Abstract
Caulobacter crescentus initiates a single round of DNA replication during each cell cycle. Following the initiation of DNA replication, the essential CckA histidine kinase is activated by phosphorylation, which (via the ChpT phosphotransferase) enables the phosphorylation and activation of the CtrA global regulator. CtrA∼P then blocks the reinitiation of replication while regulating the transcription of a large number of cell cycle-controlled genes. It has been shown that DNA replication serves as a checkpoint for flagellar biosynthesis and cell division and that this checkpoint is mediated by the availability of active CtrA. Because CckA∼P promotes the activation of CtrA, we addressed the question of what controls the temporal activation of CckA. We found that the initiation of DNA replication is a prerequisite for remodeling the new cell pole, which includes the localization of the DivL protein kinase to that pole and, consequently, the localization, autophosphorylation, and activation of CckA at that pole. Thus, CckA activation is dependent on polar remodeling and a DNA replication initiation checkpoint that is tightly integrated with the polar phospho-signaling cascade governing cell cycle progression.
The Caulobacter cell cycle progresses through a series of consecutive stages: the differentiation of a swarmer cell into a stalked cell, the initiation of chromosome replication, the onset of remodeling of the new cell pole, segregation of the newly replicated chromosomes, flagellum biogenesis, and cell division (6, 27, 32, 38). Cell cycle and polar differentiation events are interdependent processes, and checkpoints are in place to ensure that both cell cycle and differentiation processes are completed before the next stage is initiated (27, 54). The orchestrated coordination of cell cycle events results in the formation of two distinct cell types, a motile swarmer cell and a sessile stalked cell. The swarmer cell has a single polar flagellum, several polar pili, and a polar chemotaxis complex. As Caulobacter moves through the cell cycle, the cell poles undergo critical remodeling (see Fig. 6A). The flagellum and pili are lost, and a stalk grows at that pole, thereafter maintaining a stalked pole identity (6, 26). Subsequently, the pole opposite the stalk eventually acquires a flagellum, pili, and chemotaxis proteins.
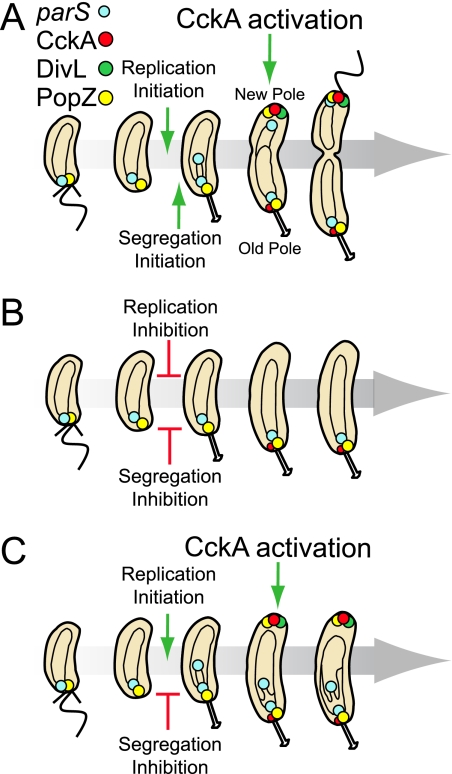
FIG. 6.
Initiation of DnaA replication controls new pole remodeling and CckA kinase activation required for cell cycle progression. (A) Schematic of the Caulobacter cell cycle in a wild-type strain. The flagellum-bearing swarmer cell possesses a single chromosome (black circle inside the cell), and its centromere-like parS sequence (blue circles) is positioned at the old cell pole. As soon as parS is replicated, a copy is moved rapidly to the new pole. Following replication initiation, DivL (green circles) localizes at the new cell pole, where it interacts with and activates the newly arrived CckA kinase (red circles). The accumulation of the PopZ polar localization factor (yellow circle) at the new pole is dependent upon replication initiation (4). (B) If DNA replication is inhibited, DivL and CckA fail to localize to the new pole, and CckA fails to be autophosphorylated and activated. (C) If the segregation of parS, but not replication initiation, is delayed in an inversion strain or if segregation, but not replication, is aborted in a strain expressing a parA(K20R) allele, PopZ and CckA still localize to the new pole, and CckA is activated by phosphorylation in a DivL-dependent manner.
In Caulobacter, if the initiation of DNA replication is inhibited, many temporally controlled events fail to occur. These include flagellum biosynthesis, synthesis of the CcrM DNA methyltransferase, and cell division (10, 11, 47, 48, 54). The essential CtrA response regulator, in its phosphorylated state, is central to the regulation of these cell cycle events. Flagellar genes, grouped in a complex regulatory hierarchy, are expressed sequentially; the order of expression corresponds to the order of assembly of the gene products into the growing flagellum. The expression of the class IV flagellar genes requires the previous expression of the class III genes, and this requires the expression of the class II flagellar genes (26, 56). CtrA functions as a class I gene controlling the transcription of class II flagellar genes (39, 57). CtrA also controls cytokinesis by regulating the expression of genes encoding essential components of cell division apparatus, including FtsZ, FtsA, and FtsQ (30, 54). In addition, CtrA activates the transcription of genes involved in chemotaxis, chromosome methylation, and pilus formation (29, 41, 45).
In the swarmer cell, CtrA is in the active phosphorylated state, where it binds to the origin of replication and blocks the initiation of replication (12, 40). Upon differentiation of a swarmer cell into a stalked cell (12), CtrA is cleared from the cell, allowing replication to begin. After DNA replication has initiated, CtrA accumulates and is activated by phosphotransfer from the essential CckA hybrid kinase through the ChpT phosphotransferase (3, 23). It has been shown that the accumulation of CtrA requires the initiation of DNA replication (54). The expression of ctrA is under the control of two promoters, P1 and P2. P1 is a weak promoter that is active only in the late stalked cell, following the initiation of DNA replication (13). In a positive feedback loop, phosphorylated CtrA activates the strong ctrA P2 promoter in the late predivisional cell (13). It was shown that if DNA replication is inhibited, CtrA is depleted due to the inhibition of the ctrA P2 promoter (54). Because the activation of the P2 promoter requires CtrA in its phosphorylated state, the activation of the CckA phospho-signaling pathway may be the primary responder to a DNA replication checkpoint.
The subcellular localization of CckA changes during the cell cycle; it accumulates at the cell poles following DNA replication initiation (2, 24). We have recently shown that CckA is autophosphorylated and activated when localized at the new cell pole (20). In addition, we demonstrated that CckA autophosphorylation, activation, and localization at the new pole is dependent on the essential DivL protein kinase, which predominantly localizes at the new pole in the same protein complex with CckA (20).
Because polar localization of DivL and CckA occurs following replication initiation, we asked if the initiation of DNA replication is required for the localization of DivL and CckA at the new cell pole and, consequently, for CckA phosphorylation and activation. Here, using three different methods to inhibit replication initiation, we demonstrate that the initiation of DNA replication is required for localizing both DivL and CckA at the new cell pole and for CckA autophosphorylation and activation. We ascertained that replication initiation, and not segregation, is the essential element for CckA new pole localization and autophosphorylation. These results argue that the initiation of DNA replication is an important cell cycle requirement for the activation of the CckA hybrid kinase and, consequently, via activated CtrA∼P, the expression of multiple cell cycle events, including flagellum and pilus formation, chemotaxis, DNA methylation, and cell division.
MATERIALS AND METHODS
Growth conditions and cell manipulations.
Caulobacter was grown at 28°C in PYE (peptone-yeast extract) rich medium, M2G medium, or M5G low-phosphate medium supplemented with glutamate (1 mM) (M5GG) (12, 16). The experiments were performed growing Caulobacter in M2G, keeping them in logarithmic phase. Where necessary, medium was supplemented with 0.3% xylose, 0.2% glucose, or 0.5 mM vanillate. When appropriate, media were supplemented with antibiotics at the following concentrations (liquid/solid media for Caulobacter; liquid/solid media for Escherichia coli [μg/ml]): kanamycin (5/25; 30/50), gentamicin (0.6/5; 15/20), oxytetracycline (1/2; 12/12), chloramphenicol (1/1; 20/30), spectinomycin (25/100; 50/50). Caulobacter transformation was performed by electroporation as previously described (22). Generalized transduction was performed with the phage ΦCr30 following a previously described procedure (16). G1-phase cell synchrony or swarmer cell isolation was performed as described previously (52). Immunoblot analysis and in vivo phosphorylation experiments are described by Iniesta et al. (20). β-Galactosidase assays were conducted as previously described (37).
Buffers and materials.
SDS-lysis buffer contains 4% SDS, 100 mM EDTA, and 50 mM Tris-HCl (pH 7). K2-low-salt buffer contains 50 mM Tris-HCl (pH 7), 100 mM NaCl, 50 mM EDTA, and 2% Triton X-100. K2-high-salt buffer contains 50 mM Tris-HCl (pH 7), 500 mM NaCl, and 50 mM EDTA. SDS loading buffer contains 125 mM Tris base, 20% glycerol, 2% SDS, and 0.01 mg/ml bromophenol blue, adjusted to pH 6.8. Quick T4 DNA ligase and endonucleases were purchased from Fermentas (Hanover, MD) and New England Biolabs (Ipswich, MA). DNA oligonucleotides were purchased from the Stanford Protein and Nucleic Acid Biotechnology Facility (Stanford, CA). One Shot Top10 chemically competent E. coli kits were purchased from Invitrogen (Carlsbad, CA) and used for cloning purposes. Electroporation cuvettes (0.1 cm) were purchased from Bio-Rad (Hercules, CA). DNA sequencing was performed by Sequetech (Mountain View, CA). KOD Hot Start DNA polymerase was purchased from Novagen (Madison, WI). DNA miniprep and gel extraction kits were purchased from Qiagen (Valencia, CA).
Fluorescence microscopy.
Strains were grown in M2G minimal medium and immobilized onto 1.0% agar in M2G before imaging with phase and epifluorescent microscopy with a Leica DM6000 microscope, using KAMS v0.8 software (7). Adobe Photoshop CS3 was utilized to scale channel intensity, maintaining a gamma of one, to maximize the dynamic display range. The histograms of monopolar CckA-CFP or DivL-mCherry partitioning between new and old poles were made by observing and quantifying monopolar fluorescence dots in cells presenting a stalk.
Construction of plasmids and strains.
Plasmids and strains used in this study are presented in Table S1 and oligonucleotides are presented in Table S2 in the supplemental material. For strain AA841, cckA::cckA-cfp was transduced with ΦCr30 lysate from a CB15N strain (17) harboring plasmid pGB024 into strain MT97. For strain AA843, cckA::cckA-cfp was transduced with ΦCr30 lysate from a CB15N strain harboring plasmid pGB024 into strain ET224. For strain AA847, cckA::cckA-cfp was transduced with ΦCr30 lysate from a CB15N strain harboring plasmid pGB024 into strain GM2471. For strain AA877, cckA::cckA-cfp was transduced with ΦCr30 lysate from a CB15N strain harboring plasmid pGB024 into strain ET165, resulting in strain AA869. Plasmid pJP69, derived from the plasmid pVCHYN-2, was integrated at the vanA locus in strain AA869, resulting in strain AA877. For strain AA878, cckA::cckA-cfp was transduced with ΦCr30 lysate from a CB15N strain harboring plasmid pGB024 into strain ET166, generating the strain AA870. The plasmid pJP69 was integrated at the vanA locus in strain AA870, resulting in strain AA878. For strain AA920, plasmid pNJH281 was introduced into the strain GM2471. For strain AA968, ccrM::Plac-ccrM was transduced with ΦCr30 lysate from strain LS1 into strain NJH530. Strain AA977 was obtained after the sequential introduction of plasmids pfixK and pNJH143 into strain AA968. For strain AA992, a fragment containing the whole coding sequence of dnaA was amplified by PCR from CB15N genomic DNA by using the oligonucleotides DnaA_NdeI.for and DnaA_NheI.rev. The dnaA fragment was digested with NdeI and NheI and ligated into the similarly digested pVCHYC-4 vector backbone, generating the plasmid pPvanA::dnaA. Plasmid pPvanA::dnaA was introduced into strain NJH530, resulting in the strain AA990. The deletion of dnaA, ΔdnaA, was transduced with ΦCr30 lysate from the strain GM2471 into the strain AA990, generating the strain AA992. Strain AA1000 was obtained after the sequential introduction of plasmids pfixK and pNJH143 into strain AA992. Strain AA1046 was obtained by introducing the pID42Δ3Ω plasmid into strain NJH602. Strain AA1059 was obtained by introducing the pID42Δ3Ω plasmid into a CB15N strain harboring plasmid pGB024. For strains NJH544 and NJH545, a fragment containing ctrA(D51E) was amplified by PCR by using plasmid pIDC42 as a template and oligonucleotides ctrARBSv2.for and ctrADown2v2.rev. This ctrA(D51E) fragment was then digested with EcoRI and BamHI and ligated into the similarly digested pNJH143 vector backbone, resulting in plasmid pNJH197. A fragment containing the vanillate promoter Pvan was amplified by PCR using plasmid pBVMCS-2 as a template and oligonucleotides NJH266 and NJH267. This Pvan fragment was then digested with EcoRI and ligated into the similarly digested pNJH197 vector backbone, resulting in plasmid pNJH210. Fragments containing ctrA(D51E)Δ3Ω or ctrA were respectively amplified by PCR using plasmids pID42Δ3Ω or pID42 as a template and oligonucleotides NJH395 and (NJH396 or NJH397). These ctrA(D51E)Δ3Ω and ctrA fragments were then digested with BglII and SpeI, and ligated into similarly digested pNJH210 vector backbone, resulting in plasmids pNJH268 and pNJH269. Strain NJH530 was sequentially transformed with plasmids pfixK and pNJH268 and plasmids pfixK and PNJH269, resulting in strains NJH544 and NJH545, respectively. For strain NJH602, the divL fragment derived from digesting plasmid pNJH281 with NdeI and AflII was ligated into the similarly digested pCHYC-2 vector backbone, resulting in plasmid pNJH286. The CB15N strain was transformed with plasmid pNJH286, resulting in strain NJH602.
RESULTS
Relative timing of the polar localization of CckA and the parS/ParB/MipZ complex.
DNA replication initiates only once per Caulobacter cell cycle from a single chromosomal origin located at the cell pole. Upon differentiation of the swarmer cell into a stalked cell, replication initiates, and a copy of the origin moves rapidly to the opposite cell pole (28, 34). The centromere-like parS sequence, located 8 kb away from the origin, forms a complex with the ParB partitioning protein and the MipZ ATPase, traveling together to the opposite pole as soon as parS is replicated (50, 52). Because it has been shown that the CckA histidine kinase localizes to the cell poles in stalked cells (2, 24), we constructed cells doubly labeled with MipZ-yellow fluorescent protein (YFP) and CckA-CFP and asked if the localization of CckA occurs after the initiation of DNA replication and segregation of the newly replicated parS/ParB/MipZ complex. We isolated swarmer cells containing fluorescently tagged MipZ and CckA and observed the relative timing of CckA and MipZ polar localization as the cells proceeded through the cell cycle (Fig. 1A). In the majority of cells, MipZ arrived at the opposite pole before CckA, confirming that the localization of CckA occurs after the initiation of DNA replication and parS segregation, as suggested previously (24) by comparison of time parallel experiments from different synchronized cultures.
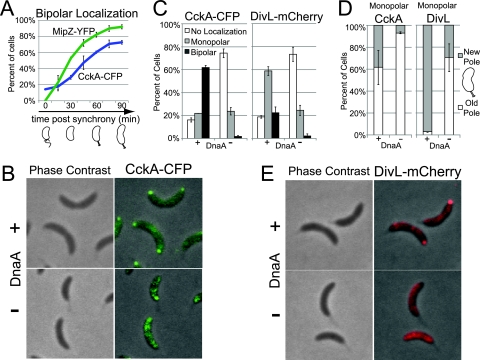
FIG. 1.
The expression of dnaA is required for CckA and DivL localization to the new cell pole. (A) Distribution of bipolar CckA-CFP and MipZ-YFP (strain AA841) throughout the cell cycle, starting with isolated swarmer cells (between 144 and 438 cells were counted at each time point and for each fluorescent marker; notated as n = 144 to 438 cells, hereafter). A cell cycle schematic is shown (below). (B) Phase contrast and fluorescence images of CckA-CFP localization 90 min after swarmer cells isolation (strain AA847) in the presence of DnaA (with 0.3% xylose) or in its absence (without xylose). (C) Histograms of CckA-CFP and DivL-mCherry localization (strains AA847 and AA920) in the presence of DnaA or in its absence (see description for panel B) (n = 213 to 421 cells). (D) Histograms of monopolar CckA-CFP and DivL-mCherry (strains AA847 and AA920) partitioning between new and old cell poles in the presence of DnaA or in its absence (see description for panel B) (n = 32 to 73 cells). (E) Phase contrast and fluorescence images of DivL-mCherry localization (strain AA920) in the presence of DnaA or in its absence (see description for panel B).
To determine if the initiation of DNA replication is required for CckA polar localization, we used three different methods to inhibit replication initiation: depletion of DnaA, treatment with novobiocin, and the use of a strain containing a dominant ctrA(D51E)Δ3Ω allele, in which the chromosomal origin is blocked (12, 40).
The expression of dnaA is required for CckA and DivL localization to the new cell pole.
We observed the pattern of CckA-CFP subcellular localization in cells depleted of DnaA, a protein required for replication initiation (18). We isolated swarmer cells from the strain AA847 containing a cckA-cfp allele, dnaA deleted from its native chromosomal site, and a copy of wild-type dnaA at the chromosomal xylX site (36) so that wild-type dnaA expression is under the control of the xylose-inducible PxylX promoter. We split the culture into aliquots that were incubated for 90 min in either the presence or absence of xylose. The inhibition of dnaA transcription rapidly blocks DNA replication (18). Only 2% of the cells exhibited bipolar CckA-CFP localization after 90 min of incubation in the absence of the xylose inducer, compared with 62% of cells in which dnaA was transcribed in the presence of the xylose inducer (Fig. 1B and C). Those cells that retained CckA polar localization in the absence of DnaA exhibited predominantly monopolar localization at the old (stalked) pole (Fig. 1D). This result suggests that the initiation of DNA replication is required for the localization of CckA to the new (opposite) pole.
The DivL protein kinase localizes predominantly to the new cell pole after initiation of DNA replication (44). We have shown previously that DivL is required for the localization and autophosphorylation of CckA at this cell pole (20). Because the localization of CckA at the new pole is dependent on dnaA expression (Fig. 1B to D) and CckA localization is also dependent on DivL, we asked if DivL localization is perturbed in the absence of dnaA expression. Swarmer cells from a strain, AA920, containing a chromosomal divL-mCherry and wild-type dnaA under the control of the xylose-inducible promoter, were incubated in the presence and in the absence of xylose. After 90 min of DnaA depletion, 74% of the cells exhibited no polar localization of DivL-mCherry, compared to 18% in cells expressing dnaA. In the absence of DnaA, 2% of the cells exhibited bipolar DivL-mCherry localization compared with 23% of cells expressing dnaA (Fig. 1C and E). Importantly, the fraction of cells that retained DivL monopolar localization in the absence of DnaA exhibited localization predominantly at the old pole, compared with cells expressing dnaA, in which DivL was almost exclusively localized at the new pole (Fig. 1D). These results suggest that the initiation of DNA replication is also required for the localization of DivL to the new pole.
In cells depleted of DnaA, the otherwise coincident G1-to-S transition and swarmer-to-stalked cell differentiation are temporally separated (18). Thus, the results shown in Fig. 1 suggest that the accumulation of DivL and CckA at the new cell pole is dependent on the G1-to-S transition and not on swarmer-to-stalked cell differentiation. However, because DnaA also functions as a transcription factor for multiple genes (19), it is possible that the loss of the proper localization of DivL and CckA in the absence of DnaA is due to the loss of specific DnaA-dependent gene expression rather than the inhibition of DNA replication initiation. We therefore turned to additional methods to inhibit replication initiation.
Proteolytically stable and active CtrA inhibits both replication initiation and CckA and DivL localization to the new cell pole.
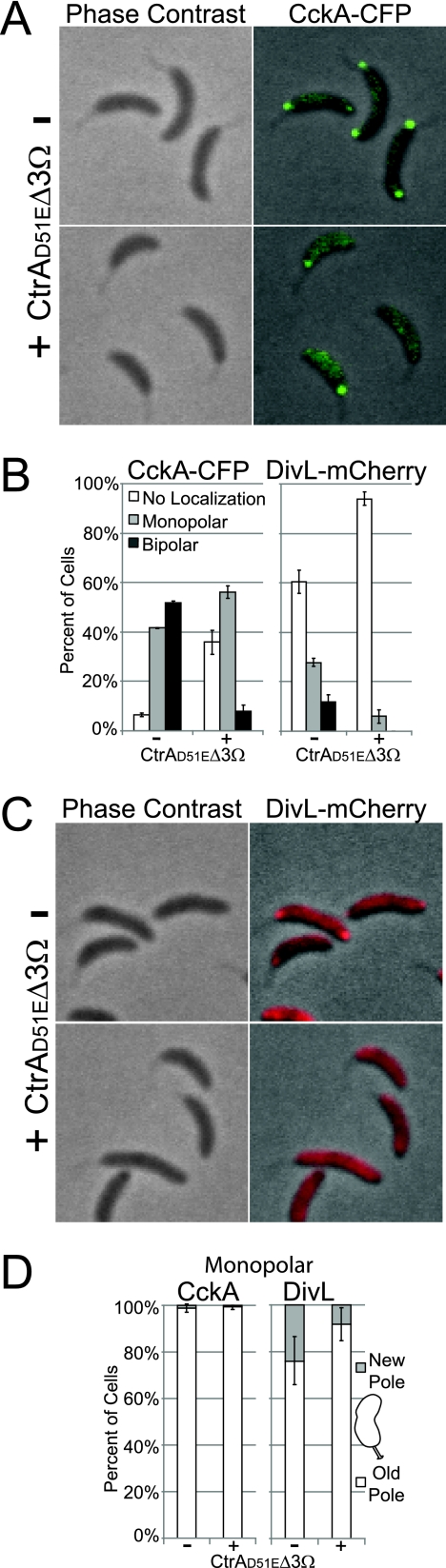
The phosphorylated (active) form of the CtrA response regulator blocks the initiation of DNA replication by binding to the chromosomal origin of replication (40). For the initiation of replication to occur, active CtrA must be eliminated by dephosphorylation or proteolysis. When both mechanisms for CtrA deactivation are blocked in a strain expressing the allele ctrA(D51E)Δ3Ω, encoding a proteolytically stable phosphomimetic variant of CtrA that is constitutively active, DNA replication is inhibited (12). We introduced a high-copy-number plasmid harboring the ctrA(D51E)Δ3Ω allele, whose expression is under the control of the PxylX promoter, into strains containing either chromosomal cckA-cfp (strain AA1059) or divL-mCherry (strain AA1046). In each case, cultures were grown in the presence of xylose for 1 h to induce the expression of ctrA(D51E)Δ3Ω. Swarmer cells were then isolated, allowed to grow in the presence of xylose for 90 min, and then visualized by fluorescence microscopy. Cells overexpressing ctrA(D51E)Δ3Ω exhibited a significant reduction in CckA-CFP and DivL-mCherry bipolar localization and an increase in the proportion of cells with no localization at the new pole (Fig. 2A to D). In the case of DivL, the expression of ctrA(D51E)Δ3Ω resulted in decreased localization of monopolar DivL (Fig. 2B), and of those cells that had monopolar DivL, 92% were at the old pole (Fig. 2D). Even without xylose induction, we observed a decrease in monopolar DivL localization at the new pole, compared to a wild-type control, and monopolar CckA was almost never observed at the new pole (compare Fig. 2D and 1D). This can be explained by background expression of ctrA(D51E)Δ3Ω from the PxylX promoter, which is not completely repressed in the absence of xylose (36). Cumulatively, these results provide additional evidence that the initiation of DNA replication is a critical element in the localization of CckA and DivL to the new cell pole and, furthermore, that the proper polar localization of CckA and DivL requires the inactivation of the CtrA master regulator.
FIG. 2.
Proteolytically stable and active CtrA inhibits CckA and DivL localization at the new cell pole. (A) Phase contrast and fluorescence images of CckA-CFP localization 90 min after swarmer cells (strain AA1059) were isolated and allowed to proceed synchronously through the cell cycle in the presence of CtrA(D51E)Δ3Ω or in its absence. A culture of strain AA1059 was incubated for 1 h in the presence of 0.3% xylose [to induce ctrA(D51E)Δ3Ω], and another culture of the same strain was incubated without inducer. Swarmer cells were then isolated from the induced and the uninduced culture, washed, and resuspended in medium containing 0.3% xylose or in the absence of xylose, respectively. (B) Histograms of CckA-CFP and DivL-mCherry (strains AA1059 and AA1046) in the presence of CtrA(D51E)Δ3Ω or in its absence (see description for panel A) (n = 319 to 637 cells). (C) Phase contrast and fluorescence images of DivL-mCherry localization (strain AA1046) in the presence of CtrA(D51E)Δ3Ω or in its absence (see description for panel A). (D) Histograms of monopolar CckA-CFP and DivL-mCherry (strains AA1059 and AA1046) partitioning between new and old cell poles in the presence of CtrA(D51E)Δ3Ω or in its absence (see description for panel A) (n = 12 to 114 cells).
CckA and DivL localization at the new cell pole fails to occur when replication initiation is inhibited by novobiocin.
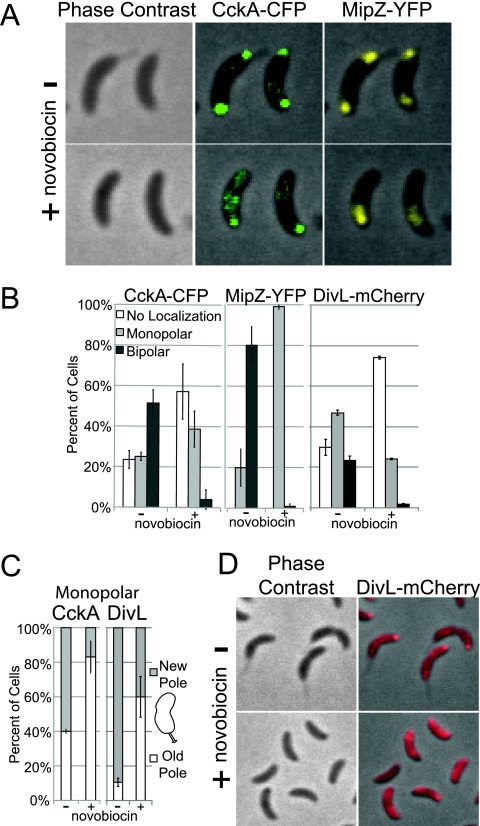
Novobiocin has been shown to inhibit the initiation of DNA replication in Caulobacter (33). To determine the effect of novobiocin on the polar localization of CckA, we isolated swarmer cells from strain AA841 that contained CckA-CFP and MipZ-YFP (to label the parS/ParB/MipZ complex) and incubated these cells in the presence or absence of novobiocin (100 μg/ml) for 90 min. In the culture of AA841 treated with novobiocin, a duplicated origin (represented by MipZ-YFP) failed to appear at the new pole (Fig. 3A and B), confirming that treatment with novobiocin inhibited the replication or the segregation of the parS region of the chromosome. In the presence of novobiocin, the bipolar localization of CckA-CFP was observed in only 4% of cells (Fig. 3A and B), compared to 52% exhibiting bipolar localization in the absence of novobiocin (Fig. 3A and B). Furthermore, of the fraction of novobiocin-treated cells that exhibited monopolar localization of CckA, it was found at the old pole at a frequency of 83%, compared with a frequency of 40% in untreated controls. In a separate experiment, we examined DivL-mCherry localization in swarmer cells of strain NJH602 incubated for 90 min in the presence or absence of novobiocin and found that 74% of the cells lost all polar accumulation of DivL-mCherry (Fig. 3B and D). Of the cells that retained monopolar localization in the presence of novobiocin, 60% exhibited accumulation at the old pole compared to 5% in untreated controls (Fig. 3B and C). These results indicate that treatment with novobiocin and its effect on replication and/or segregation of the parS/ParB/MipZ complex result in a significant decrease in the localization of CckA and DivL to the new cell pole.
FIG. 3.
The DNA replication inhibitor novobiocin inhibits CckA and DivL localization at the new cell pole. (A) Phase contrast and fluorescence images of CckA-CFP and MipZ-YFP subcellular localization (in strain AA841) with or without novobiocin treatment. Synchronized swarmer cells were incubated for 90 min without or with 100 μg/ml novobiocin. (B) Histograms of CckA-CFP and MipZ-YFP (strain AA841) and DivL-mCherry (strain NJH602) localization with or without novobiocin (see description for panel A) (n = 158 to 613 cells). (C) Histograms of monopolar CckA-CFP and DivL-mCherry (strains AA841 and NJH602) partitioning to the new and old cell poles with or without novobiocin (see description for panel A) (n = 5 to 144 cells). (D) Phase contrast and fluorescence images of DivL-mCherry localization (strain NJH602) with or without novobiocin (see description for panel A).
DNA segregation is not required for CckA localization.
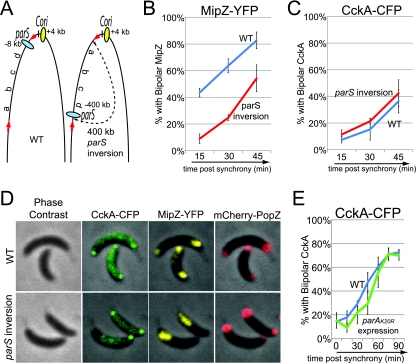
In Caulobacter, as soon as the centromere-like parS sequence is replicated, it quickly moves to the opposite pole, indicating that replication and segregation are concurrent processes (52). To address the possibility that CckA localization is dependent upon the act of parS segregation, we observed CckA-CFP localization in a strain with a parS chromosomal inversion, where the parS sequence was moved 400 kb away from the origin. In this strain, the segregation of the parS sequence is delayed without affecting the timing of replication initiation (52) (Fig. 4A). Although DNA segregation, visualized by MipZ-YFP foci marking the parS sequence, was delayed in the parS chromosomal inversion background compared with wild type (Fig. 4B), the time of the appearance of bipolar CckA-CFP localization was not significantly different in the two strains (Fig. 4C). At 45 min after cell synchronization and the initiation of replication, we could observe monopolar MipZ-YFP (parS not yet segregated), while bipolar localization of CckA-CFP had occurred (Fig. 4D), demonstrating that the polar localization of CckA is not dependent on the parS segregation process. As a control, we observed that PopZ, a polar organizing protein whose polar localization is dependent on replication initiation (4), also formed bipolar foci in the inversion strain (Fig. 4D), showing that its localization is also not dependent on parS segregation.
FIG. 4.
DNA segregation is not required for CckA localization. (A) Schematic of the relative positions of chromosomal parS and Cori sequences regions in wild-type cells and in cells with an inversion of a chromosomal region (a 400-kb inverted region is marked between red arrows and contains the parS locus and other regions arbitrarily called a, b, c, and d). (B and C) Time course of the fraction of wild-type (blue line; strain AA877) and parS chromosomal inversion (red line; strain AA878) strains that have bipolar MipZ-YFP, indicating that the origin and parS sequences have been duplicated and segregated to the two cell poles (B) and bipolar CckA-CFP (C) (n = 131 to 704 cells). Swarmer cells were isolated and imaged 15, 30, and 45 min after synchrony. (D) Phase contrast and fluorescence images of CckA-CFP, MipZ-YFP, and mCherry-PopZ localization in wild-type (strain AA877) and parS chromosomal inversion (strain AA878) strains, 45 min after synchronization. (E) Distribution of bipolar CckA-CFP localization in the parA(K20R) expression strain (green line; strain AA843), compared with the wild-type strain (blue line; strain AA841) (n = 121 to 438 cells). Swarmer cells were isolated and imaged every 15 min during 90 min of incubation. For the strain bearing a parA(K20R) allele, cells were grown in the presence of 0.03% xylose for 1 h [to induce parA(K20R)] before the isolation of swarmer cells and then incubated in the presence of 0.03% xylose.
Taking advantage of the fact that segregation of the parS sequence is dependent on the action of the ParA ATPase (52), we obtained additional evidence that the polar accumulation of CckA is independent of parS segregation. A parA(K20R) mutation was shown previously to be a dominant negative allele that blocks DNA segregation without inhibiting replication initiation (see Fig. S1A in the supplemental material) (52). The induction of parA(K20R) transcription had no effect on the extent of the bipolar accumulation of CckA-CFP (Fig. 4E; also see Fig. S1B in the supplemental material). Cumulatively, these results indicate that the segregation of the parS/ParB/MipZ complex is not required for the localization of CckA at the new pole.
Initiation of DNA replication, but not segregation, is required for CckA autophosphorylation and activation.
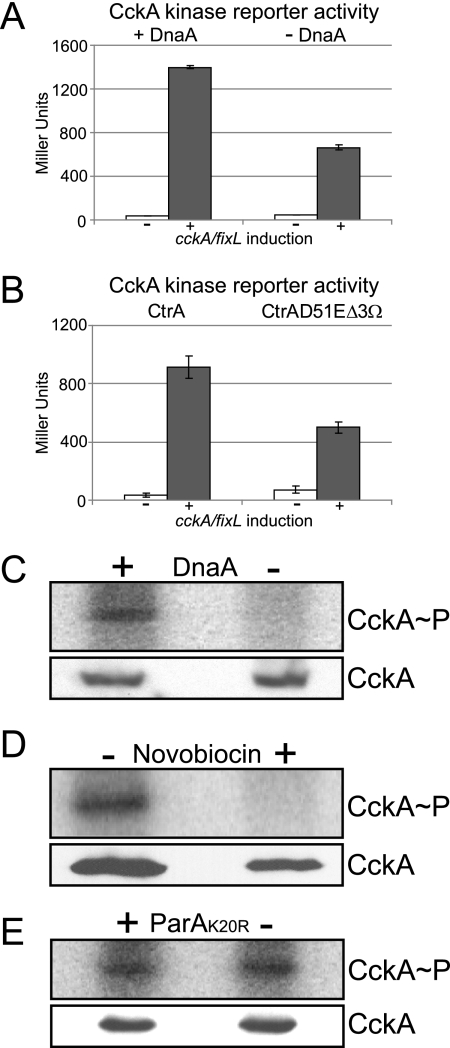
We have previously shown that the localization of CckA at the new pole correlates with its activity and phosphorylation (20). Here, we have shown by three independent methods that the localization of CckA at the new cell pole is significantly reduced when the initiation of DNA replication is blocked. To ascertain that loss of CckA localization upon inhibition of replication initiation is coupled to the loss of CckA autophosphorylation and activity, we first took advantage of the CckA-FixL chimeric kinase reporter system recently designed and validated as an assay for in vivo CckA activity (20). This chimeric kinase reporter is based on the LacZ reporter system developed for the FixL-FixJ two-component oxygen-sensing system (9). The chimeric kinase is composed of an N-terminal portion of CckA, which senses CckA-dependent signals, that is fused to a C-terminal portion of the FixL kinase. When the chimera CckA-FixL kinase is activated, phosphate is transferred to the response regulator FixJ, which in turn activates the transcription of lacZ controlled by the PfixK promoter (20). The gene encoding the chimeric kinase is transcribed from a xylose-inducible promoter. We used the CckA-FixL PfixK-lacZ reporter to measure the activity of CckA upon the depletion of DnaA in strain AA1000. This strain contained only one copy of dnaA, whose expression was under the control of the vanillate-inducible PvanA promoter (49). Cultures of AA1000 were grown to mid-log phase in the presence of vanillate (to induce dnaA), washed to remove the vanillate from the medium, and then separated into four aliquots. One aliquot was incubated with vanillate and xylose (to induce the accumulation of DnaA and the CckA-FixL chimera), another was incubated with vanillate but not with xylose (to induce the accumulation of DnaA but not the CckA-FixL chimera), a third one was incubated in the absence of vanillate and the presence of xylose (to induce accumulation of the CckA-FixL chimera but not DnaA), and the final aliquot was incubated in the absence of both vanillate and xylose. After 6 h of incubation under these conditions, we observed that the activity of the chimeric kinase (in Miller units) in the presence of DnaA was more than double that observed in the absence of DnaA (Fig. 5A). In addition, we measured the chimera kinase activity in a strain in which the overexpression of ctrA(D51E)Δ3Ω blocks the initiation of DNA replication. The strain used, NJH544, contained a high-copy-number plasmid bearing the cckA-fixL chimera under the control of the PxylX promoter, and on the same plasmid, the ctrA(D51E)Δ3Ω allele was under the control of the PvanA promoter. We also constructed an isogenic strain, NJH545, with the plasmid-borne wild-type allele ctrA instead of ctrA(D51E)Δ3Ω. We incubated both strains for 4 h in the presence of vanillate to induce either ctrA or ctrA(D51E)Δ3Ω expression and then added xylose to induce the expression of cckA-fixL. After 6 h of cckA-fixL induction, the activity of the chimeric kinase in the ctrA(D51E)Δ3Ω-expressing strain was half of that exhibited by the wild-type ctrA control (Fig. 5B).
FIG. 5.
Initiation of DNA replication is required for CckA phosphorylation and activity. (A) CckA-FixL chimera kinase reporter activity (20) in strain AA1000 incubated in the presence or absence of DnaA. A culture of strain AA1000 was incubated in the presence of vanillate (to induce dnaA) and then washed to remove the vanillate inducer. Two aliquots of the washed culture were grown in the presence of 0.3% xylose to induce cckA-fixL (one with newly added vanillate [presence of DnaA] and the other with no vanillate [absence of DnaA]) and then assayed for β-galactosidase activity after 6 h. The results of control experiments without cckA-fixL induction (no xylose) are shown. (B) CckA-FixL kinase reporter activity assays in strains overexpressing ctrA (strain NJH545) or ctrA(D51E)Δ3Ω (strain NJH544). Cells were grown in the presence of vanillate [to induce the expression of ctrA or ctrA(D51E)Δ3Ω] for 4 h before inducing the expression of cckA-fixL with 0.3% xylose and then assayed for β-galactosidase activity after 6 h of incubation. The results of control experiments without cckA-fixL induction (no xylose) are shown. (C) PhosphorImager results of in vivo phosphorylation of CckA performed with cultures of a DnaA depletion strain (strain GM2471), in the presence of DnaA or depletion of DnaA, using antibodies to CckA (upper panel). Corresponding CckA immunoblots are shown in the lower panel. Synchronized swarmer cells were grown without or with 0.3% xylose (for dnaA induction) for 90 min prior to the in vivo phosphorylation assay. (D) In vivo phosphorylation assay using anti-CckA antibodies performed with the wild-type strain (strain CB15N) in the absence or in the presence of novobiocin (100 μg/ml). Synchronized swarmer cells were grown for 90 min with or without novobiocin prior to the in vivo phosphorylation assay (upper panel) and CckA immunoblot analysis (lower panel). (E) In vivo phosphorylation assay using anti-CckA antibodies performed with the parA(K20R) expression strain (strain ET227) in the absence or in the presence of ParA(K20R). Cultures of ET227 were grown in the absence or presence of 0.03% xylose [to induce parA(K20R)-mCherry] for 1 h. Swarmer cells were then isolated and incubated for 90 min in the absence or presence of 0.03% xylose. The CckA in vivo phosphorylation assay (upper panel) and immunoblot analysis (lower panel) were carried out for each culture.
To determine directly if the initiation of DNA replication is required for the autophosphorylation of CckA, we isolated swarmer cells from a strain (GM2471) containing chromosomal dnaA under the control of the PxylX promoter, as the only copy of dnaA. Swarmer cells were isolated from this strain and divided into two aliquots, one was incubated for 90 min in the presence of xylose (to induce dnaA) and the other in the absence of the xylose inducer. In vivo CckA phosphorylation assays showed that CckA was not phosphorylated in the absence of DnaA (Fig. 5C). The absence of phosphorylated CckA in the strain that did not express dnaA was not due to a lack of CckA protein, since the levels of CckA protein were the same in the presence and absence of DnaA (Fig. 5C). Moreover, when novobiocin was added to isolated swarmer cells and incubated for 90 min, CckA did not exhibit autophosphorylation (Fig. 5D). However, in the strain bearing the mutant parA(K20R) allele, in which chromosome segregation, but not the initiation of DNA replication, was inhibited (52), CckA was autophosphorylated (Fig. 5E). Together, these results demonstrate that the initiation of DNA replication, but not DNA segregation, is required for the phosphorylation, activation, and localization of CckA at the new cell pole.
DISCUSSION
Caulobacter crescentus exhibits well-coordinated mechanisms for integrating critical cell cycle events such as the initiation of DNA replication and cell division with polar differentiation. The order of cell cycle events is controlled, in part, by the nature of the cell poles. During cell cycle progression, the cell poles undergo critical remodeling. In the swarmer cell, the chromosomal parS/ParB/MipZ complex is tethered to a ribosome-free oligomeric network of PopZ at the pole bearing the flagellum (Fig. 6A) (5, 15). The single chromosome in the swarmer cell cannot initiate DNA replication because phosphorylated (active) CtrA is bound to the origin and blocks replication initiation. During the swarmer-to-stalked cell transition, the old pole exhibits external and internal transformations. Externally, the flagellum and pili are lost, to be replaced by a stalk. Internally, the PopZ-parS/ParB/MipZ tether is broken, releasing the parS/ParB/MipZ complex from its interaction with PopZ at that pole (4). PopZ then switches function, acting to recruit factors that promote the transformation to a stalked pole by enabling the sequential localization of regulatory proteins (4). CtrA accumulates at that pole, where it is cleared from the cell by a transiently localized complex of the ClpXP protease and accessory factors (14, 21, 35, 43). Also, at that pole, the components of the chemotaxis machinery are degraded (1, 21). Once CtrA is cleared from the cell, DNA replication is initiated (12). Because CtrA represses the transcription of ftsZ, the clearance of CtrA from the cell allows the accumulation of FtsZ in preparation for divisome assembly and cell division (30).
Upon initiation of replication, a copy of the parS sequence in complex with ParB/MipZ moves rapidly to the new pole (28, 51-53). Simultaneously, PopZ accumulates at the new pole so as to be in place to tether the parS/ParB/MipZ complex newly arrived at that pole (5, 15). The CckA histidine kinase localizes to the cell poles during these polar remodeling events (2, 24), and the positioning of CckA at the new pole correlates with its autophosphorylation and activation (20). Time lapse imaging of cells doubly labeled with MipZ-YFP and CckA-CFP confirmed that the localization of CckA to the new pole follows the initiation of DNA replication and parS segregation (Fig. 1A). If the initiation of DNA replication is blocked, either by depleting DnaA, by overexpressing the phosphomimetic and stable ctrA(D51E)Δ3Ω allele, or by novobiocin treatment, neither CckA nor the DivL protein kinase can localize at the new pole (Fig. 6B). Under normal conditions, DivL predominantly localizes at the new pole following DNA replication (44), and it is required for the autophosphorylation, functional activation, and new cell pole localization of CckA (20). Consequently, a block in the initiation of DNA replication results in the loss of CckA phosphorylation and CckA function (Fig. 5). Interestingly, it has been recently shown that the initiation of DNA replication is also required for the new pole localization of PopZ (4). Therefore, the initiation of replication triggers remodeling of the new cell pole, facilitating polar localization of PopZ, which tethers the newly replicated parS sequence, and DivL, which promotes the localization and activation of CckA. This polar remodeling, in turn, prepares the cell for cell division. The tethering of the parS/ParB/MipZ complex to the new pole targets MipZ, an inhibitor of FtsZ polymerization, to both poles, thereby directing FtsZ ring formation to a site mid-way between the poles, the region of lowest MipZ concentration (51). The autophosphorylation and activation of CckA following the start of DNA replication promote CtrA activation by phosphorylation and also the inhibition of CtrA proteolysis (3, 21, 23). We propose that this chain of events, beginning with the initiation of DNA replication and ending with the activation of CtrA by phosphorylation, comprises an “off switch” for chromosome replication, limiting it to a narrow window of the cell cycle and preventing additional initiation events. In addition, CtrA∼P amplifies its own transcription (13) and activates the transcription of critical cell division genes, such as ftsQ and ftsA (54).
Because the initiation of DNA replication and parS segregation are concurrent processes, we had to rule out the possibility that the arrival of the parS/ParB/MipZ complex at the new pole is the critical event that promotes the localization of CckA at that pole. To do this, we used a strain with a chromosome inversion (52) in which parS was separated by 408 kb, instead of by 8 kb, from the origin of replication (Fig. 4A). In this strain, the segregation of parS, but not the initiation of DNA replication, is significantly delayed (52). We showed that the delay in parS segregation did not delay the localization of CckA (Fig. 4B, C, and D) or the localization of PopZ at the new pole (Fig. 4D and 6C). In addition, using a ParA mutant strain where segregation but not replication initiation is blocked (52), we found that CckA was phosphorylated (Fig. 5E) and that its localization to the new cell pole was indistinguishable from wild type (Fig. 4E). Cumulatively, these results argue that the signal that triggers remodeling of the new cell pole occurs between the initiation of DNA replication and the segregation of the parS centromere.
The mechanism that connects the initiation of DNA replication with the remodeling of the new cell pole is not known. A reasonable possibility is that the signal affecting the composition and function of the new pole is a consequence of early bidirectional DNA replication around the origin of replication. It is formally possible that the replication of origin-proximal sequences promotes the transcription of coding or noncoding RNA or the activation of a nucleotide-based second messenger that triggers new cell pole development. Possible candidates are two genes encoding validated small RNAs that are located near the origin with their peak of expression coinciding with the time of initiation of DNA replication (31). How might replication trigger the controlled transcription of a specific gene or groups of genes? Following the initiation of replication of a fully methylated chromosome, newly replicated DNA remains in a hemimethylated state until the CcrM DNA methyltransferase is synthesized upon completion of chromosome duplication (46, 55). Sequential changes in chromosomal methylation state during replication have been reported previously to modulate the transcription of genes encoding the DnaA and CtrA global cell cycle regulators (8, 42). We tested the possibility that changes in methylation state could affect the function of the new cell pole by examining a strain that constitutively expresses ccrM, thereby maintaining the DNA in a fully methylated state (58). This strain had no effect on CckA localization or activity (see Fig. S2 in the supplemental material), so the mechanism that connects replication initiation to remodeling the new cell pole remains an area of active investigation.
The use of DNA replication inhibition as a vital checkpoint appears to be universal, as it is common to all kingdoms of life. In Caulobacter, replication initiation is required to promote the physical remodeling of the pole opposite the stalk, creating a niche for the localization and activation of essential regulators, which feeds back into the control of cell cycle progression. At the opposite pole, a different remodeling process drives the initiation of replication, concurrent with the loss of the flagellum and its replacement with a stalk. Thus, the asymmetric and temporal internal remodeling of the poles is a key element for cell fate determination in this bacterial system.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants GM51426, GM32506, and GM073011 (L.S.) and Department of Energy grant DE-FG-02-05ER64136 (L.S.). N.J.H. was supported by Damon Runyon Cancer Research Foundation fellowship DRG-1880-05.
We thank G. R. Bowman, P. Mera, and E. Goley for critical reading of the manuscript; G. R. Bowman and J. L. Ptacin for providing plasmids pGB024 and pJP69; and G. Marczynski for strain GM2471.
Footnotes
Published ahead of print on 4 June 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alley, M. R., J. R. Maddock, and L. Shapiro. 1993. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science 259:1754-1757. [DOI] [PubMed] [Google Scholar]
- 2.Angelastro, P. S., O. Sliusarenko, and C. Jacobs-Wagner. 2010. Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. J. Bacteriol. 192:539-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biondi, E. G., S. J. Reisinger, J. M. Skerker, M. Arif, B. S. Perchuk, K. R. Ryan, and M. T. Laub. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899-904. [DOI] [PubMed] [Google Scholar]
- 4.Bowman, G. R., L. R. Comolli, G. M. Gaietta, M. Fero, S. H. Hong, Y. Jones, J. H. Lee, K. H. Downing, M. H. Ellisman, H. H. McAdams, and L. Shapiro. 2010. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol. Microbiol. 76:173-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, G. R., L. R. Comolli, J. Zhu, M. Eckart, M. Koenig, K. H. Downing, W. E. Moerner, T. Earnest, and L. Shapiro. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, P. J., G. G. Hardy, M. J. Trimble, and Y. V. Brun. 2009. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv. Microb. Physiol. 54:1-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christen, B., M. J. Fero, N. J. Hillson, G. Bowman, S. H. Hong, L. Shapiro, and H. H. McAdams. 2010. High-throughput identification of protein localization dependency networks. Proc. Natl. Acad. Sci. U. S. A. 107:4681-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collier, J., H. H. McAdams, and L. Shapiro. 2007. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc. Natl. Acad. Sci. U. S. A. 104:17111-17116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosson, S., P. T. McGrath, C. Stephens, H. H. McAdams, and L. Shapiro. 2005. Conserved modular design of an oxygen sensory/signaling network with species-specific output. Proc. Natl. Acad. Sci. U. S. A. 102:8018-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degnen, S. T., and A. Newton. 1972. Dependence of cell division on the completion of chromosome replication in Caulobacter. J. Bacteriol. 110:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingwall, A., W. Y. Zhuang, K. Quon, and L. Shapiro. 1992. Expression of an early gene in the flagellar regulatory hierarchy is sensitive to an interruption in DNA replication. J. Bacteriol. 174:1760-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415-424. [DOI] [PubMed] [Google Scholar]
- 13.Domian, I. J., A. Reisenauer, and L. Shapiro. 1999. Feedback control of a master bacterial cell-cycle regulator. Proc. Natl. Acad. Sci. U. S. A. 96:6648-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duerig, A., S. Abel, M. Folcher, M. Nicollier, T. Schwede, N. Amiot, B. Giese, and U. Jenal. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 23:93-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebersbach, G., A. Briegel, G. J. Jensen, and C. Jacobs-Wagner. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134:956-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 17.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbatyuk, B., and G. T. Marczynski. 2001. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol. Microbiol. 40:485-497. [DOI] [PubMed] [Google Scholar]
- 19.Hottes, A. K., L. Shapiro, and H. H. McAdams. 2005. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 58:1340-1353. [DOI] [PubMed] [Google Scholar]
- 20.Iniesta, A. A., N. J. Hillson, and L. Shapiro. 2010. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc. Natl. Acad. Sci. U. S. A. 107:7012-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iniesta, A. A., P. T. McGrath, A. Reisenauer, H. H. McAdams, and L. Shapiro. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. U. S. A. 103:10935-10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iniesta, A. A., and L. Shapiro. 2008. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc. Natl. Acad. Sci. U. S. A. 105:16602-16607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs, C., N. Ausmees, S. J. Cordwell, L. Shapiro, and M. T. Laub. 2003. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol. Microbiol. 47:1279-1290. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs, C., I. J. Domian, J. R. Maddock, and L. Shapiro. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111-120. [DOI] [PubMed] [Google Scholar]
- 25.Reference deleted.
- 26.Jenal, U. 2000. Signal transduction mechanisms in Caulobacter crescentus development and cell cycle control. FEMS Microbiol. Rev. 24:177-191. [DOI] [PubMed] [Google Scholar]
- 27.Jenal, U., and C. Stephens. 2002. The Caulobacter cell cycle: timing, spatial organization and checkpoints. Curr. Opin. Microbiol. 5:558-563. [DOI] [PubMed] [Google Scholar]
- 28.Jensen, R. B., and L. Shapiro. 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. U. S. A. 96:10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, S. E., N. L. Ferguson, and M. R. Alley. 2001. New members of the ctrA regulon: the major chemotaxis operon in Caulobacter is CtrA dependent. Microbiology 147:949-958. [DOI] [PubMed] [Google Scholar]
- 30.Kelly, A. J., M. J. Sackett, N. Din, E. Quardokus, and Y. V. Brun. 1998. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 12:880-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landt, S. G., E. Abeliuk, P. T. McGrath, J. A. Lesley, H. H. McAdams, and L. Shapiro. 2008. Small non-coding RNAs in Caulobacter crescentus. Mol. Microbiol. 68:600-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laub, M. T., L. Shapiro, and H. H. McAdams. 2007. Systems biology of Caulobacter. Annu. Rev. Genet. 41:429-441. [DOI] [PubMed] [Google Scholar]
- 33.Loewy, B., G. T. Marczynski, A. Dingwall, and L. Shapiro. 1990. Regulatory interactions between phospholipid synthesis and DNA replication in Caulobacter crescentus. J. Bacteriol. 172:5523-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marczynski, G. T. 1999. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J. Bacteriol. 181:1984-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGrath, P. T., A. A. Iniesta, K. R. Ryan, L. Shapiro, and H. H. McAdams. 2006. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell 124:535-547. [DOI] [PubMed] [Google Scholar]
- 36.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 38.Quardokus, E. M., and Y. V. Brun. 2003. Cell cycle timing and developmental checkpoints in Caulobacter crescentus. Curr. Opin. Microbiol. 6:541-549. [DOI] [PubMed] [Google Scholar]
- 39.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 40.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. U. S. A. 95:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reisenauer, A., L. S. Kahng, S. McCollum, and L. Shapiro. 1999. Bacterial DNA methylation: a cell cycle regulator? J. Bacteriol. 181:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reisenauer, A., and L. Shapiro. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 21:4969-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan, K. R., S. Huntwork, and L. Shapiro. 2004. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc. Natl. Acad. Sci. U. S. A. 101:7415-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sciochetti, S. A., N. Ohta, and A. Newton. 2005. The role of polar localization in the function of an essential Caulobacter crescentus tyrosine kinase. Mol. Microbiol. 56:1467-1480. [DOI] [PubMed] [Google Scholar]
- 45.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephens, C., A. Reisenauer, R. Wright, and L. Shapiro. 1996. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc. Natl. Acad. Sci. U. S. A. 93:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephens, C. M., and L. Shapiro. 1993. An unusual promoter controls cell-cycle regulation and dependence on DNA replication of the Caulobacter fliLM early flagellar operon. Mol. Microbiol. 9:1169-1179. [DOI] [PubMed] [Google Scholar]
- 48.Stephens, C. M., G. Zweiger, and L. Shapiro. 1995. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J. Bacteriol. 177:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thanbichler, M., A. A. Iniesta, and L. Shapiro. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thanbichler, M., and L. Shapiro. 2008. Getting organized—how bacterial cells move proteins and DNA. Nat. Rev. Microbiol. 6:28-40. [DOI] [PubMed] [Google Scholar]
- 51.Thanbichler, M., and L. Shapiro. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147-162. [DOI] [PubMed] [Google Scholar]
- 52.Toro, E., S. H. Hong, H. H. McAdams, and L. Shapiro. 2008. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc. Natl. Acad. Sci. U. S. A. 105:15435-15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Viollier, P. H., M. Thanbichler, P. T. McGrath, L. West, M. Meewan, H. H. McAdams, and L. Shapiro. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 101:9257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wortinger, M., M. J. Sackett, and Y. V. Brun. 2000. CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. EMBO J. 19:4503-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright, R., C. Stephens, and L. Shapiro. 1997. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J. Bacteriol. 179:5869-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, J., and A. Newton. 1997. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol. Microbiol. 24:233-239. [DOI] [PubMed] [Google Scholar]
- 57.Wu, J., N. Ohta, and A. Newton. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 95:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zweiger, G., G. Marczynski, and L. Shapiro. 1994. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 235:472-485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.