Abstract
Polarity is a fundamental cellular feature that is critical for generating cell diversity and maintaining organ functions during development. In C. elegans, the one-cell embryo is polarized via asymmetric localization of the PAR proteins, which in turn are required to establish the future anterior-posterior axis of the embryo. PAR-3, a conserved PDZ domain-containing protein, acts with PAR-6 and PKC-3 (atypical protein kinase; aPKC) to regulate cell polarity and junction formation in a variety of cell types. To understand how PAR-3 localizes and functions during C. elegans development, we produced targeted mutations and deletions of conserved domains of PAR-3 and examined the localization and function of the GFP-tagged proteins in C. elegans embryos and larvae. We find CR1, the PAR-3 self-oligomerization domain, is required for PAR-3 cortical distribution and function only during early embryogenesis and that PDZ2 is required for PAR-3 to accumulate stably at the cell periphery in early embryos and at the apical surface in pharyngeal and intestinal epithelial cells. We also show that phosphorylation at S863 by PKC-3 is not essential in early embryogenesis, but is important in later development. Surprisingly neither PDZ1 not nor PDZ3 are essential for localization or function. Our results indicate that the different domains and phosphorylated forms of PAR-3 can have different roles during C. elegans development.
Keywords: cell polarity, PAR-3, phosphorylation, PDZ domain
INTRODUCTION
Acquisition of cell polarity is a critical process for specifying body axis and maintaining distinct organ function in metazoan development. The PAR (partitioning defective) proteins, which are highly conserved from worms to mammals, are part of the core machinery to control cell polarization in many different cell types (Goldstein and Macara, 2007). PAR-3, a multi-PDZ domain scaffold protein, can interact with PAR-6 and PKC-3 (atypical protein kinase C; aPKC) to control cell polarization in different developmental stages and in different tissues. For example, in the Drosophila central nervous system, PAR-3 (also called Bazooka), PAR-6 and aPKC co-localize at the apical surface of neuroblasts and ensure that the neural fate determinants segregate into one of the two daughter cells (Rolls et al., 2003; Schaefer et al., 2001; Schober et al., 1999; Wodarz et al., 1999). In mammalian epithelial cells, PAR-3, PAR-6 and aPKC localize to the tight junctions to control apical-basolateral polarity (Chen and Macara, 2005; Izumi et al., 1998; Yamanaka et al., 2001).
The role of PAR-3, PAR-6 and PKC-3 as regulators of polarity was first identified in C. elegans, where they play critical roles in the establishment of embryonic polarity and organization of epithelial cells (Aono et al., 2004; Etemad-Moghadam et al., 1995; Kemphues et al., 1988; Nance et al., 2003; Tabuse et al., 1998; Totong et al., 2007; Watts et al., 1996). Early in the first embryonic cell cycle, PAR-3, PAR-6, and PKC-3 are uniformly distributed at the cell periphery of the fertilized egg (Cuenca et al., 2003; Etemad-Moghadam et al., 1995; Hung and Kemphues, 1999; Tabuse et al., 1998). In response to the polarity cue provided by the sperm centrosomes or microtubules emanating from them, localized reduction of actomyosin contractility at the posterior pole results in the cortical actin network flowing away from the sperm, carrying PAR-3, PAR-6 and PKC-3 toward the anterior (Cowan and Hyman, 2007). This restricted localization of the anterior PAR proteins is critical for the first asymmetric division, which generates two daughter cells different in size, fate, and spindle orientation (Boyd et al., 1996; Cheeks et al., 2004; Etemad-Moghadam et al., 1995; Hao et al., 2006; Munro et al., 2004; Tabuse et al., 1998; Watts et al., 1996). PAR-3 appears to act upstream to localize PAR-6 and PKC-3 at the cell periphery (Beers and Kemphues, 2006; Tabuse et al., 1998; Watts et al., 1996). However, little is known about how PAR-3 associates with the cortex in the one-cell stage worm embryo.
Recent studies in C. elegans organogenesis have revealed that PAR-3, PAR-6 and PKC-3 also play important roles in epithelial development (Aono et al., 2004; Nance et al., 2003; Totong et al., 2007). Levels of maternal PAR-3, PAR-6 and PKC-3 gradually diminish after the 26-cell stage, and zygotic expression of PAR-3 initiates when the embryo approaches 400 cells (Leung et al., 1999; McMahon et al., 2001; Nance et al., 2003). The re-expressed PAR-3, PAR-6 and PKC-3 proteins localize at the apical surface of developing pharynx, intestine, vulva, spermatheca, uterus, and male tail rays (Aono et al., 2004; Nance et al., 2003; our unpublished results). Interestingly, PAR-3 localizes basolaterally to PAR-6 and PKC-3 in fully polarized epithelial cells, suggesting that PAR-3 may act independently from the other two proteins (Totong et al., 2007), similar to results reported in flies (Harris and Peifer, 2005). Targeted degradation of maternal PAR-3 in embryonic somatic precursor cells leads to aberrant cell adhesion and cell ingression (Nance et al., 2003) and knockdown of zygotic PAR-3 protein in larvae causes defects in distal spermathecal junctions (Aono et al., 2004).
Like its homologues, C. elegans PAR-3 contains a conserved N-terminal domain called CR1, which mediates PAR-3 oligomerization both in vitro and in vivo (Benton and St Johnston, 2003a; Feng et al., 2007; Mizuno et al., 2003) three PDZ domains in tandem (PDZ1, PDZ2, PDZ3) followed by a region called CR3 containing a conserved PKC-3 binding site (Figure 1A) (Etemad-Moghadam et al., 1995) Izumi et al. 1998). There has been considerable progress in understanding PAR-3 function and localization in mammalian cultured cells (Goldstein and Macara, 2007). However, less is known about how the domains of PAR-3 contribute to its function in cells of living animals. To understand how PAR-3 localizes and functions during worm development, we have introduced targeted mutations and deletions into PAR-3::GFP and examined the localization and function of the mutated proteins in the genetic background of two different par-3 alleles that allow us to assess maternal versus zygotic requirements. Our results indicate that although the role of PAR-3 in controlling cell polarity is widely conserved, the protein acts via different mechanisms in early embryos and epithelial cells.
Figure 1. Structure of the par-3 gene and its transcripts.
(A) Schematic drawing of the par-3 gene (upper row) and PAR-3 protein structure (lower rows). par-3 exons are represented by black boxes, introns are black lines and untranslated sequences are grey boxes. Asterisk shows the location of the it71 nonsense mutation, and bracket shows the tm2010 deletion. Blue, red and green open rectangles denote the genomic regions corresponding to CR1, PDZ1, 2, 3 and CR3 domains respectively. The blue and orange open rectangle indicates the sequences encoding CR1 (blue) and coding sequences unique to F54E7.3c that disrupt the CR1 domain (orange). In the lower row, colored boxes indicate the conserved domains of PAR-3 protein; numbers denote the amino acids marking the endpoints of each conserved domain (Etemad-Moghadam et al., 1995; Izumi et al., 1998).
(B) Results of RT-PCR reactions showing diagnostic segments of F54E7.3b (Lane B) and F54E7.3c (Lane C) amplified from mRNA isolated from wild type mixed L3 and L4 stage larvae and sequenced (see text). The asterisk indicates a spuriously amplified segment of bacterial RNA.
MARTERIALS AND METHODS
Nematode strains
Caenorhabditis elegans strains were cultured under standard procedures (Brenner, 1974), except that all transgenic strains were maintained at 25°C. The Bristol N2 strain was used as wild type. Mutant strains used in this study are KK653, unc32(e189)par-3(it71)/qC1 III (Etemad-Moghadam et al., 1995), SS104, glp-4(bn2ts) (Beanan and Strome, 1992) and KK928, par-3(tm2010)/qC1 III. par-3(tm2010), generated by the National Bioresource Project (S. Mitani, Tokyo Women’s Medical University), was outcrossed 6 times, balanced, and sequenced.
Transgene construction and transformation
All par-3 transgenes were derived from plasmid pJN210 which contains genomic par-3 and 3962 base pairs of upstream regulatory sequences (gift from Dr. Jeremy Nance (Nance et al., 2003)). Mutations or deletions were constructed by site-directed mutagenesis (Quickchange kit, Stratagene) or recombinant PCR. In most cases, internal deletions could be constructed without deleting any intronic sequences, which could potentially contain regulatory elements. However, deletion of PDZ2 required that we delete all of intron 7 as well. All constructs included the wild-type unc-119 gene as a transformation marker. unc-119(ed3) worms were transformed by microparticle bombardment (Praitis et al., 2001). Only 5–10% of the Unc + transgenic lines stably express GFP both maternally and zygotically, and although we recovered lines with stable long-term maternal expression for most of our constructs, maternal expression was lost in par-3ΔNT::gfp lines within 6 weeks after the lines were generated. In all other cases, we selected for subsequent analysis only lines that expressed the mutated fusion protein at least to the same level as wild type protein as assayed by immunofluorescence.
Analysis of transgene rescue of par-3(it71) and par-3(tm2010)
We recovered integrated homozygous transgenic lines that express mutated variants of PAR-3::GFP both maternally and zygotically and tested at least two independent lines from each construct for rescue and fusion protein distribution, except for the PAR-3ΔPDZ3::GFP mutant, for which we recovered only one line. The identity of the transgene in each rescue experiment was confirmed by single-worm PCR followed by DNA sequencing (Barstead et al., 1991; Williamson et al., 1991).
To assess maternal function of the mutant constructs, we mated unc32(e189)par-3(it71)/qC1 III males to transgenic hermaphrodites. F1 outcross progeny were allowed to self individually. The recessive marker unc-32 was used to select par-3(it71) homozygotes in the F2, which were plated individually and allowed to lay eggs. Unc-32 hermaphrodites will produce viable progeny only if it71 recombines away from unc-32 or if the transgene rescues. Because recombination away from the marker is rare as determined by control crosses lacking transgenes, rescue is easily distinguishable by the high frequency of Unc-32 animals that give progeny. In addition, we confirmed rescue by showing that production of progeny correlated with GFP expression in the pharynx and developing embryos of the Unc-32 worms.
To assess the ability of mutated forms of PAR-3 to rescue the zygotic requirement for the gene, we mated par-3(tm2010)/qC1 III males to hermaphrodites from each homozygous integrated transgenic line. Offspring from the F1 worms that did not segregate qC1 homozygotes were scored for embryonic and larval lethality. If the transgene fully rescues in a single copy, we expect approximately 15/16 embryos and larvae to be viable in the F2; if the transgene fails to rescue, we expect ¾ of the embryos and larvae to be viable. Therefore we define full zygotic rescue as 93.75% survival to adult, and no rescue as 75% survival. The percentage rescue was determined by the following formula: (X-75%)/(93.75%-75%), X=scored viability. Note that due to undercounting of embryos, the level of rescue can exceed 100%. Each cross was performed in parallel with crosses using the wild-type PAR-3::GFP line as positive controls and N2 as negative controls.
To determine the localization of non-rescuing PAR-3::GFP fusion proteins in homozygous par-3(tm2010) embryos and larvae, we constructed par-3(tm2010)/qC1 strains homozygous for par-3S863A::gfp (itIs182), par-3ΔNT::gfp (itIs195), par-3ΔCT::gfp (itIs200) and par-3ΔPDZ2::gfp (itIs232) respectively. For each strain, we examined GFP distribution in the alimentary tract of a mixture of 50 or more bean, comma, 1.5-fold, 2-fold and 3-fold stage embryos from the tm2010/qC1 mothers. One fourth of the embryos are expected to be homozygous for tm2010. Counts verified that one fourth of the progeny died as embryos or arrested near the L1 to L2 molt. If the mutated fusion proteins localized normally, 100% of the examined embryos exhibited normal localization; if not, 25%showed an abnormal distribution. par-3(tm2010)/qC1 strains homozygous for wild-type par-3::gfp (itIs179) and rescuing construct par-3S863E::gfp(itIs166) served as controls.
Microscopy
Observations of live embryos were made on a Leica DM RA2 microscope with a 63× Leica HCX PL APO oil emersion lens and Hamamatsu ORCA-ER digital camera. Digital images were captured using Openlab software (Improvision). Unless indicated otherwise, for each construct images were obtained from at least two independent lines and more than 50 embryos. Confocal images were collected on a Leica TCS SP2 system with a Leica DMRE-7 microscope and an HCX PL APO 63× oil immersion lens. Images were processed using the Leica Confocal SP2 software program and Adobe PhotoShop.
RT-PCR
mRNAs were extracted from L3-L4 N2 or glp-4(bn2ts) worms using the FastTrack mRNA isolation kit (Invitrogen). RT-PCR reactions were performed using the First Strand DNA synthesis kit and the pdN(6) primer it provided (Biosciences). PCR primers used for amplification of the diagnostic fragments are: Primer (B)5′-acagttggtcaactagcagacgcagc-3′; Primer (C)5′-atgcataacggtcgtggtggtcg-3′; Primer(ctrl1)5′-gagacgcaggtggtatgcgcaatg-3′; Primer (ctrl2) 5′-acacgcatcggctataatttcagcac-3′; Primer (R) 5-gctcggcgagcttcttctcaacttc-3′. All procedures were performed according to the manufacture’s protocols. PCR products were then cloned into TOPO vector (Invitrogen) and sequenced.
Western blots
For detection of proteins in embryo extracts, embryos were collected from hypochlorite-treated adult worms and boiled in SDS-sample buffer (Etemad-Moghadam et al., 1995). Gel electrophoresis and Western blots were performed by standard procedures. anti-PAR-3 primary antibody was diluted 1:1000 and HRP-conjugated secondary antibody (Jackson ImmunoResearch) was diluted 1:5000.
Immunostaining
Embryos were fixed in methanol following previously published procedures (Guo and Kemphues, 1995). The following primary antibodies and dilutions were used: anti-PAR-3 mouse monoclonal (Nance et al., 2003) 1:70, anti-GFP goat polyclonal (Rockland Immunochemicals) 1:400, anti-PAR-2 rabbit polyclonal antibody (Boyd et al, 1996) 1:15, anti-PAR-6 rabbit polyclonal antibody (Hung and Kemphues, 1999) 1:30, anti-PKC-3 rat polyclonal antibody (Aono et al. 2004) 1:30. Primary antibodies were detected by Alexa Fluor 488 labeled goat anti-mouse (Invitrogen), Cy3 labeled goat anti-mouse at 1:200 or 1:250, Cy 3 labeled donkey anti-goat 1:400 or Cy3 labeled donkey anti-rabbit or anti-rat at 1:200 (Jackson ImmunoResearch Laboratories, Inc.). Unless indicated otherwise, immunostaining observations were based on the analysis of more than 10 embryos at the appropriate stage.
in vitro kinase assays
His-PKC-3 and His-PKC-3K266A were expressed and purified from baculovirus-infected Sf21 cells (Fujise et al., 1994). GST-PAR-3678-935, GST-PAR-31-152, GST-PAR-3153-382, GST-PAR-3759-868, and GST-PAR-3869-1379 were produced in Escherichia coli and purified by standard procedures. His-PKC-3 and His-PKC-3K266A were incubated with 10μCi[γ-32P]ATP (ICN Biomedicals, Inc.) and GST-PAR-3 fragments in 100 μl kinase buffer (25mM Tris-HCl, pH 7.4, 25ng phosphatidylserine, 5mM MgCl2, 500μM EGTA, 1mM dithiothreitol). Reactions were incubated at 30°C for 2 hours and terminated by addition of SDS sample dilution buffer. Proteins were separated by 10% SDS-PAGE, and phosphorylation was visualized by autoradiography.
RESULTS
A par-3 deletion mutant causes larval lethality
All but one previously reported par-3 alleles are strict maternal-effect-lethal mutations (Cheng et al., 1995; Kemphues et al., 1988; Kirby et al., 1990). par-3(it71), the strongest of these, contains a nonsense mutation in exon 3 and shows no detectable protein in early embryos (Etemad-Moghadam et al., 1995). However, PAR-3 accumulates normally in epithelial cells of the digestive tract and somatic gonad in embryos from homozygous it71 mothers, indicating that it71 is not a null allele (Aono et al., 2004). We obtained a par-3 deletion allele (tm2010, generously provided by the National Bioresource Project, Tokyo), which contains a 409bp internal deletion (5049–5457, start codon=1) including part of intron 6 and exon 7 (Fig. 1A). In contrast to most previously identified par-3 mutants, par-3(tm2010) homozygotes die as L1 larvae (33/47) or embryos (14/47). The mutation failed to complement par-3(it71) and was rescued by a par-3::gfp transgene (viability 95.1±3%, n=523). The larval lethality of tm2010 indicates that zygotic expression of PAR-3 is required for viability.
Previous studies suggested that the maternal-specific alleles were due to mutations within a region of the 5′ end of the mRNA (F54E7.3a or b) that were not included in a putative alternative transcript that was expressed only in late embryonic or larval stages or both (Aono et al., 2004). Indeed, a short transcript, F54E7.3c, is predicted and has been partially confirmed (Wormbase, release WS207; Fig. 1A). To test whether this transcript is expressed post-embryonically we performed RT-PCR using mRNA isolated from a mix of L3 and L4 N2 worms. Diagnostic fragments of F54E7.3b and F54E7.3c were amplified by primers B and R and C and R respectively (Fig. 1B, lanes B and C) and confirmed by sequencing. In addition, both primer ctrl2 which targets sequences immediately 5′ of the predicted start codon of F54E7.3c, ATG of F, and primer ctrl1 which targets a more 5′ region of intron 4, failed to amplify any product (Fig. 1A, lanes ctrl2 and ctrl1), indicating that it is likely that F54E7.3c reading frame initiates from the predicted start codon. The long transcript must be maternal because strictly maternal mutations affect only the long transcript. The long transcript also appears to be transcribed zygotically because L3 and L4 glp-4(bn2ts) hermaphrodites grown at restrictive temperature, which have severely reduced germ lines (Beanan and Strome, 1992), show the same levels of both transcripts as wild type (Supplemental Figure S1). We did not determine whether the short transcript is expressed maternally; if it is expressed, it is not capable of substituting for the long form.
The predicted protein product of F54E7.3c substitutes 71 novel amino acids for the first 107 amino acids of F54E7.3b and results in a disruption of the CR1 domain in the protein product of F54E7.3c, replacing the first 38 amino acids of the 83 amino acid domain. Blast search revealed no homology to the novel amino acids coded by the F54E7.3c message in Par-3 proteins other than in C. elegans and C. briggsae.
Both the N-terminal and C-terminal portions of PAR-3 contain information required for cortical accumulation
To identify the core sequences in PAR-3 important for localization and function, we tested the ability of truncated PAR-3::GFP proteins to localize and function in par-3 mutants. For these and all subsequent transgene constructs we mutated the full length genomic DNA within pJN102 (Nance et al., 2004; see materials and methods). We first made reciprocal constructs missing either C-terminal (ΔCT) or N-terminal (ΔNT) portions of PAR-3 fused to GFP: par-3ΔCT::gfp (Δ aa 809-1379) and par-3ΔNT::gfp (Δ aa 1-809), driven by its native promoter. We introduced both constructs and a control full-length par-3::gfp into worms by biolistic transformation of an unc119(−); par-3(+) strain (Praitis et al., 2001) and examined the distribution of the GFP fusion proteins in early embryos, late embryos and developing larvae. Because PAR-3 can self-oligomerize via its CR1 domain in flies and mammals (Benton and St Johnston, 2003a; Feng et al., 2007; Mizuno et al., 2003), it is possible that the endogenous wild-type PAR-3 present in the worms could recruit the mutant protein via oligomer formation and thus mask any abnormal localization. Therefore we also examined GFP distribution after crosses to replace the endogenous wild-type par-3 gene with par-3(it71) or par-3(tm2010) mutations. These crosses also enabled us to test whether the truncated transgenes could provide par-3 function in early embryogenesis (it71) and in late embryogenesis or post-embryonic development (tm2010).
In both par-3(+) and par-3(it71) worms, wild-type PAR-3::GFP displayed a weak signal but an identical distribution to endogenous PAR-3 protein as reported previously (Aono et al., 2004; Etemad-Moghadam et al., 1995; Nance et al., 2003); PAR-3::GFP distributed uniformly at the cortex early in the cell cycle, then cleared from the posterior cortex during the first mitotic prophase. After the first mitotic division, PAR-3::GFP covered the entire cortex of the anterior cell, AB, as well as the anterior cortex of the posterior cell, P1 (Fig. 2A, E; Supplementary Movie S1). In L4 larvae, PAR-3::GFP localized to apical surfaces of pharyngeal and vulval epithelial cells (Fig. 3A, B). In addition, both par-3(it71) and par-3(tm2010) were rescued by PAR-3::GFP (Fig. 4A, B), indicating that this fusion protein functions normally throughout development.
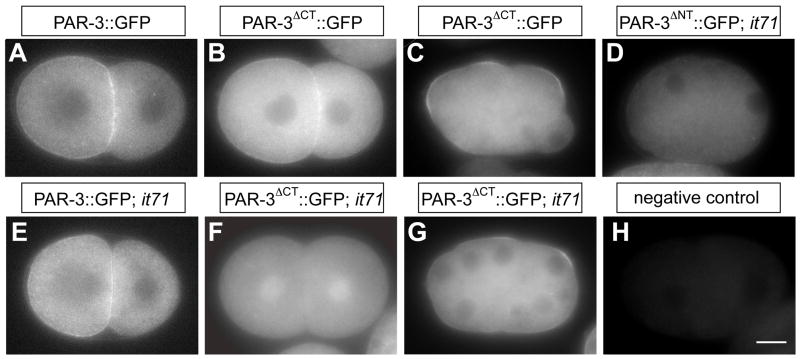
Figure 2. PAR-3ΔCT::GFP and PAR-3ΔNT::GFP in early embryos.
Fluorescence images of PAR-3::GFP, PAR-3ΔCT::GFP and PAR-3ΔNT::GFP in par-3(+) embryos (A–C) and in par-3(it71) embryos (D–G). (H) shows an embryo with no transgene under the same microscopy conditions. In this and all figures, anterior is to the left of the embryo and the scale bar is approximately 10μm. The transient enrichment of GFP signal at the time of nuclear envelope localization in (F) is a common occurrence of GFP fusion proteins.
Figure 3. PAR-3ΔCT::GFP and PAR-3ΔNT::GFP in late par-3(+) embryos, developing larvae and par-3(tm2010) embryos.
Fluorescence images of par-3(+) larvae (A, B, D, E, G, H) and representative comma stage progeny of par-3(tm2010)/qC1 embryos (C, F, I) expressing PAR-3::GFP (A–C), PAR-3ΔCT::GFP (D–F) and PAR-3ΔNT::GFP (G–I). (A, D, G), vulva; (B, E, H), pharynx. Note that we could not determine the genotypes of the embryos in C, F and I, but all embryos from the par-3(tm2010)/qC1 mothers exhibited the protein distributions shown.
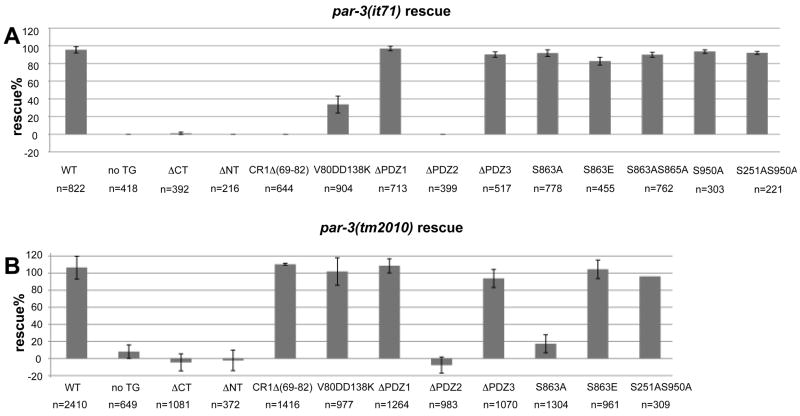
Figure 4. PAR-3 transgene rescue of par-3(it71) and par-3(tm2010).
(A) Percentage of viable embryos from it71 homozygous mothers carrying the indicated transgene. WT= wild-type PAR-3 transgene; no TG = no transgene (See text for explanation of abbreviations for transgene constructs). (B) Percentage rescue of progeny viability from par-3(tm2010)/+ mothers calculated as described in Materials and Methods. Error bars represent standard deviation of the values obtained for each experiment. n=total embryos checked for viability. Rescue can appear greater than 100% due to undercounting of laid eggs.
Neither PAR-3ΔCT::GFP nor PAR-3ΔNT::GFP behave like wild-type PAR-3. When expressed in par-3(+) embryos, PAR-3ΔCT::GFP localizes to the anterior cortex of early embryos, although high levels of protein are present in the cytoplasm (compare Fig. 2B to 2A). However, in early par-3(it71) embryos, little PAR-3ΔCT::GFP was detected at the cortex and the cytoplasmic GFP signal was higher than for the wild-type transgene (compare Fig. 2F to 2E). The truncated protein is not completely incapable of cortical accumulation, however, because cortical protein is detectable after the 16-cell stage, in both the par-3(+) and par-3(it71) backgrounds (Fig. 2C, 2G). Since par-3(it71) embryos lack maternal PAR-3, and zygotic PAR-3 does not express until the 300–400 cell stage (Nance et al., 2003), this late cortical localization is likely a result of gradual accumulation of PAR-3ΔCT::GFP rather than recruitment of the truncated protein via interaction with zygotically expressed wild-type PAR-3. It is also possible that this delayed cortical accumulation indicates that different mechanisms localize PAR-3ΔCT::GFP in one-cell and >16-cell stage embryos. In spite of this weak localization, PAR-3ΔCT::GFP failed to rescue the maternal-effect-lethality of par-3(it71) (Fig. 4A). Thus the C-terminal region of PAR-3 is required for the maternal function of the protein, and contains information necessary for robust accumulation at the cell cortex.
Zygotically expressed PAR-3ΔCT::GFP showed the same distribution as zygotically expressed wild-type PAR-3:GFP either in par-3(+) (Fig. 3D, E) or par-3(tm2010) (Fig. 3F) genetic backgrounds. In spite of this normal distribution, PAR-3ΔCT::GFP failed to rescue par-3(tm2010) (Fig. 4B), indicating a requirement for amino acids 809-1379 for PAR-3 zygotic function.
In par-3(+) embryos, maternally-expressed PAR-3ΔNT::GFP failed to localize to the cortex and was barely detectable in the cytoplasm. One-cell embryos of transgenic worms had consistently higher levels of cytoplasmic signal than the negative controls (Fig. 2D, H; 121.6±8% of background fluorescence, n=31 embryos, single-tail t-test, P<0.005). We verified that this weak signal was due to expression of the PAR-3ΔNT::GFP by Western blot (Supplementary Figure S2). Compared to wild-type PAR-3::GFP, which displayed restricted apical localization in epithelial tissues (Fig. 3A, B), zygotically expressed PAR-3ΔNT::GFP was diffuse in all epithelial tissues examined irrespective of the presence of endogenous wild type PAR-3 (Fig. 3G, H, I). Consistent with its failure to localize cortically and apically, PAR-3ΔNT::GFP also failed to rescue par-3(it71) and par-3(tm2010) (Fig. 4).
Overall, these results indicate that the first 808 amino acids of PAR-3 (ΔCT), contain information sufficient for cortical localization but not for proper function, and that amino acids 809 to 1379 (ΔNT) contribute to cortical accumulation or protein stability and are required for function.
PDZ2, but not PDZ1 or PDZ3, is necessary for PAR-3 localization and function
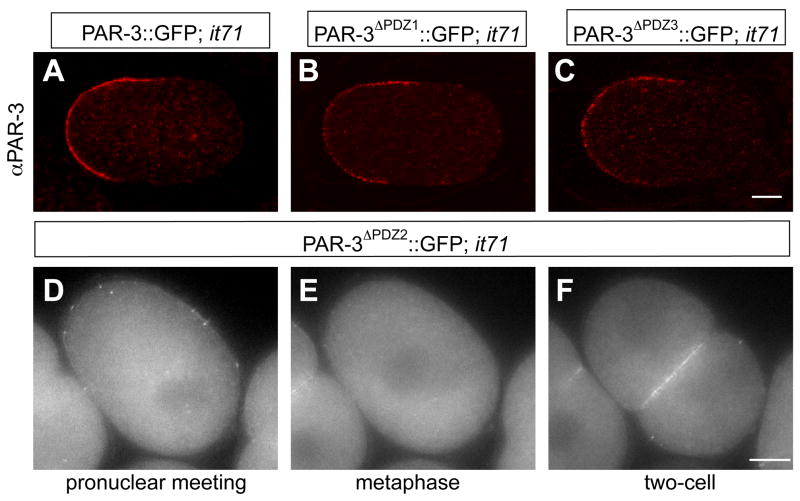
Our attempts to identify smaller fragments sufficient for localization by sequential deletion of the N-terminal fragment failed because we were unable to recover transgenic lines expressing any fragment smaller than PAR-3ΔCT::GFP. Therefore we took an alternative approach by making targeted deletions of conserved domains (summarized in Table 1). We started our analysis by deleting each of the PDZ domains. We generated lines expressing constructs PAR-3ΔPDZ1::GFP (Δ aa 383-463), PAR-3ΔPDZ2::GFP (Δ aa 515-584), PAR-3ΔPDZ3::GFP (Δ aa 659-738) and examined the expression level and distribution of the GFP-tagged transgenes. We were able to generate lines with levels of accumulation of fusion protein similar to that of wild type fusion proteins. In wild-type par-3(+) embryos, we found that deletion of any one of the three PDZ domains had no obvious effect on the cortical localization of the corresponding fusion protein (summarized in Table 1). Occasionally we observed par-3(+) embryos expressing PAR-3ΔPDZ2::GFP that showed par-3(it71)-like phenotypes indicating that PDZ2 deletion may cause some dominant-negative effects (data not shown). When endogenous maternal PAR-3 was absent, as in progeny from homozygous par-3(it71) mothers, PAR-3ΔPDZ1::GFP and PAR-3ΔPDZ3::GFP proteins showed distributions indistinguishable from PAR-3::GFP (Fig. 5A, B, C) and rescued the progeny from homozygous mothers to near wild-type viability (Fig. 4A). In contrast, in the absence of endogenous wild type PAR-3, PAR-3ΔPDZ2::GFP deviated from wild type, forming sparse and large cortical puncta (Fig. 5D). These embryos retained high levels of GFP signal in the cytoplasm indicating that failure to rescue was not likely to be due to reduced expression of the mutant protein (Fig. 5D; Fig. 6E, M). Embryos fell into two categories. In 20 of the 28 embryos we examined either via live imaging or in fixed specimens, cortical puncta were extremely sparse and showed no obvious asymmetry (Supplementary Movie S2, the right embryo). Eight of the embryos had larger numbers of cortical puncta which occasionally showed some asymmetry during the early phase of the first cell cycle (Fig. 5D; Supplementary Movie S2, the left embryo); 7 of 24 fixed embryos showed numbers of cortical puncta similar to the embryo in the movie. In both classes of embryos, the cortical puncta disappear at metaphase and return during prophase of the next cell cycle (Fig. 5, E, F; Supplementary Movie S2).
Table 1.
Embryonic localization of PAR-3::GFP protein and the mutant variants.
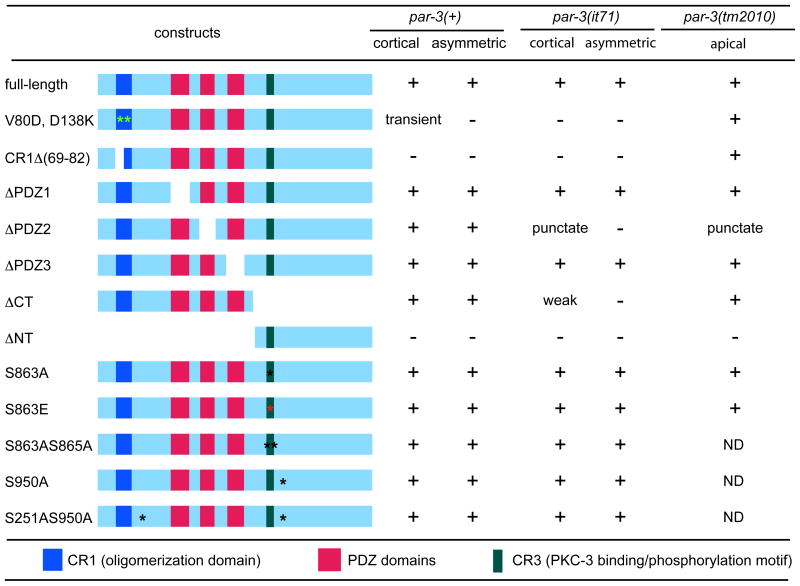 |
Summary of the localization of indicated transgenic protein in early par-3(+) embryos, early par-3(it71) embryos and late par-3(tm2010) embryos respectively. Asterisks show positions of point mutations. “+” indicates normal localization; “−” designates failure to localize. If abnormally large and sparse GFP puncta were observed, this was noted as “punctate”.
Figure 5. Effect of PDZ domain deletions on PAR-3 protein distribution.
One-cell par-3(it71) embryos that express PAR-3::GFP (A), PAR-3ΔPDZ1::GFP (B), PAR-3ΔPDZ3::GFP (C) and PAR-3ΔPDZ2::GFP (D–F) stained with anti-PAR-3 antibody only (A–C) or co-stained with anti-PAR-3 and anti-PAR-6 antibody (D–I). (A–C) are low resolution confocal images taken at the middle plane of the cell and (D–I) are mid focal plane wide-field microscope images from a live embryo (see Supplemental Movie 2).
Figure 6. PAR-3ΔPDZ2::GFP fails to associate with PAR-6 or PKC-3.
(A–H) Confocal sections of one and two-cell stage embryos of the indicated genotypes doubly labeled with anti-PAR-3 and anti-PAR-6 antibodies. The anti-PAR-6 positive larger puncta in the posterior of the embryos expressing PAR-3::GFP and clustered around the nuclei in the embryos expressing PAR-3ΔPDZ2::GFP are P granules which are stained by this rabbit polyclonal antibody. (I–P) Confocal sections of one and two-cell stage embryos of the indicated genotypes doubly labeled with anti-PAR-3 and anti-PKC-3 antibodies. Panels E-G and M-O are projections of six adjacent sections; all other panels are single sections.
We next asked how deletion of PDZ2 affects the ability of PAR-3 to co-localize with PAR-6 and PKC-3. In par-3::gfp; par-3(it71) embryos, PAR-3 puncta co-localize extensively with PAR-6 (Fig. 6A-D; supplementary Fig. S3) and PKC-3(Fig. 6I-L; supplementary Fig. S3). However, the large puncta containing PAR-3ΔPDZ2::GFP fail to co-localize with PAR-6 (n= 15 embryos) or PKC-3 (n= 15 embryos). Indeed, PAR-6 and PKC-3 are not enriched at the cortex at all in these embryos (Fig. 6F, H, N, P; supplementary Fig. S3).
Consistent with these defects, PAR-3ΔPDZ2::GFP transgenes failed to rescue it71 mutants (Figure 4A) and the embryos exhibited phenotypes typical for loss of maternal PAR-3: cytoplasmic flow was attenuated, spindle displacement failed, and the first division was equal (n=27). These observations showed that PDZ2, but not PDZ1 or PDZ3, is required for PAR-3 to localize and function properly in early embryos.
We obtained similar results when we introduced these three constructs into par-3(tm2010)/+ worms and scored progeny for viability. Based on survival rates we concluded that homozygous tm2010 embryos were rescued by PAR-3ΔPDZ1::GFP and, to a slightly lesser extent, by PAR-3ΔPDZ3::GFP, but not by PAR-3ΔPDZ2::GFP (Figure 4B). To verify the unexpected result suggesting that PDZ1 and PDZ3 were not required and to determine whether the surviving worms were sterile or exhibited any morphogical or behavior abnormality, we constructed stable lines that were homozygous for tm2010 and carried the rescuing transgene. Homozygous tm2010 worms rescued by PAR-3ΔPDZ1::GFP or by PAR-3ΔPDZ3::GFP exhibit variable amounts of embryonic and larval lethality. For example, among the progeny of 20 fourth generation tm2010 homozygotes expressing PAR-3ΔPDZ1::GFP, viability to adult ranged from 10% to 89%; among progeny of 20 fourth generation tm2010 homozygotes expressing PAR-3ΔPDZ3::GFP viability ranged from no survivors (two cases) to 96% survivors. Most worms that survive to adulthood, however, exhibit wild type morphology and behavior and produce normal broods of viable progeny. We cannot distinguish to what extent the variable level of lethality is due to compromised protein function vs. variable expression of the transgenes.
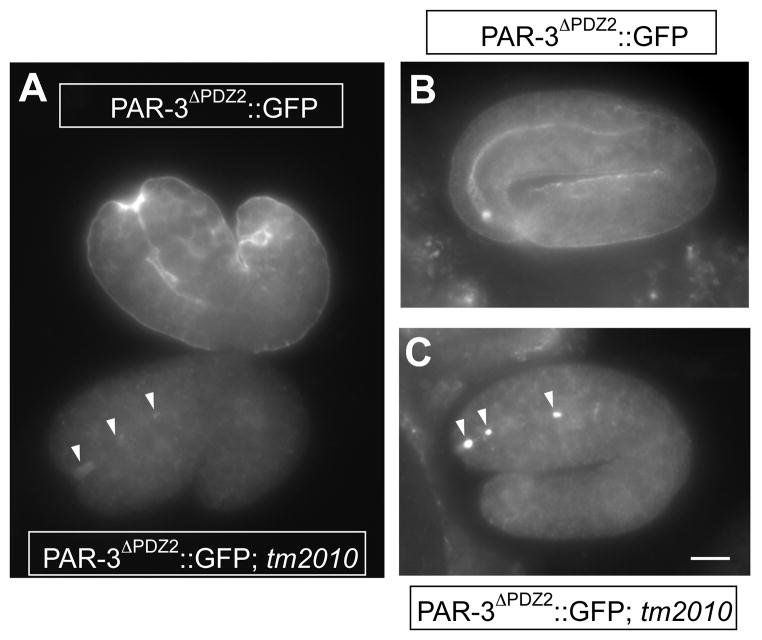
As stated above, the PAR-3ΔPDZ2::GFP fusion protein failed to rescue tm2010 mutants. We examined the localization of fusion protein among embryos of tm2010/+; par-3ΔPDZ2::gfp mothers, in which 25% of the offspring were expected to express PAR-3ΔPDZ2::GFP and be homozygous for tm2010. We found 21% of embryos lacked apical accumulation of GFP but showed accumulations of large GFP puncta adjacent to or in the lumen of the developing pharynx during morphogenesis (n=8/38; Fig. 7A, C) while the remaining embryos showed normal localization (n=30/38; Fig. 7A, B). Control embryos lacking the transgene showed no signal (data not shown). These results suggested that PDZ2, but not PDZ1 or PDZ3, is required for apical localization and function of PAR-3 in late embryogenesis or larval development.
Figure 7. PAR-3ΔPDZ2::GFP in late par-3(tm2010) embryos.
1.5-fold stage embryos (A) and 2-fold stage embryos (B, C) expressing PAR-3ΔPDZ2::GFP stained with anti-GFP antibody. Note that PAR-3ΔPDZ2::GFP shows apical localization in the developing pharynx, gut and rectum in par-3(+) embryos (A upper embryo; B), but is undetectable at the cortex of most cells of par-3(tm2010) embryos except developing pharynx where it forms aggregates (arrowheads) in or near the lumen (A lower embryo; C). Note that the tm2010 genotype is inferred because embryos of this phenotype occur as ¼ of the progeny of tm2010/+ mothers.
CR1 is necessary for PAR-3 function in early embryos but dispensable in late-embryogenesis and post-embryonic development
CR1 (conserved region 1), also called NTD (N-terminal domain), is highly conserved among PAR-3 homologues (Benton and St Johnston, 2003a; Feng et al., 2007; Mizuno et al., 2003). It has been shown to mediate PAR-3 oligomerization both in vitro and in vivo and is necessary for PAR-3 apical localization in Drosophila and in mammalian cultured cells (Benton and St Johnston, 2003a; Feng et al., 2007; Mizuno et al., 2003). To investigate the role of CR1 in C. elegans, we first tested whether CR1 of worm PAR-3 mediates oligomerization. Using the yeast-two-hybrid system, we found that CR1 of C. elegans PAR-3 was indeed capable of self–association. We found that deletion of aa 1-68, which is specific to worm PAR-3, did not block PAR-3 self-association, although three other small deletions in CR1 (Δ69-82, Δ109-119; Δ122-132) each abolished this property (Supplementary Fig. S4). The structure of the CR1 (NTD) domain of mammalian Par3 has been solved (Feng et al., 2007) and two point mutations (equivalent to V80D and D138K in C. elegans PAR-3) were identified as being able to disrupt CR1 oligomerization without significantly affecting its overall structure. We introduced the deletion Δ(69-82) and the double point mutation V80D, D138K into PAR-3::GFP (PAR-3Δ(69-82)::GFP and PAR-3V80D, D138K::GFP respectively) and generated lines expressing these constructs to assess the requirement for oligomerization of PAR-3 in vivo.
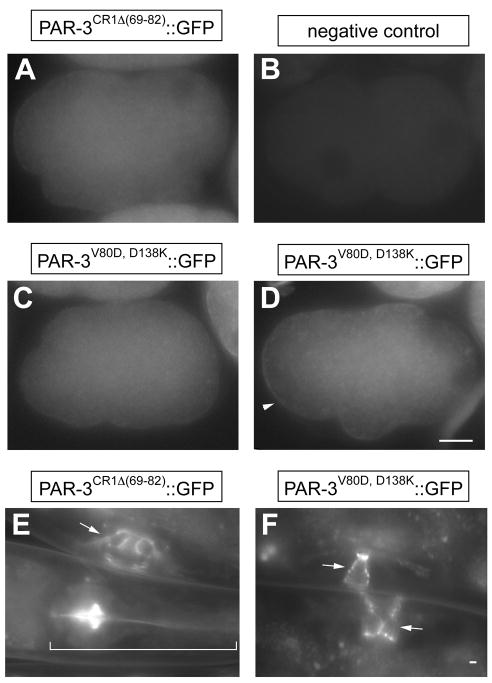
Neither PAR-3Δ(69-82)::GFP nor PAR-3V80D, D138K::GFP localized normally in early par-3(+) or par-3(it71) embryos. PAR-3Δ(69-82)::GFP displayed a diffuse and uniform signal in the cytoplasm (n>50, figure 8A). In embryos after pronuclear meeting, PAR-3V80D, D138K::GFP behaved indistinguishably from PAR-3Δ(69-82)::GFP (n>50, Fig. 8C); however, among thirty-one very early embryos expressing PAR-3V80D, D138K::GFP, five exhibited a very weak transient cortical signal which clears from the posterior pole then disappears from the cell periphery during pronuclear migration (Fig. 8D). In contrast to their behavior in early embryos, in late par-3(+) and par-3(tm2010) embryos and larvae, both PAR-3Δ(69-82)::GFP and PAR-3V80D, D138K::GFP localized similarly to wild-type PAR-3::GFP; they accumulate at apical surfaces of cells in pharynx, intestine, vulva, and somatic gonad (Fig. 8E, F and data not shown).
Figure 8. PAR-3CR1Δ(69-82)::GFP and PAR-3V80D, D138K ::GFP in par-3(+) embryos and larvae.
(A–D) Fluorescence images of par-3(+) embryo expressing PAR-3CR1Δ(69-82)::GFP (A), PAR-3V80D, D138K::GFP (C, D) and no transgene (B). Note that PAR-3V80D, D138K::GFP is cytoplasmic in most early par-3(+) embryos (C), but occasionally shows weak and transient cortical localization (D). Arrowhead points to the weak cortical signal. (E–F) Fluorescence images of par-3(+) larvae expressing PAR-3CR1Δ(69-82)::GFP (E) and PAR-3V80D, D138K::GFP (F). Arrows point to the vulva; bracket indicates the pharynx.
Consistent with its failure to localize in early embryos, PAR-3Δ(69-82)::GFP failed to rescue the maternal-effect lethality of par-3(it71) (Fig. 4A), indicating that aa 69-82 are essential for maternal PAR-3 function. PAR-3V80D, D138K::GFP showed partial and variable rescue −8.6% to 40.4% of the offspring survived and grew to fertile adults. This variability in extent of rescue occurs both between and within lines, and appears to be specific to the PAR-3V80D, D138K::GFP construct only (Fig. 4A). Two possible explanations for the weak rescue by PAR-3V80D, D138K::GFP can be drawn: the mutations do not abolish the ability of PAR-3 to form oligomers, or the CR1 domain has a function in addition to oligomer formation that monomers can facilitate.
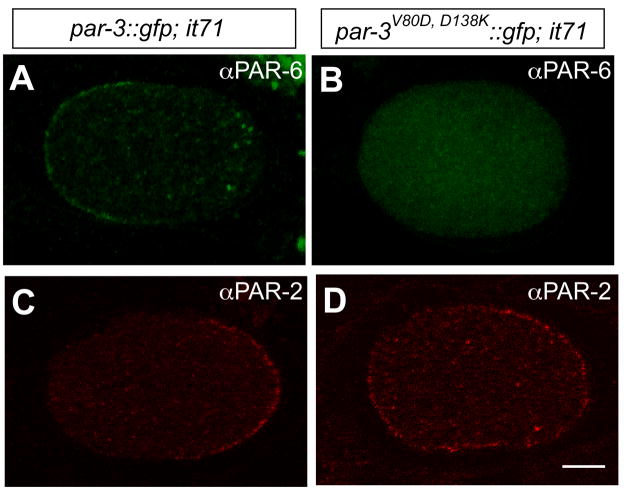
To determine the importance of PAR-3 self-oligomerization to the distribution of other PAR proteins in early embryos, we examined the localization of PAR-6 and PAR-2 in par-3(it71) embryos carrying PAR-3::GFP or PAR-3V80D, D138K::GFP. In par-3(it71); par-3::gfp embryos, PAR-6 and PAR-2 localized reciprocally at the anterior and posterior cortex (Fig. 9A, C). However, when PAR-3::GFP was substituted by PAR-3V80D, D138K::GFP, little PAR-6 was detected at the cell periphery, and PAR-2 expanded into the anterior domain (Fig. 9B, D), indicating that the oligomerization function of PAR-3 is necessary for PAR-6 localization and PAR-2 restriction in early embryos.
Figure 9. PAR-6 and PAR-2 in par-3V80D, D138K ::gfp; par-3(it71) embryos.
Fluorescence images of PAR-6 (green) and PAR-2 (red) in par-3::gfp; par-3(it71) embryos (A, C) and par-3V80D, D138K ::gfp; par-3(it71) embryos (B, D).
Surprisingly, both PAR-3Δ(69-82)::GFP and PAR-3V80D, D138K::GFP were capable of rescuing the larval-lethality of par-3(tm2010) efficiently (Fig. 4B). Together with the observation that both PAR-3Δ(69-82)::GFP and PAR-3V80D, D138K::GFP localized properly in the rescued larvae, these results suggest that CR1 is dispensable for late embryogenesis and postembryonic development.
PKC-3 phosphorylates PAR-3 at a conserved serine
In mammals, aPKC, the homologue of C. elegans PKC-3, can bind and phosphorylate mPar3 both in vitro and in vivo (Izumi et al., 1998; Joberty et al., 2000; Lin et al., 2000; Nagai-Tamai et al., 2002; Suzuki et al., 2001). The single phosphorylation target of mPar3 is serine 827, although binding to aPKC requires serine 829 (Nagai-Tamai et al., 2002); the equivalent serines in C. elegans PAR-3 are S863 and S865. To investigate whether PAR-3 is a target of C. elegans PKC-3 in vitro, we carried out kinase assays using partially purified C. elegans proteins (see Materials and Methods). We divided PAR-3 into five pieces and tested whether any of them could be phosphorylated by His-PKC-3 in vitro. Except for the fragment containing amino acids 383 to 678, which we were unable to express, we found that only the PAR-3 fragment containing amino acids 678 to 935, which includes the C. elegans region corresponding to the aPKC binding and phosphorylation site in mammals, could be phosphorylated by wild-type PKC-3 (Fig. 10A). PKC-3K266A, a kinase-dead form of PKC-3, failed to phosphorylate PAR-3, indicating that PAR-3 is specifically phosphorylated by PKC-3 in our assay (Fig. 10A).
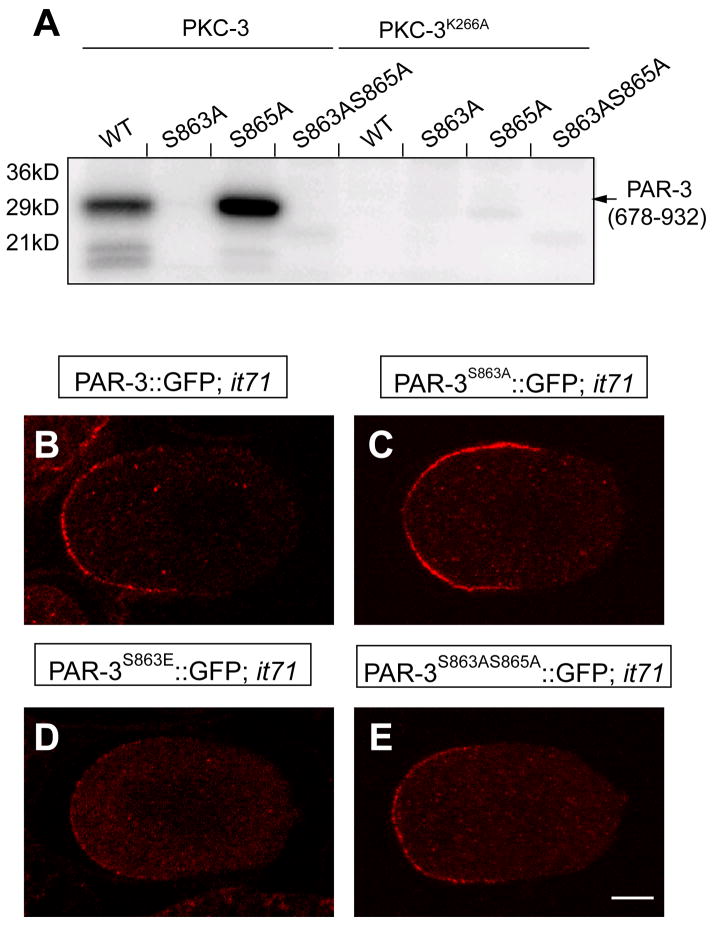
Figure 10. PAR-3 phosphorylation at S863 in par-3(it71) embryos.
(A) in vitro PKC-3 kinase assay with a portion of PAR-3(aa 678-932) and the mutated variants PAR-3S863A::GFP, PAR-3S863E::GFP and PAR-3S863AS865A::GFP as substrate. PKC-3K266A, the kinase-dead form of PKC-3, was used as negative control. (B–E) Anti-PAR-3 antibody stained par-3(it71) embryos that express PAR-3::GFP (B), PAR-3S863A::GFP (C), PAR-3S863E::GFP (D) and PAR-3S863AS865A::GFP (E).
Conversion of the putative target, S863, to alanine completely abolished the phosphorylation by His-PKC-3 but conversion of S865 to alanine had no effect. Thus C. elegans PKC-3 phosphorylates PAR-3 at conserved serine S863 in vitro.
Phosphorylation at S863 in PAR-3 is not required in early embryogenesis, but is important for later development
To investigate the in vivo significance of PKC-3 phosphorylation, we mutated S863 to alanine to block phosphorylation or to glutamic acid to mimic constitutive phosphorylation, and then generated transgenic worms expressing PAR-3S863A::GFP and PAR-3S863E::GFP. We found that in both par-3(+) and par-3(it71) embryos, PAR-3S863A::GFP and PAR-3S863E::GFP were able to localize to the anterior cortex like wild-type PAR-3::GFP (Figure 10B–D, Supplementary Movie S3). Moreover, both PAR-3S863A::GFP and PAR-3S863E::GFP exhibited robust rescue of par-3(it71) (Fig. 4A). To test if S865 could serve as a redundant phosphorylation site in vivo, we generated PAR-3S863AS865A::GFP and found that this double mutant also localized properly and rescued par-3(it71) efficiently (Fig. 4A; Fig. 10E). These results suggest that phosphorylation of PAR-3 on S863 or S865 is not essential for early embryogenesis in C. elegans. We did, however, note a difference between these two constructs. PAR-3S863A::GFP appeared to have a much stronger cortical signal than PAR-3S863E::GFP or wild-type PAR-3::GFP in the early embryos (Supplementary Movie S3). This difference is quite consistent among at least three independent lines for each construct.
To test whether there is a zygotic requirement for phosphorylation of PAR-3 by PKC-3, we crossed both PAR-3S863A::GFP and PAR-3S863E::GFP into the par-3(tm2010) strain. In contrast to the results showing rescue of par-3(it71), PAR-3S863A::GFP showed poor ability to rescue the lethality of par-3(tm2010), but PAR-3S863E:GFP was able to rescue par-3(tm2010) efficiently (single tail t-test, p<0.005; Fig. 4B). We checked offspring from the tm2010/qC1 III mothers expressing either PAR-3S863A::GFP or PAR-3S863E::GFP (n=62 and 103 respectively) and examined the localization of the transgene. All embryos (of which ¼ should be homozygous for tm2010) were indistinguishable from wild-type PAR-3::GFP staining patterns (data not shown). Furthermore we were able to isolate par-3(tm2010)/par-3(tm2010); par-3S863E::gfp lines that produce fertile progeny, confirming that PAR-3 S863E::GFP is functional during zygotic development. Normal localization of PAR-3S863A::GFP may result from perdurance of maternal PAR-3 loaded by the heterozygous mothers, or may indicate that S863A impairs PAR-3 function in some way other than by affecting its apical localization. We conclude that PKC-3 phosphorylation is required for C. elegans PAR-3 function in late embryogenesis or post-embryonic development or both, but not in early embryos.
Phosphorylation at two conserved potential 14-3-3 binding sites is not essential for PAR-3 function in C. elegans
PAR-5 is a C. elegans 14-3-3 protein and restricts PAR-3 distribution to the anterior in one-cell embryos (Cuenca et al., 2003; Morton et al., 2002). Previous studies in Drosophila and mammals suggest that PAR-3 binds to 14-3-3 proteins directly and this interaction requires the phosphorylation of a conserved serine, S950 on a potential PAR-1 target site (Benton and St Johnston, 2003b; Hurd et al., 2003a; Izaki et al., 2005). To assess the physiological significance of this phosphorylation in C. elegans, we mutated S950, singly and in combination with S251, another potential PAR-1 target that may be involved in this interaction, to alanines (Benton and St Johnston, 2003b; Hurd et al., 2003a; Izaki et al., 2005). However PAR-3S950A::GFP and PAR-3S251A, 950A::GFP localize asymmetrically throughout development and rescue it71. We did not test PAR-3S950A::GFP for rescue of tm2010, but PAR-3S251A, 950A::GFP rescues tm2010 (Figure 4 and Table 1). Because we could identify at least 10 additional putative PAR-5 binding sites, our results do not rule out a role for PAR-1 or PAR-5 in regulating PAR-3.
DISCUSSION
PAR-3 is a highly conserved scaffold protein that functions in a variety of polarized cellular events such as asymmetric cell division, epithelial polarization, directional cell migration and neuronal specification (Goldstein and Macara, 2007). In C. elegans, PAR-3 is essential for anterior-posterior polarity in the early embryo (Cuenca et al., 2003; Etemad-Moghadam et al., 1995; Kemphues et al., 1988; Tabuse et al., 1998; Watts et al., 1996) and for processes in later embryonic (Nance et al., 2003) early larval (this report) and later larval development (Aono et al., 2004). Here we report results of an analysis of the function of PAR-3’s conserved protein domains in living animals. We find that in spite of the overall structural conservation among animals, the requirements for specific PAR-3 domains appear to be stage and species-specific.
The role of PAR-3 PDZ domains
PDZ domains are 80–90 amino acid-long modules, forming a barrel-like structure consisting of 5–6 β-strands and 2 α-helices (Nourry et al., 2003; Sheng and Sala, 2001). PDZ domains can bind the C-terminus, internal peptides, other PDZ domains of their client proteins or phosphatidylinositol moieties (Roh and Margolis, 2003; Tonikian et al., 2008). PAR-3 has three PDZ domains, so it is reasonable to suppose that this protein may organize large complexes via these PDZ domains. Surprisingly, we found that although deletion of PDZ2 rendered the protein nonfunctional, deletion of either PDZ1 or PDZ3 had little or no effect on the ability of the mutated protein to rescue loss-of-function mutations of par-3.
The PDZ1 domain of mPar3 or Bazooka has been shown to bind to various proteins including mPar-6, JAM-1, nectins, Inscuteable, and p75 to regulate junction formation in epithelial cells, asymmetric division in neuroblasts and myelination in hippocampal cells (Chan et al., 2006; Ebnet et al., 2001; Itoh et al., 2001; Joberty et al., 2000; Lin et al., 2000; Schober et al., 1999; Takekuni et al., 2003; Wodarz et al., 1999). The in vitro interaction between PAR-3 and PAR-6 has been verified in many species including C. elegans (Li et al., 2010), although the consequence of this binding remains unclear (Gibson and Perrimon, 2003). In one study, overexpressed mPar6 can perturb epithelial polarity, and mutations in mPar6 that reduce mPar3-mPar6 interaction (KPLG167-170AAAA) abolished this activity (Joberty et al., 2000). However the same mutations can also abolish the interaction between mPar6 and Pals1, therefore making it difficult to interpret the results (Hurd et al., 2003b). In another study, mPar-3 binding to mPar6 is dispensable for tight junction (TJ) assembly in polarizing MDCK cells (Chen and Macara, 2005). Because of this conserved interaction and because PAR-6 and PAR-3 are mutually required for stable localization to the cell cortex of early embryos, our finding that deletion of PDZ1 had no apparent effect on PAR-3 function in C. elegans was unexpected, although it is consistent with results from our parallel analysis of the PAR-6 PDZ domain. Point mutations in the PAR-6 PDZ domain that block binding of PAR-3 PDZ1 and PAR-6 PDZ in vitro have no effect on the PAR-6 function in C. elegans (Li et al., 2010).
Limited research has been reported on PAR-3 PDZ2 and PDZ3 domains until recently, when the structure of mPar3 PDZ2 and PDZ3 domains were solved and their roles in mammalian epithelial polarization were examined (Feng et al., 2008; Wu et al., 2007). mPar3 PDZ2 shows high affinity to phosphatidylinositol lipids, but the physiological significance in epithelial polarization is still controversial (Chen and Macara, 2005; Wu et al., 2007); in one study, PDZ2 is not required for mPar3 to restore TJ assembly in mPar3-depleted MDCK cells (Chen and Macara, 2005), whereas another study showed that mPar3 with a PDZ2 deletion fails to localize and function properly in MDCK cells (Wu et al., 2007). We found that PAR-3 PDZ2 is absolutely required for C. elegans early embryogenesis and later development. Although the sequence of C. elegans PAR-3 PDZ2 domain is not strikingly similar to its mammalian homologues, it does contain a cluster of positively charged amino acids (H512, H555, K557, R597) with spacing similar to that proposed to mediate the electrostatic interaction between mPar3 PDZ2 and phospholipid membranes (K458, R504, K506, R546). It is possible then that C. elegans PAR-3 associates with the cell periphery through PDZ2-lipid interaction. Because deleting PDZ2 does not completely dissociate PAR-3 from the cell periphery in early embryos, this putative interaction with phospholipid cannot be the sole mechanism responsible for PAR-3 cortical localization. Indeed, the association of PAR-3 with the cortex in the early embryo is also dependent upon an intact actomyosin network (Severson and Bowerman, 2003).
We found that although PAR-3 lacking PDZ2 retains weak ability to become enriched at the cortex, it is unable to recruit PAR-6 or PKC-3, suggesting either that PDZ2 plays some direct role in recruiting one or both of these proteins or that proper cortical association or concentration of PAR-3 is required for formation of stable complexes.
We found that PAR-3 PDZ3 is dispensable in C. elegans in spite of its clear role in other animals. For example, in mammals, PTEN, the phosphatase that generates PtdIns(4,5)P2, binds directly to mPar3 PDZ3 and this interaction is important for membrane enrichment of PTEN and epithelial polarity (Feng et al., 2008). PDZ3 is also required for mPar3 to concentrate at TJ and to control TJ assembly in polarizing MDCK cells (Chen and Macara, 2005).
The role of PAR-3 CR1 domain
The CR1 domain of PAR-3 is highly conserved in all PAR-3 homologues and mediates PAR-3 oligomerization both in vitro and in vivo (Benton and St Johnston, 2003a; Feng et al., 2007; Mizuno et al., 2003). mPar3 lacking CR1 shows diffuse cellular distribution in MDCK cells, and overexpression of CR1 delays the formation of functional TJs (Feng et al., 2007; Mizuno et al., 2003). In Drosophila, deletion of CR1 disrupts Bazooka apical localization and strongly compromises its function in follicular epithelial cells (Benton and St Johnston, 2003a). We find that in C. elegans, intact CR1 is critical for PAR-3 function and cortical localization in early embryos, but not in late embryos and larvae, suggesting that the later function of PAR-3 is independent of CR1-mediated oligomerization. Our verification of the expression of an alternative mRNA that has a disrupted and presumably non-functional CR1 domain is consistent with our functional data and with the existence of maternal-specific mutant alleles. Mutations like it71 that affect only the large mRNA are able to support zygotic development because the short mRNA can function without an intact CR1 domain, but fail to provide the maternally provided function in early embryos because the short mRNA, even if it is produced in embryos, would generate a protein lacking a functional CR1 domain. Whether this alternative form of PAR-3 can function in a monomeric form or forms oligomers through via a mechanism other than CR1 multimerization needs further investigation.
The role of phosphorylation of PAR-3
The CR3 region of PAR-3 is highly conserved from worms to mammals (Izumi et al., 1998). In mammals, aPKC binds to the CR3 region of mPar3 directly and phosphorylates serine 827 both in vitro and in vivo (Lin et al., 2000; Nagai-Tamai et al., 2002). However, the physiological significance of this phosphorylation is not clear. One study showed that overexpression of an S827A mutant, but not wild-type mPar3, significantly inhibits TJ reformation in polarizing MDCK cells (Nagai-Tamai et al., 2002). In another study, however, mPar3 can function properly in epithelial polarization independent of aPKC (Chen and Macara, 2005). In our study, we found that in C. elegans, the phosphorylation of PAR-3 by PKC-3 does not markedly affect PAR-3 function in early embryogenesis. The phosphorylation may play a subtle role, however, because blocking the phosphorylation consistently resulted in higher levels of cortical PAR-3. In contrast, in late embryogenesis or post-embryonic development, phosphorylation at S863 is required for PAR-3 function. The phosphorylation appears to be permissive rather than regulatory because the phospho-mimetic mutation can function as well as wild-type PAR-3. A recent report from Drosophila is consistent with our results and provides insight into a possible basis for these results. Phosphorylation at S890 of Bazooka (PAR-3), the analogous position to C. elegans S863, is required in epithelial cells to exclude PAR-3 from the apical domain, but appears not be required in the Drosophila oocyte (Eurico Morais-de-Sá et al., 2010). Larval lethality in C. elegans par-3 mutants is likely to be due to defects in polarized epithelial cells (Achilleos et al., 2010). Thus, the difference in dependency on phosphorylation likely reflects differences in the precise role of PAR-3 in epithelial cells vs. its role in the early embryo.
In flies and mammals, Bazooka and mPar3 can bind to 14-3-3 proteins in a phosphorylation-dependent manner to regulate epithelial polarization (Benton and St Johnston, 2003b; Hurd et al., 2003a). We found that blocking the phosphorylation of PAR-3 at two conserved 14-3-3 (PAR-5) binding sites also had no effect on asymmetric distribution or function of the protein. It is possible that in C. elegans additional putative PAR-5 binding sites have assumed the role of the two conserved sites that we tested.
The role of PAR-3 C-terminal region
The C-terminal region of PAR-3 does not contain any recognizable domain structures, but in its mammalian homologues the region plays important roles in polarity establishment in neurons and epithelia (Chen and Macara, 2005; Nishimura et al., 2004; Nishimura et al., 2005; Zhang and Macara, 2006). Several studies have revealed that motifs in the C-terminal region are essential for mPar3 to localize properly and to recruit effectors, such as Tiam1, a RacGEF protein (Chen and Macara, 2005; Nishimura et al., 2005). We found that PAR-3 lacking the C-terminal region is still able to associate with the cell periphery in late embryos and developing larvae. These differences are consistent with the significant sequence difference between worm PAR-3 and its vertebrate homologues (Etemad-Moghadam et al., 1995; Lin et al., 2000; von Trotha et al., 2006).
In summary, our results revealed differential requirements for the conserved domains of PAR-3 in early embryogenesis and late embryonic or larval development. Although PAR-3, PAR-6 and PKC-3 function co-dependently, direct binding between PAR-3 and PAR-6 appears not to be essential, and a requirement for PKC-3 phosphorylation may be dynamic throughout worm development. Interestingly, PAR-3 may function as a monomer or oligomer at different developmental stages, since CR1, the self-association domain, is not required for zygotic development. These findings illustrate the dynamic complexity of PAR-3 interactions and regulation in different developmental contexts to control cell polarity.
Supplementary Material
mRNA isolated from L3-L4 stage glp-4(bn2ts) larvae raised from L1 at 25°C (non-permissive temperature). The reduction of germ-line was confirmed by gross morphology observation. Lane B shows RT-PCR products using primers B and R, which yield a 741 bp fragment diagnostic for F54E7.3c, the short transcript; Lane C shows RT-PCR products using primers C and R, which yield a 819 bp fragment diagnostic for F54E7.3b the long transcript. Lanes ctrl1 and 2 show additional negative controls (see text). The upper band in Lane B (asterisk) is due to spurious amplification of a contaminating bacterial sequence.
Embryo extracts were prepared from wild type (N2) and from lines expressing PAR-3 S863A::GFP and PAR-3CT::GFP, then probed with anti-PAR-3 antibody following separation by 7% SDS-PAGE. Note that because the collected embryos were somewhat asynchronous and might include late stage embryos, we cannot rule out the possibility that some of the positive signal is due to zygotic expression.
Confocal cortical sections of embryos of the indicated genotypes doubly labeled with anti-PAR-3 and anti-PAR-6 antibodies (top six panels) or with anti-PAR-3 and anti-PKC-3 (bottom six panels).
(A) Portion of a yeast growth plate showing growth when both Gal4 binding domain and activation domain fused to PAR-3 CR1 domain are co-expressed, but little or no growth when amino acids 69-82 are deleted from either of the fusion proteins. (B) Summary of yeast two-hybrid results with the indicated deletions of CR1.
Images for all movies were collected at 18-second intervals.
Acknowledgments
We thank Dr. Jeremy Nance for plasmid pJN210 and personal communications, Shohei Mitani and the National Bioresource Project for the Experimental Animal “Nematode C. elegans” for providing tm2010, Margaret M. Lee for helping construct PAR-3S863E::GFP and PAR-3S863AS865A::GFP, Wendy Hoose and Mona Hassab for technical assistance, members of the Kemphues laboratory, Jun Liu, Dave Pruyne, Sylvia Lee and Anthony Bretscher for helpful discussions, and Diane Morton for some embryo preparations and, along with Jun Liu, for editorial advice. We are grateful to Yingsong Hao and Geraldine Seydoux for teaching B.L. the bombardment technique. This research was supported by NICHD grant HD27689 and NIGMS grant GM079112 to K.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achilleos A, Wehman AM, Nance J. PAR-3 mediates the initial clustering and apical localization of junction and polarity proteins during C. elegans intestinal epithelial cell polarization. Development. 2010;137:1833–42. doi: 10.1242/dev.047647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono S, Legouis R, Hoose WA, Kemphues KJ. PAR-3 is required for epithelial cell polarity in the distal spermatheca of C. elegans. Development. 2004;131:2865–74. doi: 10.1242/dev.01146. [DOI] [PubMed] [Google Scholar]
- Barstead RJ, Kleiman L, Waterston RH. Cloning, sequencing, and mapping of an alpha-actinin gene from the nematode Caenorhabditis elegans. Cell Motil Cytoskeleton. 1991;20:69–78. doi: 10.1002/cm.970200108. [DOI] [PubMed] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–66. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- Beers M, Kemphues K. Depletion of the co-chaperone CDC-37 reveals two modes of PAR-6 cortical association in C. elegans embryos. Development. 2006;133:3745–54. doi: 10.1242/dev.02544. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol. 2003a;13:1330–4. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003b;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Boyd L, Guo S, Levitan D, Stinchcomb DT, Kemphues KJ. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JR, Jolicoeur C, Yamauchi J, Elliott J, Fawcett JP, Ng BK, Cayouette M. The polarity protein Par-3 directly interacts with p75NTR to regulate myelination. Science. 2006;314:832–6. doi: 10.1126/science.1134069. [DOI] [PubMed] [Google Scholar]
- Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr Biol. 2004;14:851–62. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–9. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- Cheng NN, Kirby CM, Kemphues KJ. Control of cleavage spindle orientation in C. elegans: the role of the genes par-2 and par-3. Genetics. 1995;139:549–559. doi: 10.1093/genetics/139.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Acto-myosin reorganization and PAR polarity in C. elegans. Development. 2007;134:1035–43. doi: 10.1242/dev.000513. [DOI] [PubMed] [Google Scholar]
- Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–65. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebnet K, Suzuki A, Horikoshi Y, Hirose T, Meyer Zu Brickwedde MK, Ohno S, Vestweber D. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM) Embo J. 2001;20:3738–48. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. Embo J. 2007;26:2786–96. doi: 10.1038/sj.emboj.7601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M. Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J Biol Chem. 2008;283:23440–9. doi: 10.1074/jbc.M802482200. [DOI] [PubMed] [Google Scholar]
- Fujise A, Mizuno K, Ueda Y, Osada S, Hirai S, Takayanagi A, Shimizu N, Owada MK, Nakajima H, Ohno S. Specificity of the high affinity interaction of protein kinase C with a physiological substrate, myristoylated alanine-rich protein kinase C substrate. J Biol Chem. 1994;269:31642–8. [PubMed] [Google Scholar]
- Gibson MC, Perrimon N. Apicobasal polarization: epithelial form and function. Curr Opin Cell Biol. 2003;15:747–52. doi: 10.1016/j.ceb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–22. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos encodes a putative ser/thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hao Y, Boyd L, Seydoux G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev Cell. 2006;10:199–208. doi: 10.1016/j.devcel.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol. 2005;170:813–23. doi: 10.1083/jcb.200505127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung TJ, Kemphues KJ. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development. 1999;126:127–35. doi: 10.1242/dev.126.1.127. [DOI] [PubMed] [Google Scholar]
- Hurd TW, Fan S, Liu CJ, Kweon HK, Hakansson K, Margolis B. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol. 2003a;13:2082–90. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003b;5:137–42. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- Itoh M, Sasaki H, Furuse M, Ozaki H, Kita T, Tsukita S. Junctional adhesion molecule (JAM) binds to PAR-3: a possible mechanism for the recruitment of PAR-3 to tight junctions. J Cell Biol. 2001;154:491–7. doi: 10.1083/jcb.200103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaki T, Kamakura S, Kohjima M, Sumimoto H. Phosphorylation-dependent binding of 14-3-3 to Par3beta, a human Par3-related cell polarity protein. Biochem Biophys Res Commun. 2005;329:211–8. doi: 10.1016/j.bbrc.2005.01.115. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Hirose T, Tamai Y, Hirai S, Nagashima Y, Fujimoto T, Tabuse Y, Kemphues KJ, Ohno S. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–9. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–20. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kirby C, Kusch M, Kemphues K. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev Biol. 1990;142:203–215. doi: 10.1016/0012-1606(90)90164-e. [DOI] [PubMed] [Google Scholar]
- Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–34. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Li J, Kim H, Aceto DG, Hung J, Aono S, Kemphues KJ. Binding to PKC-3, but not to PAR-3 or to a conventional PDZ domain ligand, is required for PAR-6 function in C. elegans. Dev Biol. 2010;340:88–98. doi: 10.1016/j.ydbio.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol. 2000;2:540–7. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- McMahon L, Legouis R, Vonesch JL, Labouesse M. Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J Cell Sci. 2001;114:2265–77. doi: 10.1242/jcs.114.12.2265. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Suzuki A, Hirose T, Kitamura K, Kutsuzawa K, Futaki M, Amano Y, Ohno S. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J Biol Chem. 2003;278:31240–50. doi: 10.1074/jbc.M303593200. [DOI] [PubMed] [Google Scholar]
- Morais-de-Sa E, Mirouse V, St Johnston D. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 2010;141:509–23. doi: 10.1016/j.cell.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DG, Shakes DC, Nugent S, Dichoso D, Wang W, Golden A, Kemphues KJ. The Caenorhabditis elegans par-5 Gene Encodes a 14-3-3 Protein Required for Cellular Asymmetry in the Early Embryo. Dev Biol. 2002;241:47–58. doi: 10.1006/dbio.2001.0489. [DOI] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–24. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7:1161–71. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- Nance J, Munro EM, Priess JR. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development. 2003;130:5339–50. doi: 10.1242/dev.00735. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol. 2004;6:328–34. doi: 10.1038/ncb1118. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, Hoshino M, Kaibuchi K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol. 2005;7:270–7. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP. PDZ domain proteins: plug and play! Sci STKE. 2003;2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–26. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh MH, Margolis B. Composition and function of PDZ protein complexes during cell polarization. Am J Physiol Renal Physiol. 2003;285:F377–87. doi: 10.1152/ajprenal.00086.2003. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163:1089–98. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–94. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–51. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- Severson AF, Bowerman B. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J Cell Biol. 2003;161:21–6. doi: 10.1083/jcb.200210171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yamanaka T, Hirose T, Manabe N, Mizuno K, Shimizu M, Akimoto K, Izumi Y, Ohnishi T, Ohno S. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J Cell Biol. 2001;152:1183–96. doi: 10.1083/jcb.152.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998 doi: 10.1242/dev.125.18.3607. (In press) [DOI] [PubMed] [Google Scholar]
- Takekuni K, Ikeda W, Fujito T, Morimoto K, Takeuchi M, Monden M, Takai Y. Direct binding of cell polarity protein PAR-3 to cell-cell adhesion molecule nectin at neuroepithelial cells of developing mouse. J Biol Chem. 2003;278:5497–500. doi: 10.1074/jbc.C200707200. [DOI] [PubMed] [Google Scholar]
- Tonikian R, Zhang Y, Sazinsky SL, Currell B, Yeh JH, Reva B, Held HA, Appleton BA, Evangelista M, Wu Y, Xin X, Chan AC, Seshagiri S, Lasky LA, Sander C, Boone C, Bader GD, Sidhu SS. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totong R, Achilleos A, Nance J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 2007;134:1259–68. doi: 10.1242/dev.02833. [DOI] [PubMed] [Google Scholar]
- von Trotha JW, Campos-Ortega JA, Reugels AM. Apical localization of ASIP/PAR-3:EGFP in zebrafish neuroepithelial cells involves the oligomerization domain CR1, the PDZ domains, and the C-terminal portion of the protein. Dev Dyn. 2006;235:967–77. doi: 10.1002/dvdy.20715. [DOI] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, Priess JR, Kemphues KJ. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- Williamson VM, Long M, Theodoris G. Isolation of caenorhabditis-elegans mutants lacking alcohol dehydrogenase activity. Biochem Genet. 1991;29:313–324. doi: 10.1007/BF00554139. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–7. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell. 2007;28:886–98. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Horikoshi Y, Suzuki A, Sugiyama Y, Kitamura K, Maniwa R, Nagai Y, Yamashita A, Hirose T, Ishikawa H, Ohno S. PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induced formation of the epithelial junctional complex. Genes Cells. 2001;6:721–31. doi: 10.1046/j.1365-2443.2001.00453.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–37. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mRNA isolated from L3-L4 stage glp-4(bn2ts) larvae raised from L1 at 25°C (non-permissive temperature). The reduction of germ-line was confirmed by gross morphology observation. Lane B shows RT-PCR products using primers B and R, which yield a 741 bp fragment diagnostic for F54E7.3c, the short transcript; Lane C shows RT-PCR products using primers C and R, which yield a 819 bp fragment diagnostic for F54E7.3b the long transcript. Lanes ctrl1 and 2 show additional negative controls (see text). The upper band in Lane B (asterisk) is due to spurious amplification of a contaminating bacterial sequence.
Embryo extracts were prepared from wild type (N2) and from lines expressing PAR-3 S863A::GFP and PAR-3CT::GFP, then probed with anti-PAR-3 antibody following separation by 7% SDS-PAGE. Note that because the collected embryos were somewhat asynchronous and might include late stage embryos, we cannot rule out the possibility that some of the positive signal is due to zygotic expression.
Confocal cortical sections of embryos of the indicated genotypes doubly labeled with anti-PAR-3 and anti-PAR-6 antibodies (top six panels) or with anti-PAR-3 and anti-PKC-3 (bottom six panels).
(A) Portion of a yeast growth plate showing growth when both Gal4 binding domain and activation domain fused to PAR-3 CR1 domain are co-expressed, but little or no growth when amino acids 69-82 are deleted from either of the fusion proteins. (B) Summary of yeast two-hybrid results with the indicated deletions of CR1.
Images for all movies were collected at 18-second intervals.