Abstract
Cellulosic biofuels represent a powerful alternative to petroleum but are currently limited by the inefficiencies of the conversion process. While Gram-positive and fungal organisms have been widely explored as sources of cellulases and hemicellulases for biomass degradation, Gram-negative organisms have received less experimental attention. We investigated the ability of Cellvibrio japonicus, a recently sequenced Gram-negative cellulolytic bacterium, to degrade bioenergy-related feedstocks. Using a newly developed biomass medium, we showed that C. japonicus is able to utilize corn stover and switchgrass as sole sources of carbon and energy for growth. We also developed tools for directed gene disruptions in C. japonicus and used this system to construct a mutant in the gspD gene, which is predicted to encode a component of the type II secretion system. The gspD::pJGG1 mutant displayed a greater-than-2-fold decrease in endoglucanase secretion compared to wild- type C. japonicus. In addition, the mutant strain showed a pronounced growth defect in medium with biomass as a carbon source, yielding 100-fold fewer viable cells than the wild type. To test the potential of C. japonicus to undergo metabolic engineering, we constructed a strain able to produce small amounts of ethanol from biomass. Collectively, these data suggest that C. japonicus is a useful platform for biomass conversion and biofuel production.
The need for renewable alternatives to petroleum has stimulated interest in many areas of research, including the production of biofuels. Strategies for the production of ethanol from corn starch are well established, but enthusiasm for these approaches is tempered by concerns about cost, as well as indirect effects on global nutrition (38). Cellulosic biofuels represent a potentially useful alternative to starch-based ethanol, in that fuel could be produced from low-value agricultural waste products (10, 39). However, the economic viability of this approach requires overcoming the recalcitrance of plant cell walls to enzymatic degradation (55). This recalcitrance results from the crystalline nature of cellulose, as well as the complexity of plant cell walls, which are a composite of cellulose, hemicellulose, and lignin. As a result, efficient degradation of plant cell walls requires the addition of large quantities of a diverse collection of enzymes, which greatly increases the cost of biomass processing (23).
A recent analysis suggested that a combination of consolidated bioprocessing and pretreatment could significantly reduce production costs associated with cellulosic biofuels (48, 52). Consolidated bioprocessing involves the use of a single organism for the degradation of biomass to its component sugars and the subsequent conversion of these sugars to biofuel. Current approaches for the production of consolidated bioprocessors (CBPs) involve the introduction of the genes necessary for ethanol production into organisms capable of deconstructing biomass or the introduction of genes encoding biomass-degrading enzymes and their cognate secretion systems into ethanologenic microorganisms.
While Saccharomyces cerevisiae is arguably the most prominent industrial ethanologen, Gram-negative bacteria, such as Zymomonas mobilis, are also used in the commercial production of ethanol (1, 45). Furthermore, studies by the Ingram group have shown that the Gram-negative bacterium Escherichia coli can be engineered for efficient production of ethanol (32). The strong ethanologenic potential of these Gram-negative organisms necessitates the development of technologies for their conversion to consolidated bioprocessors. However, due to differences in cell surface architecture, unique strategies for enzyme display and secretion are required for the construction of consolidated bioprocessors in Gram-negative microorganisms. One approach to overcoming the challenges of secretion/display is to identify Gram-negative organisms that can efficiently degrade bioenergy-relevant biomass substrates. Due to the broad conservation of secretion strategies in Gram-negative microorganisms, the cellulolytic enzymes and secretion machinery from these bacteria would be expected to be transportable to ethanologenic organisms, such as Z. mobilis and E. coli. Consistent with this notion, previous studies have shown that the introduction of the type II secretion system (TTSS) from Erwinia chrysanthemi endowed E. coli with the ability to secrete heterologously expressed E. chysanthemi pectinases (29).
Cellvibrio japonicus, originally referred to as Pseudomonas fluorescens var. cellulosa (culture no. 107, isolated in 1948 from soil in Saitama-ken, Japan), has long been known to be capable of cellulose degradation (51). The organism has been reported to produce an extracellular cellulase activity (22, 59), which is secreted into the culture supernatant and does not associate with the cell surface (28). Furthermore, the extracellular cellulases of C. japonicus are inducible, and their activities are increased in the presence of cellulose (35, 40, 60) and strongly downregulated when cellobiose is present in the medium (58).
An extensive literature involving the biochemical characterization of C. japonicus polysaccharide-degrading enzymes has been produced over the past 40 years, and in several cases, the corresponding genes have been cloned. Using C. japonicus genomic libraries expressed in E. coli, Wolff et al. described the isolation of four distinct carboxymethylcellulase genes (56). Subsequently, genes encoding endoglucanases, cellodextrinases, xylanases, mannanases, and an arabinofuranosidase have been cloned and characterized (5-8, 17-19, 21, 24). The crystal structures of many of these enzymes have also been published (43, 44). Thus, C. japonicus contains a diverse collection of cellulose-degrading enzymes. The recent analysis of the C. japonicus genome confirmed the presence of an extensive plant cell wall-degrading machinery, predicting the presence of 123 glycoside hydrolases, as well as 14 predicted carbohydrate lyases (15).
In this report, we build upon these previous studies and evaluate the ability of C. japonicus to utilize cellulose sources relevant to bioenergy. We demonstrate that C. japonicus can utilize the biomass substrates corn stover (CS) and switchgrass (SG) as sole sources of carbon and energy and show that the bacterium produces acetate, pyruvate, and lactate when grown on monosaccharides and soluble cellulosic carbon sources. We also developed a method for construction of directed gene disruptions in C. japonicus and demonstrate that efficient cellulase secretion and growth on biomass are prevented by disruption of the type II secretion system. In addition, we show that C. japonicus can be metabolically engineered using broad-host-range plasmids. Collectively, our results demonstrate that C. japonicus contains biomass-degrading enzymes and a secretion system that can be used for the engineering of Gram-negative consolidated bioprocessors.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
C. japonicus sp. nov. strain Ueda107 was obtained from the National Collections of Industrial, Marine, and Food Bacteria. Table 1 lists the genotypes of the strains of E. coli and C. japonicus used in these studies. E. coli and C. japonicus strains were grown on M9 minimal medium (41) supplemented with MgSO4 (1 mM) and CaCl2 (0.1 mM). Glucose (0.2% [wt/vol]), cellobiose (0.34% [wt/vol]), carboxymethylcellulose (CMC) (1% [wt/vol]), and Avicel (1% [wt/vol]) were used as sole carbon and energy sources for cells grown in minimal medium. Lysogenic broth (LB) was used as rich medium (3, 4). All incubations in liquid medium were performed at 30°C with high aeration (225 rpm). For small-scale preparations, 5 ml of C. japonicus cells were grown in a 30-ml culture tube. For larger-scale preparations, 15 ml cells were grown in a 125-ml baffled flask. When used, antibiotics were present in the medium at the following concentrations: ampicillin, 100 μg/ml; gentamicin, 15 μg/ml; chloramphenicol, 25 μg/ml; and kanamycin, 50 μg/ml. All chemicals were purchased from Fisher.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Laboratory collection |
| S17 λPIR | Tpr SmrrecA thi pro hsdR−M+ RP4::2-Tc::Mu::Km Tn7 λPIR | Laboratory collection |
| C. japonicus | ||
| Ueda107 | Wild type | Laboratory collection |
| MSB279 | Ueda107 gspD::pJGG1 kan+ | This study |
| MSB280 | Ueda107/pJGG2; Gmr | This study |
| Plasmids | ||
| pJGG1 | Contains 500-bp internal gspD cloned at XbaI and SphI sites of plasmid pK18mobsacB; Kmr | This study |
| pK18mobsacB | pMB1 ori mob+sacB+; Kmr | 47 |
| pJGG2 | adhB and pdc subcloned from pLOI295 into pBBR1-MCS5 at EcoRI and XbaI sites; Gmr | This study |
| pBBR1-MCS5 | pBBR oriT; Plac; Gmr | 36 |
| pRK2013 | Kmr ColE1 RK2-Mob+ RK2-Tra+ | 20 |
Preparation of feedstock medium.
A derivative of M9 medium was developed with corn stover or ammonia fiber expansion (AFEX)-treated corn stover (AFEX-CS) or switchgrass (AFEX-SG) as a sole carbon source. Briefly, the AFEX-CS was ground to a fine particle size (approximately 0.5 mm) with a porcelain mortar and pestle. The ground AFEX-CS then underwent a 15-min autoclave cycle with 30 min of drying. The sterile AFEX-CS was washed 5 times with sterile water before being used in the minimal medium. AFEX-treated corn stover, AFEX-treated switchgrass, and nonpretreated corn stover were added to M9 medium at a concentration of 1% (wt/vol).
Mobilization of plasmids.
Plasmids were introduced into E. coli strains by calcium chloride-mediated transformation (41) or electroporation (14). The replicating plasmid pJGG2, a derivative of pBBRMCS, was mobilized into C. japonicus by conjugation. Briefly, an E. coli S17 λPIR strain harboring pJGG2, an E. coli DH5α strain harboring helper plasmid pRK2013, and the C. japonicus recipient strain were streaked into a common region of an LB plate. After 2 days of incubation at 30°C, the cells were streaked on selective minimal medium containing M9 salts, 0.2% glucose, 15 μg/ml gentamicin, and 50 μg/ml valine (to counterselect against the E. coli strains). Exconjugants were visible after 3 to 5 days of incubation at 30°C. The resulting C. japonicus colonies were streaked twice more onto the same medium and then tested for the presence of the plasmid. Approximately 1/104 C. japonicus cells were found to harbor plasmids.
Construction of targeted gene disruptions.
For gene disruptions in C. japonicus, plasmid pJGG1, a derivative of plasmid pK18mobsacB, was introduced into S17 λPIR by electroporation, selecting for kanamycin resistance. The E. coli S17 λPIR strain harboring plasmid pJGG1, an E. coli DH5α strain harboring helper plasmid pRK2013, and the C. japonicus recipient strain were streaked into a common region of an LB plate. After 48 h of incubation at 30°C, the cells from the LB plate were streaked on selective minimal medium containing M9 salts, 0.2% glucose, 25 μg/ml kanamycin to select for integration of pJGG1, and 50 μg/ml valine (to counterselect against the E. coli strains). After 4 to 5 days of incubation at 30°C, the resulting C. japonicus colonies were restreaked twice on the same medium and then screened for the presence of the plasmid by PCR. Integration frequencies for pJGG1 in C. japonicus were generally low (approximately 1/106 C. japonicus cells were found to harbor integrated pJGG1).
PCR.
All amplifications used TripleMaster polymerase (Eppendorf) and were performed in an Eppendorf Mastercycler gradient PCR thermocycler (Brinkmann Instruments). Primers were purchased from Integrated DNA Technologies. Template DNA was obtained from C. japonicus by adding 5 μl of an overnight LB culture directly to the PCR mixture.
Plasmid construction.
Plasmid pJGG1 was constructed by amplifying an internal 500-bp fragment of the gspD gene using the primers 5′-GGTGGTTCTAGAGTTGTTATCCGTGGTTA-3′ and 5′-GGTGGTGCATGCAGTCTTGCTGACAG-3′. The resulting PCR product was then digested with XbaI and SphI. Plasmid pK18mobsacB was cut with enzymes XbaI and SphI. The linearized plasmid was purified and was ligated with the gspD fragment and transformed into E. coli strain DH5α, selecting for kanamycin resistance on rich medium. Plasmid DNA was recovered from the kanamycin-resistant transformants, and the plasmid was purified.
Plasmid pJGG2 was constructed from plasmid pLOI295 (61), a ColE1 derivative containing the pdc and adhB genes from Z. mobilis. Plasmid pLO1295 was digested with EcoRI and XbaI to liberate the pdc and adhB genes, which were then ligated into pBBR1-MCS5 cut with the same enzymes. The ligation products were transformed into E. coli strain DH5α, selecting for gentamicin resistance on rich medium. The resulting plasmid was conjugated into C. japonicus as described above, selecting for gentamicin resistance.
Acid and ethanol analysis.
The concentrations of organic acids and ethanol that were secreted during growth in liquid medium were determined using commercially available detection kits. Formate and acetate were detected using kits from MegaZyme (Ireland, United Kingdom), whereas pyruvate, lactate, and ethanol were detected using kits from BioVision (Mountain View, CA), according to the manufacturer's instructions. The pHs of culture supernatants were determined with an UltraBASIC pH probe (Denver Instruments).
Congo red staining.
To determine the relative amount of endoglucanase secretion, an agar plate-based assay was used as described previously (49). Carboxymethylcellulose (1% [wt/vol]) and glucose (0.1% [wt/vol]) were used as carbon sources. After 48 h of growth, the plate was flooded with 0.1% (wt/vol) Congo red solution (Ricca Chemical) and stained for 15 min at room temperature. The dye was removed, and 5 ml of water was used to wash the plate. Finally, 5 ml of a 1 M NaCl solution was applied for 15 min, and the plates were then dried and photographed.
RESULTS
Growth of C. japonicus in the presence of cellulosic substrates.
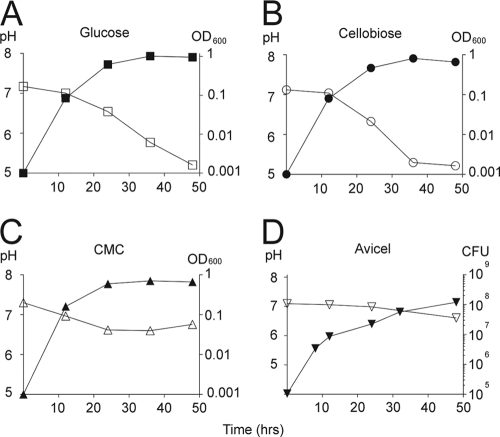
Although C. japonicus strain Ueda107 can grow in the presence of laboratory cellulosic substrates (51), little has been reported about its ability to utilize carbon sources relevant to bioenergy. We examined the ability of C. japonicus to grow in the presence of a collection of cellulosic and noncellulosic carbon sources. The growth rates of C. japonicus in M9 minimal medium at 30°C supplemented with 0.2% glucose, 0.34% cellobiose, or 1% carboxymethylcellulose were approximately the same (Fig. 1 A, B, and C), with generation times of 2.7 h, 2.8 h, and 2.4 h, respectively. Similar results were observed when 1% glucose or 1% cellobiose was provided as a carbon source (J. G. Gardner and D. H. Keating, unpublished data).
FIG. 1.
Growth and pH analysis of C. japonicus. Wild-type C. japonicus was cultured in M9 medium supplemented with 1 mM MgSO4, 0.1 mM CaCl2, and glucose (0.2% [wt/vol]) (A), cellobiose (0.34% [wt/vol]) (B), carboxymethycellulose (1% [wt/vol]) (C), or Avicel (1% [wt/vol]) (D) as the sole carbon and energy source, as described in Materials and Methods. All incubations were at 30°C with high aeration (225 rpm). Growth (closed symbols) was measured by measurement of the OD600 (A to C) or by viable-cell counting (D). The pH (open symbols) was determined with a Denver Instruments ultraBASIC probe. All experiments were performed in triplicate. The error bars, representing standard deviations, are too small to be seen. No growth was observed in media that lacked an exogenous carbon source.
We then examined the ability of C. japonicus to utilize the insoluble cellulosic substrate Avicel. Because measurement of the optical density at 600 nm (OD600) was challenging due to the insoluble Avicel present in the medium, we determined bacterial growth by viable-cell counting. Growth of C. japonicus in the presence of Avicel was biphasic, with rapid cell division (2.1-h generation time) occurring during the first 10 h, followed by a reduced growth rate (8.3-h generation time) (Fig. 1D). Although the reason for the biphasic growth is unknown, C. japonicus can clearly use soluble and insoluble cellulosic substrates as sole sources of carbon and energy.
Growth of C. japonicus in the presence of biomass.
The robust growth of C. japonicus in the presence of soluble and insoluble forms of cellulose led us to examine its ability to utilize feedstocks relevant to the bioenergy field. We chose two key biomass feedstocks for these studies: corn stover and switchgrass. Because current approaches to cellulosic biofuels typically involve the use of biomass pretreatment, we focused on corn stover and switchgrass subjected to the AFEX process (37, 50). We chose AFEX as a pretreatment because it leaves the hemicellulose fraction intact (48, 50), which allowed us to examine the ability of C. japonicus to utilize both cellulose and hemicellulose.
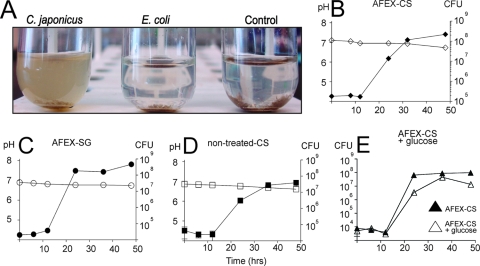
When added to minimal medium, AFEX-treated corn stover was initially found to support the growth of diverse bacteria, including E. coli (Gardner and Keating, unpublished). Because our work, as well as the work of others, had previously shown that E. coli cannot utilize carboxymethylcellulose or Avicel and lacks any detectable cellulase activity, we hypothesized that the AFEX process results in the release of soluble sugars or peptides that support the growth of noncellulolytic bacteria. We therefore developed an alternative protocol involving grinding of the AFEX-treated material with a mortar and pestle and autoclaving, followed by extensive washing with sterile water. When added to M9 minimal medium, the washed, autoclaved, AFEX-treated corn stover (or AFEX-treated switchgrass) failed to support the growth of E. coli but allowed the growth of C. japonicus (Fig. 2 A). Growth of C. japonicus in the presence of AFEX-treated corn stover was associated with release of glucose and xylose monosaccharides (Gardner and Keating, unpublished), suggesting that cellulose and hemicellulose were used as carbon sources.
FIG. 2.
Growth and degradation of biomass by C. japonicus. (A) Wild-type C. japonicus and E. coli were cultured in the presence of 1% (wt/vol) autoclaved, washed corn stover for 48 h at 30°C. The tubes containing cells and residual cellulose were then photographed. “Control” refers to mock-inoculated medium containing 1% (wt/vol) AFEX-corn stover. (B) Growth of C. japonicus in minimal medium supplemented with 1% (wt/vol) AFEX-CS. (C) Growth of C. japonicus in minimal medium supplemented with 1% (wt/vol) AFEX-SG. (D) Growth of C. japonicus in minimal medium supplemented with 1% (wt/vol) nonpretreated corn stover (nontreated-CS). (E) Growth of C. japonicus in minimal medium supplemented with 1% (wt/vol) AFEX-treated corn stover and 0.2% glucose (AFEX-CS + glucose). In panels B to E, the number of cells was determined by viable-cell counting. All experiments were performed in triplicate. The error bars (often too small to be seen) represent standard deviations.
When the growth rate was measured by viable-cell counting, C. japonicus grown in the presence of AFEX-treated corn stover or switch grass displayed growth kinetics distinct from what was observed with Avicel, glucose, carboxymethylcellulose, or cellobiose as a carbon source. No detectable increase in cell numbers was observed for approximately 10 h after addition of the cells to the medium (Fig. 2B). After the 10-hour growth lag, however, the cells displayed a growth rate similar to what was seen in other media (generation time, 1.9 h). Interestingly, the addition of 0.2% glucose to media containing AFEX-treated corn stover did not reduce the length of this lag (Fig. 2E), although it appeared to increase the growth rate after the initiation of cell division. C. japonicus was also able to utilize AFEX-treated switchgrass as a carbon source (Fig. 2C), with the cells displaying a growth lag similar to that seen with AFEX-treated corn stover but more rapid growth at later time points (generation time, 0.98 h). To determine if the lag was related to the AFEX pretreatment, we measured the rate of growth of C. japonicus with nonpretreated corn stover as a carbon source. Growth in the presence of nonpretreated corn stover led to an initial growth lag similar to that observed in the presence of pretreated corn stover and a similar postlag growth rate (1.7-h generation time) (Fig. 2D). However, the final cell yield was reduced by about 10-fold with respect to growth in the presence of AFEX-treated corn stover.
Production of organic acids during growth in the presence of cellulosic substrates.
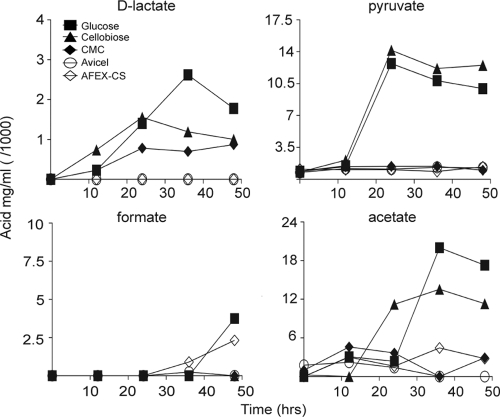
A previous study noted that C. japonicus was capable of producing acid when grown with cellobiose and mannitol as carbon sources, but acid was not produced when glucose or other mono- and disaccharides were provided as carbon sources. The surprising variability in acid production during growth with different carbon sources led us to ask whether acid was produced when C. japonicus was grown in the presence of cellulosic substrates. Similar to what was reported previously (31), we observed a decrease in medium pH when C. japonicus was grown with 0.34% cellobiose as a carbon source (Fig. 1B). However, in contrast to what has been reported previously, we also observed a decrease in pH when 0.2% glucose was included as a carbon source (Fig. 1A). When C. japonicus was grown with carboxymethylcellulose as a carbon source, we observed a decrease in pH, but it was greatly diminished with respect to what was observed in the presence of glucose or cellobiose (Fig. 1C). A similar trend was observed with insoluble forms of cellulose, which led to a negligible pH decrease (Fig. 2B, C, and D). We do not have an explanation for the reduced amount of acid production observed when cells were cultured in the presence of cellulosic carbon sources, although it could simply reflect the reduced availability of monosaccharides for the bacterium.
The decrease in medium pH during growth with glucose or cellobiose as a carbon source led us to characterize the acids excreted by C. japonicus. When cells were grown with either glucose or cellobiose as a carbon source, we observed that acetate and pyruvate were the dominant acids excreted into the culture medium, in addition to a small amount of lactate (Fig. 3). A small amount of formate was also detected at the final time points. When C. japonicus was grown in the presence of carboxymethylcellulose, small amounts of lactate and acetate were produced. However, only negligible amounts of acids were detected when cells were grown with Avicel or AFEX-treated corn stover as a carbon source, consistent with the observed stable pH.
FIG. 3.
Acid analyses of C. japonicus grown on soluble and insoluble sources of cellulose. Wild-type C. japonicus was cultured in the presence of 0.2% glucose, 0.34% cellobiose, 1% (wt/vol) carboxymethylcellulose, 1% Avicel (wt/vol), or 1% AFEX-CS (wt/vol). At various time points postinoculation, the cell supernatants were assayed for the presence of d-lactate, pyruvate, formate, and acetate using enzymatic detection assays, as described in Materials and Methods. All experiments were performed in triplicate. The error bars, representing standard deviations, are too small to be seen.
A C. japonicus gspD mutant is deficient for cellulase secretion and the use of cellulose and biomass as carbon sources.
Previous studies have shown that C. japonicus produces a diverse and extensive group of cellulases and hemicellulases (15). Furthermore, a substantial fraction (ca. 90%) of these activities have been reported to be extracellular (59, 60). In Gram-negative plant-pathogenic organisms, such as Erwinia spp., extracellular glycosylhydrolyase and pectate lyase activities are secreted via the TTSS (46). The genome of C. japonicus encodes a single predicted TTSS, but little has been reported about its role in cellulase secretion or growth on cellulosic substrates. To characterize the role of the TTSS in cellulase and hemicellulase secretion, we developed a system for construction of directed gene disruptions in C. japonicus. We cloned a 500-bp fragment corresponding to an internal region of gspD into the plasmid pK18mobsacB, which contains a pMB1 origin of replication and cannot replicate within C. japonicus. Retention of the pK18mobsacB-encoded antibiotic resistance by C. japonicus can only occur via a recombination event between the plasmid-borne gspD internal fragment and the gspD gene present in the genome, which results in integration of the plasmid into the chromosome and an inactivated form of the gene (see Fig. S1 in the supplemental material). The gspD gene is predicted to encode the outer membrane secretin of the type II system, which is essential for activity in all type II systems examined to date (46). Furthermore, gspD is the second gene in a predicted 11-gene operon and would be expected to exert a polar effect on transcription of the downstream genes. Thus, we expected the gspD::pJGG1 mutant to disrupt type II secretion.
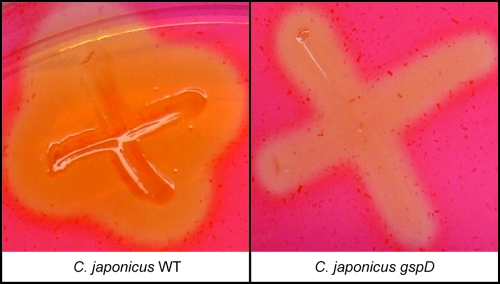
We compared the ability of the wild-type C. japonicus and the gspD::pJGG1 mutant to produce extracellular endoglucanase activity via staining with Congo red. Wild-type C. japonicus and the gspD::pJGG1 mutant were patched onto solid M9 medium containing carboxymethylcellulose and 0.1% glucose, followed by staining with Congo red to detect the presence of endoglucanase activity. The gspD::pJGG1 mutant displayed a smaller zone of clearing than the wild type, indicating reduced secretion of endoglucanases (Fig. 4). Based on cleavage of carboxymethylcellulose, we estimate that secretion of endoglucanases was reduced at least 2-fold in the gspD::pJGG1 mutant.
FIG. 4.
Endoglucanase secretion defect of the C. japonicus gspD mutant. Wild-type (WT) C. japonicus and a gspD::pJGG1 mutant were patched onto an M9 plate containing 0.1% glucose and 1% carboxymethylcellulose and incubated at 30°C for 48 h. Endoglucanase activity was then detected by staining with Congo red, as described in Materials and Methods.
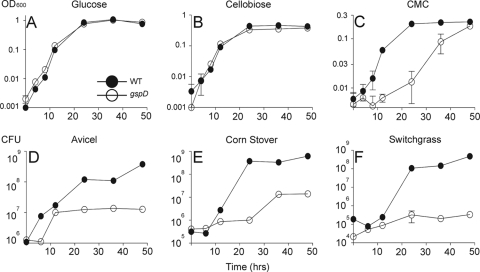
We then examined the ability of the mutant to utilize monosaccharides, disaccharides, and cellulosic substrates. The gspD::pJGG1 mutation displayed the same growth rate as the wild type on medium containing either glucose or cellobiose as a carbon source (Fig. 5 A and B). However, when carboxymethylcellulose was employed as a carbon source, the gspD::pJGG1 mutant displayed a lag in growth during the first 8 h and a reduced growth rate at later time points (Fig. 5C). Despite the reduced rate of growth, the gspD::pJGG1 mutant was able to reach the same final OD600 as the wild type. A more severe growth defect was observed in medium containing insoluble cellulose. The gspD::pJGG1 mutant showed a 6-h lag in growth in medium containing Avicel, which was followed by a brief period of rapid growth (Fig. 5D). However, growth appeared to cease at time points beyond 12 h, resulting in a greater-than-100-fold decrease in viable cells with respect to the wild type. When cultured in the presence of AFEX-treated corn stover, the gspD::pJGG1 mutant displayed a growth lag during the first 24 h (Fig. 5E), followed by a slight increase in the growth rate between 24 and 48 h and a greater-than-100-fold reduction in viable cells with respect to the wild type. Interestingly, growth of the gspD::pJGG1 mutant in the presence of AFEX-treated switchgrass resulted in a 1,000-fold reduction in viable cells with respect to the wild type (Fig. 5F).
FIG. 5.
Growth defect of the C. japonicus gspD mutant in media with insoluble cellulose. Wild-type C. japonicus and the gspD::pJGG1 mutant were cultured in M9 medium supplemented with 1 mM MgSO4, 0.1 mM CaCl2, and glucose (0.2% [wt/vol]) (A), cellobiose (0.34% [wt/vol]) (B), CMC (1% [wt/vol]) (C), Avicel (1% [wt/vol]) (D), AFEX-treated corn stover (1% [wt/vol]) (E), or AFEX-treated switchgrass (1% [wt/vol]) (F) as the sole carbon and energy sources, as described in Materials and Methods. All incubations were at 30°C with high aeration (225 rpm). Growth was determined by measurement of the OD600 (A to C) or by viable-cell counting (D to F). All experiments were performed in triplicate. The error bars (often too small to be seen) represent standard deviations.
Metabolic engineering of C. japonicus.
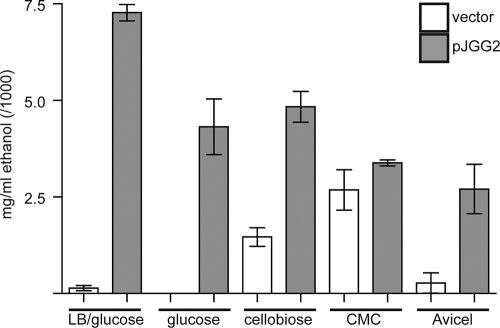
The ability of C. japonicus to utilize bioenergy-relevant forms of cellulose led us to investigate its ability to be metabolically engineered for the production of bioproducts. We constructed a version of the broad-host-range plasmid pBBR-MCS5 (referred to as pJGG2) containing the adhB and pdc genes from Z. mobilis. The pdc gene encodes pyruvate decarboxylase, which catalyzes the decarboxylation of pyruvate to form acetaldehyde, and the adhB gene encodes alcohol dehydrogenase, which catalyzes the NADH-dependent reduction of acetaldehyde to ethanol. Studies from the Ingram laboratory have shown that expression of the adhB and pdc genes from Z. mobilis can enhance the production of ethanol in E. coli (61) and other Gram-negative bacteria (32). Plasmid pJGG2 replicated efficiently within C. japonicus, as judged by antibiotic resistance and recovery of intact plasmids from C. japonicus cells (Gardner and Keating, unpublished). When cultured under aerobic conditions in LB plus 0.2% glucose, cells harboring the pJGG2 plasmid grew at a rate approximately equivalent to that of cells containing the vector control but displayed a 15-fold increase in ethanol production (Fig. 6). Although ethanol production was modest under all conditions, the greatest ethanol production was observed in cells grown in the presence of LB supplemented with glucose, with growth in minimal glucose and minimal cellobiose media producing a slightly reduced amount of ethanol. In addition, growth in carboxymethylcellulose- and Avicel-containing media resulted in a low but detectable level of ethanol production. We were unable to detect any growth of C. japonicus containing the pJGG2 plasmid or vector control under anaerobic conditions with glucose, cellobiose, carboxymethylcellulose, Avicel, or AFEX-treated corn stover as a carbon source.
FIG. 6.
Ethanol production by engineered C. japonicus. Wild-type C. japonicus harboring plasmid pJGG2 or a vector control was cultured aerobically at 30°C for 48 h in the presence of LB supplemented with 0.2% glucose or M9 medium supplemented with 1 mM MgSO4, 0.1 mM CaCl2, and either glucose (0.2% [wt/vol]), cellobiose (0.34% [wt/vol]), carboxymethylcellulose (1% [wt/vol]), or Avicel (1% [wt/vol]) as a carbon source. Ethanol was then detected enzymatically, as described in Materials and Methods. All experiments were performed in triplicate. The error bars represent standard deviations.
DISCUSSION
The C. japonicus genome is predicted to encode ca. 154 enzymes capable of cleaving glycosidic bonds. Consistent with this prediction, our studies showed that C. japonicus can utilize diverse cellulosic substrates as sources of carbon and energy, including the key bioenergy feedstocks corn stover and switchgrass. In contrast to a previous report (31), we observed that growth in the presence of glucose resulted in a reduction in medium pH, accompanied by the production of acetic acid, pyruvate, and a small amount of lactate. Using a directed gene disruption system developed as part of these studies, we demonstrated a central role for the type II secretion system in the utilization of insoluble cellulose. Collectively, our results support the notion that C. japonicus can serve as a useful source of biomass-degrading enzymes, and perhaps secretion systems, for the generation of Gram-negative consolidated bioprocessors.
Growth of C. japonicus in the presence of insoluble cellulose.
When glucose, cellobiose, or carboxymethylcellulose was supplied as a carbon source, C. japonicus showed similar generation times (Fig. 1A, B, and C). However, the kinetics of growth differed significantly when C. japonicus was cultured in the presence of insoluble forms of cellulose. In the presence of Avicel, growth of C. japonicus was biphasic, with the most rapid cell division occurring in the first 10 h, followed by a reduced growth rate (Fig. 1D). Interestingly, several groups have reported similar biphasic curves for cellulose hydrolysis in the presence of purified cellulases (2). In these cases, the initial rapid rate of degradation has been suggested to result from cleavage of amorphous cellulose, followed by a much slower hydrolysis of the crystalline cellulose (although other explanations for this phenomenon have been advanced [26, 39]). We suggest that the initial rapid growth of C. japonicus may also be supported by the hydrolysis of amorphous cellulose, whereas the lower rate of growth at later time points results from the reduced rate of hydrolysis of crystalline cellulose.
We also examined the ability of C. japonicus to use two sources of biomass relevant to the bioenergy industry: corn stover and switchgrass. In both cases, C. japonicus was able to degrade the substrates, as judged by an increase in cell number and a decrease in insoluble biomass in the culture (Fig. 2) and the release of glucose and xylose monosaccharides (Gardner and Keating, unpublished). Although the maximum growth rate of C. japonicus was comparable to that seen in the presence of purified cellulose, an approximate 8-h lag was observed when biomass was provided as a growth substrate. Interestingly, this lag in growth was observed in the absence of corn stover pretreatment (Fig. 2D) and in the presence of corn stover medium supplemented with 0.2% glucose (Fig. 2E). Because C. japonicus grows well with 0.2% glucose as a carbon source (Fig. 1A), these data strongly suggest that the growth delay observed during the first 10 h results from the presence of the biomass. Furthermore, since the growth delay was seen with biomass that had not undergone pretreatment (Fig. 2D), the lag does not result from compounds arising during the AFEX process. Plants have been reported to produce a significant number of compounds that could negatively affect the growth of C. japonicus. For example, lignin is a major substituent of plant cell walls (20 to 30% of the total) and contains phenolic substituents with known toxic effects (11, 13, 30, 42). Plants produce additional defensive compounds, such as phenylpropanoid derivatives and hydroxylated phenols with known antimicrobial activities (13). In particular, corn extracts contain 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one, which has been shown to prolong the lag phase of Erwinia species, as well as that of other Gram-negative bacteria (12, 27). Interestingly, the ability to detoxify this compound has been reported in some organisms (25, 57). We are currently employing C. japonicus microarrays to measure the transcriptional response of C. japonicus to the presence of glucose, cellobiose, Avicel, and biomass. We expect that the results of these transcriptional-profiling studies will provide insight into the nature of this growth inhibition.
Type II secretion system-dependent secretion of cellulases.
Previous studies by several groups have shown that pectinases and cellulases produced by Gram-negative organisms can be secreted via the type II secretion system (34, 46). Although C. japonicus contains a predicted type II secretion system, the roles of these genes in cellulase and hemicellulase secretion, or growth on cellulosic substrates, had not been investigated. By the use of a vector integration strategy, we constructed a gspD::pJGG1 mutant that would be expected to prevent the function of the type II secretion system. The gspD::pJGG1 mutant displayed a marked delay in growth when the soluble cellulose substrate carboxymethylcellulose was provided as a carbon source but ultimately grew to the same final OD600 as the wild-type strain (Fig. 5C). A similar phenotype was observed in measurements of endoglucanase activity, as detected by carboxymethylcellulose cleavage and Congo red staining. After 48 h of incubation, a greater-than-2-fold reduction in endoglucanase activity was observed (Fig. 4). However, prolonged incubation resulted in a detectable zone of clearing from the gspD::pJGG1 mutant (Gardner and Keating, unpublished). We suggest that the initial delay in growth in the presence of carboxymethylcellulose, and the initial lack of detectable activity observed in the Congo red assay, results from an insufficient extracellular concentration of endoglucanases required for hydrolysis of carboxymethylcellulose. In both cases, prolonged incubation results in a greater release of endoglucanases (either by alternative secretion pathways or by cell lysis), which then allow efficient carboxymethylcellulose degradation and cell growth.
When the gspD::pJGG1 mutant was cultured in the presence of insoluble cellulosic substrates, a more severe growth phenotype was observed. In the presence of Avicel, AFEX-treated corn stover, or AFEX-treated switchgrass, the growth of the gspD::pJGG1 mutant arrested prematurely, which led to a greater-than-100-fold decrease in viable cells compared to the wild type. These data indicate that the type II-independent endoglucanase release is insufficient for growth on more recalcitrant carbon sources. A recent study in the bacterium Saccharophagus degradans demonstrated the presence of a processive endoglucanase activity encoded by the cel5H gene, which was capable of the conversion of insoluble cellulose to cellobiose, as well as an apparent lack of cellobiohydrolase activity (54). The cellulose-degrading apparatus of C. japonicus bears a great deal of sequence similarity to the S. degradans system (15) and may be similarly reliant on processive endoglucanases. If this class of enzymes is poorly released in the gspD::pJGG1 mutant, this could explain the severe growth defects observed in the presence of insoluble cellulose. It remains possible that the reduced growth of the gspD::pJGG1 mutant simply results from sensitivity of the cells to components of insoluble cellulose, although this possibility is rendered less likely by the normal rate of growth of the organism on glucose or cellobiose (Fig. 5A and B). Future studies will employ proteomics to characterize the secretion of extracellular enzymes in the wild type and the gspD::pJGG1 mutant during growth in the presence of soluble and insoluble forms of cellulose. These data will not only expedite the identification of substrates for the type II secretion system, but should also identify the critical enzymes necessary for breakdown of cellulose and biomass.
A role for C. japonicus in biomass processing.
C. japonicus can utilize diverse forms of cellulose, including bioenergy-relevant biomass, demonstrating its potential as a source of enzymes for consolidated bioprocessing. Consistent with this idea, we have demonstrated functional expression in E. coli of codon-optimized genes encoding the C. japonicus β-glucosidase Cel3A, the endoglucanases Cel5B and Cel9A, and the predicted cellobiohydrolase Cel6A (Gardner and Keating, unpublished). Furthermore, the growth and cellulase secretion phenotypes of the gspD::pJGG1 mutant strongly suggest that the type II secretion system is the primary means of cellulase and hemicellulase secretion by C. japonicus. If this is confirmed by the proteomic studies ongoing in our laboratory, then introduction of the C. japonicus type II secretion system into Gram-negative bacteria, such as E. coli, may allow the secretion of diverse C. japonicus glycosylhydrolyases and carbohydrate lyases expressed within the organism. We are currently testing this idea in our laboratory.
Our demonstration that pBBR plasmids replicate within C. japonicus and development of methods for directed gene disruption, combined with previous reports of transposon-based gene knockouts (5, 17), provide the tools necessary for metabolic engineering. As a proof of concept, engineered C. japonicus was shown to produce ethanol from cellulosic substrates (Fig. 6), although at low yields. This modest amount of ethanol production may have resulted from the low expression of the pdc and adhB genes in C. japonicus, and we are currently testing alternative promoters to improve pdc and adhB expression. Alternatively, the reduced ethanol yields may have resulted from our use of aerobic growth conditions, which would be expected to greatly reduce the intracellular concentrations of NADH required for ethanol production (9, 33, 53). Growth under anaerobic conditions has been reported previously for C. japonicus (16); however, we were unable to confirm this in our laboratory. Studies are currently under way to test the utility of C. japonicus to produce next-generation biofuels that can be produced under aerobic growth conditions.
Supplementary Material
Acknowledgments
This work was funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494).
We acknowledge the laboratories of Bruce Dale for production of the AFEX-treated corn stover and switchgrass and Shawn Kaeppler for the untreated corn stover. We also acknowledge Jorge Escalante-Semerena for the gifts of pBBR1-MCS5 and pRK2013 and Laura A. Zeitler for technical assistance.
Footnotes
Published ahead of print on 11 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Bai, F. W., W. A. Anderson, and M. Moo-Young. 2008. Ethanol fermentation technologies from sugar and starch feedstocks. Biotechnol. Adv. 26:89-105. [DOI] [PubMed] [Google Scholar]
- 2.Bansal, P., M. Hall, M. J. Realff, J. H. Lee, and A. S. Bommarius. 2009. Modeling cellulase kinetics on lignocellulosic substrates. Biotechnol. Adv. 27:833-848. [DOI] [PubMed] [Google Scholar]
- 3.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beylot, M. H., V. A. McKie, A. G. Voragen, C. H. Doeswijk-Voragen, and H. J. Gilbert. 2001. The Pseudomonas cellulosa glycoside hydrolase family 51 arabinofuranosidase exhibits wide substrate specificity. Biochem. J. 358:607-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolam, D. N., N. Hughes, R. Virden, J. H. Lakey, G. P. Hazlewood, B. Henrissat, K. L. Braithwaite, and H. J. Gilbert. 1996. Mannanase A from Pseudomonas fluorescens ssp. cellulosa is a retaining glycosyl hydrolase in which E212 and E320 are the putative catalytic residues. Biochemistry 35:16195-16204. [DOI] [PubMed] [Google Scholar]
- 7.Braithwaite, K. L., T. Barna, T. D. Spurway, S. J. Charnock, G. W. Black, N. Hughes, J. H. Lakey, R. Virden, G. P. Hazlewood, B. Henrissat, and H. J. Gilbert. 1997. Evidence that galactanase A from Pseudomonas fluorescens subspecies cellulosa is a retaining family 53 glycosyl hydrolase in which E161 and E270 are the catalytic residues. Biochemistry 36:15489-15500. [DOI] [PubMed] [Google Scholar]
- 8.Braithwaite, K. L., G. W. Black, G. P. Hazlewood, B. R. Ali, and H. J. Gilbert. 1995. A non-modular endo-beta-1,4-mannanase from Pseudomonas fluorescens subspecies cellulosa. Biochem. J. 305:1005-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brambilla, L., D. Bolzani, C. Compagno, V. Carrera, J. P. van Dijken, J. T. Pronk, B. M. Ranzi, L. Alberghina, and D. Porro. 1999. NADH reoxidation does not control glycolytic flux during exposure of respiring Saccharomyces cerevisiae cultures to glucose excess. FEMS Microbiol. Lett. 171:133-140. [DOI] [PubMed] [Google Scholar]
- 10.Carere, C. R., R. Sparling, N. Cicek, and D. B. Levin. 2008. Third generation biofuels via direct cellulose fermentation. Int. J. Mol. Sci. 9:1342-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, D. H., Y. J. Lee, Y. Um, B. I. Sang, and Y. H. Kim. 2009. Detoxification of model phenolic compounds in lignocellulosic hydrolysates with peroxidase for butanol production from Clostridium beijerinckii. Appl. Microbiol. Biotechnol. 83:1035-1043. [DOI] [PubMed] [Google Scholar]
- 12.Corcuera, L. J., M. D. Woodward, J. P. Helgeson, A. Kelman, and C. D. Upper. 1978. 2,4-Dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one, an inhibitor from Zea mays with differential activity against soft rotting Erwinia species. Plant Physiol. 61:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowan, M. M. 1999. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 12:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, R. W., D. Bostein, and J. R. Roth. 1980. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.DeBoy, R. T., E. F. Mongodin, D. E. Fouts, L. E. Tailford, H. Khouri, J. B. Emerson, Y. Mohamoud, K. Watkins, B. Henrissat, H. J. Gilbert, and K. E. Nelson. 2008. Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J. Bacteriol. 190:5455-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dees, C., D. Ringelberg, T. C. Scott, and T. J. Phelps. 1995. Characterization of the cellulose-degrading bacterium NCIMB 10462. Appl. Biochem. Biotechnol. 51-52:263-274. [Google Scholar]
- 17.Emami, K., T. Nagy, C. M. Fontes, L. M. Ferreira, and H. J. Gilbert. 2002. Evidence for temporal regulation of the two Pseudomonas cellulosa xylanases belonging to glycoside hydrolase family 11. J. Bacteriol. 184:4124-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emami, K., E. Topakas, T. Nagy, J. Henshaw, K. A. Jackson, K. E. Nelson, E. F. Mongodin, J. W. Murray, R. J. Lewis, and H. J. Gilbert. 2009. Regulation of the xylan-degrading apparatus of Cellvibrio japonicus by a novel two-component system. J. Biol. Chem. 284:1086-1096. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira, L. M., G. P. Hazlewood, P. J. Barker, and H. J. Gilbert. 1991. The cellodextrinase from Pseudomonas fluorescens subsp. cellulosa consists of multiple functional domains. Biochem. J. 279:793-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert, H. J., J. Hall, G. P. Hazlewood, and L. M. Ferreira. 1990. The N-terminal region of an endoglucanase from Pseudomonas fluorescens subspecies cellulosa constitutes a cellulose-binding domain that is distinct from the catalytic centre. Mol. Microbiol. 4:759-767. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert, H. J., G. Jenkins, D. A. Sullivan, and J. Hall. 1987. Evidence for multiple carboxymethylcellulase genes in Pseudomonas fluorescens subsp. cellulosa. Mol. Gen. Genet. 210:551-556. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, H. J., H. Stalbrand, and H. Brumer. 2008. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr. Opin. Plant Biol. 11:338-348. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert, H. J., D. A. Sullivan, G. Jenkins, L. E. Kellett, N. P. Minton, and J. Hall. 1988. Molecular cloning of multiple xylanase genes from Pseudomonas fluorescens subsp. cellulosa. J. Gen. Microbiol. 134:3239-3247. [DOI] [PubMed] [Google Scholar]
- 25.Glenn, A. E., D. M. Hinton, I. E. Yates, and C. W. Bacon. 2001. Detoxification of corn antimicrobial compounds as the basis for isolating Fusarium verticillioides and some other Fusarium species from corn. Appl. Environ. Microbiol. 67:2973-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta, R., and Y. Y. Lee. 2009. Mechanism of cellulase reaction on pure cellulosic substrates. Biotechnol. Bioeng. 102:1570-1581. [DOI] [PubMed] [Google Scholar]
- 27.Hartman, J. R., A. Kelman, and C. D. Upper. 1975. Differential Inhibitory activity of a corn extract to Erwinia spp. causing soft rot. Phytopathology 65:1082-1088. [Google Scholar]
- 28.Hazlewood, G. P., J. I. Laurie, L. M. Ferreira, and H. J. Gilbert. 1992. Pseudomonas fluorescens subsp. cellulosa: an alternative model for bacterial cellulase. J. Appl. Bacteriol. 72:244-251. [DOI] [PubMed] [Google Scholar]
- 29.He, S. Y., M. Lindeberg, A. K. Chatterjee, and A. Collmer. 1991. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc. Natl. Acad. Sci. U. S. A. 88:1079-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heipieper, H. J., H. Keweloh, and H. J. Rehm. 1991. Influence of phenols on growth and membrane permeability of free and immobilized Escherichia coli. Appl. Environ. Microbiol. 57:1213-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphry, D. R., G. W. Black, and S. P. Cummings. 2003. Reclassification of ‘Pseudomonas fluorescens subsp. cellulosa’ NCIMB 10462 (Ueda et al. 1952) as Cellvibrio japonicus sp. nov. and revival of Cellvibrio vulgaris sp. nov., nom. rev. and Cellvibrio fulvus sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 53:393-400. [DOI] [PubMed] [Google Scholar]
- 32.Jarboe, L. R., T. B. Grabar, L. P. Yomano, K. T. Shanmugan, and L. O. Ingram. 2007. Development of ethanologenic bacteria. Adv. Biochem. Eng. Biotechnol. 108:237-261. [DOI] [PubMed] [Google Scholar]
- 33.Jeppsson, M., B. Johansson, B. Hahn-Hagerdal, and M. F. Gorwa-Grauslund. 2002. Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl. Environ. Microbiol. 68:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazemi-Pour, N., G. Condemine, and N. Hugouvieux-Cotte-Pattat. 2004. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics 4:3177-3186. [DOI] [PubMed] [Google Scholar]
- 35.Kellett, L. E., D. M. Poole, L. M. Ferreira, A. J. Durrant, G. P. Hazlewood, and H. J. Gilbert. 1990. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem. J. 272:369-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 37.Lau, M. W., and B. E. Dale. 2009. Cellulosic ethanol production from AFEX-treated corn stover using Saccharomyces cerevisiae 424A(LNH-ST). Proc. Natl. Acad. Sci. U. S. A. 106:1368-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, Y., and S. Tanaka. 2006. Ethanol fermentation from biomass resources: current state and prospects. Appl. Microbiol. Biotechnol. 69:627-642. [DOI] [PubMed] [Google Scholar]
- 39.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKie, V. A., G. W. Black, S. J. Millward-Sadler, G. P. Hazlewood, J. I. Laurie, and H. J. Gilbert. 1997. Arabinanase A from Pseudomonas fluorescens subsp. cellulosa exhibits both an endo- and an exo-mode of action. Biochem. J. 323:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, J. H. 1975. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Mills, T. Y., N. R. Sandoval, and R. T. Gill. 2009. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol. Biofuels 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickersgill, R. W., J. A. Jenkins, M. Scott, I. Connerton, G. P. Hazlewood, and H. J. Gilbert. 1993. Crystallization and preliminary X-ray analysis of the catalytic domain of xylanase a from Pseudomonas fluorescens subspecies cellulosa. J. Mol. Biol. 229:246-248. [DOI] [PubMed] [Google Scholar]
- 44.Pires, V. M., J. L. Henshaw, J. A. Prates, D. N. Bolam, L. M. Ferreira, C. M. Fontes, B. Henrissat, A. Planas, H. J. Gilbert, and M. Czjzek. 2004. The crystal structure of the family 6 carbohydrate binding module from Cellvibrio mixtus endoglucanase 5A in complex with oligosaccharides reveals two distinct binding sites with different ligand specificities. J. Biol. Chem. 279:21560-21568. [DOI] [PubMed] [Google Scholar]
- 45.Rogers, P. L., Y. J. Jeon, K. J. Lee, and H. G. Lawford. 2007. Zymomonas mobilis for fuel ethanol and higher value products. Adv. Biochem. Eng. Biotechnol. 108:263-288. [DOI] [PubMed] [Google Scholar]
- 46.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 47.Schäfer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 48.Sendich, E. N., M. Laser, S. Kim, H. Alizadeh, L. Laureano-Perez, B. Dale, and L. Lynd. 2008. Recent process improvements for the ammonia fiber expansion (AFEX) process and resulting reductions in minimum ethanol selling price. Bioresour. Technol. 99:8429-8435. [DOI] [PubMed] [Google Scholar]
- 49.Teather, R. M., and P. J. Wood. 1982. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43:777-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teymouri, F., L. Laureano-Perez, H. Alizadeh, and B. E. Dale. 2004. Ammonia fiber explosion treatment of corn stover. Appl. Biochem. Biotechnol. 113-116:951-963. [DOI] [PubMed] [Google Scholar]
- 51.Ueda, K., S. Ishikawa, T. Itami, and T. Asai. 1952. Studies on the aerobic mesophilic cellulose-decomposing bacteria. Taxonomical study of genus Pseudomonas. J. Agric. Chem. Soc. 26:35-41. [Google Scholar]
- 52.van Zyl, W. H., L. R. Lynd, R. den Haan, and J. E. McBride. 2007. Consolidated bioprocessing for bioethanol production using Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108:205-235. [DOI] [PubMed] [Google Scholar]
- 53.Vemuri, G. N., M. A. Eiteman, J. E. McEwen, L. Olsson, and J. Nielsen. 2007. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 104:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watson, B. J., H. Zhang, A. G. Longmire, Y. H. Moon, and S. W. Hutcheson. 2009. Processive endoglucanases mediate degradation of cellulose by Saccharophagus degradans. J. Bacteriol. 191:5697-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson, D. B. 2008. Three microbial strategies for plant cell wall degradation. Ann. N. Y. Acad. Sci. 1125:289-297. [DOI] [PubMed] [Google Scholar]
- 56.Wolff, B. R., T. A. Mudry, B. R. Glick, and J. J. Pasternak. 1986. Isolation of endoglucanase genes from Pseudomonas fluorescens subsp. cellulosa and a Pseudomonas sp. Appl. Environ. Microbiol. 51:1367-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woodward, M. D., L. J. Corcuera, J. P. Helgeson, and C. D. Upper. 1978. Decomposition of 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one in aqueous solutions. Plant Physiol. 61:796-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamane, K., H. Suzuki, M. Hirotani, H. Ozawa, and K. Nisizawa. 1970. Effect of nature and supply of carbon sources on cellulase formation in Pseudomonas fluorescens var. cellulosa. J. Biochem. 67:9-18. [DOI] [PubMed] [Google Scholar]
- 59.Yamane, K., H. Suzuki, and K. Nisizawa. 1970. Purification and properties of extracellular and cell-bound cellulase components of Pseudomonas fluorescens var. cellulosa. J. Biochem. 67:19-35. [DOI] [PubMed] [Google Scholar]
- 60.Yamane, K., T. Yoshikawa, H. Suzuki, and K. Nisizawa. 1971. Localization of cellulase components in Pseudomonas fluorescens var. cellulosa. J. Biochem. 69:771-780. [DOI] [PubMed] [Google Scholar]
- 61.Yomano, L. P., S. W. York, S. Zhou, K. T. Shanmugam, and L. O. Ingram. 2008. Re-engineering Escherichia coli for ethanol production. Biotechnol. Lett. 30:2097-2103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.