee T. S., Kobayashi S., Khosla C. (2003) Biochemistry 42, 6588–6595 [DOI] [PubMed] [Google Scholar]
- 39.Astner I., Schulze J. O., van den Heuvel J., Jahn D., Schubert W. D., Heinz D. W. (2005) EMBO J. 24, 3166–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerber R., Lou L., Du L. C. (2009) J. Am. Chem. Soc. 131, 3148–3149 [DOI] [PubMed] [Google Scholar]
- 41.Hanada K. (2003) Biochim. Biophys. Acta 1632, 16–30 [DOI] [PubMed] [Google Scholar]
- 42.Webster S. P., Alexeev D., Campopiano D. J., Watt R. M., Alexeeva M., Sawyer L., Baxter R. L. (2000) Biochemistry 39, 516–528 [DOI] [PubMed] [Google Scholar]
- 43.Pacholec M., Freel Meyers C. L., Oberthür M., Kahne D., Walsh C. T. (2005) Biochemistry 44, 4949–4956 [DOI] [PubMed] [Google Scholar]
- 44.Schmutz E., Steffensky M., Schmidt J., Porzel A., Li S. M., Heide L. (2003) Eur. J. Biochem. 270, 4413–4419 [DOI] [PubMed] [Google Scholar]
- 45.Yu T. W., Bai L., Clade D., Hoffmann D., Toelzer S., Trinh K. Q., Xu J., Moss S. J., Leistner E., Floss H. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7968–7973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vineis P., Bartsch H., Caporaso N., Harrington A. M., Kadlubar F. F., Landi M. T., Malaveille C., Shields P. G., Skipper P., Talaska G., Tannenbaum S. R. (1994) Nature 369, 154–156 [DOI] [PubMed] [Google Scholar]
- 47.Cole L. J., Gatti D. L., Entsch B., Ballou D. P. (2005) Biochemistry 44, 8047–8058 [DOI] [PubMed] [Google Scholar]
- 48.Kirchner U., Westphal A. H., Müller R., van Berkel W. J. (2003) J. Biol. Chem. 278, 47545–47553 [DOI] [PubMed] [Google Scholar]
- 49.van der Werf M. J., Swarts H. J., de Bont J. A. (1999) Appl. Environ. Microbiol. 65, 2092–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claxton H. B., Akey D. L., Silver M. K., Admiraal S. J., Smith J. L. (2009) J. Biol. Chem. 284, 5021–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koglin A., Löhr F., Bernhard F., Rogov V. V., Frueh D. P., Strieter E. R., Mofid M. R., Güntert P., Wagner G., Walsh C. T., Marahiel M. A., Dötsch V. (2008) Nature 454, 907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotowska M., Pawlik K., Smulczyk-Krawczyszyn A., Bartosz-Bechowski H., Kuczek K. (2009) Appl. Environ. Microbiol. 75, 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim B. S., Cropp T. A., Beck B. J., Sherman D. H., Reynolds K. A. (2002) J. Biol. Chem. 277, 48028–48034 [DOI] [PubMed] [Google Scholar]
- 54.Yeh E., Kohli R. M., Bruner S. D., Walsh C. T. (2004) ChemBioChem 5, 1290–1293 [DOI] [PubMed] [Google Scholar]
- 55.Song F., Zhuang Z., Finci L., Dunaway-Mariano D., Kniewel R., Buglino J. A., Solorzano V., Wu J., Lima C. D. (2006) J. Biol. Chem. 281, 11028–11038 [DOI] [PubMed] [Google Scholar]
- 56.Kotaka M., Kong R., Qureshi I., Ho Q. S., Sun H., Liew C. W., Goh L. P., Cheung P., Mu Y., Lescar J., Liang Z. X. (2009) J. Biol. Chem. 284, 15739–15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nasser W., Reverchon S. (2007) Anal. Bioanal. Chem. 387, 381–390 [DOI] [PubMed] [Google Scholar]
- 58.Ramos J. L., Martínez-Bueno M., Molina-Henares A. J., Terán W., Watanabe K., Zhang X., Gallegos M. T., Brennan R., Tobes R. (2005) Microbiol. Mol. Biol. Rev. 69, 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Groban E. S., Clarke E. J., Salis H. M., Miller S. M., Voigt C. A. (2009) J. Mol. Biol. 390, 380–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kitani S., Iida A., Izumi T. A., Maeda A., Yamada Y., Nihira T. (2008) Gene 425, 9–16 [DOI] [PubMed] [Google Scholar]
- 61.Simbahan J., Drijber R., Blum P. (2004) Int. J. Syst. Evol. Microbiol. 54, 1703–1707 [DOI] [PubMed] [Google Scholar]
- 62.Zhang W., Bolla M. L., Kahne D., Walsh C. T. (2010) J. Am. Chem. Soc. 132, 6402–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.