Abstract
We recently reported that the oral mucosal pathogen Porphyromonas gingivalis, through its 67-kDa Mfa1 (minor) fimbria, targets the C-type lectin receptor DC-SIGN for invasion and persistence within human monocyte-derived dendritic cells (DCs). The DCs respond by inducing an immunosuppressive and Th2-biased CD4+ T-cell response. We have now purified the native minor fimbria by ion-exchange chromatography and sequenced the fimbria by tandem mass spectrometry (MS/MS), confirming its identity and revealing two putative N-glycosylation motifs as well as numerous putative O-glycosylation sites. We further show that the minor fimbria is glycosylated by ProQ staining and that glycosylation is partially removed by treatment with β(1-4)-galactosidase, but not by classic N- and O-linked deglycosidases. Further monosaccharide analysis by gas chromatography-mass spectrometry (GC-MS) confirmed that the minor fimbria contains the DC-SIGN-targeting carbohydrates fucose (1.35 nmol/mg), mannose (2.68 nmol/mg), N-acetylglucosamine (2.27 nmol/mg), and N-acetylgalactosamine (0.652 nmol/mg). Analysis by transmission electron microscopy revealed that the minor fimbria forms fibers approximately 200 nm in length that could be involved in targeting or cross-linking DC-SIGN. These findings shed further light on molecular mechanisms of invasion and immunosuppression by this unique mucosal pathogen.
Porphyromonas gingivalis is one of several mucosal pathogens that have been implicated in chronic periodontitis (CP), a common oral disease that may affect 40 to 60% of the U.S. population (7). P. gingivalis utilizes a myriad of virulence factors that contribute to chronic periodontitis. Among these are a polysaccharide capsule, fimbriae, proteases for opsonins C3 and IgG, gingipains (21, 30, 43, 52), bacterial lipopolysaccharides (LPS) (22, 44), and toxins and hemagglutinins (10, 25).
The fimbriae of P. gingivalis play a crucial role in adhesion to and invasion of host cells. We have shown that optimum entry of P. gingivalis into human dendritic cells (DCs) requires the presence of two fimbriae, termed the major and minor fimbriae. The major fimbria is composed of a 41-kDa protein termed fimbrillin, encoded by the fimA gene (65). Much less is known about the minor fimbria, the focus of this paper. The minor fimbria is comprised of a 67-kDa protein (19) that is encoded by the mfa1 gene. The major and minor fimbriae are antigenically distinct, and they also differ based on amino acid composition and size (5, 19). Very little is understood about the formation and secretion of the minor fimbriae and about possible posttranslational modifications of these fimbriae. Formation and secretion of the major fimbriae is a complex reaction consisting of numerous steps required for transfer of prefimbrillin proteins from the cytoplasm to the periplasm, cleavage of the N-terminal signal peptide (24, 50), transport of prefimbrillin to the outer face of the outer membrane, and assembly into fimbria structures (23, 24, 34).
Deciphering the cellular receptors for the fimbriae is an active area of research. Evidence suggests that the cellular targets of the major fimbriae are the β-1 integrins (CD29) (32, 66). Others have proposed a role for β-2 integrins (CD18) (17, 18, 55) in the cellular response to major fimbriae. In contrast, little is known of the cellular receptors for the minor fimbriae. Lamont et al. in 2002 showed that the minor fimbria of P. gingivalis intimately interacts with the SspB protein of Streptococcus gordonii (26). This interaction might aid in P. gingivalis colonization of plaque biofilm before it invades gingival tissue (26, 41). We recently showed that the minor fimbria targets DC-SIGN on DCs for entry into DCs and that this targeting has the immunological consequence of dampening the immune response (68).
DC-SIGN is a type II membrane protein on DCs in which the extracellular domain consists of a stalk that promotes tetramerization (13). DC-SIGN contains a C-terminal carbohydrate-recognizing domain (CRD) that belongs to the C-type lectin superfamily (13). Early studies by Feinberg et al. in 2001 showed that the DC-SIGN CRD preferentially binds to the high-mannose N-linked oligosaccharides GlcNAc (N-acetylglucosamine) and Manα1-3[Manα1-6] Man (mannose) (13). Furthermore, Appelmelk et al. showed that DC-SIGN also binds to fucose-containing Lewis blood antigens (4). Guo et al. utilized an extensive glycan array and showed that DC-SIGN will bind high-mannose-containing glycans or glycans that contain terminal fucose residues (16). Previous studies showed that DC-SIGN on DCs is used by microorganisms such as Neisseria gonorrhoeae, Mycobacterium tuberculosis, Mycobacterium leprae, HIV, and Helicobacter pylori for entry into DCs and induction of immunosuppression (4, 27, 42, 51, 69). Like P. gingivalis, many of these pathogens can induce chronic life-long infections.
Our previously published work established that the minor fimbria is necessary for targeting DC-SIGN, resulting in entry of P. gingivalis into DCs (68). We were able to abrogate minor fimbria-mediated DC-SIGN ligation by using DC-SIGN-blocking agents or agonists, including fucose, mannose, and mannan (68). Additionally, we described that the minor fimbria is able to induce immunosuppression of DCs via its interaction with DC-SIGN, which was blocked by sugars (68). Further, we demonstrated that minor fimbriated strains of P. gingivalis inhibited DC maturation and suppressed proinflammatory cytokine secretion (68). Moreover, DCs that were pulsed with minor fimbriated strains of P. gingivalis and then cocultured with autologous T cells shifted the T-cell effector phenotype to a Th2 effector phenotype, as evidenced by high interleukin-4 (IL-4) production (68).
Our previous results, described above, suggested that the minor fimbria-DC-SIGN interaction was mediated by glycosylated proteins. We therefore set out to identify the carbohydrate moieties on the minor fimbria that could account for its DC-SIGN-targeting function. The intact native minor fimbria was purified and analyzed for glycosylation and for the presence of relevant monosaccharides. We show here by a combination of ProQ gel staining and gas chromatography-mass spectrometry (GC-MS) analysis that the minor fimbria is glycosylated and expresses the DC-SIGN ligands fucose, mannose, GlcNAc, and GalNAc. Use of classic N- and O-linked deglycosidases on the native minor fimbria revealed a novel glycoprotein structure. Overall, these results indicate that the minor fimbria is glycosylated with DC-SIGN-binding motifs that likely account for the reported ability of P. gingivalis to bind to and invade DCs, resulting in an immunosuppressive DC response.
MATERIALS AND METHODS
Bacterial growth conditions and minor fimbria purification.
The isogenic, major fimbria-deficient mutant DPG3, which expresses only the minor fimbria (Pg min+/maj−), was maintained anaerobically (10% H2, 10% CO2, 80% N2) in a Forma Scientific anaerobic system glove box, model 1025/1029, at 37°C in Difco anaerobe broth MIC. Erythromycin (5 μg/ml) was added according to the selection requirements of the strain (11, 20, 54, 68). Fimbriae were purified as described by Davey et al. (11). Briefly, bacterial pellets of P. gingivalis DPG3 were shattered by ultrasonication for 5 min, pulsing at 50% power, on ice. The cellular debris was removed by centrifugation, and the remaining supernatant was combined with saturated ammonium sulfate (40%) to precipitate the fimbriae. After centrifugation the resulting pellets were dialyzed in 20 mM Tris buffer (pH 7.8). The dialysate was further purified by multiple runs on a DEAE-Sepharose CL-6B column (Amersham Biosciences) equilibrated with 20 mM Tris buffer (pH 7.6 to 8.0) and eluted with a linear gradient of 0 to 1.0 M NaCl. Fractions were analyzed by 12% SDS-PAGE and silver staining (Bio-Rad) to ensure purity and quantified by Bradford assay. Fimbria preparations underwent further screening to confirm lack of LPS contamination via silver staining (20). Samples were then analyzed by tandem MS (MS/MS) to verify identity and to ensure no other protein contaminants were present.
Limulus amebocyte lysate assay.
Lack of endotoxin was verified by using the limulus amebocyte lysate (LAL) Pyrogent 03 Plus gel clot assay (catalog no. N294-03; Lonza) following the manufacturer's instructions. Briefly, an Escherichia coli LPS standard as well as 100-μg/ml, 50-μg/ml, and 25-μg/ml replicates of purified minor fimbriae were incubated with limulus amebocyte lysate for 1 h at 37°C. After 1 h the glass test tubes were inverted. A positive test was characterized by momentary formation of a firm gel when the tube was inverted. A negative test result was the absence of a solid clot. The sensitivity of this test is 0.03 endotoxin units (EU)/ml. An EU is defined by the manufacturer as the endotoxin activity of 0.2 ng of reference endotoxin standard.
Tandem mass spectrometry.
To confirm the identity of the minor fimbria, purified proteins were run on an SDS-PAGE gel and analyzed by the Proteomics Center at Stony Brook University. Gel bands were cut out, destained, reduced, alkylated, and digested with trypsin (Promega Gold, mass spectrometry grade) as described by Shevchenko et al. with minor modifications (49). The resulting concentrated peptide extract was diluted into a solution of 2% acetonitrile (ACN)-0.1% formic acid (FA) (buffer A) for analysis. For solution digest, 10 μl of purified protein was diluted in 40 μl of 50 mM ammonium bicarbonate. The proteins were reduced with 2 mM dithiothreitol and alkylated with 4 mM iodoacetamide for 30 min each. A 0.25-μg aliquot of trypsin was added, and digests were incubated overnight at 37°C. Protease reactions were stopped with 100% formic acid (final concentration, 5%). Ten microliters of the peptide mixture was analyzed by automated microcapillary liquid chromatography-tandem mass spectrometry. Fused silica capillaries (100 μm inside diameter [i.d.]) were pulled using a P-2000 CO2 laser puller (Sutter Instruments, Novato, CA) to a 5 μm i.d. tip and packed with 10 cm of 5-μm Magic C18 material (Agilent, Santa Clara, CA) by using a pressure bomb. This column was then placed in line with a Dionex 3000 high-performance liquid chromatograph (HPLC) equipped with an autosampler. The column was equilibrated in buffer A, and the peptide mixture was loaded onto the column using the autosampler. The HPLC separation at a flow rate of 300 nl/min was provided by a gradient between buffer A and buffer B (98% acetonitrile, 0.1% formic acid). The HPLC gradient was held constant at 100% buffer A for 5 min after peptide loading, followed by a 30-min gradient from 5% buffer B to 40% buffer B. Then, the gradient was switched from 40% to 80% buffer B over 5 min and held constant for 3 min. Finally, the gradient was changed from 80% buffer B to 100% buffer A over 1 min and then held constant at 100% buffer A for 15 more minutes. The application of a 1.8-kV distal voltage electrosprayed the eluted peptides directly into a Thermo LTQ ion trap mass spectrometer equipped with a custom nanoLC electrospray ionization source. Full MS spectra were recorded on the peptides over a 400 to 2,000 m/z range, followed by five MS/MS events sequentially generated in a data-dependent manner on the first, second, third, fourth, and fifth most intense ions selected from the full MS spectrum (at 35% collision energy). Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan, San Jose, CA). MS/MS spectra were extracted from the RAW file with ReAdW.exe (http://sourceforge.net/projects/sashimi). The resulting mzXML file with all the data for all MS/MS spectra was read by using the analysis software. The MS/MS data were searched with Inspect (56) against a Porphyromonas gingivalis database containing 4,251 proteins, in addition to an Escherichia coli database plus common contaminants, with the following modifications: +16 on methionine, +57 on cysteine, and +1 on asparagine and glutamine. Only peptides with a P value of ≤0.01 were analyzed further.
Detection of glycosylation.
To detect the presence of glycosylation, purified native minor fimbriae were run on SDS-PAGE and carbohydrates were stained using the ProQ Emerald glycoprotein stain kit (Molecular Probes) following the manufacturer's instructions. All ProQ gels were run with CandyCane glycoprotein molecular weight standards (Molecular Probes) provided in the ProQ Emerald glycoprotein stain kit, i.e., as positive controls for staining. Further verification of glycosylation on the minor fimbriae included treatment with the native protein deglycosylation kit (NDEGLY; Sigma) following the manufacturer's instructions. This kit is specific for N-glycosylation and utilizes three different endoglycosidase F (endo F) enzymes. According to the manufacturer's instructions, endo F1 cleaves all asparagine-linked hybrid or high-mannose oligosaccharides but not complex oligosaccharides. Endo F2 cleaves biantennary complex and to a lesser extent high-mannose oligosaccharides. Fucosylation has little effect on endo F2 cleavage of biantennary structures. Endo F2 will not cleave hybrid structures. Endo F3 cleaves biantennary and triantennary complex oligosaccharides. However, nonfucosylated biantennary and triantennary structures are hydrolyzed at a slow rate by endo F3. Core fucosylated biantennary structures are efficient substrates for endo F3 oligosaccharides. Core fucosylation of biantennary structures increases activity up to 400-fold. Endo F3 has no activity on oligomannose and hybrid molecules. The untreated and treated minor fimbriae were run under native nonreducing conditions and reducing conditions with boiling on SDS-PAGE and probed with ProQ to detect loss of glycosylation. Further glycosylation analysis was performed using the enzymatic protein deglycosylation kit (E-DEGLY; Sigma) following the manufacturer's instructions. This kit utilizes PNGase F (which cleaves N-glycosylation), α-2(3,6,8,9)-neuraminidase (which removes sialic acids), O-glycosidase (endo-α-N-acetylgalactosaminidase removes core structure with no modifications to serine or threonine residues), β(1-4)-galactosidase, and β-N-acetylglucosaminidase. Samples were then run on 12% SDS-PAGE gels and stained for ProQ. Additionally, we preincubated the minor fimbriae with α-l-fucosidase (from bovine kidney; Sigma) following the manufacturer's instructions, prior to treatment with E-DEGLY.
Monosaccharide analysis by GC-MS.
To identify the carbohydrate motifs, 2 mg of purified minor fimbriae was sent to a commercial laboratory (M-SCAN, Inc., West Chester, PA) for analysis by GC-MS. An aliquot of purified minor fimbriae (60 μl) was spiked with 10 μg arabitol (Ara) as an internal standard (IS) and lyophilized. The dried sample was hydrolyzed, re-N-acetylated, derivatized, and analyzed by GC-MS. A standard mixture containing 10 μg each of fucose (Fuc), xylose (Xyl), mannose (Man), galactose (Gal), glucose (Glc), N-acetylgalactosamine (GalNAc), and N-acetylglucosamine (GlcNAc) plus arabitol (Ara) and a tube/reagent blank containing 10 μg Ara were also hydrolyzed, re-N-acetylated, derivatized, and analyzed by GC-MS (as described below) alongside the carbohydrate sample. An aliquot (1 μl) of each derivatized carbohydrate sample dissolved in hexane (2 ml) was analyzed by GC-MS using a Perkin-Elmer Turbomass quadrupole mass spectrometer with integrated gas chromatograph under the following conditions. Samples were injected onto a DB5 column at 95°C using helium as a carrier gas. The program was run as follows: 1 min at 90°C, then 25°C/minute to 140°C, then 5°C/minute to 220°C, then 10°C/min to 300°C, and finally holding at 300°C for 5 min. The mass spectrometry ionization voltage was 70 eV, the acquisition mode was set to scanning, and mass range was 50 to 500 Da. Monitored ions were 173 for N-acetylhexosamines, 204 for hexoses, deoxyhexoses, and pentoses, and 217 for arabitol. On comparison of the data with those obtained from the standard mixtures containing known amounts of the expected monosaccharides, the sugars hydrolyzed from the sample were identified and the quantity of each monosaccharide present was estimated.
Transmission electron microscopy of native minor fimbriae.
For transmission electron microscopy (TEM), a 0.8-μg/μl solution of purified native Mfa1 protein in 20 mM Tris-HCl (pH 7.8) was adsorbed onto polyvinyl formal carbon-coated grids (Ernest F. Fullam, Latham, NY) for 2 min, washed twice with phosphate-buffered saline and twice with water, and then negatively stained with 0.5% phosphotungstic acid (Ted Pella, Inc., Redding, CA) for 30 s. All grids were viewed in a transmission electron microscope (FEI TECNAI 12 BioTwin G02) at an 80-kV accelerating voltage, and images were obtained by using an AMT XR-60 charge-coupled-device digital camera system. Direct magnification was at ×98,000.
RESULTS
Purification of native minor fimbriae.
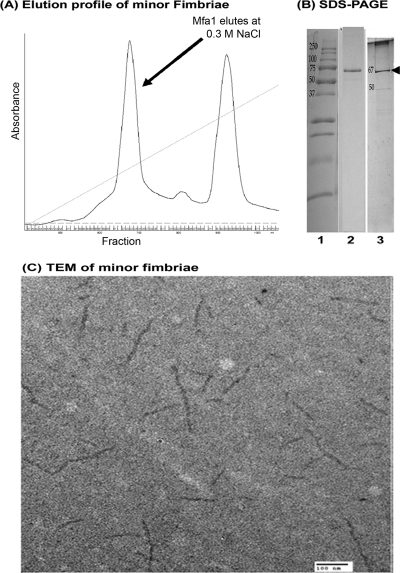
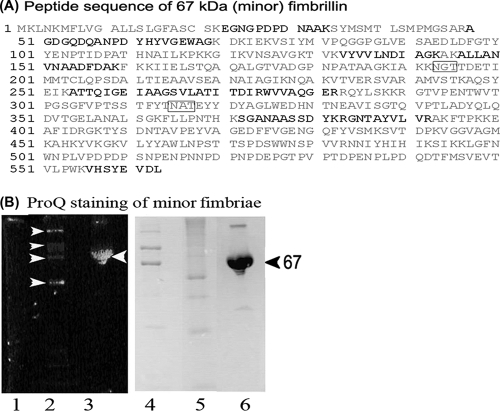
After growth of P. gingivalis DPG-3 under anaerobic conditions, the bacteria were disrupted by sonication, and minor fimbriae were purified by ion-exchange chromatography using a DEAE-Sepharose column. In Fig. 1A we show a representative elution profile of the minor fimbria on a DEAE-Sepharose column. The minor fimbria eluted at 0.3 M NaCl, whereas other proteins were still bound until 0.5 M NaCl. An aliquot of the peak corresponding to the putative minor fimbria was analyzed by SDS-PAGE and rerun on the DEAE-Sepharose column multiple times, changing either the steepness of the elution gradient or the pH of the buffers. Protein purification was continued until minor fimbria samples showed no contaminating bands (i.e., for LPS or other contaminants) by Coomassie staining and silver staining (Fig. 1B) (20). Absence of endotoxin was further confirmed by LAL assay (Lonza). None of the dilutions tested generated a positive LAL reaction at an assay sensitivity of 0.03 EU/ml (data not shown) (6, 11, 48). The native fimbria was further analyzed by TEM, demonstrating the presence of oligomeric strands approximately 100 to 200 nm in length, which is similar to previous observations (41) (Fig. 1C). The single band corresponding to the correct 67-kDa size of the minor fimbria protein was analyzed for protein purity and peptide sequence by MS/MS (Fig. 2A). MS/MS confirmed the purity and correct peptide sequence (Fig. 2A, bold letters). Examination of the peptide sequence also revealed two conserved Asn-Xaa-Ser/Thr asparagine-linked (N-linked) (putative) glycosylation motifs (Fig. 2A, gray boxes) (29, 53). The Mfa1 amino acid sequence also contained numerous serines and threonines, which can function as putative O-linked glycosylation sites (1, 53). We further analyzed the purified native minor fimbria for glycosylation by SDS-PAGE and ProQ staining (Fig. 2B), which revealed a positive staining reaction.
FIG. 1.
Purification and characterization of the minor (Mfa1) fimbria. (A) Elution profile of the 67-kDa fimbria on DEAE-Sepharose CL-6B, showing a peak that eluted with 0.3 M NaCl. (B) SDS-PAGE analysis of the minor fimbria. Lane 1, MW standard; lane 2, Coomassie blue stain showing the 67-kDa minor fimbria; lane 3, silver stain showing the single band of the minor fimbria (arrow). (C) Transmission electron micrograph of the purified minor fimbria showing 100- to 200-nm fibers. Bar, 100 nm.
FIG. 2.
Glycosylation of the minor fimbriae. (A) Peptide sequence obtained by MS/MS (shown in bold), which confirmed the identity of the minor fimbria. Boxed are putative N-X-S/T asparagine-linkage motifs. (B) Confirmation of glycosylation on the minor fimbria by ProQ (glycosylation stain). Minor fimbriae samples were run on SDS-PAGE gels and stained with ProQ (lanes 1 to 3), and then the same gel was stained with Coomassie (lanes 4 to 6). Lanes 1 and 4, nonglycosylated MW standard; lanes 2 and 5, CandyCane glycoprotein standard; lanes 3 and 6, the minor fimbria. White arrowheads highlight the CandyCane (Molecular Probes) glycoprotein standard.
The native minor fimbria is susceptible to some N- and O-linked enzymatic deglycosylation.
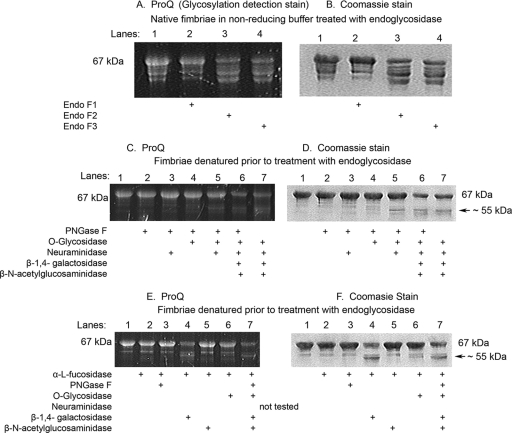
Based on the amino acid sequence, N-linked glycosylation was a distinct possibility. We therefore subjected the purified minor fimbria to treatment with endoglycosidases F1, F2, and F3. These enzymes cleave N-linked glycoproteins but differ significantly in their oligosaccharide specificity, as previously reported (37, 57-59). Briefly, endoglycosidase F1 cleaves oligomannose and hybrid oligosaccharides, but this activity is greatly reduced by core fucosylation. Endoglycosidase F1 will also not cleave any complex oligosaccharides. Endoglycosidase F2 does not cleave hybrid or triantennary complex oligosaccharide structures, but it does cleave oligomannose and biantennary complex oligosaccharides. Core fucosylation has little to no effect on endoglycosidase F2 activity. Endoglycosidase F3 can cleave biantennary and triantennary complex oligosaccharides with a preference for those oligosaccharides with core fucosylation. Endoglycosidase F3 will also cleave fucosylated trimannosyl core structures on oligosaccharides but has no activity on oligomannose or hybrid structures (37, 57-59).
The results under nonreducing (native) conditions (Fig. 3A and B, lanes 3 and 4) showed that endoglycosidases F2 and F3 have moderate effects on the native minor fimbria, as revealed by either by ProQ staining or an apparent molecular mass shift. This suggested one of several possibilities. First, the minor fimbria contains N-linked complex biantennary complex oligosaccharide glycosylation, but the glycosylation site might be inaccessible to the endoglycosidases under its oligomeric native configuration (Fig. 1C), resulting in the step ladder pattern in lanes 3 and 4 (Fig. 3A and B). Second, it is possible that the minor fimbriae are O-linked glycoproteins or contain both N- and O-linked motifs. Finally, the minor fimbria may contain a novel glycosylation structure that is resistant to classic deglycosylation. To address the first two possibilities, we employed an additional N-linked endoglycosidase, PNGase F, and denatured the minor fimbria prior to enzymatic treatment (Fig. 3C to F). PNGase F is a more potent N-linked deglycosidase that optimally works on non-core-fucosylated denatured proteins, as reported previously (37). It works by specifically recognizing an Asp-GlcNAc-oligosaccharide complex. Under these conditions we observed that the protein retained its glycosylation even when denatured and treated with PNGase F (Fig. 3C and D). To address the possibility that fucose residues interfere with PNGase F, as previously reported (37), we pretreated the minor fimbria with α-l-fucosidase. Again, pretreatment had no affect on PNGase F, ruling out classic Asp-GlcNAc-oligosaccharide linkages or the presence of other blocking carbohydrates.
FIG. 3.
Enzymatic deglycosylation of the minor fimbria observed in the presence of endoglycosidase F2, endoglycosidase F3, and β(1-4)-galactosidase. Enzymatic deglycosylation treatment on purified minor fimbria (Mfa1), as verified by lack of shift or the loss of ProQ (glycosylation detection) signal, is shown. (A, C, E) ProQ gels; (B, D, and F) the same gel after Coomassie blue staining. (A and B) Nonreduced native fimbria treated with endoglycosidase. All lanes were loaded with 5 μg of Mfa1 and digested with the indicated endoglycosidase(s). (C and D) Fimbriae denatured prior to treatment with endoglycosidase. All lanes were loaded with 7 μg of Mfa1 and digested with the indicated endoglycosidase(s). (E and F) Minor fimbria pretreated with α-l-fucosidase, then denatured and treated with endoglycosidase. All lanes were loaded with 7 μg Mfa1 and digested with the indicated endoglycosidase(s).
To examine O-linked glycosylation, the purified minor fimbriae were treated with α-2(3,6,8,9)-neuraminidase (which removes sialic acids), O-glycosidase, which cleaves serine- or threonine-linked unsubstituted Gal-β(1-3)-GalNAc-α,β(1-4)-galactosidase, which releases the terminal β(1-4)-galactose provided that it is nonreducing, and β-N-acetylglucosaminidase, which cleaves terminal nonreducing β-linked N-acetylglucosamine residues (Fig. 3C to F) (37). Intriguingly, we did observe a reduction in ProQ staining of the 67-kDa band in response to treatment with β(1-4)-galactosidase and β-N-acetylglucosaminidase (Fig. 3C and 3D, lanes 6 and 7), suggesting the presence of O-linked galactose and N-acetylglucosamine motifs in the minor fimbria. Furthermore, the samples treated with β(1-4)-galactosidase (Fig. 3E and F, lanes 4 and 7) were partially deglycosylated and contained a band with an approximate molecular mass of 55 kDa, suggesting a modest amount of β(1-4)-galactose on the minor fimbria. The 55-kDa band does not correlate to any of the known molecular masses of the enzymes we tested. It should be mentioned in this context that the predicted molecular mass of the minor fimbria is 61 kDa (8, 41), based on the complete amino acid sequence as analyzed with the ExPASy proteomics server (Expert Protein Analysis System; http://au.expasy.org/cgi-bin/protparam; Swiss Institute of Bioinformatics, Geneva, Switzerland). However, Mfa1 has two predicted signal peptidase cleavage sites. We propose that the 55-kDa band is a cleaved form of Mfa1 that has been tethered by carbohydrates, as further discussed below. The 55-kDa bands were confirmed to be the minor fimbria (lacking the first 50 amino acids) by MS/MS analysis (data not shown). Overall, these results suggest a novel pattern of glycosylation of the minor fimbria that is resistant to common methods of enzymatic deglycosylation.
Monosaccharide analysis of the minor fimbria by GC-MS.
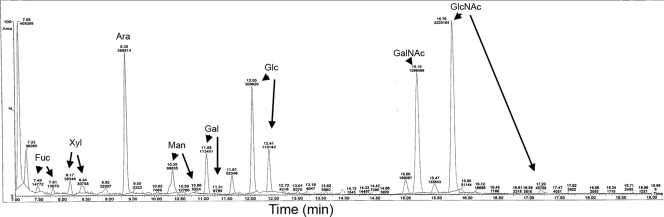
Samples of the purified native minor fimbria were analyzed by GC-MS as described in Materials and Methods. A representative chromatograph is shown (Fig. 4), and results are summarized in Table 1. We confirmed that the monosaccharides fucose, mannose, and N-acetylglucosamine were present on the purified minor fimbria (1.35 nmol/mg, 2.68 nmol/mg, and 2.27 nmol/mg, respectively) (Table 1). These carbohydrates have been implicated in ligation of DC-SIGN on dendritic cells, which promotes an immunosuppressive response (4, 68, 70). Also present in large quantities were xylose, galactose, and glucose (3.76 nmol/mg, 5.71 nmol/mg, and 14.1 nmol/mg, respectively) (Table 1). N-Acetylgalactosamine was also present in low concentrations (0.65 nmol/mg) (Table 1).
FIG. 4.
Representative chromatograph from GC-MS analysis of the purified minor fimbria. The purified minor fimbria was analyzed by GC-MS for monosaccharide content relative to monosaccharide standards for Fuc, Xyl, Man, Gal, Glc, GalNac, and GlcNAc.
TABLE 1.
Summary of monosaccharide compositional analyses by GC-MS of purified minor fimbriae
| Monosaccharide | Mfa1 (nmol/mg) | Ratio to GalNAca |
|---|---|---|
| Fuc | 1.35 | 2.07 |
| Xyl | 3.76 | 5.77 |
| Man | 2.68 | 4.11 |
| Gal | 5.71 | 8.76 |
| Glc | 14.1 | 21.6 |
| GalNAc | 0.652 | 1.00 |
| GlcNAc | 2.27 | 3.48 |
Shown are the ratios of monosaccharides found on the minor fimbriae relative to GalNAc (set as 1.0).
DISCUSSION
Our results demonstrate that the purified native minor fimbria is present as strands of 100 to 200 nm in length and is glycosylated (Fig. 1). The glycosylated minor fimbria, while susceptible to endoglycosidases F2 and F3 as well as β(1-4)-galactosidase (Fig. 3), was resistant to other classical N-linked and O-linked deglycosylation enzymes, suggesting a novel structural linkage. Finally, we showed using GC-MS that the monosaccharide composition of the minor fimbria contains moderate amounts of the DC-SIGN ligands fucose, mannose, and N-acetylglucosamine and large amounts of xylose, galactose, and glucose (Fig. 4 and Table 1).
Many mucosal pathogens exhibit glycosylation motifs on their flagella, pili, and fimbriae (53). Glycosylation reportedly plays a role in maintenance of the protein structure, protection from proteolytic degradation, immune evasion, host cell adhesion, and surface recognition (53). A role for glycosylation of the major fimbria in P. gingivalis has been previously suggested. Knockouts of gftA (a wcaE glycotransferase homolog of E. coli) in P. gingivalis fail to make mature fimbriae (35). The gingipains of P. gingivalis are apparently glycosylated, with different isoforms being differentially glycosylated (9, 15). The glycosylation activity is regulated by the vimF, vimA, and vimE glycotransferase genes (60, 61). Knocking out these genes causes a failure to glycosylate these gingipains, leading to their inactivation (9, 60, 61). Recently, it was discovered that the OMP85 protein of P. gingivalis is glycosylated (33). All of these outer membrane protein genes encode a signal peptide that gets cleaved before it exits the periplasm (31, 36, 50). Given that all of the elements for glycosylation are present in P. gingivalis and that the minor fimbria apparently targets DC-SIGN (68), this study confirms our suspicion that the minor fimbria is glycosylated.
We have shown that the minor fimbria does not contain LPS by LAL test and silver staining (Fig. 1B). Although the LPS structures of different strains of P. gingivalis may not be conserved (9, 38, 39, 47), it is worth mentioning a study by Curtis et al. in 1999 which characterized the LPS of P. gingivalis (9). This study determined that the LPS does not contain mannose or fucose, while our minor fimbria does. Furthermore, the Curtis study determined that the core region of P. gingivalis LPS lacked N-acetylglucosamine and N-acetylgalactosamine (9), while our minor fimbria contains these sugars. The Curtis study was also the first to characterize the glycosylation of the gingipains, which contain arabinose, rhamnose, fucose, mannose, galactose, glucose, N-acetylglucosamine, N-acetylgalactosamine, and N-acetylneuraminic acid (9). Again, our samples lacked the gingipains as determined by both silver staining (Fig. 1B) and tandem MS (Fig. 2A). However, the molar ratios of monosaccharides that we detected on the minor fimbria (Table 1) differed from those previously published for the gingipains and LPS (9, 15, 38, 45, 47). Finally, the fimbria was purified from a nonencapsulated P. gingivalis strain, ruling out the possibility that the monosaccharides originated from the capsule (2, 11).
The Mfa1 protein component of the minor fimbria has a predicted size of 61 kDa, but reported sizes vary starkly from 67 to 75 kDa (41, 67). These size discrepancies could be attributed to glycosylation, the extent of which may depend on purification protocols and growth conditions (3, 28, 40, 63, 64).
Although the predicted molecular mass for Mfa1 is approximately 61 kDa (8, 41), we showed that the purified native minor fimbria migrated at 67 kDa. Shoji et al. in 2004 demonstrated that the minor fimbria is processed by a lipoprotein signal peptidase (signal peptidase II), as evidenced by improper processing in the presence of globomycin (50). They also described that the precursor proteins of the minor fimbria are lipidated (50). Moreover, they described that the minor fimbria gets processed twice, first by the lipoprotein signal peptide and then again by the gingipains (50). This led us to search for signal peptide cleavage sites on Mfa1 by using the SignalP server (http://www.cbs.dtu.dk/services/SignalP) (12). Interestingly, this analysis revealed two potential signal peptide cleavage sites, at amino acid (aa) positions 21 and 22 and at 50 and 51 (12, 14). Removal of the first signal peptide (aa 21-22) by signal peptidase I would result in a protein of approximately 58 kDa (based on the ExPASy prediction). However, removal of the second signal peptide (aa 50-51) by lipoprotein signal peptidase would result in a protein of approximately 55 kDa (based on ExPASy prediction). We demonstrated that treatment with β(1-4)-galactosidase resulted in the minor fimbria migrating at 55 kDa, corresponding to the second cleavage site. Therefore, it is possible that Mfa1 is processed at this site but that the N-terminal piece is tethered to the mature protein by carbohydrates with a β(1-4)-galactose linkage.
Additionally, the minor fimbria is not the only fimbria expressed by P. gingivalis that exhibits sizes in the expected range. The major fimbria is also reported to vary in size (41 to 43 kDa) (67). Efforts are under way to determine if the major fimbria is also glycosylated. Since both fimbriae and gingipains undergo similar mechanisms of translocation to the outer membrane, it is feasible that during this process they might become glycosylated.
While nonpathogenic E. coli normally does not express the glycosylation machinery necessary to modify proteins, recent studies have transferred the pgl gene cluster of Campylobacter jejuni, enabling E. coli to perform N-linked glycosylation (1, 62). Also, Fleckenstein et al. (14) reported that EptA from a naturally occurring strain of enterotoxigenic E. coli (ETEC) is glycosylated. The report went on to demonstrate that the entire eptBAC locus is necessary for production of glycosylated EptA and that the loss of EptC results in nonglycosylated EptA (14). Moreover, they demonstrated that the eptBAC gene locus was restricted to some ETEC strains but was absent in other pathogenic and nonpathogenic strains of E. coli, confirming that E. coli can glycosylate proteins when provided with the proper genes (14). Recently Sartain and Belisle showed that expression of recombinant SodC (of M. tuberculosis) resulted in proteins that were not processed correctly, nor were they glycosylated (46). These studies suggest that E. coli normally does not possess the necessary glycosylation machinery. Our finding that the native minor fimbria is glycosylated suggests that caution should be used in interpretation of studies that use recombinant minor fimbria expressed in E. coli (41).
It is important to note that bacterial O-glycosylation makes use of unusual sugars (1). Also, the sugars are not always added in a sequential manner to the protein. There are reports that sugars are preassembled and added to a lipid carrier before being added to the protein acceptor (1). Understanding how the P. gingivalis minor fimbria becomes glycosylated and translocated would expand our understanding of this organism and how it eludes host immunity.
Acknowledgments
This study was supported by U.S. Public Health Service grants from the NIH NIDCR (R01 DE014328 to C.W.C. and F31 DE020014 to A.Z.) and the NIH NIGMS (R01 GM62987 to D.G.T.).
We thank Caroline A. Genco for providing us with the isogenic fimbria mutant (DPG-3) and for guidance in purification of minor fimbriae. We thank Susan Van Horn and the Central Microscopy Imaging Center (Stony Brook University) for assistance with electron microscopy. We thank Antonius Koller and the Proteomics Center (Stony Brook University) for assistance with tandem mass spectrometry.
Footnotes
Published ahead of print on 18 June 2010.
REFERENCES
- 1.Abu-Qarn, M., J. Eichler, and N. Sharon. 2008. Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. 18:544-550. [DOI] [PubMed] [Google Scholar]
- 2.Aduse-Opoku, J., J. M. Slaney, A. Hashim, A. Gallagher, R. P. Gallagher, M. Rangarajan, K. Boutaga, M. L. Laine, A. J. Van Winkelhoff, and M. A. Curtis. 2006. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect. Immun. 74:449-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano, A., A. Sharma, H. T. Sojar, H. K. Kuramitsu, and R. J. Genco. 1994. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect. Immun. 62:4682-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appelmelk, B. J., I. van Die, S. J. van Vliet, C. M. Vandenbroucke-Grauls, T. B. Geijtenbeek, and Y. van Kooyk. 2003. Cutting edge: carbohydrate profiling identifies new pathogens that interact with dendritic cell-specific ICAM-3-grabbing nonintegrin on dendritic cells. J. Immunol. 170:1635-1639. [DOI] [PubMed] [Google Scholar]
- 5.Arai, M., N. Hamada, and T. Umemoto. 2000. Purification and characterization of a novel secondary fimbrial protein from Porphyromonas gingivalis strain 381. FEMS Microbiol. Lett. 193:75-81. [DOI] [PubMed] [Google Scholar]
- 6.Asai, Y., M. Hashimoto, H. M. Fletcher, K. Miyake, S. Akira, and T. Ogawa. 2005. Lipopolysaccharide preparation extracted from Porphyromonas gingivalis lipoprotein-deficient mutant shows a marked decrease in toll-like receptor 2-mediated signaling. Infect. Immun. 73:2157-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burt, B. 2005. Position paper: epidemiology of periodontal diseases. J. Periodontol. 76:1406-1419. [DOI] [PubMed] [Google Scholar]
- 8.Chung, W. O., D. R. Demuth, and R. J. Lamont. 2000. Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect. Immun. 68:6758-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shepherd, N. Paramonov, and E. F. Hounsell. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutler, C. W., J. R. Kalmar, and C. A. Genco. 1995. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 3:45-51. [DOI] [PubMed] [Google Scholar]
- 11.Davey, M., X. Liu, T. Ukai, V. Jain, C. Gudino, F. C. Gibson, 3rd, D. Golenbock, A. Visintin, and C. A. Genco. 2008. Bacterial fimbriae stimulate proinflammatory activation in the endothelium through distinct TLRs. J. Immunol. 180:2187-2195. [DOI] [PubMed] [Google Scholar]
- 12.Emanuelsson, O., S. Brunak, G. von Heijne, and H. Nielsen. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2:953-971. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg, H., D. A. Mitchell, K. Drickamer, and W. I. Weis. 2001. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294:2163-2166. [DOI] [PubMed] [Google Scholar]
- 14.Fleckenstein, J. M., K. Roy, J. F. Fischer, and M. Burkitt. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74:2245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher, A., J. Aduse-Opoku, M. Rangarajan, J. M. Slaney, and M. A. Curtis. 2003. Glycosylation of the Arg-gingipains of Porphyromonas gingivalis and comparison with glycoconjugate structure and synthesis in other bacteria. Curr. Protein Pept. Sci. 4:427-441. [DOI] [PubMed] [Google Scholar]
- 16.Guo, Y., H. Feinberg, E. Conroy, D. A. Mitchell, R. Alvarez, O. Blixt, M. E. Taylor, W. I. Weis, and K. Drickamer. 2004. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11:591-598. [DOI] [PubMed] [Google Scholar]
- 17.Hajishengallis, G., and E. Harokopakis. 2007. Porphyromonas gingivalis interactions with complement receptor 3 (CR3): innate immunity or immune evasion? Front. Biosci. 12:4547-4557. [DOI] [PubMed] [Google Scholar]
- 18.Hajishengallis, G., M. Wang, E. Harokopakis, M. Triantafilou, and K. Triantafilou. 2006. Porphyromonas gingivalis fimbriae proactively modulate β2 integrin adhesive activity and promote binding to and internalization by macrophages. Infect. Immun. 74:5658-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamada, N., H. T. Sojar, M. I. Cho, and R. J. Genco. 1996. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect. Immun. 64:4788-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamada, N., K. Watanabe, M. Arai, H. Hiramine, and T. Umemoto. 2002. Cytokine production induced by a 67-kDa fimbrial protein from Porphyromonas gingivalis. Oral Microbiol. Immunol. 17:197-200. [DOI] [PubMed] [Google Scholar]
- 21.Imamura, T., J. Travis, and J. Potempa. 2003. The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr. Protein Pept. Sci. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 22.Jotwani, R., B. Pulendran, S. Agrawal, and C. W. Cutler. 2003. Human dendritic cells respond to Porphyromonas gingivalis LPS by promoting a Th2 effector response in vitro. Eur. J. Immunol. 33:2980-2986. [DOI] [PubMed] [Google Scholar]
- 23.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072-29076. [DOI] [PubMed] [Google Scholar]
- 24.Kato, T., T. Tsuda, H. Omori, T. Yoshimori, and A. Amano. 2007. Maturation of fimbria precursor protein by exogenous gingipains in Porphyromonas gingivalis gingipain-null mutant. FEMS Microbiol. Lett. 273:96-102. [DOI] [PubMed] [Google Scholar]
- 25.Lamont, R. F. 1998. New approaches in the management of preterm labour of infective aetiology. Br. J. Obstet. Gynaecol. 105:134-137. [DOI] [PubMed] [Google Scholar]
- 26.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 27.Maeda, N., J. Nigou, J. L. Herrmann, M. Jackson, A. Amara, P. H. Lagrange, G. Puzo, B. Gicquel, and O. Neyrolles. 2003. The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J. Biol. Chem. 278:5513-5516. [DOI] [PubMed] [Google Scholar]
- 28.Masuda, T., Y. Murakami, T. Noguchi, and F. Yoshimura. 2006. Effects of various growth conditions in a chemostat on expression of virulence factors in Porphyromonas gingivalis. Appl. Environ. Microbiol. 72:3458-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medzihradszky, K. F. 2008. Characterization of site-specific N-glycosylation. Methods Mol. Biol. 446:293-316. [DOI] [PubMed] [Google Scholar]
- 30.Mezyk-Kopec, R., M. Bzowska, J. Potempa, N. Jura, A. Sroka, R. A. Black, and J. Bereta. 2005. Inactivation of membrane tumor necrosis factor alpha by gingipains from Porphyromonas gingivalis. Infect. Immun. 73:1506-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagano, K., Y. Murakami, K. Nishikawa, J. Sakakibara, K. Shimozato, and F. Yoshimura. 2007. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J. Med. Microbiol. 56:1536-1548. [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa, I., A. Amano, M. Kuboniwa, T. Nakamura, S. Kawabata, and S. Hamada. 2002. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect. Immun. 70:277-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakao, R., Y. Tashiro, N. Nomura, S. Kosono, K. Ochiai, H. Yonezawa, H. Watanabe, and H. Senpuku. 2008. Glycosylation of the OMP85 homolog of Porphyromonas gingivalis and its involvement in biofilm formation. Biochem. Biophys. Res. Commun. 365:784-789. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama, K., F. Yoshimura, T. Kadowaki, and K. Yamamoto. 1996. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J. Bacteriol. 178:2818-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narimatsu, M., Y. Noiri, S. Itoh, N. Noguchi, T. Kawahara, and S. Ebisu. 2004. Essential role for the gtfA gene encoding a putative glycosyltransferase in the adherence of Porphyromonas gingivalis. Infect. Immun. 72:2698-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okamoto, K., Y. Misumi, T. Kadowaki, M. Yoneda, K. Yamamoto, and Y. Ikehara. 1995. Structural characterization of argingipain, a novel arginine-specific cysteine proteinase as a major periodontal pathogenic factor from Porphyromonas gingivalis. Arch. Biochem. Biophys. 316:917-925. [DOI] [PubMed] [Google Scholar]
- 37.O'Neill, R. A. 1996. Enzymatic release of oligosaccharides from glycoproteins for chromatographic and electrophoretic analysis. J. Chromatogr. A 720:201-215. [DOI] [PubMed] [Google Scholar]
- 38.Paramonov, N., D. Bailey, M. Rangarajan, A. Hashim, G. Kelly, M. A. Curtis, and E. F. Hounsell. 2001. Structural analysis of the polysaccharide from the lipopolysaccharide of Porphyromonas gingivalis strain W50. Eur. J. Biochem. 268:4698-4707. [DOI] [PubMed] [Google Scholar]
- 39.Paramonov, N. A., J. Aduse-Opoku, A. Hashim, M. Rangarajan, and M. A. Curtis. 2009. Structural analysis of the core region of O-lipopolysaccharide of Porphyromonas gingivalis from mutants defective in O-antigen ligase and O-antigen polymerase. J. Bacteriol. 191:5272-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, Y., C. E. James, F. Yoshimura, and R. J. Lamont. 2006. Expression of the short fimbriae of Porphyromonas gingivalis is regulated in oral bacterial consortia. FEMS Microbiol. Lett. 262:65-71. [DOI] [PubMed] [Google Scholar]
- 41.Park, Y., M. R. Simionato, K. Sekiya, Y. Murakami, D. James, W. Chen, M. Hackett, F. Yoshimura, D. R. Demuth, and R. J. Lamont. 2005. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect. Immun. 73:3983-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pohlmann, S., F. Baribaud, B. Lee, G. J. Leslie, M. D. Sanchez, K. Hiebenthal-Millow, J. Munch, F. Kirchhoff, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 75:4664-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potempa, J., J. Mikolajczyk-Pawlinska, D. Brassell, D. Nelson, I. B. Thogersen, J. J. Enghild, and J. Travis. 1998. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J. Biol. Chem. 273:21648-21657. [DOI] [PubMed] [Google Scholar]
- 44.Pulendran, B., P. Kumar, C. W. Cutler, M. Mohamadzadeh, T. Van Dyke, and J. Banchereau. 2001. Lipopolysaccharides from distinct pathogens induce different classes of immune responses in vivo. J. Immunol. 167:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rangarajan, M., A. Hashim, J. Aduse-Opoku, N. Paramonov, E. F. Hounsell, and M. A. Curtis. 2005. Expression of Arg-gingipain RgpB is required for correct glycosylation and stability of monomeric Arg-gingipain RgpA from Porphyromonas gingivalis W50. Infect. Immun. 73:4864-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sartain, M. J., and J. T. Belisle. 2009. N-terminal clustering of the O-glycosylation sites in the Mycobacterium tuberculosis lipoprotein SodC. Glycobiology 19:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schifferle, R. E., M. S. Reddy, J. J. Zambon, R. J. Genco, and M. J. Levine. 1989. Characterization of a polysaccharide antigen from Bacteroides gingivalis. J. Immunol. 143:3035-3042. [PubMed] [Google Scholar]
- 48.Seydel, U., A. B. Schromm, L. Brade, S. Gronow, J. Andra, M. Muller, M. H. Koch, K. Fukase, M. Kataoka, M. Hashimoto, S. Kusumoto, and K. Brandenburg. 2005. Physicochemical characterization of carboxymethyl lipid A derivatives in relation to biological activity. FEBS J. 272:327-340. [DOI] [PubMed] [Google Scholar]
- 49.Shevchenko, A., O. N. Jensen, A. V. Podtelejnikov, F. Sagliocco, M. Wilm, O. Vorm, P. Mortensen, H. Boucherie, and M. Mann. 1996. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. U. S. A. 93:14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shoji, M., M. Naito, H. Yukitake, K. Sato, E. Sakai, N. Ohara, and K. Nakayama. 2004. The major structural components of two cell surface filaments of Porphyromonas gingivalis are matured through lipoprotein precursors. Mol. Microbiol. 52:1513-1525. [DOI] [PubMed] [Google Scholar]
- 51.Soilleux, E. J., E. N. Sarno, M. O. Hernandez, E. Moseley, J. Horsley, U. G. Lopes, M. J. Goddard, S. L. Vowler, N. Coleman, R. J. Shattock, and E. P. Sampaio. 2006. DC-SIGN association with the Th2 environment of lepromatous lesions: cause or effect? J. Pathol. 209:182-189. [DOI] [PubMed] [Google Scholar]
- 52.Sroka, A., M. Sztukowska, J. Potempa, J. Travis, and C. A. Genco. 2001. Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J. Bacteriol. 183:5609-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi, Y., M. Davey, H. Yumoto, F. C. Gibson III, and C. A. Genco. 2006. Fimbria-dependent activation of pro-inflammatory molecules in Porphyromonas gingivalis infected human aortic endothelial cells. Cell. Microbiol. 8:738-757. [DOI] [PubMed] [Google Scholar]
- 55.Takeshita, A., Y. Murakami, Y. Yamashita, M. Ishida, S. Fujisawa, S. Kitano, and S. Hanazawa. 1998. Porphyromonas gingivalis fimbriae use β2 integrin (CD11/CD18) on mouse peritoneal macrophages as a cellular receptor, and the CD18 beta chain plays a functional role in fimbrial signaling. Infect. Immun. 66:4056-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanner, S., H. Shu, A. Frank, L. C. Wang, E. Zandi, M. Mumby, P. A. Pevzner, and V. Bafna. 2005. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77:4626-4639. [DOI] [PubMed] [Google Scholar]
- 57.Tarentino, A. L., and T. H. Plummer, Jr. 1994. Substrate specificity of Flavobacterium meningosepticum endo F2 and endo F3: purity is the name of the game. Glycobiology 4:771-773. [DOI] [PubMed] [Google Scholar]
- 58.Tarentino, A. L., G. Quinones, L. M. Changchien, and T. H. Plummer, Jr. 1993. Multiple endoglycosidase F activities expressed by Flavobacterium meningosepticum endoglycosidases F2 and F3. Molecular cloning, primary sequence, and enzyme expression. J. Biol. Chem. 268:9702-9708. [PubMed] [Google Scholar]
- 59.Tarentino, A. L., G. Quinones, W. P. Schrader, L. M. Changchien, and T. H. Plummer, Jr. 1992. Multiple endoglycosidase (Endo) F activities expressed by Flavobacterium meningosepticum. Endo F1: molecular cloning, primary sequence, and structural relationship to Endo H. J. Biol. Chem. 267:3868-3872. [PubMed] [Google Scholar]
- 60.Vanterpool, E., F. Roy, and H. M. Fletcher. 2005. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect. Immun. 73:3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanterpool, E., F. Roy, L. Sandberg, and H. M. Fletcher. 2005. Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis. Infect. Immun. 73:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wacker, M., D. Linton, P. G. Hitchen, M. Nita-Lazar, S. M. Haslam, S. J. North, M. Panico, H. R. Morris, A. Dell, B. W. Wren, and M. Aebi. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790-1793. [DOI] [PubMed] [Google Scholar]
- 63.Wu, J., X. Lin, and H. Xie. 2007. Porphyromonas gingivalis short fimbriae are regulated by a FimS/FimR two-component system. FEMS Microbiol. Lett. 271:214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xie, H., S. Cai, and R. J. Lamont. 1997. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect. Immun. 65:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie, H., and R. J. Lamont. 1999. Promoter architecture of the Porphyromonas gingivalis fimbrillin gene. Infect. Immun. 67:3227-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yilmaz, O., K. Watanabe, and R. J. Lamont. 2002. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell. Microbiol. 4:305-314. [DOI] [PubMed] [Google Scholar]
- 67.Yoshimura, F., Y. Murakami, K. Nishikawa, Y. Hasegawa, and S. Kawaminami. 2009. Surface components of Porphyromonas gingivalis. J. Periodontal Res. 44:1-12. [DOI] [PubMed] [Google Scholar]
- 68.Zeituni, A. E., R. Jotwani, J. Carrion, and C. W. Cutler. 2009. Targeting of DC-SIGN on human dendritic cells by minor fimbriated Porphyromonas gingivalis strains elicits a distinct effector T cell response. J. Immunol. 183:5694-5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, P., O. Schwartz, M. Pantelic, G. Li, Q. Knazze, C. Nobile, M. Radovich, J. He, S. C. Hong, J. Klena, and T. Chen. 2006. DC-SIGN (CD209) recognition of Neisseria gonorrhoeae is circumvented by lipooligosaccharide variation. J. Leukoc. Biol. 79:731-738. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, P., S. Snyder, P. Feng, P. Azadi, S. Zhang, S. Bulgheresi, K. E. Sanderson, J. He, J. Klena, and T. Chen. 2006. Role of N-acetylglucosamine within core lipopolysaccharide of several species of gram-negative bacteria in targeting the DC-SIGN (CD209). J. Immunol. 177:4002-4011. [DOI] [PubMed] [Google Scholar]