Abstract
Pectinolytic enzymes play an important role in cocoa fermentation. In this study, we characterized three extracellular pectate lyases (Pels) produced by bacilli isolated from fermenting cocoa beans. These enzymes, named Pel-22, Pel-66, and Pel-90, were synthesized by Bacillus pumilus BS22, Bacillus subtilis BS66, and Bacillus fusiformis BS90, respectively. The three Pels were produced under their natural conditions and purified from the supernatants using a one-step chromatography method. The purified enzymes exhibited optimum activity at 60°C, and the half-time of thermoinactivation at this temperature was approximately 30 min. Pel-22 had a low specific activity compared with the other two enzymes. However, it displayed high affinity for the substrate, about 2.5-fold higher than those of Pel-66 and Pel-90. The optimum pHs were 7.5 for Pel-22 and 8.0 for Pel-66 and Pel-90. The three enzymes trans-eliminated polygalacturonate in a random manner to generate two long oligogalacturonides, as well as trimers and dimers. A synergistic effect was observed between Pel-22 and Pel-66 and between Pel-22 and Pel-90, but not between Pel-90 and Pel-66. The Pels were also strongly active on highly methylated pectins (up to 60% for Pel-66 and Pel-90 and up to 75% for Pel-22). Fe2+ was found to be a better cofactor than Ca2+ for Pel-22 activity, while Ca2+ was the best cofactor for Pel-66 and Pel-90. The amino acid sequences deduced from the cloned genes showed the characteristics of Pels belonging to Family 1. The pel-66 and pel-90 genes appear to be very similar, but they are different from the pel-22 gene. The characterized enzymes form two groups, Pel-66/Pel-90 and Pel-22; members of the different groups might cooperate to depolymerize pectin during the fermentation of cocoa beans.
Cocoa fermentation is a key step in the technological transformation of cocoa into chocolate (6, 33, 35). The fermentation of cocoa beans occurs at two levels: the first level involves reactions that take place in the pulp, in the outer part of the beans, and the second-level reactions are located deep within the cotyledons.
Reactions occurring in the pulp mainly concern the transformation of carbohydrates into ethanol and organic acids by a microflora essentially composed of yeast, lactic acid bacteria, acetic acid bacteria, and Bacillus (35). The resulting organic acids produced by the microbial metabolism diffuse into the bean and provoke lowering of the inner pH (16). The low pH, combined with the rise in temperature of the fermenting mass, activates two acidic-pH-dependent enzymes present in the cotyledons: an aspartic endoprotease and a serine carboxypeptidase (6, 7, 43). The combined actions of these enzymes leads to the transformation of storage proteins into hydrophobic amino acids (5), which are known to be the main precursor molecules of cocoa and the eventual chocolate aroma (4, 35).
The fermentation process is also associated with the actions of various other plant cell wall-degrading enzymes, namely, pectinolytic enzymes. These enzymes, which allow the degradation of the cocoa pulp (34, 35, 36), facilitate the diffusion of microbial metabolites (essentially acetic acid) into the beans. Furthermore, the oligomers generated from the degradation of pectin polymers are used as a carbon source for the growth of the microorganisms. In view of the role they play, pectinolytic enzymes are not only essential for the normal course of cocoa fermentation, they are also key to the good quality of fermented and dried beans (3, 35).
Pectinolytic enzymes are classified into two mains groups according to their mode of attack on pectin molecules: de-esterifying enzymes (pectin methyl esterase [EC 3.1.1.11]), which remove the methoxyl group from pectin, and depolymerases, which cleave the β(1,4)glycosidic bonds between galacturonate units, either by hydrolysis (polygalacturonase [EC 3.2.1.15]) or by trans-elimination (pectin lyase [EC 4.2.2.10] and pectate lyase [Pel] [EC 4.2.2.2]). Among these enzymes, the class of pectate lyases is widely distributed in bacteria and fungi, some phytopathogenic (1, 14, 15) and others, such as members of the genus Bacillus (2, 24, 39, 40), nonpathogenic. Pectate lyases are classified into different families according to their primary amino acid sequences (11, 38). The classification can be found on the CAZy (Carbohydrate-Active EnZymes database) server (http://www.cazy.org/) (10).
Over the last 3 decades, polygalacturonase secreted by yeasts has been the sole pectinolytic enzyme identified in cocoa fermentation. However, we recently reported the involvement of pectate lyases produced by Bacillus strains in the cocoa fermentation process (26).
Here, we report the biochemical and molecular properties of purified pectate lyases from three different Bacillus strains isolated from fermenting cocoa beans and the characterization of their cloned genes.
MATERIALS AND METHODS
Organisms, culture conditions, and enzyme production.
The three strains used here had been previously screened as Pel producers from a large population of Bacillus strains isolated from fermenting cocoa beans (26) and identified as Bacillus fusiformis (BS90), Bacillus subtilis (BS66), and Bacillus pumilus (BS22). For enzyme production, the strains were grown in 500 ml of medium prepared as follows: 450 ml basal mineral medium containing 2.8 g/liter (NH4)2SO4, 6 g/liter K2HPO4, 2 g/liter KH2PO4, 0.8 g/liter citrate, 0.2 g/liter yeast extracts, 0.1 g/liter MgSO4, and 1 g/liter CaCl2, plus 50 ml of LB medium to promote bacterial growth. This medium was supplemented with polygalacturonic acid (PGA) (2 g/liter) and adjusted to pH 6.8. Cultures were incubated at 30°C for 48 h with shaking at 150 rpm. After centrifugation at 6,000 × g for 10 min, the cell supernatants were used for enzyme purification.
Enzyme purification and protein analysis.
All steps for purification were performed at a temperature below 5°C. The cell supernatants were adjusted to 1 mM dithiothreitol (DTT), and the resulting extracts were subjected to ammonium sulfate fractionation. Solid ammonium sulfate was gradually added to the clarified supernatant fluid at 20, 40, 55, 70, 90, and 100% saturation, with gentle stirring in an ice bath. The resulting fractions were collected by centrifugation at 12,000 × g for 15 min and dissolved in small volumes of buffer A (20 mM Tris-HCl, pH 7.2, 1 mM EDTA, and 1 mM DTT). The 90% and 100% ammonium sulfate fractions containing Pel activity were pooled and dialyzed overnight against buffer A, using a 14,000-Da filtration membrane. The preparations were then applied to an AP-1 carboxyl methyl cation-exchange column (100 by 10 mm; Water, France), previously equilibrated with buffer A. The column was washed extensively with the same buffer, and the retained proteins were eluted at 1.4 ml/min with an increasing NaCl linear gradient, from 0 to 1 M, using a Water high-pressure liquid chromatography (HPLC) system. Fractions (0.7 ml each) were collected and assayed for Pel activity. The purity of the fractions containing Pel activity was estimated by SDS-PAGE (22). Protein bands were detected by Coomassie blue staining. Protein concentration determination was carried out as proposed by Bradford (8).
Pel assay and thin-layer chromatography (TLC).
Pel activity was determined spectrophotometrically (Uvikon spectrophotometer; Kontron) by the release from polygalacturonate or pectin of unsaturated oligogalacturonides that absorb at 230 nm. One unit of Pel activity was defined as the amount of enzyme that liberated 1 micromole of product per min at 37°C under assay conditions. Specific activity was expressed as enzyme units per milligram of protein. Unless otherwise specified, the standard assay mixture consisted of 100 mM Tris-HCl (pH 8.0 for Pel-66 and Pel-90 and pH 7.5 for Pel-22), 0.1 mM CaCl2, and 1 g/liter of polygalacturonate in a total volume of 1 ml. The products released from polygalacturonate were continuously monitored at 37°C for 2 min. The molar extinction coefficient of unsaturated oligogalacturonides was assumed to be 5,200 (23). The influences of divalent cations (Ba2+, Ca2+, Cu2+, Fe2+, Mg2+, Mn2+, Ni2+, Se2+, and Zn2+) on Pel activity were determined with 0.1 mM concentrations of the corresponding chloride or sulfate form. The influence of Ca2+ and Fe2+ concentrations on Pel activity was investigated by the addition of CaCl2 and FeSO4 at concentrations ranging from 0 to 1 mM. To verify the Ca2+ requirement, EDTA was added to a final concentration of 1 mM to chelate the endogenous cations. The optimum pH was determined by using 100 mM (each) of the following buffers: acetate (pH 3.0 to 6.0), PIPES [piperazine-N-N′-bis(2-ethanesulfonic acid)-HCl] (pH 6.0 to 7.5), Tris-HCl (pH 6.5 to 9.5), and glycine-NaOH (pH 8.0 to 10.5). The effect of temperature was monitored within the range of 30 to 70°C. The influence of substrate methoxylation on Pel activity was tested by evaluating the activities of the purified enzyme on polygalacturonic acid and on pectins at increasing degrees of esterification (from Copenhagen Pectin).
For Km and maximum rate of metabolism (Vmax) determinations, enzymes were incubated at 60°C with polygalacturonate or pectin (45% methoxylation) at concentrations ranging from 0.025 to 1 g/liter.
TLC was performed as described by Soriano et al. (40) using an aluminum chromatogram sheet (Merk, France) and concentrations of 5 U/ml of the different enzymes.
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Escherichia coli strains were usually grown in LB liquid or solidified agar (15 g/liter) medium at 37°C. When required, ampicillin or chloramphenicol was added, as the antibiotic, at 50 μg/ml. Genomic-DNA preparation from Bacillus strains was performed as described by Rodriguez and Tait (30) and Nasser et al. (24). DNA manipulation was carried out according to the method of Sambrook et al. (32).
Construction of the genomic library and screening of clones producing pectate lyases.
Chromosomal DNA was extracted from the three Bacillus strains and partially digested with the restriction endonuclease Sau3A. The random fragments obtained were separated on 0.5% agarose gels. Fragments of 3 to 6 kb were recovered by electroelution using a biotrap apparatus (Schleicher and Schuell) and ligated to the plasmid vector pUC18, which had been digested to completion with the endonuclease BamHI and dephosphorylated. The resulting library was used to transform E. coli NM522 cells by electroporation, and the recombinant clones were selected on agar plates containing ampicillin, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and IPTG (isopropyl-β-d-thiogalactopyranoside). Recombinant clones were then screened for Pel activity as described by Keen et al. (18).
Sequencing of the pel genes.
The pel genes were sequenced on a double-stranded DNA template by MWG Eurofins Operon (Germany). The resulting data were analyzed using the free software Serial cloner 2. The Pels from B. pumilus BS22, B. subtilis BS66, and B. fusiformis BS90 were named Pel-22, Pel-66, and Pel-90, respectively.
Nucleotide sequence accession numbers.
The DNA sequences of the pel genes were deposited in DDBJ GenBank and appear under the following accession numbers: pel-90 gene, GU576909; pel-66 gene, GU576910; and pel-22 gene, GU576911.
RESULTS
Enzyme purification.
Pels were precipitated at between 90 and 100% ammonium sulfate saturation and further purified by chromatography on a cation-exchange column. The amount of Pel recovered from 500 ml of culture varied between 0.1 and 0.4 mg depending on the production level and the final optical density at 600 nm (OD600) of the culture. SDS-PAGE analysis showed satisfactory purity, and the apparent molecular masses were estimated to be 42 kDa for Pel-66 and Pel-90 and 34 kDa for Pel-22 (data not shown).
Influence of pH and temperature.
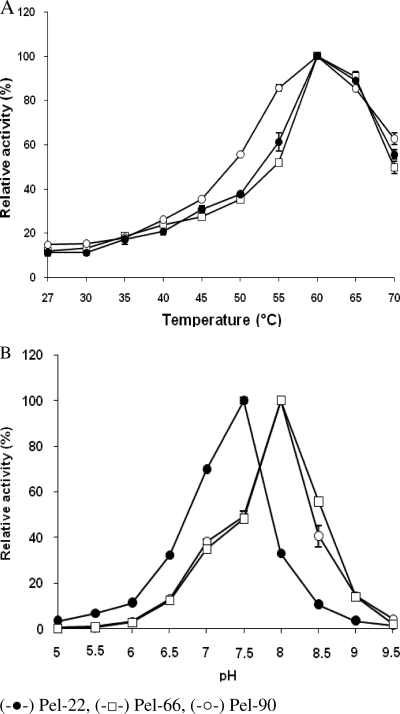
All the Pels studied showed activity over a wide range of temperatures, from 30°C (15% activity) to 70°C (50% activity), with maximum activity at 60°C (Fig. 1). These enzymes proved to be very stable at room temperature, where they lost only 5% of their activity after a 15-day incubation in their respective optimum pH buffers without PGA. Under standard conditions, the half-times of thermoinactivation at 60°C were approximately 30 min for Pel-22 and at least 27 min for Pel-66 and Pel-90. The three enzymes rapidly lost their stability above 70°C. At that temperature, Pel-22 lost 67% of its activity within 5 min, while in the same period, Pel-66 and Pel-90 lost 73 and 78% of their activities, respectively.
FIG. 1.
Effects of temperature (A) and pH (B) on Pel activity. Enzyme activities were measured using a standard reaction mixture containing 1 g/liter PGA as the substrate. The effect of pH was checked at a constant temperature of 37°C. The effect of temperature was studied at the constant optimum pH of each Pel (8 for Pel-66 and Pel-90 and 7.5 for Pel-22). Each result reported is the average of three independent experiments, and the error bars indicate the standard deviations.
Pel-22 showed an optimum level of activity at pH 7.5, while Pel-66 and Pel-90 displayed their highest activities at pH 8.0 (Fig. 1). Although the pH ranging from 3.0 to 10.0 did not significantly influence the stability of enzymes, these enzymes seem to be more stable at their optimum pHs.
Kinetic parameters.
The initial velocity for enzymatic reactions was determined using PGA and 45% esterified pectin, at different concentrations, as substrates. Pel-22 showed maximum activity at 0.5 g/liter of PGA and 0.1 g/liter of pectin. For Pel-66 and Pel-90, maximum activity occurred at 0.5 g/liter for both PGA and pectin. The kinetic parameters of purified Pels on 45% esterified pectin and PGA were determined, and the apparent Vmax and Km values are given in Table 1. The affinity of Pel-22 for both substrates was approximately 2.5-fold higher than those of Pel-66 and Pel-90. In contrast, Pel-66 and Pel-90 degraded both substrates more efficiently than Pel-22.
TABLE 1.
Km and Vmax values for each Pel with polygalacturonate and pectin (45% esterification) as substratesa
| Pel | PGA |
Pectin |
||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| Pel-22 | 0.045 | 1.41 | 0.043 | 1.45 |
| Pel-66 | 0.1 | 667 | 0.1 | 714 |
| Pel-90 | 0.125 | 909.9 | 0.111 | 1136.36 |
The initial velocity for the reactions was determined at different polygalacturonate and pectin concentrations under standard conditions. Lineweaver-Burk transformation of the data gave Km (g/liter) and Vmax (μmol/min/mg) values for each enzyme.
Effects of divalent cations.
Most of the characterized pectate lyases are known to have an absolute requirement for Ca2+ as the cofactor. The addition of 1 mM EDTA to the reaction mixture totally inhibited the three purified Pels, demonstrating their absolute requirement for cations as cofactors. The influence of Ca2+ was next investigated by adding CaCl2 at concentrations ranging from 0 to 1 mM at the optimal pHs of the different enzymes.
In the presence of PGA as a substrate, similar results were obtained for the three enzymes. The maximal activity was observed at Ca2+ concentrations of about 0.1 mM, which provoked an increase of enzyme activity of more than 100%. In the range of 0.05 to 0.2 mM Ca2+ concentrations, the rise in Pel activity was more than 75%, but enzyme activity dropped sharply below 0.05 mM and above 0.2 mM Ca2+ (Table 2 ).
TABLE 2.
Influence of calcium and iron concentrations on Pel activities using PGA and 45% esterified pectin as substrates
| Substrate | Cation concn (mM) | Enzyme activity (%) |
|||
|---|---|---|---|---|---|
| Pel-22 |
Pel-66 (Ca2+) | Pel-90 (Ca2+) | |||
| Ca2+ | Fe2+ | ||||
| PGA | 0 | 100 (9.86) | 100 (6.33) | 100 (16.18) | 100 (12.03) |
| 0.01 | 161.66 (8.33) | 163.79 (15.9) | 149.2 (7.19) | 155.47 (20.39) | |
| 0.02 | 168.33 (10.10) | 180.33 (5.18) | 143.45 (5.6) | 176.58 (0.82) | |
| 0.05 | 185 (14.50) | 193.62 (8.78) | 202.79 (7.32) | 210.33 (8.31) | |
| 0.1 | 213 (11) | 235.75 (6.34) | 231.38 (1.15) | 261.38 (13.61) | |
| 0.2 | 178.33 (13.16) | 185.63 (6.42) | 187.99 (2.53) | 193.5 (15.29) | |
| 0.5 | 141.66 (8.80) | 143.31 (12.26) | 138.42 (1.21) | 108.54 (12.11) | |
| 1 | 121.66 (4.80) | 121.36 (9.52) | 38.13 (1.5) | 32.98 (3.96) | |
| 45% esterified pectin | 0 | 100 (5.89) | 100 (12.56) | 100 (14.32) | 100 (8.12) |
| 0.01 | 149.76 (12.73) | 158.12 (7.32) | 142.36 (3.4) | 141.79 (2.94) | |
| 0.02 | 178.35 (15.39) | 185.7 (6.1) | 162.31 (3.8) | 178.64 (18.32) | |
| 0.05 | 213.54 (13.54) | 228.18 (8.18) | 211.58 (2.4) | 215.54 (1.91) | |
| 0.1 | 266.8 (9.02) | 279.67 (4.87) | 272.28 (7.99) | 251.42 (11.27) | |
| 0.2 | 271.01 (10.64) | 281.32 (4.12) | 344.71 (5.28) | 283.67 (1.93) | |
| 0.5 | 274.25 (15.74) | 272 (4.8) | 361.13 (7.88) | 304.59 (14.44) | |
| 1 | 274.34 (9.49) | 259.65 (1.25) | 375.84 (12.46) | 369.33 (14.79) | |
Activity under standard conditions (100 mM Tris-HCl, 1 g/liter PGA, and pH 7.5 or 8, or 100 mM Tris-HCl, 1 g/liter pectin [45% esterification], and pH 7.5 or 8) without any added cation was used as a reference (100%). The influence of the cation concentration was determined by using the corresponding chloride or sulfate salt from 0 to 1 mM. Each result reported is the average of three independent experiments, and the standard deviations (±) are indicated in parentheses.
When 45% methylated pectin was used as the substrate, inhibition of enzyme activity due to high Ca2+ concentration did not occur and the maximal activity was observed within a large range of Ca2+ (from 0.2 to 1 mM) for the three Pels (Table 2).
The influences of several other cations (Fe2+, Cu2+, Zn2+, Mg2+, Mn2+, Ni2+, Ba2+, and Se2+) were tested using a concentration of 0.1 mM. It appeared that all the tested cations could activate Pel-22 (Table 3). However, the best cofactors for this enzyme were Fe2+ and Ca2+, with relative activities on PGA of 231 and 208%, respectively. The best concentration of Fe2+ for Pel-22 was 0.2 mM, with 45% methylated pectin as a substrate.
TABLE 3.
Effects of divalent cations on Pel activitiesa
| Divalent cation | Enzyme activity (%) |
||
|---|---|---|---|
| Pel-22 | Pel-66 | Pel-90 | |
| None | 100 (5.53) | 100 (3.16) | 100 (4.72) |
| Ca2+ | 208.48 (0.28) | 237.16 (16.64) | 262.59 (24.52) |
| Fe2+ | 231.42 (1.91) | 94.58 (4.1) | 100.3 (0.30) |
| Cu2+ | 193.27 (1.46) | 99.84 (3.41) | 119.87 (12.51) |
| Zn2+ | 132.18 (2.6) | 1.9 (0.71) | 2.69 (0.98) |
| Mg2+ | 190.39 (16.62) | 77.22 (0.75) | 91.59 (3.86) |
| Mn2+ | 135.74 (4.6) | 15.96 (1.68) | 17.48 (2.14) |
| Ni2+ | 190.92 (7.31) | 68.71 (3.83) | 100.61 (15.95) |
| Ba2+ | 180.97 (13.76) | 62.17 (2.7) | 79.04 (7.88) |
| Se2+ | 157.52 (3.13) | 82.26 (13.81) | 89.92 (6.39) |
Activity under standard conditions (100 mM Tris-HCl, 1 g/liter PGA, and pH 7.5 or 8) without any added cation was used as a reference (100%). The influence of each divalent cation was determined by using the corresponding chloride or sulfate salt at 0.1 mM. Each result reported is the average of three independent experiments, and the standard deviations (±) are indicated in parentheses.
In contrast, Fe2+and Cu2+ did not significantly affect the activities of Pel-66 and Pel-90. Mg2+, Ni2+, Ba2+, and Se2+ all moderately inhibited these two Pels, whereas stronger inhibition of Pel-66 and Pel-90 was observed with Mn2+ and Zn2+. Among these cations, Zn2+ was the strongest inhibitor of Pel-66 and Pel-90, since it almost completely inhibited the activities of the enzymes (Table 3).
Influence of substrate methoxylation on Pel activity.
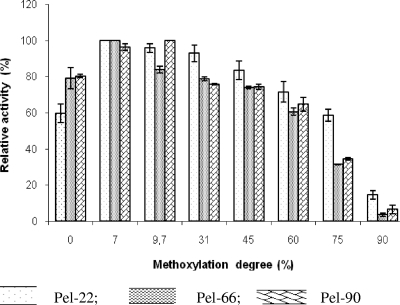
Pels were strongly active on pectin, with a degree of methoxylation up to 60% (Fig. 2). With 75% pectin methoxylation, Pel-66 and Pel-90 retained approximately 40% of their activity while Pel-22 activity remained higher (more than 75% activity). On 90% pectin esterification, the residual activities of Pel-66 and Pel-90 were around 5% and that of Pel-22 was 14%.
FIG. 2.
Influence of the degree of pectin esterification on Pel activity. The different pectins were added to the standard reaction mixture at 1 g/liter. Enzyme activities were measured at 37°C. Assays were done in triplicate, and the error bars indicate the standard deviations. Zero percent methoxylation corresponds to PGA.
Analysis of reaction products.
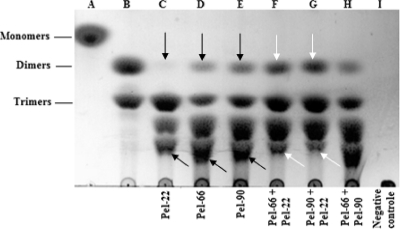
To determine precisely the modes of action of the Pels, the reaction products obtained from PGA were characterized by TLC; enzymes were used at 5 U/ml. All the Pels studied were found to release a mixture of reaction products composed of dimers, trimers, and two unsaturated oligogalacturonic acid compounds. The major products liberated by Pel-22 were trimers, while Pel-66 and Pel-90 released relatively large amounts of both oligogalacturonic acid compounds and trimers (Fig. 3). Dimers remained the minor product liberated by each Pel. However, the amount of dimers liberated by Pel-22 was extremely small compared with those produced by Pel-66 and Pel-90.
FIG. 3.
Analysis of the different end products of Pel activities. Enzymes were added to the standard reaction mixture at a concentration of 5 U/ml, and the preparations were incubated at 45°C for 10 h. For a combination of two Pels, 2.5 U/ml of each enzyme was used. Twenty microliters of reaction products was run on the chromatogram sheet (Merck, France). Lanes A and B are the standards. Lane A, galacturonic acid; lane B, di- and trigalacturonic acids. The negative control was prepared under the same conditions as the assays, except that no enzyme was added to the reaction mixture. The arrows indicate the end products of the synergistic action. Blacks arrows, individual action; white arrows, combined action.
A combination of Pel-22 and Pel-66 (each at 2.5 U/ml) provoked the degradation of the long unsaturated oligogalacturonic acid compounds, leading to an increase of dimers in the reaction product mixture compared with that observed in the presence of each enzyme individually (Fig. 3). A similar result was obtained with a combination of Pel-22 and Pel-90. The profile of the chromatogram obtained from the combined actions of Pel-90 and Pel-66 did not significantly change compared with profiles of the individual enzymes.
Cloning and characterization of the gene encoding Pel-66.
The strategy used to isolate the B. subtilis BS66 pel gene was the selection of recombinant plasmids exhibiting Pel activity on the polygalacturonate agar plates, as described by Keen et al. (18). Among at least 4,000 E. coli transformants, one clone showing Pel activity was detected. This plasmid, named pGN66, contained a DNA fragment insert of 3.8 kb. Restriction mapping of the insert enabled us to subclone the 1.8-kb HindIII-HindIII fragment harboring the pel gene (data not shown) into a pBluescript plasmid to give pGN66-1. The complete nucleotide sequence of this fragment was then determined. The sequence contains an open reading frame (ORF) of 1,260 bp that starts with an ATG codon at nucleotide 402 and ends with a TAA termination codon at nucleotide 1664. At 8 nucleotides upstream of the start codon, there is a purine-rich sequence (AGAAAA) that could be a ribosome binding site (RBS). Putative −35 (TTGCTA) and −10 (TGATAAATT) promoter signals were found upstream of the Pel-66 structural gene. An inverted-repeat sequence of 14 bp that may be a transcriptional terminator loop was found 7 nucleotides downstream of the termination codon of the pel-66 gene.
Characterization of the gene encoding Pel-90.
An attempt to clone the pel-90 gene from the genomic-DNA library of B. fusiformis was unsuccessful because of the instability of the recombinant plasmids displaying Pel activity. To avoid this problem, we PCR amplified the pel-90 gene using genomic DNA of B. fusiformis BS90 as a template and two primers (Forward, 5′-AAGCTTGGGCATAAAGCAAGG-3′; Reverse, 5′-TACTGCTGACTGTTTCCTGC-3′) designed from the pel-66 gene. This strategy was retained because Pel-66 and Pel-90 displayed similar biochemical properties and molecular weights. A 1.55-kb fragment was successfully amplified from the BS90 genome. To verify whether the 1.55-kb amplified fragment contained the pel-90 gene, the PCR products were ligated into a pGEMT vector (Promega, France), giving rise to the plasmid pGN90 (data not shown). E. coli NM522 competent cells transformed with this plasmid exhibited Pel activity. The nucleotide sequence of the pel-90 gene also revealed an ORF of 1,260 bp, starting with an ATG codon at position 152 and ending with a TAA codon at position 1414. The pel-90 gene showed 98% identity to the pel-66 gene, presenting the same purine-rich and inverted sequences. These two genes showed high identity with the nucleotide sequence of the Pel gene from B. subtilis SO113 (EMBL accession number X74880).
Characterization of the gene encoding Pel-22.
An attempt to amplify the pel-22 gene from the genomic DNA of B. pumilus BS22, using the same primers described above, was unsuccessful. Consequently, the primers F (5′-GCTTCCTAGAAAATCATGGAATACG) and Rev F (5′-TTAAGGGTTTACTTTTCCAACACCTGCATTTG-3′) were designed from the pel gene sequences of B. pumilus DKS1 and SAFR-032 (EMBL accession numbers ACD11362 and ABV64163, respectively) and of Bacillus sp. KSM-P103 (NCBI accession number AB015044.1), retrieved from the EMBL and NCBI databases. A 1,250-kb DNA fragment was successfully amplified with these two primers. The amplified fragment was ligated into a pGEMT vector (Promega, France) to give the plasmid pGN22 (data not shown). This construct was subsequently used to transform E. coli competent cells, leading to clones exhibiting Pel activity. The nucleotide sequence of the pel-22 gene showed an ORF of 1,062 bp, which starts with an ATG codon at nucleotide 142 and ends with a TGA termination codon at nucleotide 1,206.
The nucleotide sequence of the pel-22 gene displays 90% identity to that of the pel-103 gene from Bacillus sp. KSM-P103 (NCBI accession number AB015044.1) and 80% identity with the pel gene from Bacillus sp. KSM-7 (NCBI accession number AB015043.1).
Amino acid sequence analysis.
The ORFs of the Pel-66 and Pel-90 genes encode polypeptides of 420 amino acids with calculated molecular masses of 45.81 kDa and 45.52 kDa, respectively. The mature Pel-66 and Pel-90 both have a molecular mass of 42 kDa, as shown by SDS-PAGE. The difference between the calculated molecular masses and those found in SDS-PAGE experiments indicated that the three Pels are produced as precursors with a signal sequence at the NH2 extremity, which is consistent with their extracellular localization. The two proteins show 97% identity. The pel-22 gene encodes a polypeptide of 353 amino acids, with a calculated molecular mass of 38.9 kDa, while the mature protein has a molecular mass of 34 kDa. This protein exhibited 25% and 26% identity with Pel-66 and Pel-90, respectively. The deduced amino acid sequences of the cloned enzymes were compared with protein sequences in the Swissprot and EMBL databases. All the Pels studied appeared to have identity with pectate lyases belonging to the polysaccharide lyase Family 1 (see Fig. S1 in the supplemental material). However, Pel-66 and Pel-90 were closer to the Pel from B. subtilis SO113 (24), with more than 98% identity, while Pel-22 appeared to be similar to Pel-103 from Bacillus sp. KSM-P103 (12), with 95% identity, and Pel-7 from Bacillus sp. KSM-P7 (21), with 82% identity.
Among the conserved regions, Pel-66 and Pel-90 showed residues Asp-185, Asp-224, and Asp-229 in positions identical to that of the mature Pel from B. subtilis SO113. These residues were shown to be involved in the Ca2+ binding site (27). In contrast, these Asp residues do not seem to be conserved in Pel-22, which is consistent with the difference between Pel-66/Pel-90 and Pel22 in regard to the cofactors required.
DISCUSSION
Here, we report the biochemical characterization of Pels from the three main pectinolytic Bacillus strains isolated from fermenting cocoa beans: B. fusiformis (BS90), B. subtilis (BS66), and B. pumilus (BS22) (26). The genes encoding these enzymes were further cloned and characterized. Pel-22, Pel-66, and Pel-90 proved to be thermophilic, with a high level of activity at 60°C. This is not surprising, since most of the Pels from Bacillus have been reported to show maximum activity at temperatures between 60 and 70°C (12, 17, 21, 40). Furthermore, the Pels studied present strong stability at room temperature in a range of pH 3 to 10. Thus, these enzymes appear to have the same robustness as the Pel from B. subtilis SO113 (25) and PelB and PelC from Erwinia chrysanthemi 3739 (42). The high content of asparagine residues found in these Pels (15 to 18%), may be involved in the structural integrity of the proteins.
The optimum pH displayed by Pel-66 and Pel-90 (pH 8.0) is similar to those reported for the Pels from Bacillus sp. strain DT7 (17), B. subtilis SO113 (pH 8.4) (25), and B. pumilus BK2 (pH 8.5) (20). In contrast, Pel-22 showed maximum activity near neutral pH (7.5). To date, a microbial Pel has rarely been found with this feature, since most microbial Pels studied have exhibited optimum pHs in the range from 8.5 to 11.5 (2, 17, 20, 21, 25, 40, 42).
Another trait of Pel-66 and Pel-90 that is similar to the previously characterized enzymes from bacilli is their absolute requirement for Ca2+ as a cofactor. Ca2+ is known to be the relevant cofactor required by almost all Pels for catalytic activity (9, 40, 42). An exception, however, is PelZ, an extracellular pectate lyase from E. chrysanthemi, which uses Mn2+ as a cofactor more efficiently than it uses Ca2+ (28). Furthermore, the cytoplasmic enzymes PelW and Ogl from the same bacterium also use various cations (Co2+, Mn2+, or Ni2+) as cofactors (37). However, one of the atypical features of Pel-22 was that it appeared to use a wide range of divalent cations as cofactors, with iron proving to be the best. To the best of our knowledge, this is the first time that iron has acted more efficiently than Ca2+ or Mn2+ as a cofactor on a Pel produced by a bacterium. Moreover, the well-characterized calcium binding site (27) found in Pel-66 and Pel-90 was not present in Pel-22, indicating that this Pel has a cation binding site that remains to be well characterized.
One of the most notable features of the characterized Pels is their activities on both polygalacturonic acid and highly methylated pectin. Until now, most of the reported Pels have shown maximum activity on polygalacturonic acid or on low-methylated pectins (7 to 22%) (9, 19, 38, 39, 40). However, some Pels in Family 3, such as PelI from E. chrysanthemi and PelB from Erwinia carotovora, exhibit maximum activity on 45% and 68% esterified pectin, respectively (13, 37). The enzymes studied here display similar activities for PGA and pectin methylated up to 60% (for Pel-66 and Pel-90) or 75% (for Pel-22). Furthermore, on 90% methylated pectin, Pel-66 and Pel-90 retained 5% of their activity while Pel-22 retained 14%. These results regarding substrate specificity are comparable to those reported by Soriano et al. (39, 40) on PelA and PelC from Paenibacillus barcinonensis and B. subtilis, respectively. These two enzymes showed significant activity on pectin with up to 90% esterification. However, it is important to note that, in this case, the experiments were performed at pH 10 instead of at a pH near neutrality (pH 7.5 in our case). In fact, the rise in pH from 7.5 to 8.5 increased by 4-fold the activity of the studied Pels on 90% esterified pectin (see Table S2 in the supplemental material). It is known that alkaline pH de-esterifies the pectin compounds by a saponification mechanism, which leads to a considerable reduction in their degree of esterification (9). Hence, it is possible that the pH 10 used by Soriano et al. (39, 40) artificially increased the activity of Bacillus PelA and PelC on highly esterified pectins. The Pels that we have characterized, particularly Pel-22, displayed a specific trait among the pectate lyases characterized to date in regard to their actions on a large range of substrates, including highly methylated pectins.
Analysis of the mode of action on polymeric substrates showed that Pel-22, Pel-66, and Pel-90 have an endo-type mode of action, producing a mixture of degradation products. In contrast, an exo-cleaving enzyme lyase would catalyze the formation of a single product, unsaturated digalacturonate (29). This property is undoubtedly interesting, since endo-Pels, in general, display a higher maceration capacity than exo-Pels. Accordingly, the endo-type mode of action is the main mechanism used by most of the characterized Pels (1, 23, 31, 38).
The major degradation products obtained from Pel-66 and Pel-90 were unsaturated oligomers, tri- and digalacturonic acids. These products are, in part, similar to those generated by the Pels from B. subtilis WSHB04-02 (44) and Bacillus sp. RN1 (41). Indeed, these two enzymes release essentially unsaturated di- and trigalacturonic acids. In contrast, unsaturated trigalacturonic acid was the major product liberated by Pel-22, like the end product of PelA from Bacillus licheniformis 14A (2). On the other hand, the fact that the combined actions of Pel-22 and Pel-66, or Pel-22 and Pel-90, resulted in a reduced amount of the long oligomers and an increased amount of dimers strongly supports a synergistic effect between these enzymes. Further investigations should clarify whether these observed synergies on PGA are relevant to cocoa pulp.
To summarize, we have purified and characterized three pectate lyases from Bacillus strains isolated from fermented cocoa which display original features in regard to their high activity on a large range of substrates, including highly methylated pectin. Based on their amino acid sequences and their biochemical properties, the three enzymes were clustered into two groups. Pel-66 and Pel-90 are very similar to each other: they both have a relatively low affinity for substrates, a high specific activity, and an alkaline optimum pH; they absolutely require Ca2+ as a cofactor; and they have the same molecular mass. As regards these properties, they behave like most of the previously characterized Pels from the genus Bacillus. On the other hand, Pel-22 has a low specific activity, a high affinity for its substrates, and an optimum pH near neutrality, and it uses a large variety of divalent cations as cofactors, among which Fe2+ appeared to be the best. This last characteristic makes Pel-22 interesting, since it constitutes the first Pel found to use Fe2+ as the preferential cofactor. Furthermore, it appeared that the enzymes from these two groups cooperate to efficiently degrade the substrates. The synergistic effect observed is of great concern in the choice of microbial starter strains for cocoa fermentation improvement. Regarding their biochemical properties, the three Pels should be able to take important places in the process of depectinization of the pulp during cocoa fermentation. Crystallography studies on Pel-22 might give new insights into the mechanisms of activation of a Pel by various divalent cations, including Ca2+ and Fe2+.
Supplementary Material
Acknowledgments
We thank Janine Robert-Baudouy and the members of the Erwinia group, particularly Nicole Cotte-Pattat, Vladimir Shevchik, and Guy Condemine, for their helpful discussions and advice. We thank G. Effantin for technical assistance.
This research was supported by the International Foundation for Science, Sweden, under grant E/4411-1.
Footnotes
Published ahead of print on 11 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barras, F., F. Van Gijsegem, and A. K. Chatterjee. 1994. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu. Rev. Phytopathol. 32:201-234. [Google Scholar]
- 2.Berensmeier, S., S. A. Singh, J. Meens, and K. Buchholz. 2004. Cloning of the pelA gene from Bacillus licheniformis 14A and biochemical characterization of recombinant, thermostable, high alkaline pectate lyase. Appl. Microbiol. Biotechnol. 64:560-567. [DOI] [PubMed] [Google Scholar]
- 3.Bhumibhamon, O., and J. Jinda. 1997. Effect of enzymes pectinases on natural cocoa fermentation. Kasetsart J. Nat. Sci. 31:206-212. [Google Scholar]
- 4.Biehl, B., and J. Voigt. 1996. Biochemistry of cocoa flavour precursors, p. 929-938. In Proceedings of the 12th International Cocoa Research Conference, Salvador, Brazil. Cocoa Producers' Alliance, Lagos, Nigeria.
- 5.Biehl, B., J. Heinrichs, G. Voigt, G. Bytof, and P. Serrano. 1996. Nature of proteases and their action on storage proteins in cocoa seeds during germination as compared with fermentation, p. 18-23. In Proceedings of the 12th International Cocoa Research Conference, Salvador, Brazil. Cocoa Producers' Alliance, Lagos, Nigeria.
- 6.Biehl, B., J. Heinrichs, H. Ziegeler-Berghausen, S. Srivastava, Q. Xiong, D. Passern, V. I. Senyuk, and M. Hammoor. 1993. The proteases of ungerminated cocoa seeds and their role in the fermentation process. Angew. Bot. 67:59-65. [Google Scholar]
- 7.Biehl, B., J. Voigt, H. Heinrichs, V. Senjuk, and G. Bytof. 1993. pH-dependent enzymatic formation of oligopeptides and amino acids, the aroma precursors in raw cocoa beans, p. 717-722. In J. Lafforest (ed.), Proceeding of the XIth International Cocoa Research Conference. Cocoa Producers' Alliance, Yamassoukro, Ivory Coast.
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Brown, I. E., M. H. Mallen, S. J. Charnock, G. J. Davies, and G. W. Black. 2001. Pectate lyase 10A from Pseudomonas cellulosa is a modular enzyme containing a family 2a carbohydrate-binding module. Biochem. J. 355:155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. Royal Society of Chemistry, Cambridge, United Kingdom.
- 12.Hatada, Y., N. Higaki, K. Saito, A. Ogawa, K. Sawada, T. Ozawa, Y. Hakamada, T. Kobayashi, and S. Ito. 1999. Cloning and sequencing of a high alkaline pectate lyase gene from an alkaliphilic Bacillus isolate. Biosci. Biotechnol. Biochem. 63:998-1005. [DOI] [PubMed] [Google Scholar]
- 13.Heikinheimo, R., D. Flego, M. Pirhonen, M. B. Karlsson, A. Eriksson, A. Ma'e, V. Koiv, and E. T. Palva. 1995. Characterization of a novel pectate lyase from Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 8:207-217. [DOI] [PubMed] [Google Scholar]
- 14.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213-257. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins, J., V. E. Shevchik, N. Hugouvieux-Cotte-Pattat, and R. W. Pickersgill. 2004. The crystal structure of pectate lyase Pel9A from Erwinia chrysanthemi. J. Biol. Chem. 279:9139-9145. [DOI] [PubMed] [Google Scholar]
- 16.Jinap, S. 1994. Organic acids in cocoa beans: a review. ASEAN Food J. 9:3-12. [Google Scholar]
- 17.Kashyap, D. R., S. Chandra, A. Kaul, and R. Tewari. 2000. Production, purification and characterization of pectinase from a Bacillus sp. DT7. World J. Microbiol. Biotechnol. 16:277-282. [Google Scholar]
- 18.Keen, N. T., D. Dahlbeck, B. Staskawicz, and W. Belser. 1984. Molecular cloning of pectate lyase genes from Erwinia chrysanthemi and their expression in Escherichia coli. J. Bacteriol. 159:825-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi, T., H. Shibuya, T. Aikawa, and J. T. Jones. 2006. Cloning and characterization of pectate lyases expressed in the esophageal gland of the pine wood nematode Bursaphelenchus xylophilus. Mol. Plant Microbe Interact. 19:280-287. [DOI] [PubMed] [Google Scholar]
- 20.Klug-Santner, B. G., W. Schnitzhofer, M. Vrsanska, J. Weber, P. B. Agrawal, V. A. Nierstrasz, and G. M. Guebitz. 2006. Purification and characterization of a new bioscouring pectate lyase from Bacillus pumilus BK2. J. Biotechnol. 121:390-401. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, T., Y. Hatada, N. Higaki, D. D. Lusterio, T. Ozawa, K. Koike, S. Kawai, and S. Ito. 1999. Enzymatic properties and deduced amino acid sequence of a high-alkaline pectate lyase from an alkaliphilic Bacillus isolate. Biochim. Biophys. Acta 1427:145-154. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Moran, F., S. Nasuno, and M. P. Starr. 1968. Extracellular and intracellular polygalacturonic acid trans eliminase of Erwinia carotovora. Arch. Biochem. Biophys. 123:298-306. [DOI] [PubMed] [Google Scholar]
- 24.Nasser, W., A. C. Awade, S. Reverchon, and J. Robert-Baudouy. 1993. Pectate lyase from Bacillus subtilis: molecular characterization of the gene, and properties of the cloned enzyme. FEBS Lett. 335:319-326. [DOI] [PubMed] [Google Scholar]
- 25.Nasser, W., F. Chalet, and J. Robert-Baudouy. 1990. Purification and characterization of extracellular pectate lyase from Bacillus subtilis. Biochimie 72:689-695. [DOI] [PubMed] [Google Scholar]
- 26.Ouattara, H. G., L. Ban-Koffi, G. T. Karou, A. Sangare, S. L. Niamke, and J. K. Diopoh. 2008. Implication of Bacillus sp. in the production of pectinolytic enzymes during cocoa fermentation. World J. Microbiol. Biotechnol. 24:1753-1760. [Google Scholar]
- 27.Pickersgill, R., J. Jenkins, G. Harris, W. Nasser, and J. Robert-Baudouy. 1994. The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat. Struct. Biol. 1:717-723. [DOI] [PubMed] [Google Scholar]
- 28.Pissavin, C., J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1996. Regulation of pelZ, a gene of the pelB-pelC cluster encoding a new pectate lyase of Erwinia chrysanthemi 3937. J. Bacteriol. 178:7187-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preston, J. F., J. D. Rice, L. O. Ingram, and N. T. Keen. 1992. Differential depolymerization mechanisms of pectate lyases secreted by Erwinia chrysanthemi EC16. J. Bacteriol. 174:2039-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez, R. L., and R. C. Tait. 1983. Recombinant DNA techniques: an introduction, p. 37-51. Benjamin/Cummings Publishing Company, Inc., Menlo Park, CA.
- 31.Roy, C., H. Kester, J. Visser, V. Shevchik, N. Hugouvieux-Cotte-Pattat, J. Robert-Baudouy, and J. Bennen. 1999. Modes of action of five different endopectate lyases from Erwinia chrysanthemi 3937. J. Bacteriol. 181:3705-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Schwan, R. F. 1998. Cocoa fermentation conducted with a defined microbial cocktail inoculum. Appl. Environ. Microbiol. 64:1477-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwan, R. F., A. H. Rose, and R. G. Board. 1995. Microbial fermentation of cocoa beans, with emphasis on enzymatic degradation of the pulp. J. Appl. Bacterial. Symp. Suppl. 79:96-107. [Google Scholar]
- 35.Schwan, R. F., and A. E. Wheals. 2004. The microbiology of cocoa fermentation and its role in chocolate quality. Crit. Rev. Food Sci. Nutr. 44:205-221. [DOI] [PubMed] [Google Scholar]
- 36.Schwan, R. F., M. R. Cooper, and A. E. Wheals. 1997. Endopolygalacturonase secretion by Kluyveromyces marxianus and other cocoa pulp-degrading yeasts. Enzyme Microb. Technol. 21:234-244. [Google Scholar]
- 37.Shevchik, V. E., H. C. M. Kester, J. A. E. Benen, J. Visser, J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1999. Characterization of the exopolygalacturonate lyase PelX of Erwinia chrysanthemi 3937. J. Bacteriol. 181:1652-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shevchik, V. E., J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1997. Pectate lyase PelI of Erwinia chrysanthemi 3937 belongs to a new family. J. Bacteriol. 179:7321-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soriano, M., A. Blanco, P. Dıaz, and F. I. Javier-Pastor. 2000. An unusual pectate lyase from a Bacillus sp. with high activity on pectin: cloning and characterization. Microbiology 146:89-95. [DOI] [PubMed] [Google Scholar]
- 40.Soriano, M., P. Diaz, and F. I. Javier-Pastor. 2006. Pectate lyase C from Bacillus subtilis: a novel endo-cleaving enzyme with activity on highly methylated pectin. Microbiology 152:617-625. [DOI] [PubMed] [Google Scholar]
- 41.Sukhumsiirchart, W., S. Kawanishi, W. Deesukon, K. Chansiri, H. Kawasaki, and T. Sakamoto. 2009. Purification, characterization, and overexpression of thermophilic pectate lyase of Bacillus sp. RN1 isolated from a hot spring in Thailand. Biosci. Biotechnol. Biochem. 73:268-273. [DOI] [PubMed] [Google Scholar]
- 42.Tardy, F., W. Nasser, J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1997. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J. Bacteriol. 179:2503-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voigt, J., B. Biehl, H. Heinrichs, S. Kamaruddin, G. G. Marsoner, and A. Hugi. 1994. In vitro formation of cocoa specific aroma precursors: aroma-related peptides generated from cocoa seed protein by cooperation of an aspartic endoprotease and a carboxydase. Food Chem. 49:173-180. [Google Scholar]
- 44.Zhuge, B., G. Du, W. Shen, J. Zhuge, and J. Chen. 2007. Efficient secretory expression of an alkaline pectate lyase gene from Bacillus subtilis in E. coli and the purification and characterization of the protein. Biotechnol. Lett. 29:405-410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.