Abstract
Twenty-seven marine sediment- and sponge-derived actinomycetes with a preference for or dependence on seawater for growth were classified at the genus level using molecular taxonomy. Their potential to produce bioactive secondary metabolites was analyzed by PCR screening for genes involved in polyketide and nonribosomal peptide antibiotic synthesis. Using microwell cultures, conditions for the production of antibacterial and antifungal compounds were identified for 15 of the 27 isolates subjected to this screening. Nine of the 15 active extracts were also active against multiresistant Gram-positive bacterial and/or fungal indicator organisms, including vancomycin-resistant Enterococcus faecium and multidrug-resistant Candida albicans. Activity-guided fractionation of fermentation extracts of isolate TFS65-07, showing strong antibacterial activity and classified as a Nocardiopsis species, allowed the identification and purification of the active compound. Structure elucidation revealed this compound to be a new thiopeptide antibiotic with a rare aminoacetone moiety. The in vitro antibacterial activity of this thiopeptide, designated TP-1161, against a panel of bacterial strains was determined.
Natural products remain the most prolific source of new antimicrobials, and the chemical diversity of natural compounds is still unmatched by combinatorial chemistry approaches (9, 31). While the latter has been successfully applied for lead optimization, it basically failed to deliver genuinely new pharmacophores, especially in the field of antimicrobials (31), mainly due to limitations in the structural variety of compounds represented in combinatorial libraries.
Most of the antibiotics in clinical use today have been developed from compounds isolated from bacteria and fungi, with members of the actinobacteria being the dominant source (34). Traditionally, most of these antimicrobials have been isolated from soil-derived actinomycetes of the genus Streptomyces. However, isolation strategies in recent years have been directed to unexploited environments like marine sources (40). Bioprospecting efforts focusing on the isolation and screening of actinobacteria from ocean habitats (25, 27) have added new biodiversity to the order Actinomycetales and revealed a range of novel natural products of pharmacological value. The existence of marine actinobacterial species physiologically and phylogenetically distinct from their terrestrial relatives is now widely accepted, and new taxonomic groups of marine actinomycetes have been described for at least six different families within the order Actinomycetales (12). Apart from being phylogenetically distinct from their terrestrial relatives, marine isolates have been shown to possess specific physiological adaptations (e.g., to high salinity/osmolarity and pressure) to their maritime surroundings and many were found to produce novel and chemically diverse secondary metabolites (10, 13, 35).
Most streptomycetes and other filamentous actinomycetes possess numerous gene clusters for the biosynthesis of secondary metabolites (2, 32), and genome sequence studies have shown that large portions of their genomes are devoted to secondary metabolite biosynthesis. Twenty gene clusters coding for known or predicted secondary metabolites were identified in the 8.7-Mb genome of Streptomyces coelicolor A3(2) (2), and 6.4% of the 8.7-Mb genome of Streptomyces avermitilis is dedicated to gene clusters for secondary metabolite biosynthesis (32). The marine actinomycete Salinispora allocates nearly 10% of its 5.2-Mb genome to 17 diverse biosynthetic loci, including polyketide synthases (PKSs), nonribosomal peptide synthetases (NRPSs), and several hybrid clusters (4, 43). Many medicinally important natural products, including antibacterials and antifungals, are synthesized by these multimodular assembly lines (14), and genome mining for secondary metabolite gene clusters has become a common tool to assess the genetic capability of bacteria to produce novel bioactive compounds. However, even for well-studied model antibiotic producers like S. coelicolor A3(2), discrepancies between the number of known metabolites on the one hand and the number of pathways identified from genomic data on the other hand are tremendous (2). These discrepancies can only be explained by the facts that most gene clusters for secondary metabolites are silenced under standard laboratory cultivation conditions and that an expression or upregulation of these pathways is only triggered in response to certain environmental signals. It has been shown that by cultivating bacteria under a range of conditions, it is possible to obtain products of many of these “orphan” biosynthetic pathways (4). Using the OSMAC (one strain-many compounds) approach, Bode et al. were able to isolate more than 100 compounds comprising 25 structural classes from only six microorganisms (4).
In this study, marine sediment-derived actinomycete isolates were analyzed for the production of antimicrobial secondary metabolites by using microwell plate fermentations and a range of media and conditions. This approach led to the isolation of a new thiopeptide antibiotic, designated TP-1161, produced by a marine sediment-derived Nocardiopsis isolate. Here we report the isolation and structural and biological characterization of TP-1161.
MATERIALS AND METHODS
Sample collection and bacterial isolation.
Actinomycete strains were isolated from sediments and sponges collected in the Trondheim Fjord, Norway. For detailed descriptions of the sampling sites and isolation conditions for the 27 actinomycetes used in this study, see Tables S1, S2, and S3 in the supplemental material.
Molecular taxonomy and phylogenetic analyses of actinomycete bacteria.
Isolates with adaptations to the marine environment were chosen based on the results of a growth evaluation on ISP2 agar medium prepared without and with 0.5× artificial seawater. After the two conditions were compared, 27 isolates displaying a preference or obligate requirement for seawater in their growth medium (exhibited by no or only poor mycelial growth, delayed development of aerial mycelium, and later onset of sporulation on medium prepared without seawater) were selected for further studies (Table 1). A phylogenetic characterization based on molecular taxonomy was performed by sequence analysis of the 16S rRNA gene. Genomic DNA of all strains was isolated using the Qiagen DNeasy Blood and Tissue kit. The 16S rRNA gene was amplified by PCR using universal bacterial 16S rRNA gene primers F27 and R1492 (44). The PCR products obtained were cloned into the Qiagen pDrive PCR cloning vector and sequenced using standard M13 vector primers and, when necessary, primer 1100R (28). DNA sequences were assembled using the CAP3 Sequence Assembly Program (16), and the resulting partial 16S rRNA gene sequences (average length, 1,470 bp) were compared to those available in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) using nucleotide BLAST. Sequences were aligned, and a phylogenetic tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA) software version 4 (21). The tree was computed using the neighbor-joining method (38), and the resulting tree topology was tested by bootstrap analysis (11) performed with 1,000 replicates. Evolutionary distance was computed by the maximum composite likelihood method (42) and is expressed as the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 1,358 positions per sequence in the final data set. The 16S rRNA gene sequence of Bifidobacterium bifidum (GenBank accession number EF589113.1) served as an outgroup to root the tree.
TABLE 1.
Isolation sources and conditions, seawater dependency, and molecular taxonomy based on 16S rRNA gene similarity of the 27 actinomycete isolates used in this studya
| Isolate no. | Source | Sample no. | Depth (in.) | Isolation medium | Sample pretreatment | Obligatory seawater dependence | Organism with greatest 16S rRNA gene similarity (% similarity) [GenBank accession no.] |
|---|---|---|---|---|---|---|---|
| TFS59-23 | Sediment | 4 | 24.6 | 13 | None | + | Streptomyces sp. HV10 (98) [FJ222814] |
| TFS60-19 | Sediment | 9 | 102 | 15 | None | + | Saccharomonospora caesia (98) [Z38022] |
| TFS61-05 | Sediment | 4 | 24.6 | 15 | None | + | Rhodococcus marinonascens (99) [NR_026183] |
| TFS65-03 | Sediment | 4 | 24.6 | 13 | None | + | Nocardiopsis sp. TFS 806 (99) [EF216368] |
| TFS65-07 | Sediment | 4 | 24.6 | 15 | None | − | Prauseria sp. TUT1202 (99) [AB188209] |
| TFS65-13 | Sediment | 4 | 24.6 | 15 | None | − | Streptomyces sp. CNS-579_SD06 (97) [EU214954] |
| TFS65-24 | Sediment | 4 | 24.6 | 16 | None | + | Nocardiopsis sp. TFS 91 (99) [EF216363] |
| TFS66-01 | Sediment | 4 | 24.6 | 15 | None | + | Streptomycetaceae bacterium CNQ719 (98) [AY464544] |
| TFS72-17 | Sediment | 9 | 102 | 17 | Rehydration, centrifugation | − | Nocardiopsis sp. TFS 806 (99) [EF216368.1] |
| TFS73-15 | Sediment | 4 | 24.6 | 17 | Rehydration, centrifugation | + | Nocardiopsis sp. 13634A (99) [EU741113] |
| TFS74-05 | Sediment | 11 | 326 | 20 | Rehydration, centrifugation | + | Nocardiopsis sp. TFS 806 (99) [EF216368] |
| TFS76-05 | Sediment | 9 | 102 | 15 | Filter method | + | Micromonospora auratinigra (98) [AB159779] |
| TFS77-24 | Sediment | 9 | 102 | 16 | Filter method | + | Nocardiopsis sp. TFS 91 (99) [EF216363] |
| TFS79-01 | Sediment | 9 | 102 | 15 | Filter method | − | Streptomyces sp. CNQ-027_SD01 (98) [EU214912] |
| TFS79-06 | Sediment | 2A | 10 | 15 | Filter method | − | Saccharomonospora caesia (99) [Z38022] |
| TFS79-15 | Sediment | 2A | 10 | 17 | Rehydration, centrifugation | − | Actinomycetales bacterium HPA72 (99) [DQ144231] |
| TFS83-14 | Sediment | 2A | 10 | 15 | Filter method | + | Micromonospora auratinigra (98) [AB159779] |
| TFS84-03 | Sediment | 4 | 24.6 | 7b | None | + | Micromonospora sp. HBUM80369 (99) [EU119228] |
| TSI115-07 | Sponge | 3 | 57.2 | 17 | None | + | Micromonospora chokoriensis (99) [AB241454] |
| TSI116-04 | Sponge | 3 | 57.2 | 4 | None | + | Streptomyces sp. CNQ-027_SD01 (98) [EU214912] |
| TSI116-13 | Sponge | 4 | 121.4 | 4 | None | + | Pseudonocardia petroleophila IMSNU 22072T (99) [AJ252828] |
| TSI119-11 | Sponge | 4 | 121.6 | 19 | None | + | Micromonospora chokoriensis 173803 (100) [EU570342] |
| TSI124-17 | Sponge | 6 | 62.9 | 15 | Filter method | + | Streptosporangium sp. TFS 224 (99) [EF212016] |
| TSI125-23 | Sponge | 4 | 121.4 | 17 | None | + | Micromonospora matsumotoense IMSNU 22003 (99) [NR_025015] |
| TSI127-17 | Sponge | 5 | 60 | 18 | None | + | Actinoalloteichus hymeniacidonis HPA177 (98) [DQ144222] |
| TSI129-12 | Sponge | 3 | 57.2 | 18 | None | + | Streptosporangium amethystogenes DSM43179T (99) [X89935] |
| TSI129-23 | Sponge | 6 | 62.9 | 15 | None | + | Streptosporangium amethystogenes DSM43179T (99) [X89935] |
For detailed information about sampling locations and origins (Tables S1 and S2), isolation procedures (Table S3), and isolation media, see the supplemental material.
PCR screening for PKS/NRPS genes.
To assess the potential of isolated bacteria to synthesize polyketide- and nonribosomal peptide-derived secondary metabolites, their DNA was subjected to the following analysis. Three different degenerate primer pairs were used to screen the genomes of the 27 actinomycetes for the presence of modular/iterative PKS and NRPS genes. The primers and PCR amplification conditions used were as described previously (6). All PCRs were performed with 10 to 20 ng genomic template DNA isolated using the Qiagen DNeasy Blood and Tissue kit. PCR products were purified using the Qiagen gel extraction kit, cloned into the Qiagen pDrive vector, and transformed into Escherichia coli DH5α by standard methods. Plasmids were isolated using the Wizard Plus SV Minipreps DNA purification system (Promega), and inserts were sequenced using standard vector primers.
Screening of isolates for production of antimicrobial compounds.
Fermentations of the 27 marine actinomycete isolates were performed in 800-μl microcultures in 2-ml deep-well plates (Greiner Bio One 96-well polypropylene Masterblocks with 2-ml deep square wells). To prevent excessive pellet formation, 3-mm-diameter glass beads were added (one bead per well). Fermentation cultures were inoculated 5% from frozen suspensions of densely grown seed cultures and incubated at 25°C, 800 rpm, and 85% humidity on a rotary shaker. M3 (1% glucose, 0.2% yeast extract, 0.1% l-asparagine, and 0.05% K2HPO4 per liter of artificial seawater, pH 7.8), ISP2 (Difco), 0.5× ISP2 prepared with 0.5× artificial seawater, and tryptic soy broth prepared with 0.5× artificial seawater served as seed culture media. For the fermentation media for metabolite production used in this screening (PM1 to PM14), see the supplemental material. For extraction, entire fermentation cultures were freeze-dried and extracted with 600 μl dimethyl sulfoxide (DMSO) for 2 h at 800 rpm at room temperature. Cell extracts were obtained by centrifugation (8,000 × g, 10 min) and stored at −20°C.
Agar diffusion assay against Micrococcus luteus ATCC 9341 and Candida albicans ATCC 10231.
Fermentation extracts were screened in two stages for antibacterial and/or antifungal activity. The first step consisted of agar diffusion assays using M. luteus ATCC 9341 and C. albicans ATCC 10231 as bacterial and fungal indicator organisms. Inoculums of M. luteus were prepared by growing the bacteria in AM1 medium (17) to an optical density at 600 nm (OD600) of 5.0 (SpectraMaxPlus384; Molecular Devices). Sterile glycerol was added to a final concentration of 10%, and the culture was stored in aliquots at −80°C. C. albicans was grown in AM19 medium (18) to an OD660 of 10.0 and stored as described above. M1 or M19 medium prepared with 1% agarose was inoculated with 0.5 to 1% (vol/vol) indicator organism and dispensed into Nunc OmniTray plates. DMSO extracts (3 μl) were applied on top of the solidified agarose, and the plates were incubated at 4°C for 30 min to allow diffusion of the applied extract into the surrounding medium. Afterwards, the plates were incubated for 13 h at 34°C to allow microbial growth. Extracts were identified as antibacterial and/or antifungal based on the observed growth inhibition of the corresponding indicator strains (visible as inhibition zones in agarose medium seeded with test organisms).
Bioassay of selected extracts against Enterococcus faecium CCUG 37832 and C. albicans CCUG 39343.
DMSO extracts with strong antibacterial and/or antifungal activity identified in agar diffusion assays were assayed in addition for antagonistic activity against E. faecium CCUG 37832 harboring multiple antibiotic resistances and the yeast C. albicans CCUG 39343 (for a description of the antibiotic resistance phenotype, see Table S4 in the supplemental material) by using robotic bioassay procedures as described by Jørgensen et al. (19). As the assay medium for E. faecium, Difco brain heart infusion broth was used. The purpose of this second screening stage was to limit the probability of rediscovery of known antibiotics.
Identification of thiopeptide TP-1161.
Whole fermentation broth of the isolate TFS65-07 was extracted with methanol (equal volume) and fractionated on an Agilent 1100 series high-performance liquid chromatography (HPLC) system with a Zorbax Bonus-RP column (2.1 by 50 mm, 3.5 μm) connected to a diode array detector (DAD) and a fraction collector system. Methanol and 10 mM ammonium acetate (pH 4) were used as the mobile phase, and the methanol gradient was linearly increased from 10 to 90% for 24 min. Fractions were sampled every minute for the whole run. Drying of the samples and agar diffusion assays to point out fractions with antibacterial or antifungal activity were performed essentially as described above, with M. luteus ATCC 9341 and C. albicans CCUG 39343 as the indicator organisms. Bioactive fractions were analyzed using an Agilent HPLC system with a Zorbax Bonus-RP column (2.1 by 50 mm, 3.5 μm) connected to a DAD and a time-of-flight (TOF) apparatus to determine the accurate mass of the bioactive compound. Ten millimolar ammonium acetate (pH 7) and acetonitrile were used as the mobile phase, and electrospray ionization was performed in the negative mode.
Production of TP-1161.
For small-scale production of TP-1161, TFS65-07 was grown in 500-ml baffled flasks with 100 ml PM4 medium prepared with 0.5× artificial seawater (see the supplemental material). Five grams of glass beads (3-mm diameter) was added to each flask to increase the shear force and avoid extensive pellet formation. Production cultures were incubated for 14 days at 25°C and 225 rpm (orbital movement, 2.5-cm amplitude) before harvesting. Production cultures were inoculated (3%, vol/vol) from seed cultures cultivated in shake flasks for 3 days at 25°C and 225 rpm in 0.5× tryptone soya broth supplemented with 20 g/liter glucose. Fermentation of TFS65-07 for production of TP-1161 for structure elucidation was performed in 3-liter Applikon fermentors with 1.5 liters BPS-4_KD medium (soy flour at 30 g/liter, corn flour at 20 g/liter, malt extract at 7 g/liter, yeast extract at 4 g/liter, CaCO3 at 5 g/liter, and glucose at 50 g/liter [pH 7.8] prepared with 0.5× artificial seawater and supplemented with 3 ml/liter trace mineral solution TMS1 [39] and 0.6 ml/liter Symperonic PEl/61 antifoam).
Fermentations were run for 14 days at 25°C with constant aeration (gas volume flow per unit of liquid volume per minute of 0.25) and agitation (1,000 rpm). The pH was controlled at 7.5 with 2 M NaOH. Seed cultures for the fermentations were prepared in 500-ml baffled shake flasks with 100 ml 2× ISP2 medium (yeast extract at 8 g/liter, malt extract at 10 g/liter, glucose at 4 g/liter [pH 7.8]). The medium was prepared with 1× artificial seawater. Seed cultures were incubated for 2 days at 25°C and 225 rpm prior to inoculation of the fermentations (3 vol%).
Purification of TP-1161.
TP-1161 was purified in a two-step purification process. Culture broth was centrifuged, and the mycelial pellet was frozen at −20°C. After thawing, the mycelium was washed twice with deionized water and then with a 1:1 mixture of deionized water and isopropanol. TP-1161 was extracted with methanol (1 ml methanol/g wet pellet) for 1 h, and the supernatant was collected by centrifugation. The extract was concentrated to approximately half of the volume on a rotational evaporator. The extract was stored overnight at −20°C to precipitate impurities. The precipitate was then removed by centrifugation (15 min, 120,000 rpm, 4°C), followed by filtration. The concentrated extract was purified using an Agilent 1100 series preparative HPLC system with an Agilent Prep-C18 column (50 by 250 mm, 10 μm) connected to a DAD and a fraction collector. Methanol and 10 mM ammonium acetate (pH 4) were used as the mobile phase. The methanol concentration was linearly increased from 70 to 80% for the first 4 min, kept at 80% for the next 6 min, and then kept at 100% for the next 2 min. The fractions were adjusted to approximately pH 7 with NH3 solution. The methanol in the fractions was evaporated, and the aqueous phase was concentrated on an Oasis HLB solid-phase extraction cartridge.
TP-1161 structure elucidation.
Optical rotation was measured on a Perkin-Elmer 341 polarimeter. Nuclear magnetic resonance (NMR) experiments were carried out on a Bruker AVANCE III instrument at 500 MHz using a 1-mm TXI probe. Spectra were analyzed utilizing Bruker Topspin 2.1 or MestRe Nova 5.25 software using the residual solvent signals as an internal standard.
TP-1161 is a pale yellow amorphous solid: [α]20D −103 (c 0.10, CHCl3); UV (MeOH) λmax (log ɛ) 244 (3.87) nm; for 1H and 13C NMR and heteronuclear multiple bond correlation (HMBC) data, see Table 4; high-resolution mass spectrometry (HRMS) m/z [M + H + Na]2+ 592.63009 (calculated for C50H47N15O13S3 592.63024, Δ = 0.25 ppm).
TABLE 4.
MICs of TP-1161 and vancomycin against a panel of Gram-positive bacterial strains
| Gram-positive bacterial strain | MIC (μg/ml) |
|
|---|---|---|
| TP-1161 | Vancomycin | |
| S. aureus 10 | 1.0 | 1.0 |
| S. aureus 3797 | >32.0 | 8.0 |
| S. aureus 3798 | >32.0 | 8.0 |
| S. aureus 5 | 0.5 | 1.0 |
| S. aureus 100 | 2.0 | 1.0 |
| S. aureus ATCC 25923 | 4.0 | 0.5 |
| S. haemolyticus 161 | 1.0 | 2.0 |
| S. haemolyticus 1025 | 0.5 | 1.0 |
| S. haemolyticus 602 | 1.0 | 1.0 |
| S. haemolyticus 585 | 0.5 | 1.0 |
| S. epidermidis 533 | 0.5 | 1.0 |
| S. epidermidis 4603 | 4.0 | 1.0 |
| S. epidermidis 22 | 0.5 | 1.0 |
| S. epidermidis 9 | 0.5 | 1.0 |
| E. faecalis 559 | 1.0 | 1.0 |
| E. faecium 568 | 0.5 | 2.0 |
| E. faecalis 560 | 1.0 | >32.0 |
| E. faecium 569 | 1.0 | >32.0 |
| E. faecalis ATCC 29212 | 0.25 | 1.0 |
| S. pneumoniae ATCC 49619 | 0.5 | 0.25 |
| Streptococcus B 52 | 0.5 | 0.5 |
| Streptococcus B 209 | 0.5 | 0.5 |
TP-1161 in vitro antibacterial activity (MIC determination).
TP-1161 was tested against a panel of Gram-negative and Gram-positive pathogens. MICs for all Gram-positive and Gram-negative bacterial strains were determined by standardized microdilution tests using Mueller-Hinton broth (Acumedia). Bacterial inoculums containing 5 × 105 CFU/ml were incubated for 24 h at 36°C in the presence of different antibiotic concentrations according to Clinical and Laboratory Standards Institute protocols (document M100-S15 [8]). The majority of the bacterial strains used were clinical isolates, while some were obtained from ATCC (American Type Culture Collection).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the 16S rRNA gene sequences of the different species are as follows: Streptomyces sp. TFS59-23, HM001271; Saccharomonospora sp. TFS60-19, HM001272; Rhodococcus sp. TFS61-05, HM001273; Nocardiopsis sp. TFS65-03, HM001274; Nocardiopsis sp. TFS65-07, HM001275; Streptomyces sp. TFS65-13, HM001276; Nocardiopsis sp. TFS65-24, HM001277; Streptomyces sp. TFS66-01, HM001278; Nocardiopsis sp. TFS72-17, HM001279; Nocardiopsis sp. TFS73-15, HM001280; Nocardiopsis sp. TFS74-05, HM001281; Micromonospora sp. TFS76-05, HM001282; Nocardiopsis sp. TFS77-24, HM001283; Streptomyces sp. TFS79-01, HM001284; Saccharomonospora sp. TFS79-06, HM001285; Streptomyces sp. TFS79-15, HM001286; Micromonospora sp. TFS83-14, HM001287; Micromonospora sp. TFS84-03, HM001288; Micromonospora sp. TSI115-07, HM001289; Streptomyces sp. TSI116-04, HM001290; Pseudonocardia sp. TSI116-13, HM001291; Micromonospora sp. TSI119-11, HM001292; Streptosporangium sp. TSI124-17, HM001293; Micromonospora sp. TSI125-23, HM001294; Actinoalloteichus sp. TSI127-17, HM001295; Streptosporangium sp. TSI129-12, HM001296; and Streptosporangium sp. TSI129-23, HM001297.
RESULTS
Molecular taxonomy and phylogeny of marine actinomycete isolates.
Twenty-seven bacterial isolates from marine sediments and sponges, preliminarily classified as actinobacteria based on their growth morphology, were selected for further studies. An additional criterion for selection was the isolates' dependence on or preference for seawater for growth. The results of the 16S rRNA gene analysis of these isolates are summarized in Table 1, presenting the 16S rRNA gene sequences of the closest phylogenetic neighbors with the highest similarity values obtained from a BLAST search. For a description of the evolutionary relationships of the 27 isolates and closely related taxa, including the closest actinobacterial type strains in the form of a neighbor-joining tree, see Fig. S1 in the supplemental material. The phylogenetic tree divides the 27 isolates into five clades comprising eight different genera within the Actinomycetales: Nocardiopsis, Streptosporangium, Streptomyces, Actinoalloteichus, Pseudonocardia, Rhodococcus, Saccharomonospora, and Micromonospora. The largest clade consists of seven Nocardiopsis isolates and is divided into four subclusters around the closest type strains, Nocardiopsis exhalans, N. alba, N. listeri, and N. umidischolae. All seven isolates exhibited the highest level of 16S rRNA gene similarity (99%) to Nocardiopsis sp. isolated from marine sediments. Six of these strains had previously been isolated from sediments in the Trondheim Fjord (6), whereas one isolate (TFS73-15) had the highest similarity (99%) to Nocardiopsis sp. 13634A isolated from marine sediments collected in the Caribbean.
The two largest clades consist of six Streptomyces spp. and six Micromonospora spp. Five Streptomyces isolates exhibited the highest 16S rRNA gene similarity (97 to 99%) to Streptomyces spp. isolated from marine sediments (TFS65-13, TFS66-01, TFS79-01, and TFS116-04) or sponges (TFS79-15). The six Micromonospora isolates are divided into four subclusters, and the 16S rRNA gene analysis yielded the highest similarity values for all isolates to Micromonospora spp. from soil samples. The genus Streptosporangium is represented by three isolates, all originating from sponges. Two of these (TSI129-12 and TSI129-23) show the highest similarity (99%) to Streptosporangium spp. from terrestrial sources, whereas the TSI124-17 16S rRNA gene displays the highest similarity to that of Streptosporangium sp. TFS224 isolated from sediment samples from the Trondheim Fjord (6). The Saccharomonospora clade consists of two isolates which are divided into two subclusters in the phylogenetic tree and show the highest 16 rRNA gene sequence similarity (98 and 99%) to Saccharomonospora sp. isolated from soil samples. TSI127-17 was classified as a member of the genus Actinoalloteichus with the highest similarity (98%) to Actinoalloteichus hymeniacidonis cultivated from a marine sponge. The genus Rhodococcus is represented by one isolate showing the highest similarity (99%) to the marine sediment-derived Rhodococcus marinonascens. Isolate TSI116-13 was found to be the most closely (99%) related to Pseudonocardia petrophila isolated from soil.
PCR screening for PKS and NRPS genes.
Members of the Actinomycetales are known as prolific producers of secondary metabolites. To assess the genetic potential of the 27 selected isolates to produce polyketide and peptide antibiotics, a PCR screening for genes encoding PKS type I and II ketosynthase and NRPS adenylation domains was performed using degenerate primers. Amplification products for gene fragments for all three biosynthetic systems were obtained from the majority of the isolates (Table 2). To confirm that the amplified sequences in fact represented the targeted gene segments, amplicons of selected strains were subcloned and sequenced. In all cases, amplification of the target sequences was confirmed by sequence analysis (data not shown).
TABLE 2.
Results of PCR screening of 27 actinomycete isolates for the presence of genes encoding domains in modular PKS type I, iterative PKS type II, and NRPSs
| Strain | PKS Ia | PKS IIa | NRPSsa | Activity(ies)b |
|---|---|---|---|---|
| TFS59-23 | + | + | + | AB/AF |
| TFS60-19 | − | + | + | − |
| TFS61-05 | + | + | + | AB/AF |
| TFS65-03 | + | + | + | − |
| TFS65-07 | + | + | + | AB |
| TFS65-13 | + | − | + | AB |
| TFS65-24 | + | − | + | AB/AF |
| TFS66-01 | + | + | + | AB/AF |
| TFS72-17 | + | + | + | − |
| TFS73-15 | + | + | + | − |
| TFS74-05 | + | + | + | − |
| TFS76-05 | + | + | + | − |
| TFS77-24 | + | − | − | AB |
| TFS79-01 | − | − | + | − |
| TFS79-06 | + | + | + | AB |
| TFS79-15 | − | + | + | AB |
| TFS83-14 | − | + | + | AB |
| TFS84-03 | + | + | + | − |
| TSI115-07 | − | + | − | − |
| TSI116-04 | − | − | − | − |
| TSI116-13 | + | − | + | − |
| TSI119-11 | + | + | + | − |
| TSI124-17 | + | − | + | AB/AF |
| TSI125-23 | + | + | + | AB/AF |
| TSI127-17 | + | + | + | AB/AF |
| TSI129-12 | + | − | + | AB/AF |
| TSI129-23 | + | + | + | AF |
+, activity present; −, no activity present.
AB, antibacterial activity present; AF, antifungal activity present; −, no activity present.
Screening for new antimicrobials in microwell cultures.
In order to reveal the phenotypic potential of the selected isolates for production of antimicrobial compounds, they were subjected to a series of microwell fermentations using a variety of media. In the first series, 12 isolates classified as obligatorily seawater dependent (Table 1) were cultivated for 7 to 19 days in 14 different liquid production media (PM1 to PM14) prepared with 1× artificial seawater (for the composition of production media and seawater, see the supplemental material). Entire fermentation cultures were extracted with DMSO, and the extracts were analyzed for activity against M. luteus ATCC 9341 and C. albicans ATCC 10231 in agar diffusion assays. Based on the results of this first bioassay, the number of production media was scaled down to six, representing those which yielded most of the antibacterial and antifungal activity (PM2, PM3, PM4, PM6, PM7, and PM14), for screening of the remaining isolates. Isolates displaying a dependency on seawater in their growth medium were cultivated in these six media prepared with 0.5× and 1× artificial seawater, while isolates displaying a seawater preference during growth were cultivated in media prepared with deionized and 0.5× artificial seawater, respectively. All fermentations were performed at 25°C and 800 rpm, and the cultures were harvested for extraction after 10 to 21 days.
Fifteen of the 27 isolates subjected to this screening were found to produce compounds inhibiting the growth of M. luteus ATCC 9341 and/or C. albicans ATCC 10231 (Table 2). To reduce the possibility of rediscovery of commonly produced antibiotics, a second screening was performed by using multiply resistant bacterial and fungal indicator organisms. Extracts of 9 of the 15 strains displaying strong antibacterial and/or antifungal activity in the agar diffusion assay were thus analyzed for activity against vancomycin-resistant E. faecium CCUG 37832 and C. albicans CCUG 39343 (for a description of the resistance phenotype, see the supplemental material) in an automated bioassay. This second bioassay identified extracts of nine strains with activity against one or both of the multidrug-resistant indicator strains (Table 3). Based on the potent antibacterial inhibition profile of extracts from Nocardiopsis sp. TFS65-07, this strain was chosen for further analyses and identification and purification of the bioactive compound.
TABLE 3.
Antibacterial and antifungal activities of fermentation extracts of 15 isolates identified in a screening of 27 marine sediment-derived actinomycetes
| Strain | Antibacterial activitya |
Antifungal activitya |
||
|---|---|---|---|---|
| M. luteus ATCC 9341 | E. faecium CCUG 37832 | C. albicans ATCC 10231 | C. albicans CCUG 39343 | |
| TFS59-23 | + | − | + | − |
| TFS61-05 | + | NT | + | NT |
| TFS65-07 | + | + | − | − |
| TFS65-13 | + | NT | − | NT |
| TFS65-24 | + | + | + | + |
| TFS66-01 | + | NT | + | NT |
| TFS77-24 | + | + | − | − |
| TFS79-06 | + | + | − | − |
| TFS79-15 | + | NT | − | NT |
| TFS83-14 | + | + | − | − |
| TSI124-17 | + | NT | + | NT |
| TSI125-23 | + | + | + | + |
| TSI127-17 | + | + | + | + |
| TSI129-12 | + | NT | + | NT |
| TSI129-23 | − | − | + | + |
+, activity present; −, no activity present; NT, not tested.
Identification of the bioactive compound produced by isolate TFS65-07.
Fermentation extracts of TFS65-07 causing strong growth inhibition of E. faecium CCUG 37832 were subjected to liquid chromatography (LC) fractionation to identify the bioactive compound. Bioactivity against M. luteus ATCC 9341 was identified in two neighboring fractions. These fractions were not active against C. albicans CCUG 3934. LC-DAD-TOF analysis of the bioactive fractions showed a significant UV absorption peak with a maximum at 250 nm and a significant mass spectrometry peak which corresponded well to the UV absorption peak (see Fig. S2 in the supplemental material). To further characterize the putative bioactive compound, scaled-up production in both shake flasks and batch fermentors and purification (see Materials and Methods) were performed. The production levels achieved in shake flasks and fermentors were relatively low (<20 mg/liter). The pure product was analyzed by LC-DAD-TOF (see Fig. S2 in the supplemental material). The measured accurate mass, [M − H]−, of the compound was 1,160.2564. Assuming that the measured mass is within 2 ppm of the correct mass of the molecule, the accurate mass (M) was calculated to be between 1,161.2619 and 1,161.2665. The molecule is further referred to as TP-1161.
Elucidation of TP-1161 structure.
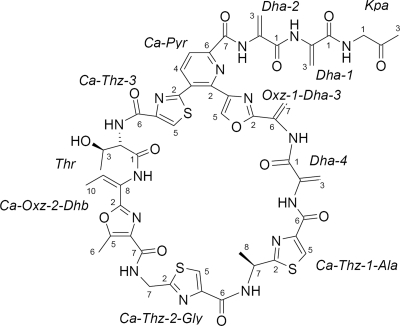
TP-1161 possesses a molecular formula of C50H47N15O13S3, as determined on the basis of HRMS and NMR data. Phase-sensitive heteronuclear single quantum coherence (HSQC) experiments revealed the peptidic nature of the molecule. From the HSQC experiments, further structural features could be derived indicating the presence of four dehydroalanine groups, i.e., three 2,4-disubstituted thiazoles and one 2,4-disubstituted oxazole (Oxz) moiety. The presence of these structural features strongly suggested the molecule to be a representative of the thiopeptide antibiotics (1). This assumption was also supported by the molecular weight of 1,161 and the UV/VIS spectrum of the molecule showing an absorption maximum at 250 nm. The structural data also strongly indicated the presence of a trisubstitued pyridine (Pyr) functional unit, thus assigning the compound to the d series of thiopeptide antibiotics (1). Further analysis of the 13C NMR, HSQC, and HMBC data led to the identification of 11 partial structures (assigned in Fig. 1). The planar structure of TP-1161 was obtained from the partial structures by further interpretation of the HMBC data, taking advantage of the correlations of the nonexchanging amide protons. A literature search of the d series of thiopeptide antibiotics (more than 49 members have been reported to date [1]) revealed that TP-1161 shows a close resemblance to the A10255 series of antibiotics published by Lilly and company in 1992 (5). The distinct structural differences are found at the methyl group attached to the C-5 position of Oxz-2 in the Ca-Oxz-2-Dhb substructure and at the C-terminal TP-1161 bearing an unusual aminoacetone moiety. The NMR data for TP-1161 (see Table S5 in the supplemental material), compared to the values reported in the literature, confirm the close similarity of the compounds mentioned.
FIG. 1.
Molecular structure of thiopeptide antibiotic TP-1161 with partial structures assigned. Kpa, 2-ketopropylamine; Dha, dehydroalanine; Dhb, dehydrobutyrine; Thz, thiazole.
As it has recently been shown that the thiopeptide antibiotics are derived from ribosomally synthesized peptide precursors with no epimerization domains encoded in the biosynthetic gene clusters (23), we assume that TP-1161 also occurs in its natural stereoconfiguration (Fig. 1).
TP-1161 in vitro antibacterial activity.
The in vitro antibacterial activity of TP-1161 was assessed against a panel of Gram-negative and Gram-positive bacteria, mostly represented by clinical isolates. All of the Gram-negative strains tested (six of E. coli, two of Klebsiella pneumoniae, one of Salmonella enterica serovar Choleraesuis, and four of Pseudomonas aeruginosa) were not susceptible to TP-1161 (data not shown). The MICs of TP-1161, ranging from 0.25 to 4 μg/ml for most Gram-positive strains, were comparable to or lower than those of the reference antibiotic vancomycin (Table 4). TP-1161 also inhibited the growth of vancomycin-resistant bacterial strains represented by Enterococcus faecalis 560 and E. faecium 569, with a MIC of 1 μg/ml.
DISCUSSION
Marine sediment-derived actinomycetes as a source of new antimicrobials.
Most of the antibiotics in clinical use today were discovered more than 5 decades ago, with only two new antibacterial agents with new mechanisms of action (the synthetic oxazolidinone linezolid and the natural-product-based lipopeptide daptomycin) approved over the last 10 years (41). Loss of efficacy of existing drugs due to emerging multidrug-resistant pathogens threatens to outpace the development of new antimicrobials. The majority of all anti-infective drugs were either derived from or inspired by natural products (9), and new antibiotics are most likely to come from natural-product-based research since neither genomics-derived target-based research nor combinatorial chemistry has so far provided drugs that have actually entered the market (33). Thus, mining microbial diversity represents the most promising source for obtaining new and diverse antimicrobial leads.
The results of this study support previous findings that fjord-derived actinomycetes are a promising resource for drug discovery (6, 7). PCR screening indicated that the overall majority of the 27 isolates studied have the genetic capability to produce representatives of more than one antibiotic class. Using different cultivation media, conditions for the production of these antibiotics could be determined for more than half of the isolates, with 30% of the isolates producing compounds with antagonistic activity against multidrug-resistant indicator organisms. The screening and further focused production/isolation resulted in the identification of the new antibacterial thiopeptide antibiotic TP-1161.
TP-1161 structure and activity.
The macrocyclic peptide scaffold of all thiopeptide antibiotics contains a six-membered tri- or tetrasubstituted nitrogen heterocycle. According to the structure and oxidation state of this Pyr ring, thiopeptides can be divided into five classes (series a to e) encompassing 29 different antibiotic families with nearly 80 structurally distinct compounds (1). TP-1161 belongs to the series d thiopeptides, which make up the most prolific class and are characterized by a 2,3,6-trisubstituted Pyr domain located at the center of a single peptide macrocycle. The amino acid residues constituting the peptide macrocycle are typically highly modified, and common modifications include cyclodehydration of serine, threonine, and cysteine side chains to oxazoline and thiazoline heterocycles, which can then be further oxidized to aromatic Oxz and thiazole systems. Series d thiopeptides typically carry a side chain of dehydrated or heterocyclic amino acids attached to the Pyr 6 position (1).
Members of this complex group of naturally occurring peptide antibiotics have been isolated predominantly from Gram-positive bacteria, mainly terrestrial actinomycetes of the genus Streptomyces (1) but also from marine sediment-derived bacteria (30). Thiopeptide antibiotics possess no activity against Gram-negative bacteria but are potent inhibitors of protein synthesis in Gram-positive bacteria, including multidrug-resistant pathogens like methicillin-resistant Staphylococcus aureus and vancomycin-resistant E. faecium. In addition to interfering with bacterial protein synthesis, antibiotics like thiostrepton and micrococcin possess activity against the human malaria parasite Plasmodium falciparum (26, 36); others have potential applications as neostatic agents (3, 22). The biosynthetic origin of thiopeptides has been elucidated only recently (20, 24, 45) and revealed these compounds to be derived from ribosomally synthesized precursor peptides which are converted to their final structures by a series of posttranslational enzymatic modifications (23).
The in vitro activity profile of TP-1161 mirrors the characteristics described for other thiopeptide antibiotics. It was not active against Gram-negative bacteria, while displaying good activity against a panel of Gram-positive clinical isolates, including two multidrug-resistant Enterococcus strains (Table 4). Thiopeptides exert their antibacterial activity through at least three different mechanisms of action. Antibiotics like thiostrepton and micrococcin interfere with peptide elongation by binding to the 23S region of the large ribosomal subunit at the ribosomal protein L11 binding domain, a region that also interacts with bacterial elongation factor G (37). The thiomuracins, the amythiamycins, or the GE2270 factors inhibit protein synthesis by binding to bacterial elongation factor Tu and preventing it from delivering incoming aminoacyl-tRNAs to the ribosome (1, 23, 29). A third mechanism is employed by the cyclothiazomycins which target DNA-dependent RNA synthesis by inhibition of RNA polymerase (15). Further studies are necessary to determine the mechanism of action and molecular target(s) of TP-1161.
Supplementary Material
Acknowledgments
We are thankful to E. P. Mirchink for performing the MIC assays.
This work was supported by the Norwegian University of Science and Technology, Research Council of Norway, and Sinvent AS.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bagley, M. C., J. W. Dale, E. A. Merritt, and X. Xiong. 2005. Thiopeptide antibiotics. Chem. Rev. 105:685-714. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bhat, U. G., M. Halasi, and A. L. Gartel. 2009. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One 4:e5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bode, H. B., B. Bethe, R. Hofs, and A. Zeeck. 2002. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3:619-627. [DOI] [PubMed] [Google Scholar]
- 5.Boeck, L. D., D. M. Berry, F. P. Mertz, and R. W. Wetzel. 1992. A10255, a complex of novel growth-promoting thiopeptide antibiotics produced by a strain of Streptomyces gardneri. Taxonomy and fermentation studies. J. Antibiot. (Tokyo) 45:1222-1230. [DOI] [PubMed] [Google Scholar]
- 6.Bredholdt, H., O. A. Galatenko, K. Engelhardt, E. Fjaervik, L. P. Terekhova, and S. B. Zotchev. 2007. Rare actinomycete bacteria from the shallow water sediments of the Trondheim Fjord, Norway: isolation, diversity and biological activity. Environ. Microbiol. 9:2756-2764. [DOI] [PubMed] [Google Scholar]
- 7.Bredholt, H., E. Fjaervik, G. Johnsen, and S. B. Zotchev. 2008. Actinomycetes from sediments in the Trondheim Fjord, Norway: diversity and biological activity. Mar. Drugs 6:12-24. [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing, 15th information supplement M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Cragg, G. M., P. G. Grothaus, and D. J. Newman. 2009. Impact of natural products on developing new anti-cancer agents. Chem. Rev. 109:3012-3043. [DOI] [PubMed] [Google Scholar]
- 10.Feling, R. H., G. O. Buchanan, T. J. Mincer, C. A. Kauffman, P. R. Jensen, and W. Fenical. 2003. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. Engl. 42:355-357. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 12.Fenical, W., and P. R. Jensen. 2006. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol. 2:666-673. [DOI] [PubMed] [Google Scholar]
- 13.Fiedler, H. P., C. Bruntner, J. Riedlinger, A. T. Bull, G. Knutsen, M. Goodfellow, A. Jones, L. Maldonado, W. Pathom-aree, W. Beil, K. Schneider, S. Keller, and R. D. Sussmuth. 2008. Proximicin A, B and C, novel aminofuran antibiotic and anticancer compounds isolated from marine strains of the actinomycete Verrucosispora. J. Antibiot. (Tokyo) 61:158-163. [DOI] [PubMed] [Google Scholar]
- 14.Fischbach, M. A., and C. T. Walsh. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106:3468-3496. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto, M., T. Murakami, K. Funahashi, T. Tokunaga, K.-I. Nihei, T. Okuno, T. Kimura, H. Naoki, and H. Himeno. 2006. An RNA polymerase inhibitor, cyclothiazomycin B1, and its isomer. Bioorg. Med. Chem. 14:8259-8270. [DOI] [PubMed] [Google Scholar]
- 16.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jørgensen, H., K. F. Degnes, A. Dikiy, E. Fjaervik, G. Klinkenberg, and S. B. Zotchev. 2010. Insights into the evolution of macrolactam biosynthesis through cloning and comparative analysis of the biosynthetic gene cluster for a novel macrocyclic lactam, ML-449. Appl. Environ. Microbiol. 76:283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jørgensen, H., K. F. Degnes, H. Sletta, E. Fjaervik, A. Dikiy, L. Herfindal, P. Bruheim, G. Klinkenberg, H. Bredholt, G. Nygard, S. O. Doskeland, T. E. Ellingsen, and S. B. Zotchev. 2009. Biosynthesis of macrolactam BE-14106 involves two distinct PKS systems and amino acid processing enzymes for generation of the aminoacyl starter unit. Chem. Biol. 16:1109-1121. [DOI] [PubMed] [Google Scholar]
- 19.Jørgensen, H., E. Fjaervik, S. Hakvag, P. Bruheim, H. Bredholt, G. Klinkenberg, T. E. Ellingsen, and S. B. Zotchev. 2009. Candicidin biosynthesis gene cluster is widely distributed among Streptomyces spp. isolated from the sediments and the neuston layer of the Trondheim Fjord, Norway. Appl. Environ. Microbiol. 75:3296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly, W. L., L. Pan, and C. Li. 2009. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J. Am. Chem. Soc. 131:4327-4334. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., M. Nei, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok, J. M.-M., S. S. Myatt, C. M. Marson, R. C. Coombes, D. Constantinidou, and E. W.-F. Lam. 2008. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol. Cancer Ther. 7:2022-2032. [DOI] [PubMed] [Google Scholar]
- 23.Li, C., and W. L. Kelly. 2010. Recent advances in thiopeptide antibiotic biosynthesis. Nat. Prod. Rep. 27:153-164. [DOI] [PubMed] [Google Scholar]
- 24.Liao, R., L. Duan, C. Lei, H. Pan, Y. Ding, Q. Zhang, D. Chen, B. Shen, Y. Yu, and W. Liu. 2009. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 16:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magarvey, N. A., J. M. Keller, V. Bernan, M. Dworkin, and D. H. Sherman. 2004. Isolation and characterization of novel marine-derived actinomycete taxa rich in bioactive metabolites. Appl. Environ. Microbiol. 70:7520-7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McConkey, G. A., M. J. Rogers, and T. F. McCutchan. 1997. Inhibition of Plasmodium falciparum protein synthesis. Targeting the plastid-like organelle with thiostrepton. J. Biol. Chem. 272:2046-2049. [DOI] [PubMed] [Google Scholar]
- 27.Mincer, T. J., P. R. Jensen, C. A. Kauffman, and W. Fenical. 2002. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 68:5005-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno, C., J. Romero, and R. T. Espejo. 2002. Polymorphism in repeated 16S rRNA genes is a common property of type strains and environmental isolates of the genus Vibrio. Microbiology 148:1233-1239. [DOI] [PubMed] [Google Scholar]
- 29.Morris, R. P., J. A. Leeds, H. U. Naegeli, L. Oberer, K. Memmert, E. Weber, M. J. LaMarche, C. N. Parker, N. Burrer, S. Esterow, A. E. Hein, E. K. Schmitt, and P. Krastel. 2009. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J. Am. Chem. Soc. 131:5946-5955. [DOI] [PubMed] [Google Scholar]
- 30.Nagai, K., K. Kamigiri, N. Arao, K. Suzumura, Y. Kawano, M. Yamaoka, H. Zhang, M. Watanabe, and K. Suzuki. 2003. YM-266183 and YM-266184, novel thiopeptide antibiotics produced by Bacillus cereus isolated from a marine sponge. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological properties. J. Antibiot. (Tokyo) 56:123-128. [DOI] [PubMed] [Google Scholar]
- 31.Newman, D. J., and G. M. Cragg. 2007. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70:461-477. [DOI] [PubMed] [Google Scholar]
- 32.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. U. S. A. 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Payne, D. J., M. N. Gwynn, D. J. Holmes, and D. L. Pompliano. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 34.Peláez, F. 2006. The historical delivery of antibiotics from microbial natural products—can history repeat? Biochem. Pharmacol. 71:981-990. [DOI] [PubMed] [Google Scholar]
- 35.Riedlinger, J., A. Reicke, H. Zahner, B. Krismer, A. T. Bull, L. A. Maldonado, A. C. Ward, M. Goodfellow, B. Bister, D. Bischoff, R. D. Sussmuth, and H. P. Fiedler. 2004. Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J. Antibiot. (Tokyo) 57:271-279. [DOI] [PubMed] [Google Scholar]
- 36.Rogers, M. J., E. Cundliffe, and T. F. McCutchan. 1998. The antibiotic micrococcin is a potent inhibitor of growth and protein synthesis in the malaria parasite. Antimicrob. Agents Chemother. 42:715-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosendahl, G., and S. Douthwaite. 1994. The antibiotics micrococcin and thiostrepton interact directly with 23S rRNA nucleotides 1067A and 1095A. Nucleic Acids Res. 22:357-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 39.Sekurova, O., H. Sletta, T. E. Ellingsen, S. Valla, and S. Zotchev. 1999. Molecular cloning and analysis of a pleiotropic regulatory gene locus from the nystatin producer Streptomyces noursei ATCC11455. FEMS Microbiol. Lett. 177:297-304. [DOI] [PubMed] [Google Scholar]
- 40.Sheridan, C. 2006. Antibiotics au naturel. Nat. Biotechnol. 24:1494-1496. [DOI] [PubMed] [Google Scholar]
- 41.Spellberg, B., J. H. Powers, E. P. Brass, L. G. Miller, and J. E. Edwards, Jr. 2004. Trends in antimicrobial drug development: implications for the future. Clin. Infect. Dis. 38:1279-1286. [DOI] [PubMed] [Google Scholar]
- 42.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Udwary, D. W., L. Zeigler, R. N. Asolkar, V. Singan, A. Lapidus, W. Fenical, P. R. Jensen, and B. S. Moore. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U. S. A. 104:10376-10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wieland Brown, L. C., M. G. Acker, J. Clardy, C. T. Walsh, and M. A. Fischbach. 2009. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc. Natl. Acad. Sci. U. S. A. 106:2549-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.