Abstract
Oxidative stress is a prominent feature of Huntington's disease (HD) due to mitochondrial dysfunction and the ensuing overproduction of reactive oxygen species (ROS). This phenomenon ultimately contributes to cognitive and motor impairment, as well as brain pathology, especially in the striatum. Targeting the transcription of the endogenous antioxidant machinery could be a promising therapeutic approach. The NF-E2-related factor-2 (Nrf2)/antioxidant response element (ARE) signaling pathway is an important pathway involved in antioxidant and anti-inflammatory responses. Synthetic triterpenoids, which are derived from 2-Cyano-3,12-Dioxooleana-1,9-Dien-28-Oic acid (CDDO) activate the Nrf2/ARE pathway and reduce oxidative stress in animal models of neurodegenerative diseases. We investigated the effects of CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) in N171-82Q mice, a transgenic mouse model of HD. CDDO-EA or CDDO-TFEA were administered in the diet at various concentrations, starting at 30 days of age. CDDO-EA and CDDO-TFEA upregulated Nrf2/ARE induced genes in the brain and peripheral tissues, reduced oxidative stress, improved motor impairment and increased longevity. They also rescued striatal atrophy in the brain and vacuolation in the brown adipose tissue. Therefore compounds targeting the Nrf2/ARE pathway show great promise for the treatment of HD.
Keywords: Huntington's disease, Oxidative stress, Nitrosative stress, Nrf2/ARE pathway, Triterpenoids, N171-82Q mice, Survival, Motor performances, Striatal atrophy
Introduction
Huntington's disease (HD) is a neurodegenerative disorder caused by a polyglutamine expansion in the huntingtin protein. It is characterized by motor impairment, cognitive decline and psychiatric symptoms that worsen as the disease progresses. Neostriatal (caudate and putamen) degeneration is characteristic of the pathology. Both neostriatal and cerebral cortex atrophy are widely believed to contribute to cognitive dysfunction and motor impairment, such as choreiform movements [1, 2]. The degree of neostriatal atrophy is indicative of disease severity [2]. Striatal medium spiny neurons, which are involved in movement regulation, are particularly affected in HD [3].
A number of studies suggest that oxidative stress is prominent in the neostriatum of HD brains [4]and contributes to degeneration of the neostriatum. In HD patients and animal models, mitochondrial dysfunction results in overproduction of reactive oxygen species (ROS), which leads to oxidative and nitrosative stress [5-7]. This stress contributes to neuronal dysfunction by damaging DNA, proteins, and lipids.
DNA oxidation, which is contributes to DNA fragmentation, is marked by increased concentrations of oxidized nucleotides, such as 8-hydroxy-2’-deoxyguanosine (8-OHdG) [8, 9]. DNA damage occurs in the striatum and cerebral cortex of HD patients [10-12]. Furthermore, levels of 8-OHdG in blood and postmortem brain tissue of HD patients are markedly increased [13-15]. Similarly, in HD transgenic mouse models, 8-OHdG levels are significantly increased in urine, blood, striatal DNA and striatal microdialysates [16]. In the R6/2 transgenic mouse model of HD, striatal and cortical mtDNA damage were increased eight fold as compared to nuclear genes [17].
Protein oxidation results from overproduction of peroxynitrite, a highly reactive product of nitric oxide and superoxide free radicals, as well as other oxidants. Peroxynitrite can also inhibit mitochondrial respiration and reduce antioxidant defenses in the cell [5, 7, 18]. Peroxynitrite formation is marked by increased 3-nitrotyrosine (3-NT) levels [5, 7]. In postmortem HD brain tissue, immunoreactivity of 3-NT is increased [13]. Moreover, peroxynitrite formation is associated with striatal damage in HD animal models [19, 20]. There are also increased levels of protein carbonyls in HD striatum and cerebral cortex [4]. The oxidized proteins include aconitase, enolase, creatine kinase B and glial fibrillary acidic protein.
Oxidation of lipids is associated with impaired lysosomal function and increased levels of lipofuscin, which are consistently observed in HD postmortem brain tissue [5]. The lipid peroxidation products 4-hydroxynonenal and malondialdehyde are increased eight fold in HD human plasma [21] and postmortem brain tissue [13]. F2-isoprotane levels are also augmented in HD human cerebrospinal fluid [22]. In addition, glutathione levels in HD plasma are significantly reduced [23]. There is also increased immunostaining for malondialdehyde, 4-hydroxynonenal and 8-iso-PGF2a, in transgenic HD mice [5]. Elevated lipid peroxidation was correlated with worsening neurological phenotype [24]. Because oxidation is injurious to DNA, protein, and lipids, it can lead to cellular dysfunction and ultimately cell death.
In normal cells, oxidative stress is combated by endogenous antioxidant pathways and free radical scavengers. In HD neurons, ROS accumulate and produce oxidative stress [5, 7]. There is also a history of inflammation in HD pathology. In HD patients, microglia activation is evident in the neostriatum and cortex, and the level of microglial activation increases with increased neuronal loss and correlates with the severity of the disease [25-27]. Therefore, stimulation of the endogenous antioxidant machinery could be a potential therapeutic intervention for the treatment of HD. The NF-E2-related factor-2 (Nrf2)/antioxidant response element (ARE) signaling pathway is an important pathway implicated in antioxidant and anti-inflammatory activities. Nrf2 binds to Keap1 in the cytoplasm which prevents its degradation by the ubiquitin proteosome pathway. Following exposure to oxidative stressors or electrophiles [28, 29], Nrf2 disassociates from Keap1 and enters the nucleus where it interacts with Maf proteins and binds to the AREs on a large number of genes with antioxidant and anti-inflammatory activity, including NAD(P)H-quinone oxidoreductase 1 (NQO1), heme oxygenase, glutamate cysteine ligase, gluthatione S-tranferase, thioredoxin, and periredoxins [30].
Triterpenoids (TPs), particularly those which are analogues of 2-Cyano-3,12-Dioxooleana-1,9-Dien-28-Oic acid (CDDO), have been investigated for their antioxidant and anti-inflammatory properties for the treatment of many diseases. CDDO analogues induced the Nrf2/ARE pathway in both cell culture and in mice [31-35]. In studies on cancer, CDDO analogues reduced nitric oxide levels in mouse macrophages [36, 37]. In an Alzheimer's disease (AD) transgenic mouse model, CDDO-methyl amide (CDDO-MA) reduced protein carbonyl levels, a biomarker of protein oxidation, improved memory and reduced amyloid pathology [38]. We showed that CDDO-MA was neuroprotective against acute and chronic MPTP administration, malonate lesions and 3-nitropropionic acid toxicity, which has been used to model HD. CDDO-MA blocked the decreases in reduced glutathione and the increases in 8-OHdG, 3-NT, malondialdehyde and isoprostanes [34]. In rodents, 2-Cyano-3,12-Dioxooleana-1,9-Dien-28-Oic acid-trifluoroethyl amide (CDDO-TFEA) upregulated both NQO1 and sulfiredoxin in the lung, which are antioxidant genes induced by the Nrf2/ARE pathway [39]. 2-Cyano-3,12-Dioxooleana-1,9-Dien-28-Oic acid-ethyl amide (CDDO-EA) exhibited cytoprotective and anti-inflammatory properties in lung of mice administered vinyl carbamate [40]. Both CDDO-EA and CDDO-TFEA show higher brain concentrations as compared to both CDDO and CDDO-MA. Therefore, CDDO-EA and CDDO-TFEA are of particular interest for the reduction oxidative stress in the central nervous system, and for the prevention of neurodegeneration in HD.
In this study, we investigated the effects of CDDO-EA and CDDO-TFEA in the N171-82Q transgenic mouse model of Huntington's disease. The N171-82Q mice express a N-terminal fragment of human huntingtin of 171 amino acids with 82 CAG repeats [41]. They become symptomatic from 60 days of age onward, when they stop gaining weight and exhibit progressive behavioral impairments, including loss of motor coordination [41]. They also exhibit brain pathology with striatal atrophy [42]. We assessed motor coordination, neuropathology, expression of Nrf2/ARE regulated genes, and oxidative damage in N171-82Q mice treated with either CDDO-EA or CDDO-TFEA. We found that CDDO-EA and CDDO-TFEA increased the transcription of genes regulated by Nrf2/ARE, resulting in decreased oxidative stress, improved motor performance, increased survival, and rescue of striatal atrophy in the brain and vacuolation in the brown adipose tissue.
Materials and methods
Animals and treatment
Male N171-82Q mice, which express a human huntingtin cDNA with a N-terminal fragment of 171 amino acids with 82 CAG repeats, were obtained from Jackson Laboratories (Bar Harbor, ME, USA). They were bred with B6C3 F1 females (Jackson Laboratories) and offspring were genotyped by PCR assay of tail DNA. At 30 days of age, mice were assigned to control diet (Lab Diet #5002, Purina-Mills, Richmond, IN, USA), diet containing 2-Cyano-3,12-Dioxooleana-1,9-Dien-28-Oic acid-trifluoroethyl amide (CDDO-TFEA, 100mg/kg diet, 200mg/kg diet, or 400mg/kg diet), or diet containing 2-Cyano-3,12-Dioxooleana-1,9-Dien-28-Oic acid-ethyl amide (CDDO-EA, 100mg/kg diet or 200mg/kg diet). Diets were formulated by Purina-Mills. CDDO-EA and CDDO-TFEA were provided by Reata Pharmaceuticals (Dallas, TX, USA). Briefly, CDDO-EA and CDDO-TFEA were synthesized by the condensation of either ethyl amine or 2,2,2-trifluoroethyl amine, respectively, with CDDO acid chloride [36]. The purity was determined to be greater than 95% by NMR and HPLC analysis.
Mice were separated into three cohorts for behavioral analysis, histological analysis, and gene expression assays respectively. Behavioral analysis was assessed from 60 days of age until endstage. Histological analysis and gene expression assays were performed on mice at 120 days of age. All experiments were carried out under the guidelines and approval of the Institutional Animal Care and Use Committee (IACUC) of Weill Cornell Medical College (New York, NY, USA).
Behavioral assessment
Motor skills were assessed once per week from 60 days of age until endstage using an accelerating rotorod apparatus (Economex, Columbus Instruments, Columbus, OH, USA). Each behavioral session consisted of three 5-minute trials on the accelerating beam of the rotorod, with 15-minute inter-trial intervals. The rotorod was set to a motor speed of 0 rpm and an acceleration of 0.1 rpm/sec. Latencies to fall were used as the measure of motor performance, and averaged for all three trials. Age of death was recorded for this cohort of mice and used to assess the effects of CDDO-EA and CDDO-TFEA on survival.
Levels of CDDO in brain and skeletal muscle
CDDO-EA and CDDO-TFEA levels were measured by liquid chromatography-mass spectrometry (LC-MS). For extraction, 300 μl acetonitrile was added to 100 mg of tissue before homogenization on ice using a Tissue-Tearor (Fisher Scientific, Pittsburgh, PA, USA). Samples were then centrifuged at 20,000 g for 10 min. The acetonitrile extracts were diluted 1:1 with 20 mM ammonium acetate pH 7.4 and centrifuged at 20,000 g for 5 min. The diluted samples (100μl injection volume) were loaded on Waters XTerra column and eluate analyzed using a Waters single-quadrapole ZQ mass spectrometer (Water macromass ZQ) in electrospray positive ionization mode for CDDO-TFEA, electrospray negative ionization mode for CDDO-EA.
Analysis was carried out using Waters MassLynx 4.1 software. Standard curves were generated by spiking control tissue extracts of both brain and muscle with at least 4 different concentrations of each CDDO. All calculated values were within the limits of the spiked standards.
Immunohistochemistry and antibodies
Mice intended for neuropathologic analysis were sacrificed at 120 days of age. Mice were anesthetized by intraperitoneal injection of sodium pentobarbital and perfused with 0.9% sodium chloride. Brains were removed and fixed in 4% paraformaldehyde in 1 mM phosphate buffer (pH 7.4). After fixation for 24 hours, brains were transferred to cryoprotectant (30% glycerol, 30% ethylene glycol in 20mM phosphate buffer, pH 7.4) at 4°C and frozen before sectioning. Sections were cut coronally at 50 μm thickness.
Sections were processed for immunohistochemistry using a modified avidin-biotin-peroxidase technique. The following antibodies were used: mouse anti-NeuN, (1:1,000, Chemicon, Temecula, CA, USA); rabbit anti-calbindin (1:1,000, Chemicon, Temecula, CA, USA); rabbit anti-3-nitrotyrosine (1:100, Millipore, Billerica, MA, USA); rabbit anti-malondialdehyde modified protein (1:1,000, gift from Dr. Craig Thomas); goat anti-8-hydroxy-2’-deoxyguanosine (1:100, Chemicon, Temecula, CA, USA). The immunoreaction was visualized using 3,3' -diaminobenzidine tetrahydrochloride dihydrate (DAB).
Analysis of striatal volume
NeuN-immunostained sections were used for analyzing striatal volume. For each animal, we analyzed five serial coronal tissue sections stained through the striatum (300 μm apart) from the level of interaural 4.9/bregma 1.1 to interaural 3.7/bregma –0.1. Stereological measurement of striatal volume was carried out using the Cavalieri estimator probe of the Stereo Investigator software (Microbrightfield, Colchester, VT, USA). Each section was examined using a Nikon Eclipse E600 microscope (Melville, NY, USA) and viewed at a magnification of 4×. The striatum was identified as being bounded by the corpus callosum dorsally, the external capsule dorsolaterally, and the border of the lateral ventricle medially. A series of grid points (100 μm × 100 μm grid spacing) was then overlaid on the region of interest, and the number of points in the lattice that lie within the striatum was counted.
Analysis of striatal neuron count and medium spiny neuron area
NeuN-immunostained sections were also used for striatal neuron counts. Unbiased stereological counts of striatal NeuN-immunoreactive neurons were obtained using the Stereo Investigator software (Microbrightfield, Colchester, VT, USA). The optical fractionator probe was used to quantify the total number of NeuN-positive neurons in a defined volume of the striatum, as described above (section 2.5). The size of the x-y sampling grid was 600 μm × 600 μm, and the counting frame thickness was 14 μm with 3 μm guard zones.
Adjacent sections were immunostained with an antibody against calbindin, a marker of medium sized spiny neurons. The size of striatal calbindin positive neurons was quantified using the nucleator probe (Stereo Investigator software). The nucleus was used as the reference point. The average neuron area for at least 100 neurons per animal was calculated.
Histological analysis of vacuolation in the brown adipose tissue
Mice intended for vacuolation analysis were sacrificed at 120 days of age. Brown adipose tissues were dissected, snap frozen in liquid nitrogen and stored at -80°C prior to sectioning. Fresh frozen sections were cut at 50μM thickness and mounted on slides.
Sections were post-fixed in 4% paraformaldehyde and processed for both hematoxylin and eosin staining. Additionally, oil red O staining was performed in semi-adjacent sections in order to verify the presence of neutral lipid vacuoles.
Gene expression by qRT-PCR
Mice intended for gene expression assay were sacrificed at 120 days of age. Brains were removed and striatum were dissected. Skeletal muscles of the hind limb and brown adipose tissue were also dissected. All tissues were snap frozen in liquid nitrogen and stored at -80°C prior to Trizol extraction.
Quantitative real-time PCR (qRT-PCR) was performed with the ABI Prism 7000HT sequence detection system (Applied Biosystems, Foster City, CA, USA). CDDO-TFEA samples were processed at the Weill Cornell Medical College Core Facility whereas CDDO-EA samples were processed in our laboratory. The following representative genes were analyzed: glutathione S-transferase 3 alpha (GST3a), heme oxygenase 1 (HO1), and NAD(P)H-quinone oxidoreductase 1 (NQO1), and actin as control.
Statistical analysis
ANOVA was used to compare mice fed with control, CDDO-EA and CDDO-TFEA diets. Post hoc Fisher's PLSD was used for comparison between groups.
Kaplan-Meier survival curve was used to compare the average age of death (Statview 5.0.1, SAS Institute Inc., Cary, NC, USA).
Results
Levels of CDDO-EA and CDDO-TFEA in brain and skeletal muscle of N171-82Q mice
Detection of CDDO analogues was performed by mass spectrometry in tissues after feeding with the compounds (Supplementary figure 1). CDDO-MA was previously developed for use in mouse models of other neurodegenerative diseases [34, 38]. In this new study, we now show that CDDO-EA and CDDO-TEFA have even better pharmacokinetics than CDDO-MA for use in treatment of neurodegenerative diseases. Thus, in a short-term feeding experiment with wildtype mice (3 days), we compared the brain levels of CDDO-MA to the newer compounds CDDO-EA (Supplementary figure 1B) and CDDO-TFEA (Supplementary figure 1C). We found that CDDO-EA and CDDO-TFEA produced higher concentrations in brain as compared to CDDO-MA (Supplementary figure 1). Furthermore, in a long-term feeding experiment with wildtype mice (10 weeks), we found that brain levels of CDDO-TFEA were higher than CDDO-EA (data not shown). Therefore, we used these two newer compounds in our study.
In our experiment with transgenic N171-82Q mice, supplementary figure 2 shows chromatograms of CDDO-EA and CDDO-TFEA spiked in mouse tissue homogenates at different concentrations, which were used to form a standard curve. CDDO-EA in brains, fed in chow, was below levels of detection at the 100mg/kg diet, and was present at 33.4 ± 2.1 nmoles/kg at the 200mg/kg diet (Supplementary figure 3). CDDO-EA in skeletal muscle was present at 92.6 ± 28.0 at the 100mg/kg diet and 256.0 ± 92.3 nmoles/kg at the 200mg/kg diet (Supplementary figure 4). The ratio of brain to plasma (skeletal muscle) for CDDO-EA at 200mg/kg diet was 1:7.66. CDDO-TFEA in brains, also fed in chow, was present at 190.2 ± 70.5 nmoles/kg at the 100mg/kg diet, 191.5 ± 39.7 nmoles/kg at the 200mg/kg diet, and 174.2 ± 36.2 nmoles/kg at the 400mg/kg diet (Supplementary figure 5). Brain concentration of the latter dose is consistent with the previous feeding study of wildtype mice mentioned above (Supplementary figure 1). CDDO-TFEA was present in skeletal muscle at 1,012.4 ± 56.8 nmoles/kg at the 100mg/kg diet, 1,008.7 ± 173.5 nmoles/kg at the 200mg/kg diet, and 1,126.7 ± 202.7 at the 400mg/kg diet (Supplementary figure 6). The ratio of brain to plasma (skeletal muscle) for CDDO-TFEA at 200mg/kg diet was 1:5.27. Both CDDO-EA and CDDO-TFEA therefore crossed the blood brain barrier in transgenic N171-82Q mice, although concentrations achieved in brain were much less than those in skeletal muscle (cf. ratios of brain to plasma).
CDDO-EA and CDDO-TFEA did not affect food intake in N171-82Q mice
Body weight was assessed and analyzed at 119 days of age to ensure that the CDDO diets did not affect food intake. We found that CDDO-EA and CDDO-TFEA did not change the body weight of N171-82Q mice as compared to mice fed the control diet, indicating first that the food intake was unchanged, and second that the compounds did not improve weight loss (Supplementary figure 7).
CDDO-EA and CDDO-TFEA improved rotorod performance in N171-82Q mice
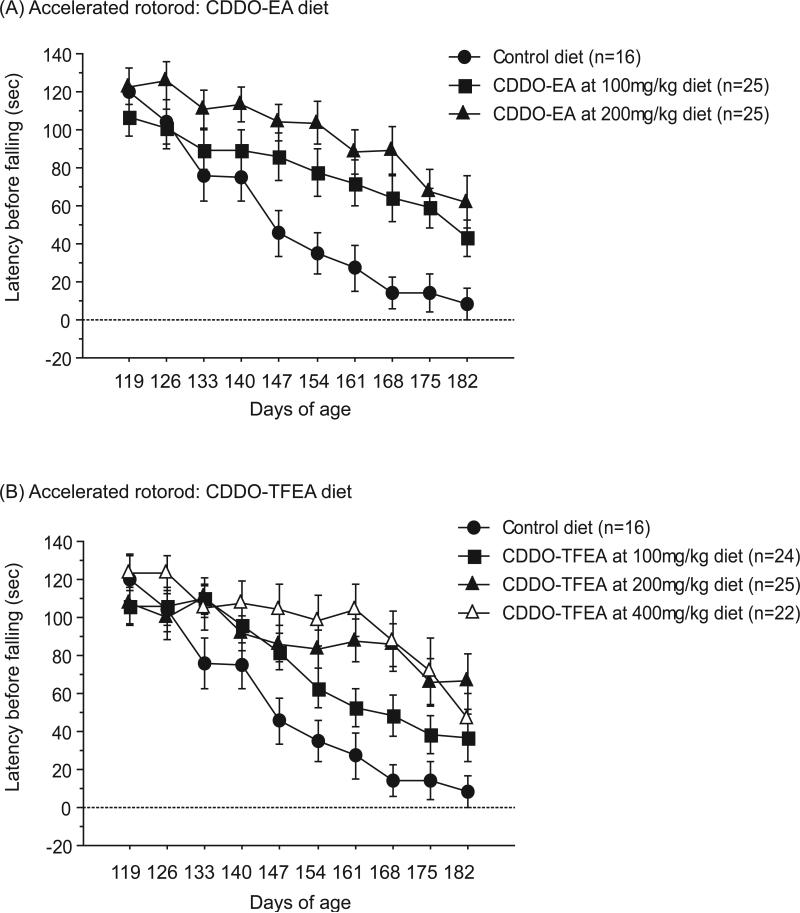
Motor coordination was assessed by performance on an accelerated rotorod apparatus. Latency to fall was recorded for 3 trials per weekly assessment and scores were averaged. Figure 1 shows rotorod performance of N171-82Q mice treated with the following diets: control, CDDO-EA 100mg/kg diet and 200mg/kg diet, and CDDO-TFEA 100mg/kg, 200mg/kg, and 400mg/kg diets from 119 days of age until 182 days of age, an age that N171-82Q mice can no longer be tested. During this symptomatic period, CDDO-EA at 100mg/kg diet and 200mg/kg diet both significantly improved motor performance in N171-82Q mice compared to N171-82Q mice fed control diet (Figure 1A; Fischer's PLSD, p=0.04, p=0.0006). CDDO-TFEA at 200mg/kg diet and 400mg/kg diet also significantly improved motor performance in N171-82Q mice as compared to N171-82Q mice fed control diet (Figure 1B; Fischer's PLSD, p=0.005, p=0.0007), however, improvement with CDDO-TFEA at 100mg/kg diet did not quite reach significance (Figure 1B; Fischer's PLSD, p=0.12).
Figure 1. CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) improved motor performance in N171-82Q mice.
(A) Latency to fall during rotorod trials of N171-82Q mice treated with control diet, CDDO-EA at 100mg/kg diet and 200mg/kg diet. Data were expressed as means and standard errors. N171-82Q mice treated with CDDO-EA 100mg/kg diet and CDDO-EA 200mg/kg diet both showed significantly improved motor coordination compared to N171-82Q mice fed with control diet (p=0.04, p=0.0006). (B) Latency to fall during rotorod trials of N171-82Q mice treated with control diet, CDDO-TFEA at 100mg/kg diet, 200mg/kg diet and 400mg/kg diet. Data were expressed as means and standard errors. N171-82Q mice treated with CDDO-TFEA at 200mg/kg diet and CDDO-TFEA 400mg/kg diet both showed significantly improved motor coordination compared to N171-82Q mice fed control diet (p=0.005, p=0.0007).
CDDO-EA and CDDO-TFEA increased survival of N171-82Q mice
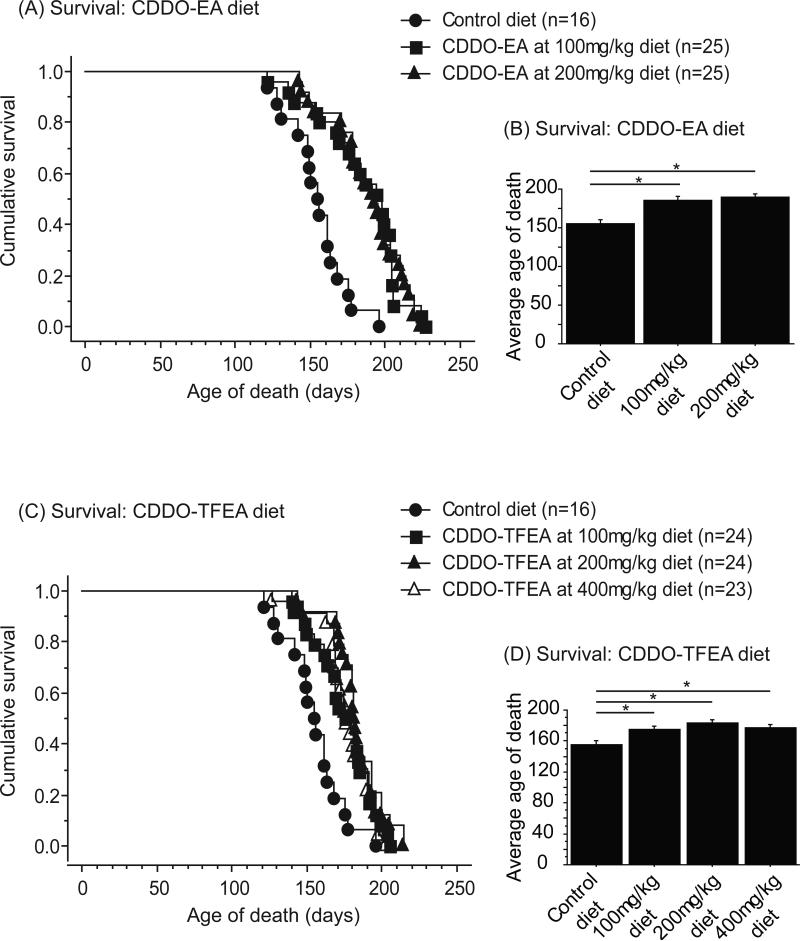
The cohort of mice used for behavioral analysis was also used for assessment of survival. Figure 2 shows a Kaplan-Meier plot of the survival of N171-82Q mice treated with control, CDDO-EA 100mg/kg and 200mg/kg diets and CDDO-TFEA 100mg/kg, 200mg/kg, and 400mg/kg diets. The average survival of N171-82Q mice fed control diet was 155 ± 4.8 days. The average survival of N171-82Q mice fed CDDO-EA was 185 ± 5.5 days for the 100mg/kg diet (19.4% increase from mice fed control diet), and 189 ± 4.8 days for the 200mg/kg diet (21.9% increase from mice fed control diet). Dietary supplementation of CDDO-EA at both doses significantly improved survival (Figure 2A, B; Fischer's PLSD, p=0.0002, p<0.0001). The average survival of N171-82Q mice fed CDDO-TFEA was 175 ± 3.9 days for the 100mg/kg diet (12.9% increase from mice fed control diet), 184 ± 3.5 days for the 200mg/kg diet (18.7% increase from mice fed control diet), and 177 ± 3.6 days for the 400mg/kg diet (14.2% increase from mice fed control diet). Each CDDO-TFEA diet significantly improved survival as compared to N171-82Q mice fed control diet (Figure 2C, D; Fischer's PLSD, p=0.001, p<0.0001, p=0.0003).
Figure 2. CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) improved survival in N171-82Q mice.
(A) Cumulative survival and (B) average age of death of N171-82Q mice treated with control diet, CDDO-EA at 100mg/kg diet and 200mg/kg diet. Data were expressed as means and standard errors. N171-82Q mice treated with CDDO-EA 100mg/kg diet and CDDO-EA 200mg/kg diet both showed significantly improved survival compared to mice fed control diet (p=0.0002, p<0.0001). (C) Cumulative survival and (D) average age of death of N171-82Q mice treated with control diet, CDDO-TFEA at 100mg/kg diet, 200mg/kg diet and 400mg/kg diet. Data were expressed as means and standard errors. N171-82Q mice treated with CDDO-TFEA at 100mg/kg diet, CDDO-TFEA 200mg/kg diet and CDDO-TFEA 400mg/kg diet each showed significantly improved survival compared to control (p=0.001, p=<0.0001, p=0.0003).
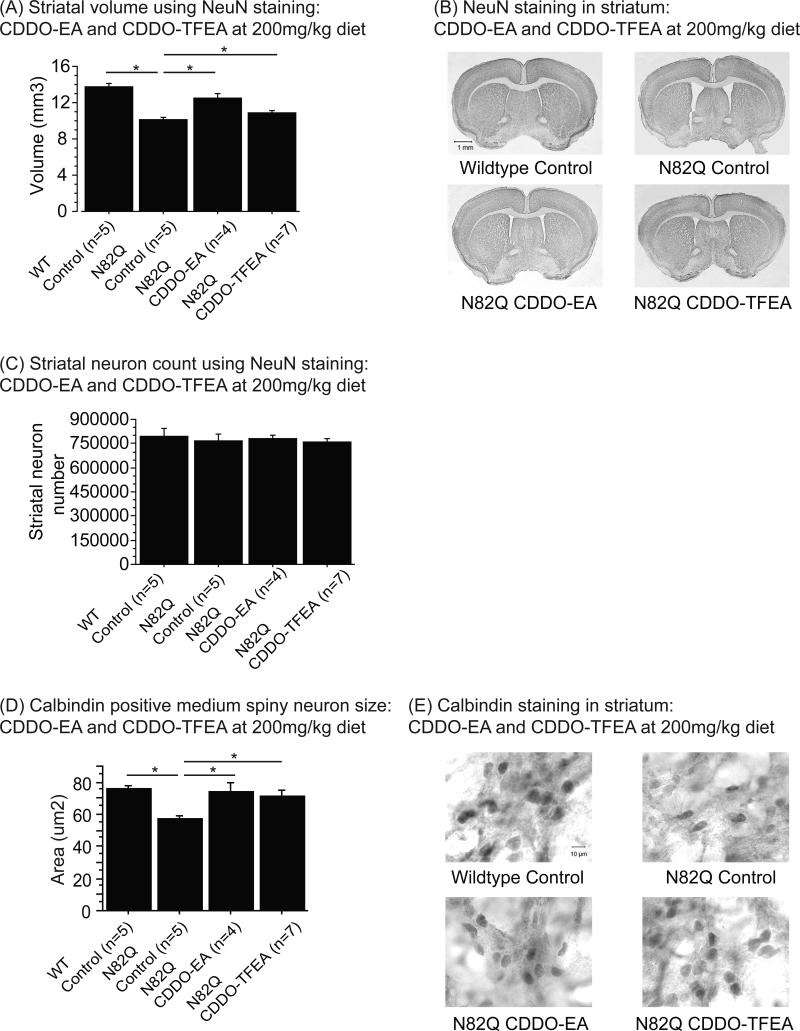
CDDO-EA and CDDO-TFEA at 200mg/kg in the diet attenuated striatal atrophy and reduction of neuron size in N171-82Q mice
Mice treated with CDDO-EA and CDDO-TFEA at 200mg/kg diet were used for brain pathology, because their effects on motor behavior were maximal at this dose. Since striatal degeneration is characteristic of N171-82Q mice, we analyzed striatal volume after staining with a NeuN antibody (Figure 3A, B). Striatal volume was significantly reduced in N171-82Q mice fed control diet compared to wildtype mice fed control diet (Figure 3A, B; Fischer's PLSD, p<0.0001). With CDDO-EA and CDDO-TFEA, there was a marked increase in striatal volume in N171-82Q mice compared to N171-82Q mice fed control diet (Figure 3A, B; Fisher's PLSD, p<0.0001, p=0.04). Figure 3B also demonstrates the increased ventricle size of N171-82Q mice fed control diet as compared to wildtype mice fed control diet, and the reduction of striatal atrophy in N171-82Q mice fed CDDO-EA and CDDO-TFEA. However, there was no change in striatal neuron count amongst any of the groups, consistent with previous observations (Figure 3C).
Figure 3. CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) attenuated striatal atrophy and reduction of medium spiny neuron size in N171-82Q mice.
(A) Striatal volume of NeuN labeled tissues of N171-82Q mice treated with control diet, CDDO-EA and CDDO-TFEA at 200mg/kg diet. Data were expressed as means and standard errors. (B) Photographs of brain sections labeled with NeuN antibody.
N171-82Q mice fed control diet showed a significant reduction in striatal volume compared to wildtype mice fed control diet (p<0.0001). However, treatment with both CDDO-EA and CDDO-TFEA significantly increased striatal volume in N171-82Q mice compared to N171-82Q mice fed control diet (p<0.0001, p=0.002).
(C) Striatal neuron count in NeuN labeled tissues of N171-82Q mice treated with control diet, CDDO-EA and CDDO-TFEA at 200mg/kg diet. Data were expressed as means and standard errors. There was no difference in striatal neuron count between any of the groups.
(D) Calbindin positive medium spiny neuron size of N171-82Q mice treated with control diet, CDDO-EA and CDDO-TFEA at 200mg/kg diet. Data were expressed as means and standard errors. (E) Photographs of brain sections labeled with calbindin antibody. N171-82Q mice fed control diet showed a significant reduction in medium spiny neuron size compared to wildtype mice fed control diet (p=0.001). N171-82Q mice treated with CDDO-EA and CDDO-TFEA showed significantly larger medium spiny neuron size compared to N171-82Q mice fed control diet (p=0.004, p=0.006).
Since there were no significant differences in the NeuN-positive neuron counts between groups, we determined whether alterations in striatal volume were associated with changes in neuron size. We analyzed the size of medium spiny neurons (Figure 3D, E) which are known to be preferentially vulnerable in HD transgenic mice [3]. Figure 3E shows calbindin immunoreactivity in the dorsolateral striatum. At baseline, calbindin positive medium spiny neurons of N171-82Q mice fed control diet were significantly smaller than those of wildtype mice fed control diet (Figure 3D, E; Fischer's PLSD, p=0.001). The average size of medium spiny neurons in N171-82Q mice fed control diet was 57.4 um2 while the average size of medium spiny neurons in wildtype mice fed control diet was 75.8 um2. CDDO-EA and CDDO-TFEA treatment at 200mg/kg diet markedly attenuated the reduction of medium spiny neuron size as compared to N171-82Q mice fed control diet (Figure 3D, E; Fischer's PLSD, p=0.004, p=0.006). Neuronal size in triterpenoid treated N171-82Q mice was not significantly different from those in wildtype mice fed control diet (average medium spiny neuron size was 74.3 um2 for N171-82Q mice treated with CDDO-EA at 200mg/kg diet, 71.4 um2 for N171-82Q mice treated with CDDO-TFEA, and 75.8 um2 for wildtype mice fed control diet).
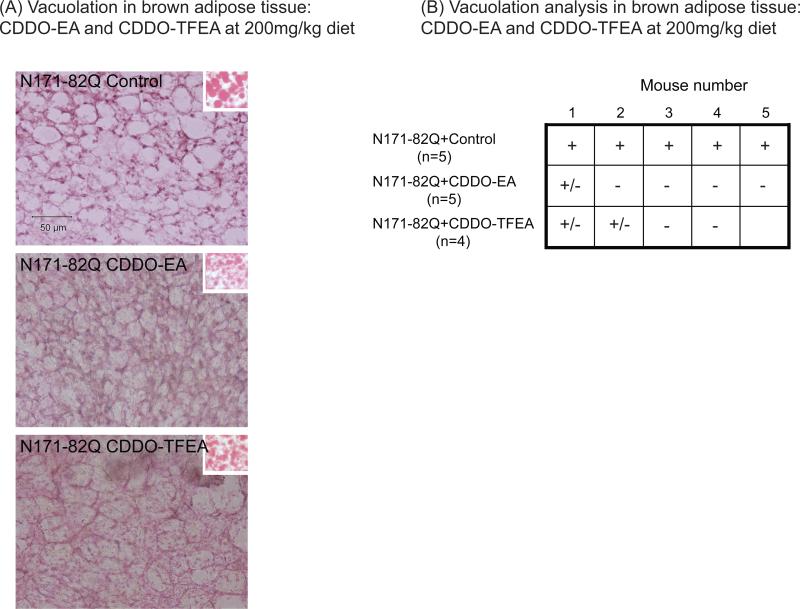
CDDO-EA and CDDO-TFEA at 200mg/kg in the diet attenuated vacuolation in the brown adipose tissue of N171-82Q mice
Mice treated with CDDO-EA and CDDO-TFEA at 200mg/kg diet were used for vacuolation analysis in the brown adipose tissue, because their effects on motor behavior were maximal at this dose. Sections were stained with both hematoxylin and eosin staining. Additionally, oil red O staining was performed in semi-adjacent sections in order to verify the presence of neutral lipid vacuoles.
CDDO-EA and CDDO-TFEA reduced vacuolation in the brown adipose tissue in N171-82Q transgenic mice as compared to N171-82Q mice fed control diet (Figure 4). CDDO-EA and CDDO-TFEA at 200mg/kg in the diet reduced oxidative and nitrosative stress in N171-82Q mice
Figure 4. CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) reduced vacuolation in the brown adipose tissue of N171-82Q mice.
(A) Photographs of brown adipose tissue stained by hematoxylin and eosin in N171-82Q transgenic mice fed control diet, CDDO-EA and CDDO-TFEA at 200mg/kg diet. Right insets show the red oil staining for lipids. (B) Vacuolation analysis of the brown adipose tissue in N171-82Q transgenic mice fed control diet, CDDO-EA and CDDO-TFEA at 200mg/kg diet. Data were expressed as “-” for the lack of extensive vacuolation, “+” for the presence of extensive vacuolation, and “+/-” for the presence of vacuolation in few area.
Oxidative and nitrosative stress are characteristic features of mouse models of Huntington's disease. Immunohistochemical analyses of oxidative and nitrosative stress were performed in striatal tissues using 8-hydroxy-2’-deoxyguanosine (8-OHdG), malondialdehyde (MDA), and 3-nitrotyrosine (3-NT) antibodies (Figure 5). Figure 5A shows 8-OHdG immunoreactivity, a marker of DNA damage, in the striatum. In N171-82Q mice fed control diet, there was increased staining for 8-OHdG as compared to wildtype mice fed control diet (Figure 5A). However, CDDO-EA and CDDO-TFEA markedly reduced 8-OHdG immunoreactivity in the striatum of the N171-82Q mice as compared to N171-82Q mice fed control diet (Figure 5A).
Figure 5. CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) reduces oxidative damage in N171-82Q mice.
(A) 8-hydroxy-2’-deoxyguanosine staining in striatal tissue of mice treated with control diet, CDDO-EA and CDDO-TFEA at 200mg/kg diet. (B) Malondialdehyde staining in striatal tissue of mice treated with control diet, CDDO-EA and CDDO-TFEA at 200mg/kg diet. (C) 3-Nitrotyrosine staining in striatal tissue of mice treated with control diet, CDDO-EA and CDDO-TFEA at 200mg/kg diet. Oxidative stress was increased in N171-82Q mice fed control diet compared to wildtype mice fed control diet. Administration of CDDO-EA and CDDO-TFEA in N171-82Q mice reduced oxidative stress compared to N171-82Q mice fed control diet.
Figure 5B shows immunoreactivity to MDA in the striatum, a marker of lipid peroxidation. In N171-82Q mice fed control diet, there was increased MDA staining in neurons and neuropil as compared to wildtype mice fed control diet (Figure 5B). CDDO-EA and CDDO-TFEA treatments substantially reduced MDA staining in neurons and neuropil as compared to N171-82Q mice fed control diet (Figure 5B).
Figure 5C shows immunoreactivity to 3-NT in the striatum, a marker of peroxynitrite formation. In N171-82Q mice fed control diet, there was increased 3-NT staining as compared to wildtype mice fed control diet (Figure 5C). CDDO-EA and CDDO-TFEA treatments markedly decreased 3-NT immunostaining in N171-82Q mice as compared to N171-82Q mice fed control diet (Figure 5C).
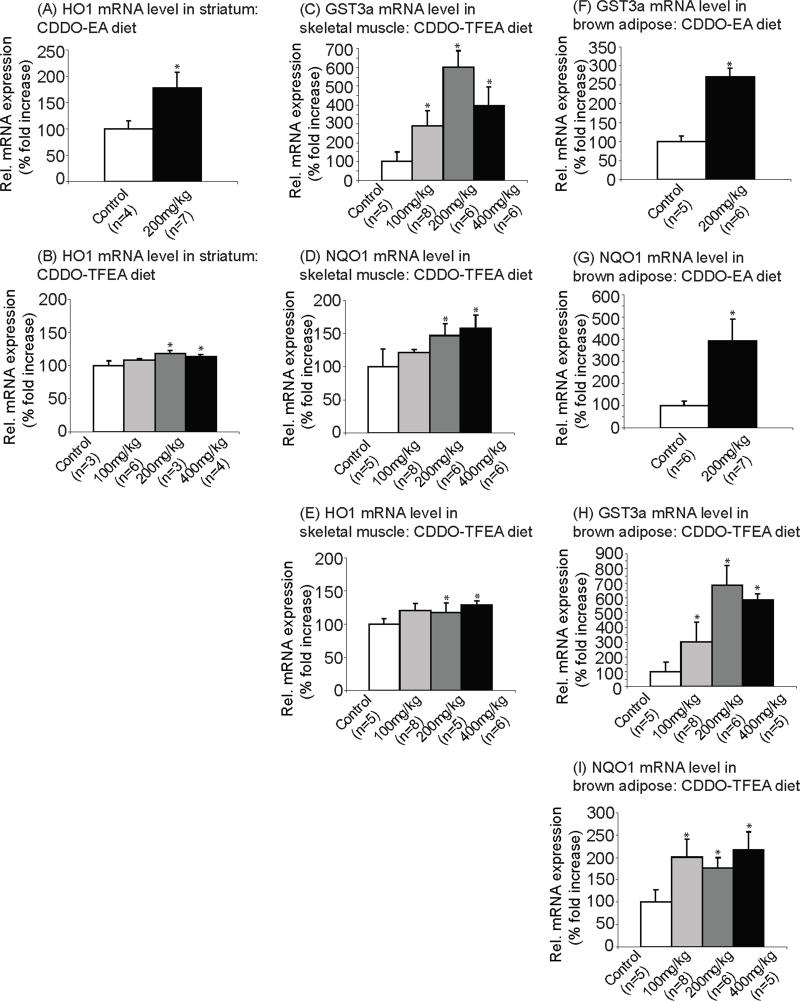
CDDO-EA and CDDO-TFEA upregulated expression of genes specifically involved in the Nrf2/ARE pathway in N171-82Q mice
For both CDDO-EA and CDDO-TFEA treated N171-82Q mice, striatum, hindlimb skeletal muscle, and brown adipose tissue were analyzed for the following three Nrf2/ARE induced genes: GST3a, NQO1 and HO1. Because of the multiple analyses, we report only the significant upregulations of these genes. It should be assumed that for the data not included in this section, we did not find significant differences in mRNA levels.
In the striatum, the mRNA level of HO1 was significantly increased in N171-82Q mice treated with CDDO-EA (Figure 6A; Fisher's PLSD, p=0.04 at 200mg/kg diet) and CDDO-TFEA (Figure 6B; Fisher's PLSD, p=0.009 at 200mg/kg diet, p=0.03 at 400mg/kg diet) as compared to N171-82Q mice fed control diet.
Figure 6. CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) upregulated expression of genes specifically involved in the Nrf2/ARE pathway in N171-82Q mice.
mRNA levels of HO1 in striatum of (A) CDDO-EA treated and (B) CDDO-TFEA treated N171-82Q mice. Data were expressed as means and standard errors. HO1 gene expression was significantly increased in striatum of N171-82Q mice treated with CDDO-EA (p=0.04 at 200mg/kg diet) and CDDO-TFEA (p=0.009 at 200mg/kg diet, p=0.03 at 400mg/kg diet) compared to N171-82Q mice fed control diet.
mRNA levels of (C) GST3a (D) NQO1 and (E) HO1 in skeletal muscle of CDDO-TFEA treated N171-82Q mice. Data were expressed as means and standard errors. mRNA level of GST3a (p=0.007 at 100mg/kg diet, p=0.0001 at 200mg/kg diet, p=0.002 at 400mg/kg diet), NQO1 (p=0.04 at 200mg/kg diet, p=0.02 at 400mg/kg diet) and HO1 (p=0.02 at 200mg/kg diet, p=0.03 at 400mg/kg diet) were significantly increased compared to mice fed control diet.
mRNA levels of (F) GST3a and (G) NQO1 in brown adipose tissue of CDDO-EA treated N171-82Q mice. Data were expressed as means and standard errors. GST3a gene expression was significantly increased in brown adipose tissue of CDDO-EA treated mice compared to control (p=0.005 at 200mg/kg diet). NQO1 gene expression was also significantly increased in brown adipose tissue of CDDO-EA treated mice compared to control (p=0.0002 at 200mg/kg diet).
mRNA levels of (H) GST3a and (I) NQO1 in brown adipose tissue of CDDO-TFEA treated N171-82Q mice. Data were expressed as means and standard errors. Both GST3a gene expression (p=0.003 at 100mg/kg diet, p<0.0001 at 200mg/kg diet, p<0.0001 at 400mg/kg diet) and NQO1 gene expression (p=0.005 at 100mg/kg diet, p=0.03 at 200mg/kg diet, p=0.005 at 400mg/kg diet) were significantly increased in CDDO-TFEA treated mice.
We also found that in skeletal muscle, after CDDO-TFEA administration, the mRNA levels of the following genes were significantly increased as compared to control diet: GST3a (Figure 6C; Fisher's PLSD, p=0.007 at 100mg/kg diet, p=0.0001 at 200mg/kg diet, p=0.001 at 400mg/kg diet), NQO1 (Figure 6D; Fisher's PLSD, p=0.04 at 200mg/kg diet, p=0.02 at 400mg/kg diet) and HO1 (Figure 6E; Fisher's PLSD, p=0.02 at 200mg/kg diet, p=0.03 at 400mg/kg diet).
In brown adipose tissue of the N171-82Q mice, we found that the mRNA level of GST3a was significantly increased after both CDDO-EA (Figure 6F; Fisher's PLSD, p=0.005 at 200mg/kg diet) and CDDO-TFEA administration (Figure 6H; Fisher's PLSD, p=0.003 at 100mg/kg diet, p<0.0001 at 200mg/kg diet, p<0.0001 at 400mg/kg diet) as compared to control diet. Similarly, we found that the mRNA level of NQO1 was significantly increased after both CDDO-EA (Figure 6G; Fisher's PLSD, p=0.0002 at 200mg/kg diet) and CDDO-TFEA (Figure 6I; Fisher's PLSD, p=0.005 at 100mg/kg diet, p=0.03 at 200mg/kg diet, p=0.005 at 400mg/kg diet) administration as compared to control diet.
Discussion
Many studies have demonstrated that increased oxidative stress and inflammation occurs in HD and thus may play a role in its pathogenesis. In HD patients and animal models, accumulation of ROS triggers oxidative and nitrosative stress in neurons [5, 7]. Oxidative and nitrosative stress are injurious to DNA, proteins, and lipids leading to cell dysfunction and death. Oxidative damage is prominent in the striatum of HD patients and animal models, and ultimately contributes to brain and behavioral pathology, including motor impairment and cognitive dysfunction [11, 19, 24].
Many therapeutic approaches target exogenous antioxidants as a way to prevent oxidative damage. A study in a Drosophila model of oxidative stress suggested that dietary supplementation with the antioxidant vitamin E increased survival [43]. Other studies have investigated the effect of ascorbate in the R6/2 mouse model of HD, and have shown that ascorbate improved motor signs in these mice [44]. We previously showed that the antioxidants lipoic acid and BN82451 both exerted modest neuroprotective effects and increased survival in transgenic mouse models of HD [42, 45]. We also showed that coenzyme Q10, which acts as an antioxidant and also improves the efficiency of the electron transport chain, was effective in transgenic mouse models of HD [46]. Coenzyme Q10 also lowered increased lactate levels in occipital cortex of HD patients [47]. In a randomized clinical trial coenzyme Q10 at 600mg/day showed a trend to slow decline in total functional capacity, and improved measures of cognition [48]. A further trial using coenzyme Q10 at a dose of 2400mg daily is now underway (CARE-HD).
Another important pathway in combating oxidative stress is the Nrf2/ARE signaling pathway, which responds by activating Phase II detoxification [49, 50]. It has been shown that in neurodegenerative diseases, Nrf2 expression is altered in both neurons and astrocytes [51]. It should be noted that ARE regulated genes are preferentially activated in astrocytes [52] and that astrocytes can play a role in Nrf2 mediated neuroprotection during neurodegeneration. After both malonate or 6-hydroxydopamine lesioning in mice, transplanting Nrf2-overexpressing astrocytes into the striatum protected against malonate- or 6-hydroxydopamine-induced damage [49, 53]. In addition, Nrf2 deficiency increased sensitivity to MPTP or 6-hydroxydopamine administration in mice whereas overexpression of GFAP-Nrf2 abolished MPTP or 6-hydroxydopamine toxicity [53, 54]. Also, crossing a mouse with Nrf2-overexpressing astrocytes with the G93A SOD transgenic mouse model of ALS produced delayed onset of symptoms and improved survival [55].
Tert-butylhydroquinone (tBHQ) and sulforaphane are two potent pharmacological inducers of the Phase II detoxification pathway, the latter increasing glutathione-S-transferase and quinone reductase activity. These two compounds have been widely used as Nrf2/ARE activators to protect against oxidative and cellular damage, especially in in vitro and in vivo models of Parkinson's disease (PD). Activation of the Nrf2/ARE pathway in astrocytes after tBHQ or sulforaphane treatment protected neurons against oxidative stress-induced cell death in cultures [56, 57], and increased NQO1 activity [56]. Both drugs protected against 6-hydroxydopamine toxicity in rat organotypic nigrostriatal cocultures, once again increasing NQO1 expression [58], and against MPTP toxicity in mice [59]. Sulforaphane also reduced toxicity in human dopaminergic neuroblastoma SH-SY5Y cells induced by arsenic and dopamine treatment [60]. Furthermore, induction of the Phase II detoxification pathway by sulforaphane suppressed neuronal loss in Drosophila Parkin mutants, a model of PD [61]. These findings suggest that the activation of Nrf2/ARE play a key role in the protection against neurodegeneration, especially in PD. Another study showed that DJ-1, a protein in which mutations cause PD, stabilizes Nrf2 by preventing its association with its inhibitor protein Keap1 and its subsequent ubiquitination [62]. Indeed, in cells, deficiency of DJ-1 affected expression of NQO1 in an Nrf2-dependent manner [62].
Synthetic triterpenoids, which are analogues of 2-Cyano-3,12-Dioxooleana-1,9-Dien-28-Oic acid (CDDO), have been of great interest because of their antioxidant and anti-inflammatory properties. Such agents induced Nrf2/ARE regulated genes, such as increasing NQO1 [35] and repressing iNOS expression [35, 63, 64]. More specifically, CDDO-methyl ester reduced nitric oxide levels produced in response to interferon-gamma in mouse macrophages [36, 37]. CDDO-methyl amide improved memory, reduced plaque burden, Aβ42 levels, and protein oxidation in a transgenic mouse model of Alzheimer's disease [38]. CDDO-imidazolide was effective in decreasing DNA damage produced by hypoxia in mouse lungs [65]. It also increased the expression level of HO1 and reduced ROS production in cells treated with tert-butyl hydroperoxide [31]. CDDO-TFEA induced sulfiredoxin in rodents [39] and upregulated expression of NQO1 in ARPE-19 retinal pigment epithelial cells, as well as in retinas of BALB/c mice following photo-oxidative stress [66].
Because of their potency in stimulating endogenous antioxidant and anti-inflammatory pathways, we examined whether the TPs could improve behavioral deficits and brain pathology of N171-82Q mice, a transgenic HD mouse model. These mice exhibit progressive behavioral impairment including a loss of motor skills in rotorod testing and impaired survival [41]. We found that CDDO-EA and CDDO-TFEA produced higher concentrations in brains as compared to CDDO itself, and other CDDO analogues such as CDDO-MA. Thus, in our study, we administered both CDDO-EA at 100mg/kg diet and 200mg/kg diet and CDDO-TFEA at 100mg/kg diet, 200mg/kg diet and 400mg/kg diet.
Both compounds significantly ameliorated motor impairment of N171-82Q mice in the accelerated rotorod test. This improvement was seen from 119 to 182 days of age, when the mice became symptomatic. Both CDDO-EA and CDDO-TFEA increased survival without affecting the body weight of N171-82Q mice at all doses. More precisely, CDDO-EA increased survival by 19.4% at 100mg/kg diet and 21.9% at 200mg/kg diet while CDDO-TFEA increased survival by 12.9% at 100mg/kg diet, 18.7% at 200mg/kg diet, and 14.2% at 400mg/kg diet. These increases in survival are comparable to the highest range of percent increases in survival seen in other therapeutic trials in mouse models of HD. For example, in both the R6/2 and N171-82Q mouse models of HD, there have been survival increases of 6.8% with dichloroacetate, 7.1% with lipoic acid, 15.5% with remacemide, and 17.4% with creatine [67]. We also showed that administration of creatine increased survival by 19% in N171-82Q mice [42] and that administration of coenzyme Q10 with remacemide or administration of mithramycin increased survival by 31.8% or by 29.1% in R6/2 mice, respectively [68, 69]. This indicates that CDDO-EA and CDDO-TFEA both improve the behavioral phenotype and survival of N171-82Q mice in a comparable range to the best therapeutic interventions thus far tested.
CDDO-EA and CDDO-TFEA exerted neuroprotective effects. Striatal atrophy is believed to play a role in HD symptoms such as motor impairment. Neostriatal atrophy, which is a consequence of loss or atrophy of striatal neurons, is a pathologic hallmark of HD. The severity of striatal degeneration correlates with the severity of the disease [1, 2]. In mice treated with MPTP and 3-nitropropionic acid, models of PD and HD, respectively, CDDO-MA protected against neuronal loss [34]. In our study, we found that CDDO-EA and CDDO-TFEA attenuated striatal atrophy in N171-82Q mice at 120 days of age. Interestingly, CDDO-EA exhibited similar neuroprotective effects as compared to CDDO-TFEA despite having lower brain concentrations. This suggests that CDDO-EA may be more active than CDDO-TFEA.
To investigate the mechanism by which CDDO-EA and CDDO-TFEA were improving striatal volumes, we assessed both striatal neuron counts and striatal neuron size. Our data showed that striatal atrophy in N171-82Q transgenic mice was not caused by a general loss of striatal neurons, but rather was due to a reduction in the size of a specific subtype of striatal neurons, the medium spiny neurons. These neurons are particularly affected in HD [3]. This finding is consistent with previous reports [42]. CDDO-EA and CDDO-TFEA prevented the atrophy of the medium spiny neurons in N171-82Q mice, and the size of the neurons in the treated mice was similar to the size of the neurons in the wildtype mice. Our data therefore show that CDDO-EA and CDDO-TFEA preserved striatal volume in the N171-82Q mice by preventing atrophy of medium spiny neurons. In this study we cannot dismiss the possibility of triterpenoids affecting Nrf2 mediated neuroprotection from astrocytes. As aforementioned, astrocytes have a particularly robust detoxification and anti-inflammatory effect associated with increased Nrf2 expression. Future investigations should explore this possibility.
In addition to their action in the brain, CDDO-EA and CDDO-TFEA were also beneficial in peripheral tissue. In HD transgenic mouse models, it is well established that mice develop peripheral pathology, including vacuolation of the brown adipose tissue [70-72]. PGC1-α is involved with thermoregulation in brown adipose tissue, however, overexpression of the mutant huntingtin interferes with PGC1- α transcription and leads to abnormal thermoregulation in HD mice [72]. In the brown adipose tissue of N171-82Q transgenic mice, defective PGC1-alpha transcription, mitochondria dysfunction, and blunted uncoupling protein 1 (UCP-1) response contribute to impaired thermoregulation in the periphery [73]. In our experiments, both CDDO-EA and CDDO-TFEA reduced vacuolation in the brown adipose tissue of N171-82Q transgenic mice as compared to mice fed control diet.
Because oxidative and nitrosative stress can cause cell damage and CDDO analogues have been shown to activate Nrf2/ARE related genes, we examined whether the neuroprotection is associated with a reduction of oxidative damage markers in brain tissue of N171-82Q mice treated with control, CDDO-EA, and CDDO-TFEA diets. CDDO-EA and CDDO-TFEA reduced 8-OHdG, MDA and 3-NT immunoreactivity in the striatal tissues. This is consistent with other reports using animal models of neurodegenerative diseases. In Tg19959 mouse brains, a model of Alzheimer's disease, CDDO-MA reduced protein carbonyl levels [38]. CDDO-MA also reduced 8-OHdG, MDA, and isoprostane levels in MPTP and 3-nitropropionic acid treated rodents [34]. In addition, CDDO-EA and CDDO-TFEA rescued retinal cells from oxidative stress induced cell death [66].
Reduced oxidative damage markers indicated that CDDO-EA and CDDO-TFEA may reduce oxidative stress by activating the Nrf2/ARE pathway. CDDO analogues induced Nrf2/ARE regulated genes such as NQO1, HO1, and GST3a [35, 63, 64]. To determine whether upregulation of Nrf2/ARE related genes was a possible mechanism whereby CDDO-EA and CDDO-TFEA exert their beneficial effects, we examined the gene expression of NQO1, HO1 and GST3a in brain and in peripheral tissues of the N171-82Q mice. We focused our efforts on these 3 genes that CDDO analogues have previously been shown to upregulate, and which play a crucial role in the endogenous antioxidant system. NQO1 is an important enzyme that detoxifies protein-bound quinones, and maintains alpha-tocopherol and coenzyme Q10 in their reduced antioxidant states [74]. HO1 is an inducible isoform of heme oxygenase that is highly responsive to oxidative stress. HO1 is implicated in the metabolism of the prooxidant heme to the antioxidant pigment biliverdin, ferrous iron and carbon monoxide [75]. Finally, we examined the mRNA levels of GST3a. Glutathione, which is utilized by GST3a [74, 76], serves as a major endogenous antioxidant in cells, and scavenges ROS.
In our study, we found that CDDO-EA and CDDO-TFEA upregulated HO1 mRNA level in the striatum, consistent with our previous report in Tg19959 mouse brain [38]. In peripheral tissues, CDDO-EA and CDDO-TFEA upregulated mRNA levels of GST3a, NQO1 and HO1. Therefore, CDDO-EA and CDDO-TFEA upregulate Nrf2/ARE induced genes both in the brain and in the periphery.
In conclusion, our study showed that both CDDO-EA and CDDO-TFEA rescued behavioral deficits, extended survival, and attenuated brain and peripheral pathology in the N171-82Q transgenic mouse model of HD. CDDO-EA and CDDO-TFEA upregulated the Nrf2/ARE pathway, leading to increased expression of important antioxidant genes, and reduction of several indices of oxidative stress. Because Nrf2 is affected in human neurodegenerative diseases such as Alzheimer's disease [51], therapeutics that target Nrf2 are of great interest for human patients. In order to restore the oxidative stress response pathways and prevent further neurodegeneration, CDDOs could be given at early stages of human disease. This novel therapeutic approach therefore holds promise for the treatment and prevention of HD, and other neurodegenerative diseases.
Supplementary Material
Supplementary figure 1: Levels of CDDO analogues in brain of wildtype mice after feeding.
(A) Chemical structures of CDDO analogues. (B) Comparison of CDDO-MA and CDDO-EA brain levels after feeding at 800mg/kg diet for 2 days. (C) Comparison of CDDO-EA and CDDO-TFEA brain levels after feeding at 200mg/kg diet and 400mg/kg diet for 3 days. Data were expressed as means and standard errors. In wildtype mice, brain levels of CDDO-EA were higher than CDDO-MA, and brain levels of CDDO-TFEA were higher than CDDO-EA.
Supplementary figure 2: Chromatograms of standard levels of CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) in N171-82Q mice. Chromatograms of standard dilution series for (A) CDDO-EA and (B) CDDO-TFEA added to mouse tissue homogenates at various concentrations. Areas under the curve increase with increasing concentrations.
Supplementary figure 3: Levels of CDDO-ethyl amide (CDDO-EA) in the brain of N171-82Q mice.
Chromatograms of (A) CDDO-EA at 100mg/kg diet and (B) at 200mg/kg diet in N171-82Q mouse brain.
Supplementary figure 4: Levels of CDDO-ethyl amide (CDDO-EA) in the skeletal muscle of N171-82Q mice.
Chromatograms of (A) CDDO-EA at 100mg/kg diet and (B) at 200mg/kg diet in N171-82Q mouse muscle. (C) Levels of CDDO-EA in the muscle of N171-82Q mice. Data were expressed as means and standard errors.
Supplementary figure 5: Levels of CDDO- trifluoroethyl amide (CDDO-TFEA) in the brain of N171-82Q mice.
Chromatograms of (A) CDDO-TFEA at 100mg/kg diet and (B) at 200mg/kg diet, and (C) at 400mg/kg diet in N171-82Q mouse brain. (D) Levels of CDDO-TFEA in the brain of N171-82Q mice. Data were expressed as means and standard errors.
Supplementary figure 6: Levels of CDDO-trifluoroethyl amide (CDDO-TFEA) in the skeletal muscle of N171-82Q mice.
Chromatograms of (A) CDDO-TFEA at 100mg/kg diet and (B) at 200mg/kg diet, and (C) at 400mg/kg diet in N171-82Q mouse muscle. (D) Levels of CDDO-TFEA in the muscle of N171-82Q mice. Data were expressed as means and standard errors.
Supplementary figure 7: CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) did not affect food intake in N171-82Q mice.
Body weights of N171-82Q mice fed with CDDO-EA (A for males and B for females) and CDDO-TFEA diets (C for males and D for females). Data were expressed as means and standard errors. CDDO-EA and CDDO-TFEA did not affect food intake in N171-82Q mice.
Acknowledgments
We thank Reata Pharmaceuticals (Dallas, TX, USA) for providing the CDDO-EA and CDDO-TFEA. We thank the Weill Cornell Medical College Microarray Core Facility for performing qRT-PCR. We also thank Lichuan Yang and Michael T. Lin for their help in the study. This work was supported by the National Institute of Health grant NS39258 and U01NS49077, and by the Huntington's Disease Society of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Halliday GM, McRitchie DA, Macdonald V, Double KL, Trent RJ, McCusker E. Regional specificity of brain atrophy in Huntington's disease. Exp Neurol. 1998;154(2):663–672. doi: 10.1006/exnr.1998.6919. [DOI] [PubMed] [Google Scholar]
- 2.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44(6):559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Ferrante RJ, Kowall NW, Richardson EP., Jr Proliferative and degenerative changes in striatal spiny neurons in Huntington's disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J Neurosci. 1991;11(12):3877–3887. doi: 10.1523/JNEUROSCI.11-12-03877.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008;45(5):667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Browne SE, Beal MF. Oxidative damage in Huntington's disease pathogenesis. Antioxid Redox Signal. 2006;8(11-12):2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 6.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 7.Stack EC, Matson WR, Ferrante RJ. Evidence of oxidant damage in Huntington's disease: translational strategies using antioxidants. Ann N Y Acad Sci. 2008;1147:79–92. doi: 10.1196/annals.1427.008. [DOI] [PubMed] [Google Scholar]
- 8.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327(6117):77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 9.Valavanidis A, Vlachogianni T, Fiotakis C. 8-hydroxy-2' -deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2009;27(2):120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 10.Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, Bird ED, Beal MF. Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41(5):646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 11.Dragunow M, Faull RL, Lawlor P, Beilharz EJ, Singleton K, Walker EB, Mee E. In situ evidence for DNA fragmentation in Huntington's disease striatum and Alzheimer's disease temporal lobes. Neuroreport. 1995;6(7):1053–1057. doi: 10.1097/00001756-199505090-00026. [DOI] [PubMed] [Google Scholar]
- 12.Polidori MC, Mecocci P, Browne SE, Senin U, Beal MF. Oxidative damage to mitochondrial DNA in Huntington's disease parietal cortex. Neurosci Lett. 1999;272(1):53–56. doi: 10.1016/s0304-3940(99)00578-9. [DOI] [PubMed] [Google Scholar]
- 13.Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington's disease. Brain Pathol. 1999;9(1):147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CM, Wu YR, Cheng ML, Liu JL, Lee YM, Lee PW, Soong BW, Chiu DT. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington's disease patients. Biochem Biophys Res Commun. 2007;359(2):335–340. doi: 10.1016/j.bbrc.2007.05.093. [DOI] [PubMed] [Google Scholar]
- 15.Hersch SM, Gevorkian S, Marder K, Moskowitz C, Feigin A, Cox M, Como P, Zimmerman C, Lin M, Zhang L, Ulug AM, Beal MF, Matson W, Bogdanov M, Ebbel E, Zaleta A, Kaneko Y, Jenkins B, Hevelone N, Zhang H, Yu H, Schoenfeld D, Ferrante R, Rosas HD. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2'dG. Neurology. 2006;66(2):250–252. doi: 10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- 16.Bogdanov MB, Andreassen OA, Dedeoglu A, Ferrante RJ, Beal MF. Increased oxidative damage to DNA in a transgenic mouse model of Huntington's disease. J Neurochem. 2001;79(6):1246–1249. doi: 10.1046/j.1471-4159.2001.00689.x. [DOI] [PubMed] [Google Scholar]
- 17.Acevedo-Torres K, Berrios L, Rosario N, Dufault V, Skatchkov S, Eaton MJ, Torres-Ramos CA, Ayala-Torres S. Mitochondrial DNA damage is a hallmark of chemically induced and the R6/2 transgenic model of Huntington's disease. DNA Repair (Amst) 2009;8(1):126–136. doi: 10.1016/j.dnarep.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson VL, Dawson TM. Nitric oxide neurotoxicity. J Chem Neuroanat. 1996;10(3-4):179–190. doi: 10.1016/0891-0618(96)00148-2. [DOI] [PubMed] [Google Scholar]
- 19.Perez-De La Cruz V, Gonzalez-Cortes C, Galvan-Arzate S, Medina-Campos ON, Perez-Severiano F, Ali SF, Pedraza-Chaverri J, Santamaria A. Excitotoxic brain damage involves early peroxynitrite formation in a model of Huntington's disease in rats: protective role of iron porphyrinate 5,10,15,20-tetrakis (4-sulfonatophenyl)porphyrinate iron (III). Neuroscience. 2005;135(2):463–474. doi: 10.1016/j.neuroscience.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 20.Tabrizi SJ, Schapira AH. Secondary abnormalities of mitochondrial DNA associated with neurodegeneration. Biochem Soc Symp. 1999;66:99–110. doi: 10.1042/bss0660099. [DOI] [PubMed] [Google Scholar]
- 21.Stoy N, Mackay GM, Forrest CM, Christofides J, Egerton M, Stone TW, Darlington LG. Tryptophan metabolism and oxidative stress in patients with Huntington's disease. J Neurochem. 2005;93(3):611–623. doi: 10.1111/j.1471-4159.2005.03070.x. [DOI] [PubMed] [Google Scholar]
- 22.Montine TJ, Beal MF, Robertson D, Cudkowicz ME, Biaggioni I, O'Donnell H, Zackert WE, Roberts LJ, Morrow JD. Cerebrospinal fluid F2-isoprostanes are elevated in Huntington's disease. Neurology. 1999;52(5):1104–1105. doi: 10.1212/wnl.52.5.1104. [DOI] [PubMed] [Google Scholar]
- 23.Klepac N, Relja M, Klepac R, Hecimovic S, Babic T, Trkulja V. Oxidative stress parameters in plasma of Huntington's disease patients, asymptomatic Huntington's disease gene carriers and healthy subjects : a cross-sectional study. J Neurol. 2007;254(12):1676–1683. doi: 10.1007/s00415-007-0611-y. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Severiano F, Rios C, Segovia J. Striatal oxidative damage parallels the expression of a neurological phenotype in mice transgenic for the mutation of Huntington's disease. Brain Res. 2000;862(1-2):234–237. doi: 10.1016/s0006-8993(00)02082-5. [DOI] [PubMed] [Google Scholar]
- 25.Pavese N, Gerhard A, Tai YF, Ho AK, Turkheimer F, Barker RA, Brooks DJ, Piccini P. Microglial activation correlates with severity in Huntington disease: a clinical and PET study. Neurology. 2006;66(11):1638–1643. doi: 10.1212/01.wnl.0000222734.56412.17. [DOI] [PubMed] [Google Scholar]
- 26.Sapp E, Kegel KB, Aronin N, Hashikawa T, Uchiyama Y, Tohyama K, Bhide PG, Vonsattel JP, DiFiglia M. Early and progressive accumulation of reactive microglia in the Huntington disease brain. J Neuropathol Exp Neurol. 2001;60(2):161–172. doi: 10.1093/jnen/60.2.161. [DOI] [PubMed] [Google Scholar]
- 27.Tai YF, Pavese N, Gerhard A, Tabrizi SJ, Barker RA, Brooks DJ, Piccini P. Microglial activation in presymptomatic Huntington's disease gene carriers. Brain. 2007;130(Pt 7):1759–1766. doi: 10.1093/brain/awm044. [DOI] [PubMed] [Google Scholar]
- 28.Holtzclaw WD, Dinkova-Kostova AT, Talalay P. Protection against electrophile and oxidative stress by induction of phase 2 genes: the quest for the elusive sensor that responds to inducers. Adv Enzyme Regul. 2004;44:335–367. doi: 10.1016/j.advenzreg.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26(1):221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278(14):12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 31.Liby K, Hock T, Yore MM, Suh N, Place AE, Risingsong R, Williams CR, Royce DB, Honda T, Honda Y, Gribble GW, Hill-Kapturczak N, Agarwal A, Sporn MB. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65(11):4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- 32.Nichols DP, Ziady AG, Shank SL, Eastman JF, Davis PB. The triterpenoid CDDO limits inflammation in preclinical models of cystic fibrosis lung disease. Am J Physiol Lung Cell Mol Physiol. 2009;297(5):L828–836. doi: 10.1152/ajplung.00171.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thimmulappa RK, Scollick C, Traore K, Yates M, Trush MA, Liby KT, Sporn MB, Yamamoto M, Kensler TW, Biswal S. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDOImidazolide. Biochem Biophys Res Commun. 2006;351(4):883–889. doi: 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L, Calingasan NY, Thomas B, Chaturvedi RK, Kiaei M, Wille EJ, Liby KT, Williams C, Royce D, Risingsong R, Musiek ES, Morrow JD, Sporn M, Beal MF. Neuroprotective effects of the triterpenoid, CDDO methyl amide, a potent inducer of Nrf2-mediated transcription. PLoS One. 2009;4(6):e5757. doi: 10.1371/journal.pone.0005757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yates MS, Tauchi M, Katsuoka F, Flanders KC, Liby KT, Honda T, Gribble GW, Johnson DA, Johnson JA, Burton NC, Guilarte TR, Yamamoto M, Sporn MB, Kensler TW. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol Cancer Ther. 2007;6(1):154–162. doi: 10.1158/1535-7163.MCT-06-0516. [DOI] [PubMed] [Google Scholar]
- 36.Honda T, Honda Y, Favaloro FG, Jr., Gribble GW, Suh N, Place AE, Rendi MH, Sporn MB. A novel dicyanotriterpenoid, 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-onitrile, active at picomolar concentrations for inhibition of nitric oxide production. Bioorg Med Chem Lett. 2002;12(7):1027–1030. doi: 10.1016/s0960-894x(02)00105-1. [DOI] [PubMed] [Google Scholar]
- 37.Honda T, Rounds BV, Bore L, Finlay HJ, Favaloro FG, Jr., Suh N, Wang Y, Sporn MB, Gribble GW. Synthetic oleanane and ursane triterpenoids with modified rings A and C: a series of highly active inhibitors of nitric oxide production in mouse macrophages. J Med Chem. 2000;43(22):4233–4246. doi: 10.1021/jm0002230. [DOI] [PubMed] [Google Scholar]
- 38.Dumont M, Wille E, Calingasan NY, Tampellini D, Williams C, Gouras GK, Liby K, Sporn M, Nathan C, Flint Beal M, Lin MT. Triterpenoid CDDO-methylamide improves memory and decreases amyloid plaques in a transgenic mouse model of Alzheimer's disease. J Neurochem. 2009;109(2):502–512. doi: 10.1111/j.1471-4159.2009.05970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soriano FX, Baxter P, Murray LM, Sporn MB, Gillingwater TH, Hardingham GE. Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol Cells. 2009;27(3):279–282. doi: 10.1007/s10059-009-0050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liby K, Royce DB, Williams CR, Risingsong R, Yore MM, Honda T, Gribble GW, Dmitrovsky E, Sporn TA, Sporn MB. The synthetic triterpenoids CDDO-methyl ester and CDDO-ethyl amide prevent lung cancer induced by vinyl carbamate in A/J mice. Cancer Res. 2007;67(6):2414–2419. doi: 10.1158/0008-5472.CAN-06-4534. [DOI] [PubMed] [Google Scholar]
- 41.Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, Copeland NG, Price DL, Ross CA, Borchelt DR. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8(3):397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 42.Andreassen OA, Dedeoglu A, Ferrante RJ, Jenkins BG, Ferrante KL, Thomas M, Friedlich A, Browne SE, Schilling G, Borchelt DR, Hersch SM, Ross CA, Beal MF. Creatine increase survival and delays motor symptoms in a transgenic animal model of Huntington's disease. Neurobiol Dis. 2001;8(3):479–491. doi: 10.1006/nbdi.2001.0406. [DOI] [PubMed] [Google Scholar]
- 43.Bahadorani S, Bahadorani P, Phillips JP, Hilliker AJ. The effects of vitamin supplementation on Drosophila life span under normoxia and under oxidative stress. J Gerontol A Biol Sci Med Sci. 2008;63(1):35–42. doi: 10.1093/gerona/63.1.35. [DOI] [PubMed] [Google Scholar]
- 44.Rebec GV, Barton SJ, Marseilles AM, Collins K. Ascorbate treatment attenuates the Huntington behavioral phenotype in mice. Neuroreport. 2003;14(9):1263–1265. doi: 10.1097/00001756-200307010-00015. [DOI] [PubMed] [Google Scholar]
- 45.Klivenyi P, Gardian G, Calingasan NY, Yang L, Beal MF. Additive neuroprotective effects of creatine and a cyclooxygenase 2 inhibitor against dopamine depletion in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) mouse model of Parkinson's disease. J Mol Neurosci. 2003;21(3):191–198. doi: 10.1385/jmn:21:3:191. [DOI] [PubMed] [Google Scholar]
- 46.Ferrante RJ, Andreassen OA, Dedeoglu A, Ferrante KL, Jenkins BG, Hersch SM, Beal MF. Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington's disease. J Neurosci. 2002;22(5):1592–1599. doi: 10.1523/JNEUROSCI.22-05-01592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koroshetz WJ, Jenkins BG, Rosen BR, Beal MF. Energy metabolism defects in Huntington's disease and effects of coenzyme Q10. Ann Neurol. 1997;41(2):160–165. doi: 10.1002/ana.410410206. [DOI] [PubMed] [Google Scholar]
- 48.A randomized, placebo-controlled trial of coenzyme Q10 and remacemide in Huntington's disease. Neurology. 2001;57(3):397–404. doi: 10.1212/wnl.57.3.397. [DOI] [PubMed] [Google Scholar]
- 49.Johnson JA, Johnson DA, Kraft AD, Calkins MJ, Jakel RJ, Vargas MR, Chen PC. The Nrf2-ARE pathway: an indicator and modulator of oxidative stress in neurodegeneration. Ann N Y Acad Sci. 2008;1147:61–69. doi: 10.1196/annals.1427.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14(1):76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramsey CP, Glass CA, Montgomery MB, Lindl KA, Ritson GP, Chia LA, Hamilton RL, Chu CT, Jordan-Sciutto KL. Expression of Nrf2 in neurodegenerative diseases. J Neuropathol Exp Neurol. 2007;66(1):75–85. doi: 10.1097/nen.0b013e31802d6da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vargas MR, Johnson JA. The Nrf2-ARE cytoprotective pathway in astrocytes. Expert Rev Mol Med. 2009;11:e17. doi: 10.1017/S1462399409001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jakel RJ, Townsend JA, Kraft AD, Johnson JA. Nrf2-mediated protection against 6-hydroxydopamine. Brain Res. 2007;1144:192–201. doi: 10.1016/j.brainres.2007.01.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen PC, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, Kan YW, Johnson JA. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson's disease: Critical role for the astrocyte. Proc Natl Acad Sci U S A. 2009;106(8):2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vargas MR, Johnson DA, Sirkis DW, Messing A, Johnson JA. Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J Neurosci. 2008;28(50):13574–13581. doi: 10.1523/JNEUROSCI.4099-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danilov CA, Chandrasekaran K, Racz J, Soane L, Zielke C, Fiskum G. Sulforaphane protects astrocytes against oxidative stress and delayed death caused by oxygen and glucose deprivation. Glia. 2009;57(6):645–656. doi: 10.1002/glia.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci. 2004;24(5):1101–1112. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siebert A, Desai V, Chandrasekaran K, Fiskum G, Jafri MS. Nrf2 activators provide neuroprotection against 6-hydroxydopamine toxicity in rat organotypic nigrostriatal cocultures. J Neurosci Res. 2009;87(7):1659–1669. doi: 10.1002/jnr.21975. [DOI] [PubMed] [Google Scholar]
- 59.Abdel-Wahab MH. Potential neuroprotective effect of t-butylhydroquinone against neurotoxicity-induced by 1-methyl-4-(2'-methylphenyl)-1,2,3,6-tetrahydropyridine (2'-methyl-MPTP) in mice. J Biochem Mol Toxicol. 2005;19(1):32–41. doi: 10.1002/jbt.20053. [DOI] [PubMed] [Google Scholar]
- 60.Shavali S, Sens DA. Synergistic neurotoxic effects of arsenic and dopamine in human dopaminergic neuroblastoma SH-SY5Y cells. Toxicol Sci. 2008;102(2):254–261. doi: 10.1093/toxsci/kfm302. [DOI] [PubMed] [Google Scholar]
- 61.Trinh K, Moore K, Wes PD, Muchowski PJ, Dey J, Andrews L, Pallanck LJ. Induction of the phase II detoxification pathway suppresses neuron loss in Drosophila models of Parkinson's disease. J Neurosci. 2008;28(2):465–472. doi: 10.1523/JNEUROSCI.4778-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A. 2006;103(41):15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dinkova-Kostova AT, Liby KT, Stephenson KK, Holtzclaw WD, Gao X, Suh N, Williams C, Risingsong R, Honda T, Gribble GW, Sporn MB, Talalay P. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc Natl Acad Sci U S A. 2005;102(12):4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liby KT, Yore MM, Sporn MB. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat Rev Cancer. 2007;7(5):357–369. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- 65.Reddy NM, Suryanaraya V, Yates MS, Kleeberger SR, Hassoun PM, Yamamoto M, Liby KT, Sporn MB, Kensler TW, Reddy SP. The Triterpenoid CDDO-Imidazolide Confers Potent Protection against Hyperoxic Acute Lung Injury in Mice. Am J Respir Crit Care Med. 2009;180(9):867–874. doi: 10.1164/rccm.200905-0670OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pitha-Rowe I, Liby KT, Royce D, Sporn MB. Synthetic Triterpenoids Attenuate Cytotoxic Retinal Injury: Crosstalk Between Nrf2 and PI3K/AKT Signaling Through Inhibition of the Lipid Phosphatase PTEN. Invest Ophthalmol Vis Sci. 2009 doi: 10.1167/iovs.09-3648. [DOI] [PubMed] [Google Scholar]
- 67.Hersch SM, Ferrante RJ. Translating therapies for Huntington's disease from genetic animal models to clinical trials. NeuroRx. 2004;1(3):298–306. doi: 10.1602/neurorx.1.3.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beal MF, Ferrante RJ. Experimental therapeutics in transgenic mouse models of Huntington's disease. Nat Rev Neurosci. 2004;5(5):373–384. doi: 10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- 69.Ferrante RJ, Ryu H, Kubilus JK, D'Mello S, Sugars KL, Lee J, Lu P, Smith K, Browne S, Beal MF, Kristal BS, Stavrovskaya IG, Hewett S, Rubinsztein DC, Langley B, Ratan RR. Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington's disease. J Neurosci. 2004;24(46):10335–10342. doi: 10.1523/JNEUROSCI.2599-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chaturvedi RK, Adhihetty P, Shukla S, Hennessy T, Calingasan N, Yang L, Starkov A, Kiaei M, Cannella M, Sassone J, Ciammola A, Squitieri F, Beal MF. Impaired PGC-1alpha function in muscle in Huntington's disease. Hum Mol Genet. 2009;18(16):3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119(1):121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 72.Ross CA, Thompson LM. Transcription meets metabolism in neurodegeneration. Nat Med. 2006;12(11):1239–1241. doi: 10.1038/nm1106-1239. [DOI] [PubMed] [Google Scholar]
- 73.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD, Cui L, Beyer RP, Easley CN, Smith AC, Krainc D, Luquet S, Sweet IR, Schwartz MW, La Spada AR. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4(5):349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 74.Watanabe N, Dickinson DA, Liu RM, Forman HJ. Quinones and glutathione metabolism. Methods Enzymol. 2004;378:319–340. doi: 10.1016/S0076-6879(04)78024-6. [DOI] [PubMed] [Google Scholar]
- 75.Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev. 2006;86(2):583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 76.Wilce MC, Parker MW. Structure and function of glutathione S-transferases. Biochim Biophys Acta. 1994;1205(1):1–18. doi: 10.1016/0167-4838(94)90086-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1: Levels of CDDO analogues in brain of wildtype mice after feeding.
(A) Chemical structures of CDDO analogues. (B) Comparison of CDDO-MA and CDDO-EA brain levels after feeding at 800mg/kg diet for 2 days. (C) Comparison of CDDO-EA and CDDO-TFEA brain levels after feeding at 200mg/kg diet and 400mg/kg diet for 3 days. Data were expressed as means and standard errors. In wildtype mice, brain levels of CDDO-EA were higher than CDDO-MA, and brain levels of CDDO-TFEA were higher than CDDO-EA.
Supplementary figure 2: Chromatograms of standard levels of CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) in N171-82Q mice. Chromatograms of standard dilution series for (A) CDDO-EA and (B) CDDO-TFEA added to mouse tissue homogenates at various concentrations. Areas under the curve increase with increasing concentrations.
Supplementary figure 3: Levels of CDDO-ethyl amide (CDDO-EA) in the brain of N171-82Q mice.
Chromatograms of (A) CDDO-EA at 100mg/kg diet and (B) at 200mg/kg diet in N171-82Q mouse brain.
Supplementary figure 4: Levels of CDDO-ethyl amide (CDDO-EA) in the skeletal muscle of N171-82Q mice.
Chromatograms of (A) CDDO-EA at 100mg/kg diet and (B) at 200mg/kg diet in N171-82Q mouse muscle. (C) Levels of CDDO-EA in the muscle of N171-82Q mice. Data were expressed as means and standard errors.
Supplementary figure 5: Levels of CDDO- trifluoroethyl amide (CDDO-TFEA) in the brain of N171-82Q mice.
Chromatograms of (A) CDDO-TFEA at 100mg/kg diet and (B) at 200mg/kg diet, and (C) at 400mg/kg diet in N171-82Q mouse brain. (D) Levels of CDDO-TFEA in the brain of N171-82Q mice. Data were expressed as means and standard errors.
Supplementary figure 6: Levels of CDDO-trifluoroethyl amide (CDDO-TFEA) in the skeletal muscle of N171-82Q mice.
Chromatograms of (A) CDDO-TFEA at 100mg/kg diet and (B) at 200mg/kg diet, and (C) at 400mg/kg diet in N171-82Q mouse muscle. (D) Levels of CDDO-TFEA in the muscle of N171-82Q mice. Data were expressed as means and standard errors.
Supplementary figure 7: CDDO-ethyl amide (CDDO-EA) and CDDO-trifluoroethyl amide (CDDO-TFEA) did not affect food intake in N171-82Q mice.
Body weights of N171-82Q mice fed with CDDO-EA (A for males and B for females) and CDDO-TFEA diets (C for males and D for females). Data were expressed as means and standard errors. CDDO-EA and CDDO-TFEA did not affect food intake in N171-82Q mice.