Abstract
Binding of the platelet-derived growth factor (PDGF)-B to its receptor PDGFRβ promotes proliferation, migration, and recruitment of pericytes and smooth muscle cells to endothelial cells, serving to stabilize developing blood vessels. The main goals of this study were to determine whether the extracellular domain of the PDGFRβ can be proteolytically released from cell membranes and, if so, to identify the responsible sheddase and determine whether activation of the PDGFRβ stimulates its shedding and potentially that of other membrane proteins. We found that the PDGFRβ is shed from cells by a metalloproteinase and used loss-of-function experiments to identify ADAM10 as the sheddase responsible for constitutive and ionomycin-stimulated processing of the PDGFRβ. Moreover, we showed that ligand-dependent activation of the PDGFRβ does not trigger its own shedding by ADAM10, but instead it stimulates ADAM17 and shedding of substrates of ADAM17, including tumor necrosis factor α and transforming growth factor α. Finally, we demonstrated that treatment of mouse embryonic fibroblasts with PDGF-B triggers a metalloproteinase-dependent cross-talk between the PDGFRβ and the epidermal growth factor receptor (EGFR)/ERK1/2 signaling axis that is also critical for PDGF-B-stimulated cell migration, most likely via ADAM17-dependent release and activation of ligands of the EGFR. This study identifies the principal sheddase for the PDGFRβ and provides new insights into the mechanism of PDGFRβ-dependent signal transduction and cross-talk with the EGFR.
Keywords: ADAM/ADAMTS, MAPKs, Metalloprotease, Shedding, Signal Transduction, Cross-talk, EGFR, PDGFRbeta
Introduction
Signaling activated by the platelet-derived growth factor (PDGF)3-B and its receptor on pericytes, the PDGF receptor β (PDGFRβ), is important for blood vessel development. PDGF-B is secreted by endothelial cells and promotes proliferation, migration, and recruitment of pericytes and smooth muscle cells to endothelial cells, which in turn stabilizes the developing vasculature (1, 2). Mice lacking PDGF-B do not have microvascular pericytes, resulting in microaneurysm, hemorrhage, edema, and ultimately death (1). The signal transduction pathways activated by the PDGFRβ are well characterized and resemble those of other receptor tyrosine kinases such as epidermal growth factor receptor (EGFR) and the vascular endothelial growth factor receptor (VEGFR) in that PDGF-B binding to PDGFRβ induces receptor dimerization and subsequent autophosphorylation on tyrosine residues in its intracellular domain (3). This, in turn, activates the tyrosine kinase and provides docking sites for downstream signaling molecules (4). Following its activation by PDGF-B, the PDGFRβ stimulates intracellular signaling proteins that include Ras-MAPK, phosphatidylinositol 3-kinase, phospholipase Cγ, and ERK1/2 (5).
This study was initiated to assess the role of protein ectodomain shedding in regulating the function of the PDGFRβ. Protein ectodomain shedding is a regulated process, and members of the ADAM (a disintegrin and metalloproteinase) family of membrane-anchored metalloproteinases (6, 7) have often been identified as the enzymes involved in the constitutive and stimulated release of integral membrane substrate proteins such as the pro-inflammatory cytokine TNFα (8–10), ligands of the EGFR including EGF, HB-EGF, and TGFα (11–15) and growth factor receptors such as VEGFR2 (16) or the p75NTR (17). We were therefore interested in determining whether the PDGFRβ is shed and, if so, to identify the responsible enzyme and assess the potential functional consequences of PDGFRβ shedding.
Recent studies on a different receptor tyrosine kinase, the VEGFR2, demonstrated that stimulation of the VEGFR2 with VEGF-A activates ADAM17, thereby triggering the release of the VEGFR2 as well as other substrates of ADAM17, including ligands of the EGFR (16). The results from this study also indicated that ADAM17-dependent processing of EGFR ligands in response to activation of the VEGFR2 extends the duration of VEGF-A-stimulated ERK1/2 phosphorylation, which can be separated into two distinct components. The first component is represented by an early stimulation of ERK1/2 via the VEGFR2 that lasts about 15–30 min and is not sensitive to metalloproteinase inhibitors, whereas the second component of ERK1/2 phosphorylation is most evident 30 and 60 min after addition of VEGF-A and is sensitive to metalloproteinase inhibitors. The second component likely depends on stimulation of ADAM17 and the release of EGFR ligands that can activate ERK1/2 by binding to the EGFR (16). These findings prompted us to test whether a similar two-stage mechanism of ERK1/2 activation might also be triggered by stimulation of the PDGFRβ with PDGF-B.
Taken together, the overall goals of this study were to identify the sheddase responsible for PDGFRβ cleavage and to determine whether shedding affects the activation or turnover of the PDGFRβ. Moreover, we wished to establish whether PDGF-B/PDGFRβ signaling also stimulates ADAM17, and if so, whether the activation of ERK1/2 elicited by PDGF-B could depend, at least in part, on stimulation of ADAM17 through the PDGFRβ. Finally, we assessed whether metalloproteinase-dependent activation of the EGFR could have a role in PDGFR-β-dependent cell migration.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
COS-7 cells and mouse embryonic fibroblasts (mEFs) from wild type, Adam10−/−, or Adam17−/− mice were cultured and transfected as described previously (14, 18). All immortalized cells were grown in Dulbecco's modified Eagle's medium with antibiotics and 5% fetal calf serum, whereas primary mEFs were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum. Cells were incubated in Opti-MEM for transfection and shedding assays. Reagents were from Sigma unless indicated otherwise. GenJet (SignaGen Laboratories, Gaithersburg, MD) and Lipofectamine 2000 (Invitrogen) were used for transient transfections of COS-7 and mEFs, respectively. Ionomycin was purchased from Calbiochem. Recombinant murine platelet-derived growth factor-B (PDGF-B) was obtained from PeproTech (Rocky Hill, NJ); rabbit anti-ERK2 was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); and mouse monoclonal anti-phosphotyrosine (clone 4G10) was from Millipore (Temecula, CA). Rabbit anti-phospho-ERK1/2, rabbit monoclonal anti-phospho-EGFR (Tyr-1068), and rabbit anti-EGFR were from Cell Signaling Technology, Inc. (Danvers, MA), and the mouse monoclonal anti-c-Myc antibody (9E10) was from Roche Applied Science. Concanavalin A-Sepharose was purchased from GE Healthcare. Marimastat was kindly provided by Dr. Ouathek Ouerfelli, Memorial Sloan-Kettering Cancer Center, New York.
Expression Vectors
The expression constructs for ADAM10 and -17, for the inactive ADAM10Glu→Ala or ADAM17Glu→Ala mutants, and for alkaline phosphatase (AP)-tagged TNFα, HB-EGF, TGFα, and EGF were described previously (14, 19). The expression construct for murine AP-tagged PDGFRβ was generated as follows. Full-length cDNA was obtained from ATCC (number 9890909, Manassas, VA), and the PCR product that included the coding sequence for the PDGFRβ that was C-terminal from the third Ig region within the extracellular domain and included the juxtamembrane region, the transmembrane domain, and the cytotail (amino acid residues Leu410–Leu1099) was generated and cloned in-frame downstream of human alkaline phosphatase in the pAP-TAG mammalian expression vector (Genehunter, Nashville, TN). To generate the full-length PDGFRβ construct, the entire coding sequence of PDGFRβ was cloned into pcDNA 3.1A− (Invitrogen). The cDNA for the oncogenic fusion protein between the first 154 amino acids of the transcription factor TEL and the transmembrane and cytoplasmic domains of the PDGFRβ (TelPDGFRβ (20, 21)), kindly provided by Dr. Michael Tomasson (Washington University School of Medicine, St. Louis), was subcloned into pcDNA 3.1A−. The gatekeeper mutant T681M PDGFRβ (22) was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Cell Culture, Transfection, and Ectodomain Shedding Assays
COS-7 cells and mEFs were transiently transfected with GenJet and Lipofectamine 2000, respectively, as described previously (14, 23). Shedding assays were performed in Opti-MEM the day following transfection, and AP activity in the supernatant and lysates was measured colorimetrically as described previously (14, 23). The ratio between the AP activity in the supernatant and the total AP activity in the cell lysate plus supernatant was calculated from two identically transfected wells and averaged. This ratio represents the relative amount of shedding by a given sheddase toward a given AP-tagged substrate. No AP activity was present in conditioned media of nontransfected cells. Each experiment was repeated at least three separate times.
Western Blot Analysis
Western blots were performed as described previously (24). Briefly, cells were lysed in lysis buffer containing phosphate-buffered saline, 1% Triton X-100, protease inhibitor mixture, 0.1 mm vanadate, 0.2 mm phenylmethylsulfonyl fluoride, and 10 mm NaF. Following cell lysis, nuclei and cell debris were removed from the sample by spinning at 13,000 rpm for 10 min at 4 °C in an Eppendorf tabletop microcentrifuge. For the pEGFR and EGFR blots in Fig. 5B, the supernatant was removed into a separate tube containing 50 μl of a 50% slurry of concanavalin A beads and rotated at 4 °C for 2 h. The samples were then spun at 13,000 rpm for 2 min; the supernatant was discarded, and the concanavalin A beads were mixed with Laemmli sample loading buffer containing 100 mm 2-mercaptoethanol and boiled for 5 min to remove the bound glycoproteins. For all other blots, the cleared supernatants were removed into a separate tube and mixed with Laemmli sample loading buffer containing 100 mm 2-mercaptoethanol and boiled for 5 min. Samples were separated by SDS-PAGE on a 10% SDS-polyacrylamide gel and transferred to nitrocellulose membranes (Millipore, Temecula, CA). Membranes were blocked for 1 h with 3% milk reconstituted from dry powder in Tris-buffered saline, pH 7.4, containing 0.1% Tween 20 (TBS-T). Blots were incubated by shaking overnight at 4 °C in 1:5000 mouse monoclonal anti-phosphotyrosine, 1:5000 mouse monoclonal anti-c-Myc, 1:2000 rabbit anti-phospho-ERK1/2, or 1:1500 rabbit anti-phospho-EGFR antibodies diluted in 3% milk in TBS-T, as indicated. Subsequently, blots were washed three times in TBS-T for 10 min each and incubated with the appropriate secondary horseradish peroxidase-coupled antibody, either anti-mouse (1:5000) or anti-rabbit (1:2500) (Promega, Madison, WI) for 1 h. After three additional washes, the membranes were incubated with ECL developer (GE Healthcare), and chemiluminescence was detected and photographed using a Bio-Rad Molecular Imager Gel Doc XR System. To obtain a loading control, blots were stripped with 0.1 m glycine, pH 2.2, and reprobed with rabbit anti-ERK2 (1:2500) or rabbit anti-EGFR (1:1500).
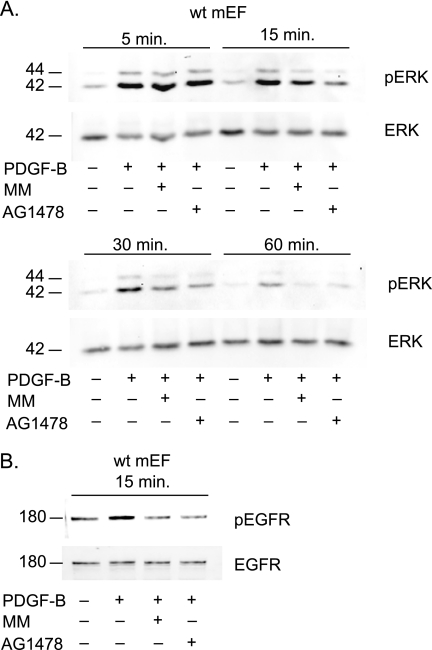
FIGURE 5.
PDGFRβ stimulates metalloproteinase and EGFR-dependent cross-talk with ERK1/2. A, Western blot of ERK1/2 phosphorylation in primary mEFs treated with or without PDGF-B (25 ng/ml) or PDGF-B and MM (4 μm) or PDGF-B and the EGFR inhibitor tyrphostin AG1478 (500 nm). A time course was performed to examine ERK1/2 phosphorylation (pERK) at different time points ranging from 5, 15, 30, and 60 min after addition of PDGF-B. Total ERK2 levels served as a loading control (ERK). Inhibition of metalloproteinases by MM and of the EGFR by AG1478 did not affect ERK1/2 phosphorylation 5 min after addition of PDGF-B, but both inhibitors slightly reduced ERK1/2 phosphorylation after 15 min, and a strong reduction was evident 30 and 60 min after addition of PDGF-B compared with cells treated only with PDGF-B. A representative sample of seven experiments with nearly identical results is shown. B, Western blot analysis of EGFR phosphorylation in mEFs treated with or without PDGF-B (25 ng/ml) or PDGF-B and MM (4 μm) or PDGF-B and AG1478 (500 nm) for 15 min (top panel). Treatment of wild type mEFs with PDGF-B stimulates phosphorylation of the EGFR, and this can be blocked by treatment with AG1478 or MM. A Western blot of total EGFR levels is included as a control (lower panel). These results, which are a representative example for three separate experiments, provide evidence for a metalloproteinase-dependent cross-talk between the PDGFRβ and EGFR/ERK1/2 signaling pathways.
Cell Migration Assays
Primary wild type mEFs were generated from E13.5 embryos as previously described (14, 23), and seeded in 6-well tissue culture dishes such that they would be confluent upon attachment to the dish. Prior to cell seeding, the bottom of the culture dishes had been labeled with a marker to ensure that pictures would be taken in exactly the same area at the beginning and end of the assay. Once cells had firmly attached to the tissue culture plates (∼8 h later), the media were replaced with Opti-MEM containing 0.4% fetal bovine serum, and the cells were incubated overnight at 37 °C. The next morning, the cell monolayer was scraped in a straight line using a p200 pipette tip (25). The cells were washed twice with Opti-MEM to remove cell debris, and 10 ng/ml PDGF-B was added to Opti-MEM containing 0.1% fetal bovine serum in the presence or absence of 4 μm marimastat or 1 μm tyrphostin AG1478. Some wells were left untreated as a negative control. Pictures of the scratches were taken immediately after they had been made (at 0 h) and after 10 h, which was the earliest time point when the primary mEF cells had completely grown over and covered the scratched area following PDGF-B stimulation. Three pictures were taken for each scratch at 0 and 10 h, and within one experiment, 3 or 4 wells were evaluated for each condition. To analyze the data, each image taken at 10 h was matched to the corresponding image taken immediately after introduction of the scratch (0 h), and vertical lines were added to the 10-h image that corresponded with the exact width of the initial scratch (shown in Fig. 6A). Using NIH Image J software to mark cell nuclei, all cells that had migrated into the wound for each image were counted in a blinded fashion. Each experiment was repeated three separate times. Statistical analysis was performed with Prism GraphPad software.
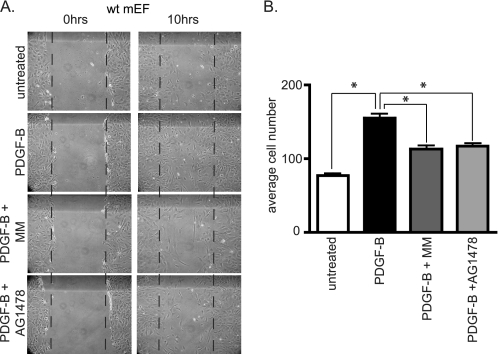
FIGURE 6.
PDGF-B-induced scratch wound healing in primary mouse embryonic fibroblasts is sensitive to the metalloproteinase inhibitor MM and the EGFR inhibitor AG1478. A, scratch wounds were introduced in confluent cultures of primary wild type mEFs and the cells were then treated with or without PDGF-B (10 ng/ml) or PDGF-B and MM (4 μm) or PDGF-B and the EGFR-inhibitor AG1478 (1 μm) for 10 h. Treatment with PDGF-B significantly increased the number of cells that were present in the scratch-wounded area (between the vertical lines) compared with untreated cells, and this increase was inhibited by MM or AG1478. B, results of three independent experiments were quantified by counting the number of cells that had entered the scratch wound under various conditions, as described under “Experimental Procedures” (n = 3 separate experiments ± S.E.). Following analysis of variance with Bonferroni post hoc analysis, p values were calculated as <0.001 between unstimulated and PDGF-B-treated samples, between PDGF-B and PDGF-B + MM samples, and between PDGF-B and PDGF-B + AG1478 samples (as denoted by asterisks). These results provide evidence for the functional relevance of the metalloproteinase-dependent cross-talk between PDGFRβ and EGFR signaling pathways.
RESULTS
To identify the ADAM responsible for shedding of the PDGFRβ, we transfected COS-7 cells with cDNA encoding an AP-tagged PDGFRβ (PDGFRβ-AP) and then quantified the shedding of the tagged receptor by measuring AP activity released into the culture supernatant (see “Experimental Procedures” for details). We found that the ectodomain of the PDGFRβ-AP expressed in COS-7 cells was shed constitutively and that stimulation of these cells with the calcium ionophore, ionomycin (IM, 2.5 μm), for 30 min increased shedding of the PDGFRβ ectodomain, whereas stimulation with 25 ng/ml phorbol 12-myristate 13-acetate (PMA) for 30 min did not (Fig. 1A). Constitutive and IM-induced shedding was reduced by the hydroxamic acid-type metalloproteinase inhibitor marimastat (MM, 4 μm) (Fig. 1A). Based on our previous studies, the stimulation of PDGFRβ-AP shedding by IM but not PMA was indicative of a role for ADAM10 in this process, because ADAM10 responds to short term stimulation (∼30–60 min) with 2.5 μm of IM but not to short term stimulation with 25 ng/ml PMA (26, 27), which activates the metalloproteinase ADAM17.
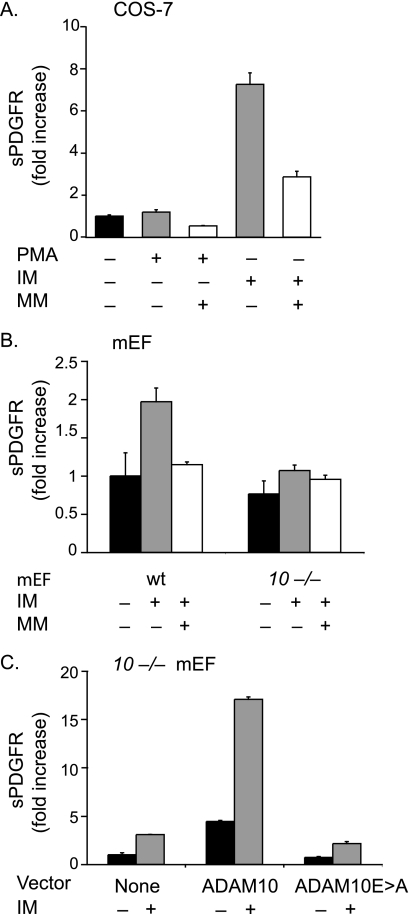
FIGURE 1.
Ectodomain shedding of PDGFRβ by ADAM10. A, shedding of PDGFRβ-AP transfected into COS-7 cells following stimulation with PMA (25 ng/ml) or ionomycin (2.5 μm) in the presence or absence of the hydroxamate-type metalloproteinase inhibitor marimastat (4 μm). Shedding of PDGFRβ-AP is stimulated by IM, but not by PMA, and the constitutive and IM-dependent shedding can be partially reduced by MM (n = 4 ± S.D.). The constitutive PDGFRβ-AP shedding was set to 1 and used as a reference point to determine the fold increase shedding of all samples. B, shedding of PDGFRβ-AP transfected into wild type (wt) or Adam10−/− (10−/−) mEFs (n = 3 ± S.D.) following stimulation with 2.5 μm IM in the presence or absence of 4 μm MM shows that the IM-stimulated component of PDGFRβ-AP shedding is strongly decreased in the absence of ADAM10. The constitutive PDGFRβ-AP shedding in wild type mEFs was set to 1 and used as a reference point to determine the relative fold increase shedding of all samples. C, shedding of PDGFRβ-AP from Adam10−/− mEFs co-transfected with or without ADAM10 or the ADAM10Glu→Ala mutant, which carries an inactivating Glu→Ala point mutation in its catalytic site, in the presence or absence of 2.5 μm IM (n = 4 ± S.D.). Please note that the differences in fold increase shedding in IM-treated samples are due to different responses of the various cell lines to these stimuli. The low constitutive shedding in Adam10−/− cells was set to 1 in C, hence the relatively high numbers for the fold increase in this panel.
To provide additional evidence for a role of ADAM10 in shedding the PDGFRβ, we compared the release of the transfected AP-tagged PDGFRβ receptor from wild type mEFs with Adam10−/− mEFs under constitutive and stimulated conditions. Shedding of PDGFRβ-AP transfected into wild type mEFs was stimulated by 2.5 μm IM and inhibited by 4 μm MM, whereas shedding of PDGFRβ-AP transfected into Adam10−/− mEFs was only weakly induced by IM (Fig. 1B). Additionally, constitutive shedding of PDGFRβ-AP from Adam10−/− mEFs could be increased by co-transfection with ADAM10 cDNA, and this could be further enhanced by addition of 2.5 μm IM (Fig. 1C). By contrast, when cDNA for the catalytically inactive ADAM10Glu→Ala mutant was co-transfected with PDGFRβ-AP in Adam10−/− mEFs, there was no increase in constitutive or stimulated shedding (Fig. 1C). These experiments in Adam10−/− mEFs further corroborate that constitutive and IM-stimulated shedding of PDGFRβ-AP depends on ADAM10.
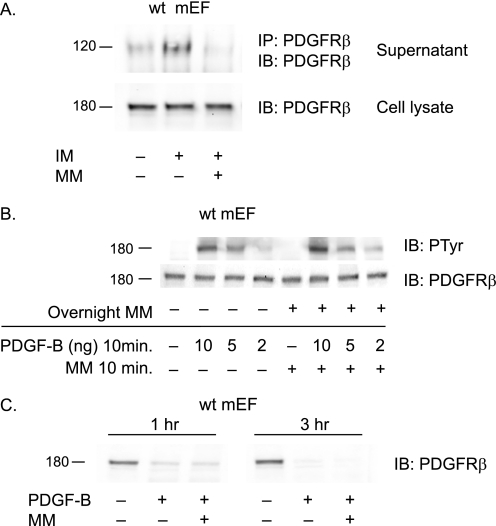
Next, we were interested in determining whether shedding had a detectable effect on the function of the PDGFRβ receptor itself. To corroborate that the endogenous PDGFRβ is shed from wild type mEFs, we performed an immunoprecipitation followed by Western blot analysis of soluble PDGFRβ released from mEFs (Fig. 2A). Shedding of the endogenous PDGFRβ from wild type mEFs was enhanced by treatment with 2.5 μm IM, and this could be blocked by addition of 4 μm MM (Fig. 2A, upper panel). However, a Western blot analysis of the full-length PDGFRβ present in the corresponding cell lysates showed that IM-stimulated shedding did not detectably reduce the levels of the cell-associated form of this receptor (Fig. 2A, lower panel). This suggests that only a relatively small percentage of the total amount of PDGFRβ is shed from mEFs treated with IM. In addition, overnight preincubation of wild type mEFs with 4 μm MM to block ADAM10-dependent constitutive release of the PDGFRβ did not detectably increase the levels of the endogenous receptor compared with untreated cells (Fig. 2B, lower panel). Moreover, overnight treatment with 4 μm MM also did not detectably affect the response of the PDGFRβ to treatment with varying amounts of PDGF-B (10, 5, and 2 ng/ml) for 10 min compared with cells that had not been treated with MM, as measured by phosphorylation of the PDGFRβ (Fig. 2B, upper panel). Finally, to assess whether ectodomain shedding contributes to the previously reported down-regulation of the PDGFRβ that is seen between 30 min and 1 h after addition of PDGF-B (28, 29), wild type mEFs were treated with or without 50 ng/ml of PDGF-B in the presence or absence of 4 μm MM for 1 or 3 h. No evidence for an effect of MM on ligand-induced receptor down-regulation was found under these conditions (Fig. 2C). Taken together, these findings argue against a role for ectodomain shedding of the PDGFRβ in regulating its turnover or response to stimulation with PDGF-B in mEF cells.
FIGURE 2.
Ectodomain shedding does not detectably affect PDGFRβ protein levels or its phosphorylation in response to PDGF-B. A, stimulation of wild type mEFs with 2.5 μm IM increased shedding of the endogenously expressed PDGFRβ into the culture supernatant, as detected by Western blot (IB) analysis of the immunoprecipitated (IP) receptor, and the increased shedding could be inhibited by 4 μm MM (top panel). However, there was no corresponding decrease in the levels of the endogenous PDGFRβ in the cell lysate of wild type mEFs stimulated with IM in the presence or absence of MM. B, to test whether long term inhibition of constitutive PDGFRβ shedding could increase receptor levels in mEF cells, thereby perhaps rendering these cells more sensitive to stimulation with PDGF-B, wild type mEFs were incubated overnight with 4 μm MM, then treated with or without different concentrations of PDGF-B, followed by Western blot analysis with antibodies against the PDGFRβ (lower panel) or against phosphotyrosine (PTyr) (upper panel). Overnight incubation with 4 μm MM did not lead to a detectable increase in the levels of the PDGFRβ (lower panel) and also did not change PDGFRβ phosphorylation in response to the addition of varying amounts of PDGF-B (10, 5, and 2 ng/ml). C, to test whether shedding of the PDGFRβ has a role in down-regulation of this receptor following stimulation with PDGF-B, wild type mEFs were treated with or without 50 ng/ml of PDGF-B in the presence or absence of 4 μm MM for 1 or 3 h. Western blot analysis of the PDGFRβ in cell lysates showed down-regulation at 1 and 3 h following the addition of PDGF-B, but this receptor down-regulation was not inhibited by MM, arguing against a role for ectodomain shedding in the ligand-dependent down-regulation of PDGFRβ. Each Western blot is a representative example of the results of at least three separate experiments.
Because we had previously observed that stimulation of the VEGFR2 by its ligand VEGF-A activated ADAM17, thereby causing shedding of the VEGFR2 and other substrates of ADAM17 (16), we tested whether activation of the PDGFRβ by PDGF-B could perhaps stimulate ADAM10-dependent shedding of the alkaline phosphatase-tagged PDGFRβ-AP or of other substrates of ADAM10. However, when COS-7 cells were transfected with the full-length wild type PDGFRβ and PDGFRβ-AP and stimulated with PDGF-B (50 ng/ml) for 30 min, no increase in the shedding of PDGFRβ-AP was observed (Fig. 3A). Moreover, when full-length wild type PDGFRβ was transfected into COS-7 cells together with another substrate for ADAM10, EGF-AP (14, 26), no increase in the shedding of this substrate was seen in cells stimulated with PDGF-B as compared with the corresponding unstimulated controls (Fig. 3A). Finally, stimulation of wild type mEFs with PDGF-B did not increase the shedding of the endogenous PDGFRβ (data not shown). Thus, there was no evidence for activation of ADAM10 by ligand-induced stimulation of the PDGFRβ, at least in COS-7 and mEF cells under the conditions used here.
FIGURE 3.
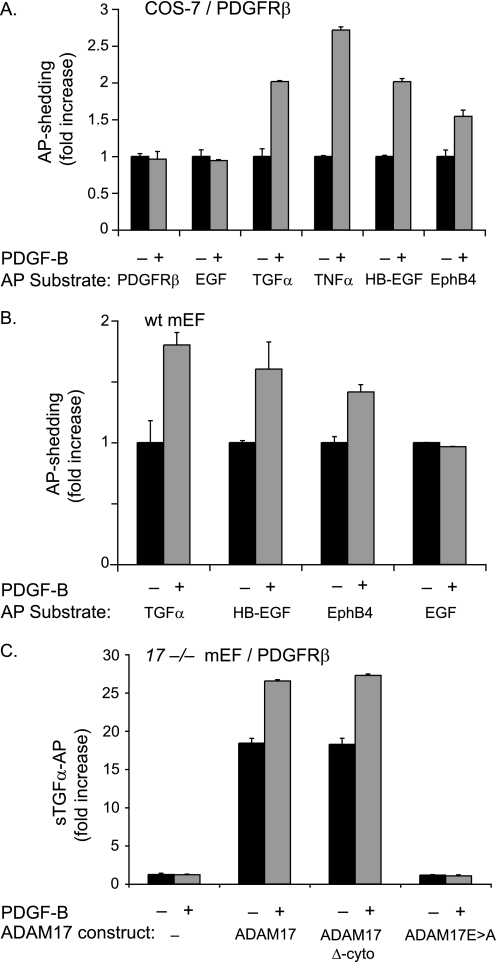
PDGF-B binding to its receptor PDGFRβ does not activate shedding of PDGFRβ-AP or of the ADAM10 substrate EGF but causes shedding of substrates of ADAM17. A, COS-7 cells were co-transfected with full-length PDGFRβ and the following alkaline phosphatase-tagged membrane-anchored substrate proteins: PDGFRβ, EGF, TGFα, TNFα, HB-EGF, and EphB4. The day following transfection, cells were starved by incubation in serum-free medium (Opti-MEM) for 6 h and then incubated for 30 min in the presence or absence of PDGF-B (50 ng/ml). PDGF-B did not stimulate the shedding of the ADAM10 substrates PDGFRβ or EGF but stimulated shedding of all ADAM17 substrates tested here (TGFα, TNFα, HB-EGF, and EphB4) (n = 6 ± S.D.). B, wild type (wt) mEFs were transfected with alkaline phosphatase-tagged TGFα, HB-EGF, EphB4, or EGF and treated with or without 50 ng/ml PDGF-B for 30 min (as in A). PDGF-B stimulation of wild type mEFs, which express the PDGFRβ endogenously, activated shedding of the ADAM17 substrates TGFα, HB-EGF, and EphB4 but not of the ADAM10 substrate EGF (n = 5 ± S.D.). The constitutive shedding (for 30 min) for each AP-tagged substrate in A and B was set to 1 to provide a reference point for the shedding in the presence of PDGF-B (30 min). C, Adam17−/− mEFs (17−/− mEF) transfected with full-length PDGFRβ and TGFα either alone or with wild type ADAM17, ADAM17 lacking its cytoplasmic domain (ADAM17 Δ-cyto), or the catalytically inactive ADAM17Glu→Ala. Cells were stimulated with 50 ng/ml PDGF-B for 30 min or left untreated (n = 6 ± S.D.). The stimulation of TGFα shedding by PDGF-B/PDGFRβ requires a functional catalytic site of ADAM17 but not its cytoplasmic domain. The low level of constitutive shedding of TGFα in untreated Adam17−/− mEFs was set to 1 and used as a reference in C.
To test whether stimulation of the PDGFRβ activates ADAM17, we co-transfected COS-7 cells with wild type full-length PDGFRβ and AP-tagged substrates for ADAM17 and stimulated these cells with PDGF-B (50 ng/ml) for 30 min. The addition of PDGF-B induced shedding of all AP-tagged ADAM17 substrates tested here, including transforming growth factor α (TGFα-AP), tumor necrosis factor α (TNFα-AP), heparin-binding epidermal growth factor (HB-EGF-AP), and EphB4-AP (Fig. 3A). The increase in shedding of the ADAM17 AP substrates following PDGF-B stimulation was comparable with that observed following activation of ADAM17 by 25 ng/ml PMA (data not shown). To extend our analysis, wild type mEFs, which endogenously express PDGFRβ (see Fig. 2), were transfected with the ADAM17 substrates TGFα-AP, HB-EGF-AP, and EphB4-AP or the ADAM10 substrate EGF-AP. Stimulation of wild type mEFs with PDGF-B induced shedding of all the ADAM17 substrates but not of the ADAM10 substrate EGF-AP (Fig. 3B). Thus, ligand-dependent activation of the endogenous PDGFRβ in mEFs also activates ADAM17.
To further confirm that PDGF-B/PDGFRβ-stimulated shedding of TGFα depends on activation of ADAM17, experiments were performed in Adam17−/− mEFs. After co-transfection of the wild type PDGFRβ (to optimize the response to stimulation by PDGF-B) and TGFα, used as a representative ADAM17 substrate, and stimulation with PDGF-B, no changes were observed in the low levels of TGFα shedding from these cells (Fig. 3C). However, when wild type ADAM17 was co-transfected with the wild type PDGFRβ and TGFα into Adam17−/− mEFs, constitutive shedding was strongly increased, and PDGF-B induction of TGFα shedding was restored (Fig. 3C). Because activation of ADAM17 by PMA does not require its cytoplasmic domain (26, 27, 30), we tested whether the PDGF-B/PDGFRβ-dependent stimulation of TGFα shedding requires the cytoplasmic domain of ADAM17. We found that transfection of Adam17−/− mEFs with ADAM17 lacking its cytoplasmic domain (ADAM17 Δ-cyto) along with PDGFRβ and TGFα was able to restore constitutive and PDGF-B-stimulated shedding of TGFα-AP to a similar extent as rescue with wild type ADAM17. As a control, transfection of Adam17−/− mEFs with the catalytically inactive ADAM17Glu→Ala mutant and PDGFRβ and TGFα did not rescue PDGF-B-stimulated shedding of TGFα (Fig. 3C).
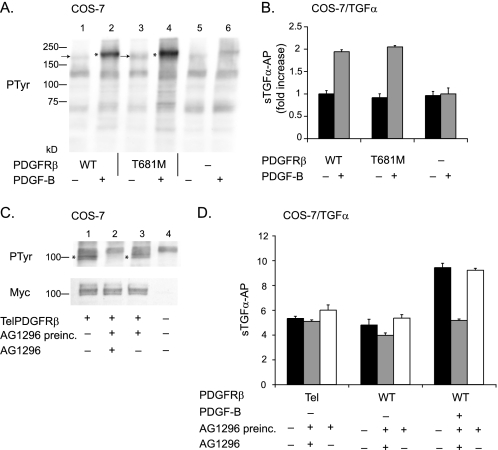
The ability of PDGF-B to activate ADAM17 raised questions about whether constitutively active forms of the PDGFRβ might chronically stimulate ADAM17. To test this possibility, we first examined whether a form of the PDGFRβ with a mutation of the gatekeeper threonine (T681M), which has been shown to activate its tyrosine kinase domain (22), can stimulate ADAM17. However, we only found a small increase in constitutive phosphorylation of this receptor by Western blot analysis with anti-phosphotyrosine antibodies compared with the wild type receptor in unstimulated cells that had been starved by incubation in serum-free medium (Opti-MEM) for 6 h (Fig. 4A, compare the bands marked with an arrow in lanes 1 and 3). Moreover, the increase in phosphorylation of the PDGFRβ T681M gatekeeper mutant in response to PDGF-B (50 ng/ml) was similar to that observed for the wild type receptor (Fig. 4A, the phosphorylated receptors are marked with an asterisk). Untransfected COS-7 cells treated with or without PDGF-B are shown as controls in Fig. 4A, lanes 5 and 6. Finally, in shedding experiments with TGFα, we observed similar levels of constitutive and PDGF-B-stimulated shedding from cells transfected with the wild type PDGFRβ or PDGFRβ T681M (Fig. 4B), suggesting that this mutation does not detectably activate ADAM17. COS-7 cells transfected with only TGFα showed similar levels of constitutive shedding but lacked the PDGF-B-stimulated component of shedding, demonstrating that the response to PDGF-B required co-transfection with the wild type or T681M mutant PDGFRβ.
FIGURE 4.
Constitutively active forms of PDGFRβ, one of which is highly phosphorylated, do not stimulate ADAM17. Two constitutively active forms of the PDGFRβ, the gatekeeper mutant (T681M) (A and B), and the TelPDGFRβ (C and D) were tested for their ability to activate ADAM17-dependent shedding of TGFα. A, Western blot of tyrosine phosphorylation (PTyr) of the wild type (WT) receptor and gatekeeper mutant in the presence or absence of PDGF-B, as indicated. Lanes 1–4 show samples of COS-7 cells transfected with wild type PDGFRβ (lanes 1 and 2) or the PDGFRβ T681M (lanes 3 and 4) and left unstimulated (lanes 1 and 3) or stimulated with 50 ng/ml PDGF-B (lanes 2 and 4). Samples from untransfected COS-7 cells that were not treated (lane 5) or treated with 50 ng/ml PDGF-B (lane 6) are shown as controls. The arrows next to lanes 1 and 3 point to the unstimulated wild type PDGFRβ and PDGFRβ T681M mutant, and the asterisks next to lanes 2 and 4 indicate the position of the phosphorylated forms of these receptors after treatment with 50 ng/ml PDGF-B. The phosphorylation of the PDGFRβ T681M mutant in unstimulated cells is only slightly increased compared with the wild type receptor, and the relative increase in phosphorylation upon stimulation with PDGF-B is similar for the wild type and PDGFRβ T681M mutant. B, comparison of TGFα shedding from COS-7 cells transfected with wild type PDGFRβ or PDGFRβ T681M showed no significant difference in constitutive or PDGF-B-stimulated conditions (n = 6 ± S.D.). There was no PDGF-B stimulation of TGFα shedding in COS-7 cells that were not co-transfected with the wild type PDGFRβ. In this graph, the unstimulated shedding of TGFα in cells expressing the wild type receptor was set to 1 and used as a reference to calculate the fold increase in shedding for the other samples. C, Western blot analysis of COS-7 cells transfected with the Myc-tagged TelPDGFRβ probed with an anti-Myc antibody (lower panel) shows comparable expression of this receptor in transfected cells (lanes 1–3) and no expression in untransfected control cells (lane 4). The band detected by the anti-Myc antibodies migrates as a doublet of ∼100 kDa, as described previously for the TelPDGRFβ (20, 21). Tyrosine phosphorylation of the TelPDGFRβ was detected with an anti-phosphotyrosine antibody in samples from transfected but untreated cells (upper panel, lane 1), and this was inhibited following 1 h of preincubation with 20 μm of the PDGFRβ-selective tyrosine kinase inhibitor AG1296 (upper panel, lane 2). When the cells were first incubated with 20 μm AG1296 followed by washing to remove the inhibitor, TelPDGFRβ phosphorylation was restored (top panel, lane 3). D, functional analysis of the ability of the TelPDGFRβ to activate ADAM17 using TGFα shedding assays in COS-7 cells co-transfected with TelPDGFRβ or the wild type PDGFRβ. There was no significant difference in the constitutive shedding of TGFα in COS-7 cells expressing TelPDGFRβ compared with the wild type PDGFRβ in the presence (gray bar) or absence of AG1296 (black bar) or following washout of AG1296 after preincubation for 1 h (white bar). The addition of PDGF-B to COS-7 cells expressing the wild type PDGFRβ and TGFα resulted in a strong increase in TGFα shedding that was blocked by AG1296 but was restored following washout of AG1296. The experiments with the ligand-stimulated wild type PDGFRβ provide a positive control for the ability of AG1296 to block the function of the PDGFRβ and of the reversibility of this inhibition when AG1296 is washed out following preincubation.
Next, we examined TelPDGFRβ, an oncogenic fusion protein found in patients with chronic myelomonocytic leukemia, which contains the N-terminal 154 amino acids of the Tel transcription factor fused to the transmembrane and cytoplasmic regions of the PDGFRβ (20, 21). The ligand-binding domain of PDGFRβ is missing in this fusion protein, but the entire tyrosine kinase domain is present and constitutively tyrosine-phosphorylated, without the presence of its ligand (20, 21). A Western blot analysis corroborated that TelPDGFRβ was constitutively phosphorylated in transfected COS-7 cells (Fig. 4C, band marked with an asterisk in lane 1). Nevertheless, there was no increase in the constitutive shedding of the ADAM17 substrate TGFα in COS-7 cells expressing TelPDGFRβ compared with cells expressing the wild type receptor without added PDGF-B (Fig. 4D, black bars). Because the TelPDGFRβ is constitutively active, we preincubated cells expressing TelPDGFRβ with a selective inhibitor of PDGFRβ kinase activity, AG1296, to generate conditions that would more closely resemble unstimulated cells. Then the AG1269 inhibitor was washed out to restore phosphorylation of the TelPDGFRβ, with the goal of replicating the relative increase in phosphorylation that is observed when the wild type PDGFRβ is stimulated with PDGF-B. COS-7 cells transfected with TelPDGFRβ and TGFα were preincubated in 20 μm AG1296 in Opti-MEM for 1 h. Following a brief wash to remove the inhibitor, the culture supernatant was collected for 1 h either in the continued presence of 20 μm AG1296 or in the absence of this inhibitor. A Western blot analysis showed that tyrosine phosphorylation of the TelPDGFRβ was completely inhibited by preincubation with AG1296 (Fig. 4C, lane 2), and washing out AG1296 restored phosphorylation of the TelPDGFRβ to normal levels (Fig. 4C, lane 3). Despite the efficient inhibition of TelPDGFRβ by AG1296, we did not see a significant decrease in shedding of TGFα in the continued presence of AG1296 (Fig. 4D, gray bar) or an increase in shedding after the inhibitor was removed and phosphorylation of the mutant receptor was restored (Fig. 4D, white bar). Similar results were obtained when COS-7 cells were transfected with the wild type PDGFRβ and TGFα but not stimulated with PDGF-B. When PDGF-B (50 ng/ml) was added to COS-7 cells expressing the wild type PDGFRβ and TGFα, there was an increase in TGFα shedding, which could be blocked by 20 μm AG1296 but restored following washing out of this inhibitor (Fig. 4D). These results provide a positive control that AG1296 effectively blocked activation of ADAM17 by the wild type PDGFRβ and that the inhibitor could be rapidly removed by washing. Thus, we found no evidence for the ability of transforming mutants of the PDGFRβ to activate ADAM17 in a ligand-independent manner.
Because VEGF-A/VEGFR2 signaling activates ADAM17-dependent stimulation of the ERK and MAPK pathways (16), we tested whether stimulation of the endogenous PDGFRβ in wild type mEFs, which also activates ADAM17 (see Fig. 2B), activates ERK through metalloproteinase-dependent cross-talk with the EGFR. A time course was performed where primary wild type mEFs were stimulated with 25 ng/ml PDGF-B in the presence or absence of 4 μm MM or 500 nm of the EGFR-specific tyrosine kinase inhibitor AG1478, and then the relative levels of ERK1/2 phosphorylation were determined by Western blot analysis. A strong activation of ERK1/2 was observed within 5 min of addition of PDGF-B, and this initial rapid response was not inhibited by MM or AG1478 (Fig. 5A, top panel). However, at later time points, starting at 15 min after addition of PDGF-B, and most evident 30 and 60 min thereafter, the phosphorylation of ERK1/2 was significantly reduced by both MM and AG1478, suggesting that the PDGFRβ also activates metalloproteinase-dependent cross-talk with ERK1/2 via activation of the EGFR. To further corroborate that this metalloproteinase-dependent cross-talk involves stimulation of the EGFR, wild type mEFs were treated with PDGF-B for 15 min, and then probed with an antibody against the phosphorylated tyrosine residue 1068 of the EGFR, which represents a binding site for Grb2 and is involved in activation of the MAPK signaling cascade. Stimulation of mEFs with PDGF-B elicited increased phosphorylation of Tyr1068 on the EGFR, which could be blocked by addition of MM or AG1478 (Fig. 5B).
To assess the potential physiological role for the metalloproteinase-dependent cross-talk between the PDGFRβ and the EGFR/ERK1/2 signaling pathways, we performed an in vitro scratch wound healing assay using primary wild type mEFs (25). A scratch wound was introduced into a confluent monolayer of wild type mEFs, and the number of cells that had entered a defined surface area within the scratch wound after treatment with or without PDGF-B in the presence or absence of MM or AG1478 was counted. Treatment with 10 ng/ml PDGF-B led to an almost complete healing of the scratch wound after 10 h, whereas untreated cells had only partially healed the scratch wound at this time point (Fig. 6A, quantification of three independent experiments is shown in Fig. 6B). The addition of 4 μm MM or 1 μm AG1478 to PDGF-B-treated cells significantly decreased the number of cells that entered the scratch wound in response to treatment with PDGF-B (Fig. 6). These results provide evidence for the functional relevance of the metalloproteinase-dependent cross-talk between the PDGFRβ and the EGFR for scratch wound healing assays with primary mEF cells.
DISCUSSION
Here, we show that the PDGFRβ, which has an important role in recruitment of pericytes to stabilize endothelial cells, is shed from the plasma membrane, and we identify ADAM10 as the responsible enzyme in mEF cells. Moreover, we show that PDGF-B-dependent stimulation of the PDGFRβ activates ADAM17, triggers metalloproteinase-dependent cross-talk between the PDGFRβ and the EGFR/ERK1/2 signaling pathway, and is critical for PDGF-B-stimulated cell migration of primary mEF cells. However, stimulation of the PDGFRβ with PDGF-B does not appear to activate ADAM10 under the conditions tested here, suggesting that shedding of the PDGFRβ itself is not regulated by ligand binding. Nevertheless, shedding of the PDGFRβ by ADAM10 could be important for regulating the levels of this receptor in cells and tissues where it is expressed in vivo, such as in pericytes. Future studies in conditional knock-out mice lacking ADAM10 in pericytes could address this issue, as could the generation of mice harboring an uncleavable form of the PDGFRβ.
Although we did not find evidence for an activation of ADAM10 by the PDGFRβ, we observed a significant stimulation of ADAM17 upon activation of the PDGFRβ. These results were similar to the activation of ADAM17 in response to VEGF-A/VEGFR2 signaling (16). The cytoplasmic domain of ADAM17 is dispensable for its activation by the PDGFRβ, further corroborating that the activation of ADAM17 does not require cytoplasmic phosphorylation, as previously shown for stimulation with the phorbol ester PMA (26, 30). Additional studies will be necessary to understand how the PDGFRβ and other cellular signaling pathways activate ADAM17 in the absence of its cytoplasmic domain.
The ability of the PDGF-B/PDGFRβ signaling axis to stimulate ADAM17 raised questions about whether constitutively active forms of the PDGFRβ can also enhance ADAM17-dependent shedding. We found that a constitutively active form of the PDGFRβ containing a mutation in the gatekeeper threonine (T681M) (22) did not activate shedding of the ADAM17 substrate TGFα compared with the unstimulated wild type receptor. However, the phosphorylation of the unstimulated PDGFRβ T681M mutant was only slightly increased compared with the unstimulated wild type PDGFRβ. Moreover, phosphorylation of the mutant and wild type receptor could be similarly enhanced upon ligand binding, and this stimulated ADAM17 to a comparable degree. These results suggest that the gatekeeper mutation does not sufficiently activate the PDGFRβ to affect the activity of ADAM17, at least under the conditions used in this study. When we examined COS-7 cells transfected with the constitutively active TelPDGFRβ, an oncogenic fusion protein found in patients with chronic myelomonocytic leukemia (20, 21), we also found no increase in the shedding of the ADAM17 substrate TGFα compared with unstimulated cells expressing the wild type PDGFRβ. It is important to note that the TelPDGFRβ fusion protein lacks an N-terminal signal sequence and thus is not properly anchored in the plasma membrane (31), which could provide an explanation for its inability to activate ADAM17. Nevertheless, these results do not rule out that a more subtle stimulation of ADAM17 by these mutants could occur in vivo, which cannot be measured in our assays, but could nevertheless possibly contribute to cancer initiation or progression in vivo.
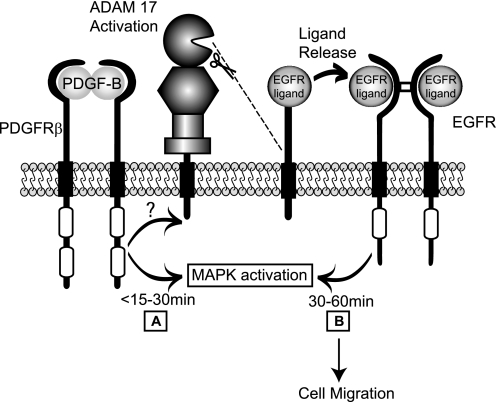
The results presented here also provide the first evidence for metalloproteinase-dependent cross-talk between the PDGFRβ and the EGFR/ERK1/2 signaling pathway, which is most likely due to the activation of ADAM17. Because this cross-talk resembles that initiated by VEGF-A/VEGFR2 (16), it is tempting to speculate that ADAM17-dependent cross-talk with the EGFR could be a more general aspect of receptor tyrosine kinase signaling pathways. A conceptually similar cross-talk between G protein-coupled receptors (GPCRs) and the EGFR also relies on activation of ADAMs (32–35). This well characterized GPCR/EGFR cross-talk is referred to as a “triple membrane-passing signal” because it occurs when a GPCR activates an ADAM, which releases membrane-anchored EGFR ligands that then stimulate the EGFR and ERK1/2 (6, 34, 35). The main difference between GPCR-stimulated cross-talk with ERK1/2 and the cross-talk initiated by the PDGFRβ and the VEGFR2 is that the latter two tyrosine kinase receptors are able to activate ERK1/2 in an initially metalloproteinase-independent manner, most likely through direct intracellular signaling pathways, whereas GPCR-dependent cross-talk with ERK1/2 depends entirely on metalloproteinases. For the PDGFRβ and VEGFR2, only the second phase of ERK1/2 activation between about 30 and 60 min depends on metalloproteinases (Fig. 7).
FIGURE 7.
Model for metalloproteinase-dependent cross-talk between the PDGFRβ and the EGFR/ERK1/2 signaling pathway. Based on the results of this study, we propose a model in which binding of PDGF-B to the PDGFRβ causes a biphasic activation of ERK1/2 with an initial response triggered via PDGFRβ-dependent stimulation of intracellular signaling pathways (A) and the second response by activation of ADAM17 (B). This second response results in the processing and release of membrane-anchored EGFR ligands, allowing them to bind to and activate the EGFR. The metalloproteinase-dependent activation of the EGFR is responsible for the extended duration of ERK1/2 phosphorylation between about 30 and 60 min after addition of PDGF-B and for ligand-induced cell migration (B).
To assess potential functional consequences of the cross-talk between the PDGFRβ and the EGFR/ERK, we tested whether PDGF-B-stimulated migration of primary mouse embryonic fibroblasts was affected by the metalloproteinase inhibitor marimastat or the EGFR-selective tyrosine kinase inhibitor AG1478. We found that both inhibitors significantly reduced the PDGF-B-stimulated cell migration, providing evidence for the functional relevance of a triple membrane-passing cross-talk that depends on the activation of a metalloproteinase (most likely ADAM17) and the EGFR.
Taken together, these results provide new insights into the regulation of PDGFRβ signaling by ectodomain shedding. ADAM10 emerged as the major sheddase of the PDGFRβ, but we did not find evidence for an activation of ADAM10 by the PDGFRβ or for a role of shedding in regulating turnover of the PDGFRβ or its response to PDGF-B. Instead, we found that the PDGFRβ activates ADAM17, thereby stimulating the release of EGFR ligands and other substrates of ADAM17, which in turn is likely responsible for the cross-talk between the PDGFRβ and EGFR/ERK and for PDGF-B-stimulated cell migration. Collectively, our results provide new information on the mechanism underlying PDGFRβ-dependent activation of ERK1/2 and demonstrate that the PDGFRβ signaling pathway is a physiological activator of ADAM17. These results suggest that inhibitors of ADAM17, which are being developed for treatment of cancer (36), are likely to attenuate activation of EGFR/ERK1/2 signaling by PDGFRβ in addition to the VEGFR2 (16) and possibly also other tyrosine kinase receptors.
Acknowledgment
The TelPDGFRβ cDNA was kindly provided by Dr. Michael Tomasson from Washington University, St. Louis.
Note Added in Proof
Previous studies by Lehti et al. ((2005) Genes Dev. 19, 979–991) have identified the membrane-type 1 matrix metalloproteinase (MT1-MMP) as a proteolytic modifier of PDGF-B/PDGFRβ signal transduction and activation of ERK1/2 in vascular smooth muscle cells so it will be interesting to further dissect the mechanisms underlying the contribution of MT1-MMP and ADAM17 to this process in different cell types.
This work was supported, in whole or in part, by National Institutes of Health Grants GM64750 and EY015719 (to C. P. B.) and the Deutsche Forschungsgemeinschaft SFB 877 and the European Union-DeZenit (to P. S.).
- PDGF-B
- platelet-derived growth factor-B
- AP
- alkaline phosphatase
- EGF
- epidermal growth factor
- EGFR
- epidermal growth factor receptor
- HB-EGF
- heparin-binding epidermal growth factor
- IM
- ionomycin
- mEF
- mouse embryonic fibroblast
- MM
- marimastat
- PDGFRβ
- platelet-derived growth factor receptor β
- PMA
- phorbol 12-myristate 13-acetate
- TGFα
- transforming growth factor α
- TNFα
- tumor necrosis factor α
- VEGF-A
- vascular endothelial growth factor-A
- VEGFR2
- vascular endothelial growth factor receptor 2
- GPCR
- G protein-coupled receptor
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Lindahl P., Karlsson L., Hellström M., Gebre-Medhin S., Willetts K., Heath J. K., Betsholtz C. (1997) Development 124, 3943–3953 [DOI] [PubMed] [Google Scholar]
- 2.Hellström M., Kalén M., Lindahl P., Abramsson A., Betsholtz C. (1999) Development 126, 3047–3055 [DOI] [PubMed] [Google Scholar]
- 3.Kelly J. D., Haldeman B. A., Grant F. J., Murray M. J., Seifert R. A., Bowen-Pope D. F., Cooper J. A., Kazlauskas A. (1991) J. Biol. Chem. 266, 8987–8992 [PubMed] [Google Scholar]
- 4.Kazlauskas A., Cooper J. A. (1989) Cell 58, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 5.Tallquist M., Kazlauskas A. (2004) Cytokine Growth Factor Rev. 15, 205–213 [DOI] [PubMed] [Google Scholar]
- 6.Blobel C. P. (2005) Nat. Rev. Mol. Cell. Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 7.Murphy G. (2008) Nat. Rev. Cancer 8, 929–941 [DOI] [PubMed] [Google Scholar]
- 8.Moss M. L., Jin S. L., Milla M. E., Bickett D. M., Burkhart W., Carter H. L., Chen W. J., Clay W. C., Didsbury J. R., Hassler D., Hoffman C. R., Kost T. A., Lambert M. H., Leesnitzer M. A., McCauley P., McGeehan G., Mitchell J., Moyer M., Pahel G., Rocque W., Overton L. K., Schoenen F., Seaton T., Su J. L., Becherer J. D. (1997) Nature 385, 733–736 [DOI] [PubMed] [Google Scholar]
- 9.Black R. A., Rauch C. T., Kozlosky C. J., Peschon J. J., Slack J. L., Wolfson M. F., Castner B. J., Stocking K. L., Reddy P., Srinivasan S., Nelson N., Boiani N., Schooley K. A., Gerhart M., Davis R., Fitzner J. N., Johnson R. S., Paxton R. J., March C. J., Cerretti D. P. (1997) Nature 385, 729–733 [DOI] [PubMed] [Google Scholar]
- 10.Horiuchi K., Kimura T., Miyamoto T., Takaishi H., Okada Y., Toyama Y., Blobel C. P. (2007) J. Immunol. 179, 2686–2689 [DOI] [PubMed] [Google Scholar]
- 11.Jackson L. F., Qiu T. H., Sunnarborg S. W., Chang A., Zhang C., Patterson C., Lee D. C. (2003) EMBO J. 22, 2704–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peschon J. J., Slack J. L., Reddy P., Stocking K. L., Sunnarborg S. W., Lee D. C., Russell W. E., Castner B. J., Johnson R. S., Fitzner J. N., Boyce R. W., Nelson N., Kozlosky C. J., Wolfson M. F., Rauch C. T., Cerretti D. P., Paxton R. J., March C. J., Black R. A. (1998) Science 282, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 13.Sahin U., Blobel C. P. (2007) FEBS Lett. 581, 41–44 [DOI] [PubMed] [Google Scholar]
- 14.Sahin U., Weskamp G., Kelly K., Zhou H. M., Higashiyama S., Peschon J., Hartmann D., Saftig P., Blobel C. P. (2004) J. Cell Biol. 164, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunnarborg S. W., Hinkle C. L., Stevenson M., Russell W. E., Raska C. S., Peschon J. J., Castner B. J., Gerhart M. J., Paxton R. J., Black R. A., Lee D. C. (2002) J. Biol. Chem. 277, 12838–12845 [DOI] [PubMed] [Google Scholar]
- 16.Swendeman S., Mendelson K., Weskamp G., Horiuchi K., Deutsch U., Scherle P., Hooper A., Rafii S., Blobel C. P. (2008) Circ. Res. 103, 916–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weskamp G., Schlöndorff J., Lum L., Becherer J. D., Kim T. W., Saftig P., Hartmann D., Murphy G., Blobel C. P. (2004) J. Biol. Chem. 279, 4241–4249 [DOI] [PubMed] [Google Scholar]
- 18.Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., von Figura K., Saftig P. (2002) Hum. Mol. Genet. 11, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 19.Zheng Y., Schlondorff J., Blobel C. P. (2002) J. Biol. Chem. 277, 42463–42470 [DOI] [PubMed] [Google Scholar]
- 20.Carroll M., Tomasson M. H., Barker G. F., Golub T. R., Gilliland D. G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14845–14850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cain J. A., Grisolano J. L., Laird A. D., Tomasson M. H. (2004) Blood 104, 561–564 [DOI] [PubMed] [Google Scholar]
- 22.Azam M., Seeliger M. A., Gray N. S., Kuriyan J., Daley G. Q. (2008) Nat. Struct. Mol. Biol. 15, 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahin U., Weskamp G., Zheng Y., Chesneau V., Horiuchi K., Blobel C. P. (2006) in Epidermal Growth Factor: Methods and Protocols (Patel T. B., Bertics P. J. eds) Vol. 327, pp. 99–113, Humana Press Inc., Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 24.Weskamp G., Krätzschmar J., Reid M. S., Blobel C. P. (1996) J. Cell Biol. 132, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang C., Park A., Guan J. (2007) Nat. Protocols 2, 329–333 [DOI] [PubMed] [Google Scholar]
- 26.Horiuchi K., Le Gall S., Schulte M., Yamaguchi T., Reiss K., Murphy G., Toyama Y., Hartmann D., Saftig P., Blobel C. P. (2007) Mol. Biol. Cell 18, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Gall S. M., Bobé P., Reiss K., Horiuchi K., Niu X. D., Lundell D., Gibb D. R., Conrad D., Saftig P., Blobel C. P. (2009) Mol. Biol. Cell 20, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claesson-Welsh L., Rönnstrand L., Heldin C. H. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 8796–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keating M. T., Williams L. T. (1987) J. Biol. Chem. 262, 7932–7937 [PubMed] [Google Scholar]
- 30.Reddy P., Slack J. L., Davis R., Cerretti D. P., Kozlosky C. J., Blanton R. A., Shows D., Peschon J. J., Black R. A. (2000) J. Biol. Chem. 275, 14608–14614 [DOI] [PubMed] [Google Scholar]
- 31.Toffalini F., Hellberg C., Demoulin J. B. (2010) J. Biol. Chem. 285, 12268–12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prenzel N., Zwick E., Daub H., Leserer M., Abraham R., Wallasch C., Ullrich A. (1999) Nature 402, 884–888 [DOI] [PubMed] [Google Scholar]
- 33.Gschwind A., Hart S., Fischer O. M., Ullrich A. (2003) EMBO J. 22, 2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schäfer B., Gschwind A., Ullrich A. (2004) Oncogene 23, 991–999 [DOI] [PubMed] [Google Scholar]
- 35.Fischer O. M., Hart S., Gschwind A., Ullrich A. (2003) Biochem. Soc. Trans. 31, 1203–1208 [DOI] [PubMed] [Google Scholar]
- 36.Zhou B. B., Peyton M., He B., Liu C., Girard L., Caudler E., Lo Y., Baribaud F., Mikami I., Reguart N., Yang G., Li Y., Yao W., Vaddi K., Gazdar A. F., Friedman S. M., Jablons D. M., Newton R. C., Fridman J. S., Minna J. D., Scherle P. A. (2006) Cancer Cell 10, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]