Abstract
Retroviral integrases associate during the early viral life cycle with preintegration complexes that catalyze the integration of reverse-transcribed viral cDNA into the host chromosomes. Several cellular and viral proteins have been reported to be incorporated in the preintegration complex. This study demonstrates that transcription factor Yin Yang 1 binds to Moloney murine leukemia virus, human immunodeficiency virus type 1, and avian sarcoma virus integrases. The results of coimmunoprecipitation and in vitro pulldown assays revealed that Yin Yang 1 interacted with the catalytic core and C-terminal domains of Moloney murine leukemia virus and human immunodeficiency virus type 1 integrases, while the transcriptional repression and DNA-binding domains of the Yin Yang 1 molecule interacted with Moloney murine leukemia virus integrase. Immunoprecipitation of the cytoplasmic fraction of virus-infected cells followed by Southern blotting and chromatin immunoprecipitation demonstrated that Yin Yang 1 associated with Moloney murine leukemia virus cDNA in virus-infected cells. Yin Yang 1 enhanced the in vitro integrase activity of Moloney murine leukemia virus, human immunodeficiency virus type 1, and avian sarcoma virus integrases. Furthermore, knockdown of Yin Yang 1 in host cells by small interfering RNA reduced Moloney murine leukemia virus cDNA integration in vivo, although viral cDNA synthesis was increased, suggesting that Yin Yang 1 facilitates integration events in vivo. Taking these results together, Yin Yang 1 appears to be involved in integration events during the early viral life cycle, possibly as an enhancer of integration.
Following retroviral infection, reverse-transcribed viral cDNA is incorporated into preintegration complexes (PICs) with viral proteins. Some examples include the human immunodeficiency virus type 1 (HIV-1) PIC, with reverse transcriptase, integrase (IN), matrix, nucleocapsid, and Vpr proteins, and the Moloney murine leukemia virus (MoMLV) PIC, with reverse transcriptase, IN, and capsid proteins. PICs are high-molecular-mass nucleoprotein complexes (approximately 5,000 kDa) that are required for viral cDNA integration into the host genome (reviewed in reference 51). Depending on the virus type and cellular context, PICs or INs associate with several cellular proteins, such as barrier-to-autointegration factor (35; reviewed in reference 44), lamina-associated polypeptide 2α (52), lens epithelium-derived growth factor/transcription coactivator p75 (LEDGF/p75) (3, 5, 34), the SET complex (64), Ku80 (32), the human Polycomb group protein EED (embryonic ectoderm development gene product) (56), and Daxx (18). In addition, the HIV-1 virion contains integrase interactor 1 (INI1) (24, 69). Among these factors, a physical interaction has been reported between HIV-1 IN and INI1 (24, 69), EED (56), and LEDGF/p75 (3, 5, 34), as well as between avian sarcoma virus (ASV) IN and Daxx (18). It has recently been reported that numerous cellular proteins, including transcription factors, in addition to chromatin and RNA-binding proteins, potentially interact with MoMLV and HIV-1 INs (50).
Yin Yang 1 (YY1) is a sequence-specific DNA-binding transcription factor that is an ortholog of Drosophila melanogaster Pleiohomeotic (Pho) and Pho like (reviewed in reference 53). YY1 is ubiquitously expressed in all tissues and highly conserved between Xenopus laevis and humans (reviewed in reference 17). It is able to activate and repress gene expression under different cellular contexts (reviewed in reference 46) and interact with a wide variety of regulator proteins, including retinoblastoma protein (40), histone acetyltransferase (p300/CBP) (28), histone deacetylase (HDAC1 to -3) (66-68), Sp1 (29), TATA box binding protein (TBP), transcription factor IIB (TFIIB) (1), YY1AP (57), and RYBP (16). In particular, YY1 can directly and indirectly bind to HIV-1 and MoMLV long terminal repeat (LTR) sequences, thereby repressing viral expression (7, 15, 19; reviewed in reference 22); for derepression of HIV-1 expression, HIV-1 Tat protein counteracts HDAC1 recruitment by YY1 and LSF to its LTR (19).
In this study, we demonstrate that YY1 is able to physically interact with MoMLV, HIV-1, and ASV INs and that it may associate with the MoMLV PIC. In vitro assays of IN activity and in vivo assessment of viral cDNA integration efficiency in YY1-knockdown cells revealed that YY1 facilitates the events required for viral cDNA integration into the host chromosomes.
MATERIALS AND METHODS
Vectors.
Based on the NCBI database (GenBank accession number AF033811), the coding sequence of MoMLV IN (amino acids 2 to 408) was amplified from the gag-pol gene sequence in the pGP plasmid of a retrovirus packaging kit (Takara) by PCR with primers 5′-AATGGATCCGGAAAATTCATCACCCTACACC-3′ and 5′-AATCTCGAGGGGGGCCTCGC-3′ and then subcloned into pETBlue-2 (Novagen). ASV and HIV-1 INs were amplified by PCR from the pSRA2 (9) and pLP1 (Invitrogen) plasmids, respectively, as templates, with primers 5′-AAACCATGGCGCCCTTGAGAGAGGCTAAAGA-3′ and 5′-AAACTCGAGTGCAAAAAGAGGGCTCG-3′ for ASV and primers 5′-AAACCATGGCGTTTTTAGATGGAATAGATAAGGC-3′ and 5′-AAACTCGAGATCCTCATCCTGTCTACTTGC-3′ for HIV-1; the underlined sequences show the restriction enzyme recognition sites used for subcloning. The PCR fragments were subcloned into pETBlue-2.
For construction of the mammalian expression plasmid for MoMLV IN fused with the Flag epitope, MoMLV IN DNA from pETBlue2-MoMLV IN was amplified by PCR and ligated into the pFlag-CMV-2 plasmid (Sigma) at the EcoRI/BamHI site. Deletion mutants of Flag-tagged MoMLV IN were also constructed by PCR, and the amplified DNA fragments were inserted into the pFlag-CMV-2 plasmid. For construction of expression plasmids for glutathione S-transferase (GST) fusion proteins of MoMLV, HIV-1, and ASV INs, PCR-amplified IN DNA fragments were inserted into pGEX-6P-2 or -5X-2 (GE Healthcare). The C209A MoMLV IN mutant (55) was generated by PCR-based site mutagenesis using pETBlue-2-MoMLV IN as the template with primers 5′-TGGAAATTACATGCTGCATACAGACCC-3′ and 5′-GGGTCTGTATGCAGCATGTAATTTCCA-3′; the underlined sequences show the mutated nucleotides.
Human YY1 was amplified from cDNA of C33A cells, and its coding sequence was inserted into pGEX-6P-2 for expression as a GST fusion protein and into pcDNA4 (Invitrogen). His-tagged deletion mutants were generated from full-length human YY1 DNA by inserting the PCR-amplified fragments into pcDNA4/His C. The mutant DNA fragments were also subcloned into pGEX-6P-2.
Purification of GST-INs and preparation of antibodies.
Escherichia coli BL21(DE3) containing the GST-MoMLV or GST-HIV-1 (amino acids 213 to 288) IN expression plasmid was grown in LB medium to an optical density at 600 nm (OD600) of 0.5 for MoMLV IN and an OD600 of 0.6 for HIV-1 IN. Then, 1 mM isopropyl-β-d-thiogalactoside was added, and E. coli was cultured at 25°C for 6 h for MoMLV and at 30°C for 6 h for HIV-1 INs. For the purification of MoMLV IN, the cells were suspended in phosphate-buffered saline containing 1 μg/ml each of aprotinin, pepstatin, and leupeptin and 100 μg/ml phenylmethylsulfonyl fluoride before being sonicated. For HIV-1 IN, the cells were harvested and suspended in phosphate-buffered saline containing 1 mg/ml lysozyme. The cells were disrupted by sonication, and Triton X-100 was added at a final concentration of 1%. IN samples of MoMLV and HIV-1 were applied to a glutathione-Sepharose 4B (GE Healthcare) column equilibrated with phosphate-buffered saline. After extensive washing with phosphate-buffered saline, the GST fusion proteins were eluted with 50 mM Tris-HCl (pH 8.0) containing 10 mM glutathione. The anti-MoMLV and anti-HIV-1 IN antisera were raised against purified GST-IN preparations.
Immunoprecipitation.
To determine the YY1-interacting regions of MoMLV IN, expression plasmids for Flag-MoMLV IN and deletion mutants were cotransfected with His-YY1 expression plasmid into 293FT cells using Lipofectamine 2000 (Invitrogen). Cells were then harvested between 36 and 48 h posttransfection and lysed in buffer A (10 mM Tris-HCl [pH 7.4], 5 mM EDTA, 150 mM NaCl, 10% glycerol, 1% sodium deoxycholate, and 1% Triton X-100) or buffer B (10 mM Tris-HCl [pH 7.4], 5 mM EDTA, 300 mM NaCl, 10% glycerol, 1% sodium deoxycholate, 1% Triton X-100, and 0.1% sodium dodecyl sulfate [SDS]). After the cell lysate had been incubated with Anti-FLAG M2 affinity gel (Sigma) for 4 h, the gel was washed twice with buffer A or B and then an additional two times with Benzonase (a broad-spectrum endonuclease from Serratia marcescens; Takara) buffer containing 20 mM Tris-HCl (pH 7.3), 50 mM KCl, and 10 mM MgCl2. The gel was treated twice with 200 U/ml Benzonase, 100 U/ml DNase I, and 5 μg/ml RNase A in Benzonase buffer at 37°C for 10 min and washed twice with buffer A or B. The samples were analyzed by Western blotting using anti-His or anti-YY1 antibody. For identification of the IN-interacting regions in YY1, Flag-MoMLV IN and His-tagged derivatives of YY1 were expressed in 293FT cells, followed by immunoprecipitation and Western blotting. The anti-Flag and anti-His antibodies were purchased from Sigma and Bethyl Laboratories, respectively. Anti-YY1 antibody (C-20) was obtained from Santa Cruz.
Preparation and analyses of PICs.
To analyze whether YY1 associates with viral cDNA of MoMLV, NIH 3T3 cells and MoMLV-producer clone no. 4 (a kind gift from Y. Suzuki, Kyoto University) (47) were cocultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum for 16 h. NIH 3T3 and producer cells were inoculated at a 4:1 ratio. PICs were prepared essentially as previously described (14). Briefly, infected cells were lysed with 20 mM HEPES-KOH (pH 7.5), 5 mM MgCl2, 150 mM KCl, and 1 mM dithiothreitol (DTT) containing 20 μg/ml aprotinin and 0.025% digitonin (Sigma), and the nuclei were removed by centrifugation. The cytoplasmic fraction was treated with 20 μg/ml of RNase A for 30 min at room temperature and subjected to gel filtration through a spin column of Sephacryl S-1000 (GE Healthcare). The partially purified PICs were incubated with anti-MoMLV IN antiserum, anti-YY1 antibody, or control rabbit preimmune serum for 4 h, followed by absorption of the immunocomplex onto salmon sperm DNA-protein A agarose (Millipore) for 1 h. The beads were washed with buffer containing 20 mM HEPES-NaOH (pH 7.5), 5 mM MgCl2, 150 mM KCl, 6% sucrose, 5 mM DTT, 6 mM EDTA, and 0.1% NP-40. The resin was suspended in 10 mM Tris-HCl (pH 7.4) containing 1 mM EDTA, 1% SDS, and 200 μg/ml proteinase K and then incubated at 55°C for 1 h. Viral DNA was purified by phenol-chloroform extraction and isopropanol precipitation and detected by Southern blotting using the LTR fragment of wild-type MoMLV (ClaI-NdeI fragment of the pNCA plasmid kindly provided by Y. Suzuki) as a probe.
The cytoplasmic fraction was also prepared from NIH 3T3 cells infected with the pQEGFP plasmid and its mutant at 4 h postinfection. After immunoprecipitation with anti-YY1 antibody, the enhanced green fluorescent protein (eGFP) sequence of the viral vector was detected by PCR with primers 5′-CGGCAACTACAAGACCCGC-3′ and 5′-GAAGTTCACCTTGATGCCGTTC-3′.
In vitro binding assay.
Various fragments of YY1 and of the MoMLV, HIV-1, and ASV INs were produced in E. coli BL21 or DH5α as GST fusions. A 1- to 2-μg aliquot of GST-MoMLV, GST-HIV-1, or GST-ASV IN or their fragments was incubated with glutathione-Sepharose 4B in binding buffer (20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl, 6% glycerol, and 0.2% NP-40). The samples were treated with a mixture of Benzonase, DNase, and RNase. His-tagged full-length YY1 treated with these nucleases was then mixed with each GST-IN fusion protein immobilized on the glutathione-Sepharose beads in binding buffer and allowed to stand for 2 h at 4°C. The beads were washed three times with binding buffer and mixed with SDS sample buffer (62.5 mM Tris-HCl [pH 6.8], 5% 2-mercaptoethanol, 2% SDS, 5% sucrose and 0.002% bromophenol blue). The bound proteins underwent SDS-polyacrylamide gel electrophoresis (PAGE) followed by Western blotting using anti-YY1 antibody. To determine the IN-interacting regions in YY1, similar pulldown experiments were performed with GST-YY1 fragments immobilized on the glutathione-Sepharose 4B beads. Bound MoMLV IN was detected by Western blotting using anti-MoMLV IN antiserum. His-tagged human YY1 was purchased from Santa Cruz and used for the in vitro integration and in vitro pulldown assays.
In vitro integration assay.
His-tagged INs of MoMLV and its C209A mutant, ASV, and HIV-1 were produced in E. coli BL21(DE3) and purified using a Ni2+-nitrilotriacetic acid agarose column as reported by Villanueva et al. (55). Stocks of INs were diluted for the assay in 20 mM HEPES-KOH (pH 7.4) containing 100 mM KCl, 1.5 mM DTT, and 10% glycerol. The following donor oligonucleotides were purchased from Hokkaido System Science: 5′-TTGACTACCCGTCAGCGGGGGTCTTTCA-3′ (28 mer) and its complementary strand 5′-AATGAAAGACCCCCGCTGACGGGTAGTCAA-3′ (30 mer), corresponding to the end of the U5 region of the MoMLV LTR; 5′-GTGTGGAAAATCTCTAGCA-3′ (19 mer) and 5′-ACTGCTAGAGATTTTCCACAC-3′ (21 mer), corresponding to the end of the U5 region of the HIV-1 LTR; and 5′-CTACAAGAGTATTGCATAAGACTACA-3′ (26 mer) and 5′-AATGTAGTCTTATGCAATACTCTTGTAG-3′ (28 mer), corresponding to the end of the U3 region of the ASV LTR. The shorter DNA fragments (100 pmol) were labeled with [γ-32P]ATP using T4 polynucleotide kinase (Takara), purified on a column (Probe Quant G-50 microcolumn; GE Healthcare), and annealed with an equivalent amount of the complementary strand. For the MoMLV IN assay, YY1, GST, and IN (10 or 20 pmol) were preincubated in 20 μl of IN dilution buffer at 4°C for 2 h before adding 2 pmol (1 μl) labeled substrate. It is possible that the addition of proteins can nonspecifically stimulate in vitro integration reactions. Thus, to eliminate the nonspecific effects, GST was added to ensure that the molar concentration of protein remained constant. After standing at 4°C for 30 min, 1.2 μg of target DNA plasmid (1 to 2 μl, pBluescript II KS−; Stratagene) was added, and the reaction was started by the addition of concentrated buffer solution at a final concentration of 20 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6.2), 165 mM KCl, 10 mM MnCl2, 10 mM DTT, and 10% dimethyl sulfoxide (DMSO) (total volume, 35 μl). The molar ratios of preincubated IN/YY1 varied between 1:0.5 and 1:4. The same reaction buffer but with a lower KCl concentration (120 mM) was used for the ASV IN assay, and a reaction buffer containing 20 mM HEPES-KOH (pH 7.2), 70 mM KCl, 10 mM MnCl2, 5 mM DTT, 10% DMSO, and 10% polyethylene glycol (PEG) 6000 was used for the HIV-1 IN assay. For the enzymatic reaction of ASV and HIV-1 INs, various amounts of YY1, GST, and IN (10 pmol) were preincubated in the reaction buffer at 4°C for 2 h. The 32P-labeled substrates were then added, and the mixtures were allowed to stand for 30 min before the target DNA addition and enzymatic reaction. Reactions were performed at 37°C for 30 min. Reaction products were digested by protease (10 mM EDTA [pH 8.0], 0.5% SDS, and 100 μg/ml proteinase K). After electrophoresis on a 1% agarose gel, integration products were detected by autoradiography.
The reaction product of MoMLV IN that showed mobility corresponding to the linear form of the target plasmid DNA was eluted from the gel, and the DNA was isolated. The DNA was amplified by PCR using KOD-Plus-Neo (Toyobo) and phosphorylated 28-mer donor DNA for the IN reaction as a primer. The enzyme in PCR buffer containing deoxynucleoside triphosphates was activated at 94°C for 2 min, and then the DNA sample was added. The mixture was stood at 68°C for 10 min in order to repair the gap resulting from the integration reaction. After the primer was added, PCR was performed using the following protocol: 94°C for 15 s, 55°C for 30 s, and 68°C for 90 s for a total of 35 cycles. The resultant PCR product was ethanol precipitated, ligated using T4 DNA ligase, and then transformed into E. coli DH5α. The donor DNA for the IN reaction was designed so that it would form a MunI site when the DNA fragments were inserted by two-end integration, and the target plasmid did not contain the restriction enzyme site. Thus, to confirm which clones had been produced by two-end integration, DNA prepared from each colony was cut with the enzyme. After rough estimation of the integration site by cutting the plasmid with SspI or PvuII/EcoRI, the DNA sequence around the junction was determined. For analyses of the linear DNA product of the HIV-1 IN reaction, the following sequence was used for PCR: 5′-AAACCATGGTGTGGAAAATCTCTAGCA-3′; the underlined sequence shows the NcoI site. The target plasmid DNA did not contain an NcoI site.
Viral vectors.
For the construction of the mouse stem cell virus (MSCV) vector pQEGFP, the eGFP DNA fragment was ligated into the EcoRI/ClaI site of pMSCVneo (Clontech), and the KpnI fragment of the recombinant plasmid containing the viral sequence was isolated and ligated to pQCXIX (Clontech). For the construction of MoMLV-based viral vector pLNΔAG, the chicken actin promoter of the pMiwZ plasmid was ligated into the pLNRβ plasmid (25). The plasmid was then cut with NotI and ligated with the GFP fragment of the pGREEN LANTERN-1 plasmid (Invitrogen). From this plasmid, the intron sequence in the actin promoter was deleted by PCR (25).
The YY1-binding-site mutant of MSCV-derived viral vector pQEGFP was constructed by PCR with primers 5′-AGCTTAAGTAACGCACGTTTGCAAGGCATG-3′ and 5′-CATGCCTTGCAAACGTGCGTTACTTAAGCT-3′; the underlined sequences show the mutated nucleotides.
The packaging cell line of viral vector pLNΔAG was established by transfecting the pLNΔAG plasmid to GP293 cells, followed by cloning. Viruses were produced by transfection of pVSV-G to packaging cells or cotransfection of pQEGFP and pVSV-G to GP293 cells (21). Both the packaging and GP293 cells express the MoMLV gag-pol gene. Supernatants were collected 48 h posttransfection, and viruses were concentrated by centrifugation as previously described (21, 25, 38). GP293 and its derivative packaging cells were cultured in DMEM (Gibco) containing 10% fetal calf serum, 1 mM sodium pyruvate, and nonessential amino acids (Gibco). All virus preparations were passed through a 0.45-μm filter, and Polybrene (8 μg/ml) was added for infection. The virus titer was measured using NIH 3T3 cells as described previously (21, 25, 38).
ChIP analysis.
NIH 3T3 cells infected with viral vector pLNΔAG were fixed with 1% formaldehyde at 4 h postinfection. Chromatin immunoprecipitation (ChIP) assays using anti-YY1 antibody or anti-MoMLV antiserum were performed using a standard procedure with salmon sperm DNA-protein A agarose beads. DNA was fragmented by sonication to a mean size of 1 kb. The GFP sequence of pLNΔAG was detected by PCR with primers 5′-TTTTTCAAAGATGACGGGAACTACA-3′ and 5′-ATGCCATTCTTTTGCTTGTCGG-3′.
In vivo integration assay.
NIH 3T3 cells (4.0 × 104 cells) were seeded on 35-mm dishes and cultured for 24 h in DMEM containing 10% fetal calf serum. Mouse YY1 small interfering RNA (siRNA) (no. 1006, 5′-GGCUGCACAAAGAUGUUCAGGGAUA-3′, and no. 1099, 5′-GCGUUCGUUGAGAGCUCAAAGCUAA-3′; 100 pmol each) or universal negative-control siRNA (B-bridge) was introduced into the cells using Lipofectamine 2000. Cells were transfected again with siRNA 24 h after the first transfection. At 24 h after the second siRNA transfection, the medium was changed to remove siRNA, and cells were infected with viral vector pQEGFP (multiplicity of infection [MOI] of 0.1) or pLNΔAG (MOI of 1.0). At 0, 4, 10, 24, and 48 h, cells were harvested for DNA extraction using a QIAamp DNA mini kit (Qiagen). The amount of total viral cDNA and two-LTR junction molecule (two-LTR circle) was measured by quantitative PCR (qPCR) with the following primers for two-LTR circle DNA: 5′-GCGCGCCAGTCCTCCGATAGAC-3′ and 5′-TACTTAAGCTAGCTTGCCAAACCTACAGG-3′ for pQEGFP and 5′-GGAGGGTCTCCTCTGAGTGAT-3′ and 5′-CTCAGTTATGTATTTTTCCATGCCTT-3′ for pLNΔAG. For the detection of total viral cDNA for pLNΔAG and pQEGFP, the primers were those used for the ChIP assay and immunoprecipitation of PICs, respectively. Genomic DNA was also purified 14 days postinfection using a MagExtractor Genome kit (Toyobo). The inserted viral DNA was quantified by qPCR using the same primers as those used for total viral cDNA amplification.
The integrated viral DNA was also assessed by B1 nested PCR. For a first round of the qPCR, the primer for the B1 sequence (5′-CTTTAATCCCAGCACTTGGGAGGC-3′; the underlined C was biotinylated) and that for eGFP (5′-GAAGTTCACCTTGATGCCGTTC-3′) or GFP (5′-ATGCCATTCTTTTGCTTGTCGG-3′) were used. The PCR was performed under the following condition: 35 cycles of denaturation at 96°C for 15 s, annealing at 55°C for 30 s, and extension at 68°C for 3 min. The amplified DNA samples were absorbed onto streptavidin-coated magnetic beads (Roche) and washed two times with 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 1 M NaCl and two times with Tris-HCl (pH 7.5), 1 mM EDTA. After elution of the DNA from the beads with 0.1 M NaOH, sodium acetate was added for neutralization and the DNA was collected by ethanol precipitation. The recovered DNA was subjected to a second round of qPCR with primers for detection of the eGFP or GFP sequences.
RESULTS
Physical interaction of INs with YY1.
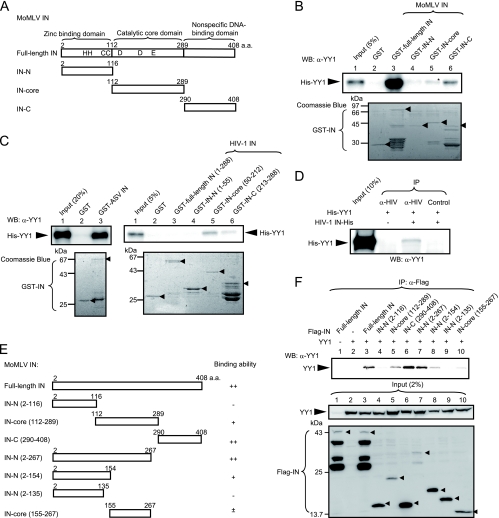
We searched for IN-interacting proteins, especially those involved in gene silencing, in order to elucidate the mechanism of retroviral silencing in undifferentiated cells. By preliminary screening involving immunoprecipitation of the extracts of cells coexpressing IN and silencing proteins, we found that MoMLV IN physically interacted with YY1. Thus, we expressed full-length MoMLV IN and its fragments as GST fusion proteins in E. coli (Fig. 1 A) and then subjected them to a pulldown assay with His-YY1 to confirm their physical association. Analysis with anti-YY1 antibody demonstrated that full-length YY1 bound to the C-terminal and catalytic core domains of MoMLV IN (Fig. 1B); GST protein alone was used as a negative control. Comparison of band densities to that of the input band showed that full-length IN bound tightly to YY1, while the C-terminal and catalytic core domains of MoMLV IN showed weaker interaction with YY1. Especially, the catalytic core domain bound weakly to YY1. In these experiments, degradation was evident in all GST-IN preparations; however, the expression levels of intact fragments were similar for all IN derivatives.
FIG. 1.
YY1 directly interacts with INs of MoMLV, ASV, and HIV-1. (A) Schematic drawing of the GST-IN of MoMLV and its deletion mutants. HHCC, zinc finger; DDE, catalytic center; a.a., amino acid. (B) Results of in vitro binding assay of GST-MoMLV IN derivatives and His-YY1. His-YY1 was detected with the anti-YY1 antibody (upper panel). The GST fusion proteins used for the pulldown assay were subjected to SDS-PAGE and detected by Coomassie brilliant blue staining (lower panel). Arrowheads indicate the intact fragments of the GST-IN derivatives. A result representative of several experiments is presented. Band intensity was measured using a densitometer, and the faint bands of lanes 4 and 5 were analyzed by paired t test. The asterisk indicates significance for GST-IN-core (lane 5) values versus GST (lane 2) values (*, P = 0.018). WB, Western blotting. (C) Results of in vitro binding assays of GST-ASV IN and GST-HIV-1 IN with His-YY1. The amino acid number of HIV IN is taken from the NCBI database (GenBank accession number NC_001802). (D) Results of immunoprecipitation (IP) of purified His-HIV-1 IN and His-YY1. Five hundred nanograms each of His-HIV-1 IN and His-YY1 were mixed in 300 μl of the binding buffer used for the pulldown assay, and the immunocomplex was precipitated with anti-HIV-1 IN antiserum and analyzed with the anti-YY1 antibody. For a negative control, mouse preimmune serum was used. (E) Schematic drawing of the Flag-tagged MoMLV IN and its deletion mutants. Full-length MoMLV IN and the mutants were tagged with Flag at the N terminus. The results of the immunoprecipitation experiment whose results are shown in panel F are summarized on the right side of the panel. (F) Human YY1 interacts with the catalytic core and C-terminal domain of MoMLV IN. Cells were lysed with buffer B. YY1 precipitated with Anti-FLAG M2 affinity beads was detected with anti-YY1 antibody after extensive washing with buffer B. Two percent of each sample was used to confirm the expression of YY1 or Flag-INs (input, lower panels). The intact bands of the Flag-IN fragments are indicated by the arrowheads. A result representative of several experiments is presented.
To determine whether ASV and HIV-1 INs also physically interact with YY1, we next analyzed their GST-IN fusion proteins by pulldown assays. ASV IN could interact with YY1 (Fig. 1C), and based on the band density, the interaction appeared to be strong. For HIV-1 IN, its catalytic core and C-terminal domains bound weakly to YY1, but its N-terminal domain showed no interaction, as found for MoMLV IN. Obvious interaction could not be detected between full-length IN and YY1 under this experimental condition. Although the reason is not fully clear, a form of steric hindrance may cause this weakened interaction between full-length IN and YY1. Immunoprecipitation of purified His-tagged full-length HIV-1 IN and His-YY1 demonstrated a weak interaction (Fig. 1D). These results for HIV-1 IN appeared to be in contrast to the results for the MoMLV and ASV INs that showed tight binding between full-length INs and YY1. Taken together, the binding assay results demonstrated that YY1 is able to physically interact with the three different INs, and thus, the interactions might have a physiological significance. In these experiments, GST-INs, His-IN, and His-YY1 were extensively treated with Benzonase, and it was confirmed by agarose gel electrophoresis that a detectable level of nucleic acids were not contaminated (data not shown). This may rule out the possibility that contaminating DNA or RNA bridged the two proteins and mediated the physical interaction (50).
To provide further evidence that the catalytic core and C-terminal domains of MoMLV IN interact with YY1, we also performed a detailed study on its YY1-binding regions by overexpressing YY1 and Flag-tagged MoMLV IN fragments (Fig. 1E) in 293FT cells, followed by immunoprecipitation using anti-Flag antibody and Western blotting. As shown by the results presented in Fig. 1F, YY1 bound to the C-terminal (amino acids 290 to 408 [290-408]) but not to the N-terminal (2-116 and 2-135) fragments. Similar expression levels were found for these truncated IN constructs. IN fragments containing the whole or a part of the catalytic core domain (fragments 112-289, 2-267, and 2-154) also bound to YY1, but the band density was dependent on the fragment. The expression levels of these truncated INs also varied. Although fragment 155-267 contained a part of the catalytic core domain, the band density was almost background level (Fig. 1F). These results are summarized in Fig. 1E. The immunocomplexes were treated with nucleases in all of these experiments. Although the interaction between the catalytic core domain of IN and YY1 seemed weak by pulldown assay, the results of immunoprecipitation demonstrated that a part of the catalytic core domain, as well as the C-terminal domain, of MoMLV IN interacted with YY1.
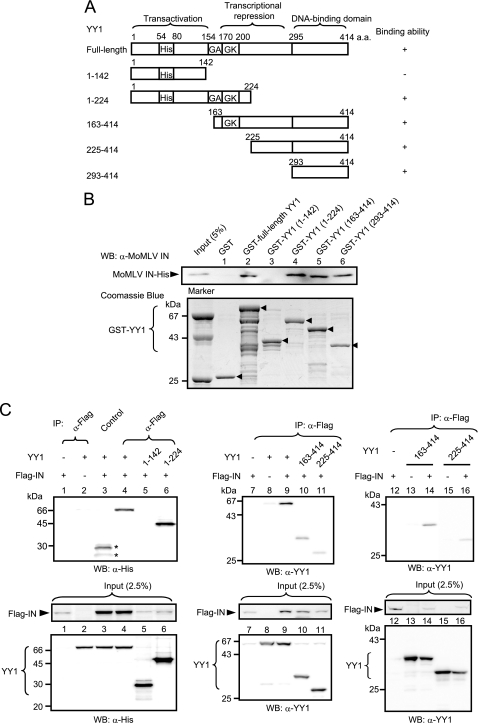
We next investigated which region of YY1 interacts with IN using in vitro pulldown assays. For these assays, we constructed several GST fusion proteins that were based on the domain structure of YY1 (Fig. 2 A). YY1 comprises three domains, including an N-terminal activation domain with a histidine-rich region, a central repressive domain with a glycine-rich region, and a C-terminal zinc finger DNA-binding domain (reviewed in reference 53). As shown in Fig. 2B, YY1 fragments 1-224, 163-414, and 293-414 but not 1-142 bound to MoMLV IN, suggesting that it bound to the central repression and C-terminal domains of YY1. We also confirmed the location of the IN-binding regions with 293FT cells overexpressing Flag-MoMLV IN and truncated YY1 fragments. The results in Fig. 2C show that YY1 fragments 1-224, 163-414, and 225-414 were coimmunoprecipitated with IN but fragment 1-142 was not precipitated. Since bands of fragments 163-414 and 225-414 were weak in intensity (Fig. 2C, center panel), immunoprecipitation was carried out under different conditions (high stringency) with controls without IN (Fig. 2C, right panel). Fragments 163-414 and 225-414 bound to IN. The results of the pulldown and immunoprecipitation experiments consistently showed that the central repressive domain of YY1, possibly including the GA and GK regions, bound to IN, while the results for the C-terminal DNA-binding domain were slightly different. That is, the interaction appeared weak by immunoprecipitation compared to the results for pulldown assays. The reason for this slight discrepancy remains unclear. However, binding of the other protein factors or posttranslational modifications may have hampered the physical interaction between the DNA-binding domain and IN in the immunoprecipitation experiments, as protein factors such as p300/CBP (28) and HDAC (66-68) bind to this domain, and amino acid residues 261 to 333, in addition to amino acids 170 to 200, contain acetylation sites (68). Another possible cause of the discrepancy is differences in the stringency of the binding assays.
FIG. 2.
Identification of the IN-binding region in YY1 in vitro and in vivo. (A) Schematic drawing of full-length YY1 and its fragments used for pulldown and coimmunoprecipitation experiments. His, histidine-rich domain; GA, glycine-alanine-rich domain; GK, glycine-lysine-rich domain. The results of in vitro pulldown and immunoprecipitation experiments are summarized on the right side of the panel. (B) Results of pulldown assay of MoMLV IN with GST fusion proteins formed with various portions of human YY1. His-MoMLV IN was detected by Western blotting using anti-MoMLV IN antiserum. GST and GST-YY1 fragments were subjected to SDS-PAGE, and the gel was stained with Coomassie brilliant blue. Arrowheads show the intact fragments of GST-YY1 derivatives (lower panel). A result representative of several experiments is presented. (C) Results of coimmunoprecipitation of human YY1 and MoMLV IN expressed in 293FT cells (upper panels). Normal mouse IgG was used in the control, and nonspecific bands are indicated by asterisks. Cells were lysed with buffer A. IN complex was precipitated and washed with buffer A (left and central panels) or buffer B (right panel). In the right panel, controls without IN were included to estimate a nonspecific interaction between YY1 and anti-Flag antibody observed in lane 8, since interaction between IN and some YY1 derivatives seemed to be weak. His-tagged fragments of human YY1 were detected with anti-YY1 or anti-His antibody. These antibodies were also used for detection of input YY1 derivatives (2.5%). Flag-IN was detected with anti-Flag antibody. A result representative of several experiments is presented.
In vivo association of YY1 with MoMLV cDNA.
As YY1 can potentially form physical interactions with three different INs, we next examined whether YY1 associates with viral DNA in the cytoplasm of infected cells. The cytoplasmic fraction of NIH 3T3 cells infected with wild-type MoMLV was subjected to gel filtration and immunoprecipitation, and the viral sequence was detected by Southern blotting using a viral cDNA fragment as a probe. As presented in Fig. 3 A, a viral sequence of the expected size was detected in the precipitates of both anti-YY1 antibody and anti-MoMLV IN antiserum, demonstrating that YY1, as well as IN, associated with viral cDNA in the cytoplasm of the infected cells. As judged by qPCR, 0.35% (YY1) and 1% (IN) of the input DNA was recovered by immunoprecipitation.
FIG. 3.
YY1 associates with MoMLV cDNA in infected cells. (A) The cytoplasmic fraction containing PICs was prepared from cocultures of NIH 3T3 and clone 4 cells, and PICs were partially purified by gel filtration and immunoprecipitated with anti-MoMLV IN antiserum or anti-YY1 antibody. Preimmune rabbit serum was used as a control antibody, and the viral sequence was detected by Southern blotting using a probe for the LTR of MoMLV. The right panel shows the percentage of immunoprecipitated viral DNA; error bars show standard deviations. Immunoprecipitated viral DNA was quantitated by qPCR using primers 5′-CGCTCACAACCAGTCGGTAGAT-3′ and 5′-AGGTCACGATGTAGGGGACCT-3′ for the packaging signal sequence. Asterisks indicate that values of anti-IN and anti-YY1 compared to those of preimmune serum were significantly different (*, P = 0.009; **, P = 0.046). (B) Results of ChIP assay of NIH 3T3 cells infected with the VSV-G-pseudotyped MoMLV vector pLNΔAG (MOI of 0.8). Infected cells were harvested 4 h postinfection. Whole-cell lysates were subjected to the assay. (C) Schematic representation of the YY1-binding site mutation in the LTR. Primers used in the experiment whose results are shown in Fig. 3D are shown by the arrowheads. (D) The YY1-binding sequences in the LTRs do not affect the association of YY1 to viral DNA. GP293 cells were cotransfected with pVSV-G and the YY1-binding site mutant pQEGFP to obtain the viral vector. NIH 3T3 cells were infected with the wild-type or mutant viral vector at the same MOI (1.6), and the cells were collected 4 h postinfection. PICs were immunoprecipitated from the cytoplasmic fraction with anti-MoMLV IN serum or anti-YY1 antibody. Preimmune rabbit serum was used as a control antibody. PCR was performed to detect the eGFP sequence of the viral vector.
We then also examined the association of YY1 with viral DNA in NIH 3T3 cells that had been infected with a replication-defective viral vector. At 4 h postinfection, the cells were collected and the association between YY1 and viral DNA was studied by ChIP; anti-MoMLV IN antiserum was used as a positive control. As shown in Fig. 3B, viral DNA could be detected with both anti-YY1 antibody and anti-MoMLV IN antiserum.
It has been reported previously that both MoMLV (15) and HIV-1 (7) contain YY1-binding motifs in their LTR sequences, suggesting that YY1 may be recruited specifically to this sequence and associated with the PIC. To verify that the association of YY1 with viral DNA is independent of the LTR, a mutated viral vector based on MSCV (pQEGFP) that contains a mutation in the YY1-binding site of the LTR sequence was constructed as reported previously (Fig. 3C) (reviewed in reference 22). The LTR sequence of nonmutated pQEGFP differed from that of MoMLV by two nucleotides; however, these changes were found outside the YY1-binding site. NIH 3T3 cells were infected with the vesicular stomatitis virus G glycoprotein (VSV-G)-pseudotyped viral vector with or without this mutation. At 4 h postinfection, the cytoplasmic fractions were immunoprecipitated with anti-YY1 antibody, and the viral DNA in the precipitate was amplified by PCR. As indicated by the results in Fig. 3D, similar levels of viral DNA could be detected for both wild-type and mutated viruses by PCR. This finding demonstrated that the association of YY1 with viral DNA was mainly mediated by physical interaction with MoMLV IN and not through binding to the YY1-binding sequence in the LTR. As the cellular distribution of YY1 is cell cycle dependent and YY1 localizes to the cytoplasm during the G1 and early S phase (39), it seems reasonable that cytoplasmic YY1 associates with viral DNA.
YY1 activates in vitro integration.
We next investigated whether the physical interaction between YY1 and INs modulates retroviral integration. We first analyzed the in vitro IN activity in both the presence and absence of YY1 using His-tagged MoMLV, HIV-1, and ASV INs produced in E. coli. The MoMLV IN activity (strand transfer activity) was measured using a short LTR sequence that was labeled with 32P at the 5′ end as the donor and plasmid pBluescript II KS− as the target. IN and YY1 were premixed at 4°C for 2 h, followed by incubation with donor DNA at 4°C for 30 min, and the reaction was initiated upon the addition of target DNA and concentrated reaction buffer (protocol 1 in Fig. 4 A). Under the assay conditions, two forms of integration products were observed. Band 1 in Fig. 4A, the mobility of which on agarose gel electrophoresis was similar to that of the open circular target plasmid, was supposed to be donor-tagged open circular target DNA, e.g., a one-end integration product. The mobility of band 2 corresponded to that of the linear form of the target DNA. From this band, the DNA was isolated, PCR amplified, self-ligated, and introduced into E. coli. After confirming that the cloned DNA had resulted from the two-end integration by MunI digestion, the sequences of the DNA were determined. Sequence analysis of the donor-target junctions of the DNA revealed that around 70 to 75% of the products contained the correct 4-bp duplication, irrespective of the presence of YY1, as shown in Table 1. From a comparison of the amounts of integration products, we found that YY1 activated integration (Fig. 4A). Four-times excess amounts of YY1 did not result in any further activation and partially impaired the enzyme activity (data not shown). To confirm this activation, IN activity was also assessed using a different preincubation protocol (protocol 2), which resulted in similar activation (Fig. 4B). This finding suggested that YY1 may not exhibit significant effects on IN-donor DNA interaction. Furthermore, activation of IN mutant C209A that shows a higher oligomerization state (equilibrium between the dimer and tetramer) and has an increased solubility (55) was also observed (data not shown). Taken together, these results indicated that MoMLV IN can be activated by a relatively low concentration of YY1.
FIG. 4.
YY1 enhances the integration activity of MoMLV and HIV-1 INs. The purity of the IN used is presented in the photographs on the right side. (A) Following protocol 1, His-YY1 was mixed with MoMLV IN first at a molar ratio as shown above the panel. The upper panel shows the products from the 32P-labeled LTR donor and the target plasmid DNA by integration. The lower bar graph indicates the normalized band intensities of the integration products compared to that of the IN plus GST (lane 2) as 1. The band density was measured using an image analyzer (BAS-5000; Fujifilm). Error bars show the standard deviations of the results of three independent assays. (B) Comparison of the activation of MoMLV IN by YY1 following protocols 1 and 2 at a ratio of IN and YY1 of 1:2. The lower bar graph indicates the normalized band intensities of the integration products compared to that of the IN plus GST (lanes 1 and 2) as 1. (C) YY1 also activated HIV-1 IN. In lane 6, GST (20 pmol) was included to quantify nonspecific activation. The bar graph shows the normalized band intensities of the products (band 1).
TABLE 1.
Sequencing results of LTR-target junctions
| Duplication | No. (%) of products obtained with: |
|
|---|---|---|
| IN | IN + YY1 | |
| 4 bp | 13 (77) | 10 (71) |
| 5 bp | 2 (12) | 2 (14) |
| 3 bp | 1 (6) | 0 (0) |
| Others | 1 (6) | 2 (14) |
| Total | 17 | 14 |
With HIV-1 IN, a band with mobility that corresponded to the linear form of the target DNA (band 2 in Fig. 4C) appeared in the presence of YY1. The mobility of band 1 matched that of the donor-tagged open circular target plasmid DNA. From band 2, DNA was isolated, PCR amplified, and introduced into E. coli. The cloned DNA from almost all colonies did not contain a two-end concerted integration product. This result agrees with the previous observation that two-end concerted integration occurred at very low frequencies when short donor fragments were used (33). Activation was also observed for HIV-1 IN, in which YY1 activated the IN reaction by more than 10-fold at an IN/YY1 ratio of 1:2 compared to the density of lane 2 (Fig. 4C). GST protein slightly activated the IN reaction (<1.5-fold) (Fig. 4C, lane 6). This result suggests that relatively weak interaction between HIV-1 IN and YY1 had a substantial effect on IN activity (Fig. 1C and D). In contrast to the results for MoMLV, excess amounts of YY1 versus IN (approximately 8 times) activated HIV-1 IN in a dose-dependent manner, possibly due to the weak interaction between the two molecules (data not shown). In addition to HIV-1 IN, ASV IN showed 4- to 5-fold activation in the presence of YY1, judging from the density of the band corresponding to the donor-tagged open circular target DNA (data not shown). The purity of the IN preparations in these experiments was high, although a small amount of degradation was observed (Fig. 4A and C). These results demonstrated that the three INs are activated by YY1, most likely through their physical interactions.
In vivo integration assay.
To investigate the physiological importance of the IN-YY1 interaction, we assessed whether YY1 affects in vivo integration events by using siRNA-mediated knockdown of YY1. VSV-G-pseudotyped MoMLV and MSCV viral vectors were prepared using cell lines that express wild-type MoMLV gag-pol. YY1-knockdown or control siRNA-transfected NIH 3T3 cells were infected with these viral vectors for analysis of total viral cDNA, two-LTR circles, and the integrated form of viral DNA. Following knockdown of YY1, the amount of YY1 in NIH 3T3 cells was reduced to less than 20% of the amount in control cells (Fig. 5 A). The amount of viral cDNA peaked at approximately 10 h postinfection for both viral vectors, irrespective of the presence of siRNA, and then gradually decreased (Fig. 5B). The total amount of synthesized viral cDNA in knockdown cells with both viral vectors was increased by 1.8- to 2-fold the amount in control cells at 10 h, suggesting that reverse transcription was partly abrogated by YY1. Previously, it was shown that the C-terminal region of HIV-1 IN binds to reverse transcriptase, and mutations in the C-terminal region of HIV-1 and MoMLV INs hamper the reverse transcriptase activity (8, 27, 58, 62). Since YY1 binds to the C-terminal region of the MoMLV IN, it is possible that YY1 may affect reverse transcriptase activity. Alternatively, we cannot rule out the possibility that YY1 directly affects the enzymatic activity. In this regard, it is noteworthy that YY1 binds to single- and double-stranded RNA with a certain sequence preference for the oligo(U) stretch of single-stranded and the A·U stretch of double-stranded RNA, based on the analysis of messenger ribonucleoprotein particle synthesis (2). Thus, it is possible that YY1 may be able to bind to viral RNA and hamper reverse transcription. So far, we have not obtained any clear result that elucidates the mechanism of the reduction of cDNA synthesis by YY1. Further extensive studies are necessary to clarify the mechanism and physiological meaning of this effect of YY1.
FIG. 5.
YY1 knockdown reduces viral integration into the host genome. (A) Knockdown of YY1 by siRNA was confirmed by Western blotting. (B and C) The amount of total viral cDNA (B) and two-LTR circle (C) of pQEGFP (siRNA no. 1006) and pLNΔAG (siRNA no. 1099) viral vectors is shown. NIH 3T3 cells were transfected twice with YY1 siRNA no. 1006 or no. 1099 and then infected with pQEGFP (MOI of 0.1) or pLNΔAG (MOI of 1). Cellular DNA was extracted at 0, 4, 10, 24, and 48 h and subjected to qPCR analysis. Copy number per cell was calculated by normalizing the value of the viral sequence with that of GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Error bars represent the standard deviations of the results of at least three experiments. Primers used for PCR for GAPDH were 5′-TGTGATGGGTGTGAACCACGAGAA-3′ and 5′-GAGCCCTTCCACAATGCCAAAGTT-3′. Asterisks indicate that values of YY1 siRNA compared to those of scrambled RNA were significantly different (*, P = 0.038, and **, P = 0.031 in panel B; *, P = 0.022, **, P = 0.041, and ***, P = 0.047 in panel C). (D) The integrated form of viral DNA was quantified 14 days postinfection by qPCR and normalized as described for the results presented in panel C. Asterisks indicate significance of values of YY1 siRNA versus those of scrambled RNA (*, P = 0.014, **, P = 0.016, and ***, P < 0.0001). (E) The integrated viral DNA was quantified by nested PCR 24 h postinfection (pQEGFP, n = 2; pLNΔAG, n = 3). The asterisk indicates that values of YY1 siRNA compared to those of scrambled RNA were significantly different (*, P = 0.027). (F) Triplicate cultures of NIH 3T3 cells (YY1 siRNA and scrambled RNA treated) infected with pLNΔAG (MOI of 1) were analyzed by fluorescence-activated cell sorting in order to quantify the mean GFP intensity. The asterisk indicates that values of YY1 siRNA compared to those of scrambled RNA were significantly different (*, P = 0.008).
We next examined the effects of YY1 on the two-LTR circle, which is thought to be the result of direct end-to-end joining of the viral DNA and is a dead-end product (45). For PCR, primers annealing to the U5 and U3 regions of the LTR of pLNΔAG or the R and U3 regions of pQEGFP were used. The levels of two-LTR circle increased by 24 h, after which they either gradually increased or decreased depending on the viral vector used (Fig. 5C). The mean copy number of the two-LTR circle was less than 1% of total viral cDNA at 24 h postinfection in both knockdown and control cells. siRNA facilitated the two-LTR-circle formation by 1.3- to 2.9-fold compared to the level in control cells at 24 h. Two-LTR-circle formation has been reported to prevent apoptosis that is induced in infected cells by nonintegrated linear viral DNA, and this formation can be used as an index for nuclear transport of PICs (31, 42). However, whether two-LTR-circle formation serves as a true reflection of nuclear entry of PICs remains controversial, as two-LTR circle was detected in the cytoplasm of MLV-infected aphidicolin-arrested NIH 3T3 cells (45). Although the physiological mechanisms for this reduction in two-LTR-circle formation by YY1 remain to be elucidated, the effects of YY1 may be secondary to the reduction in viral cDNA synthesis.
As shown by the results presented in Fig. 5D, the amount of viral sequence measured by qPCR 14 days after infection, which probably corresponded to the amount of the integrated form of viral DNA, decreased 3- to 6-fold by knockdown of YY1 with both the MoMLV- and MSCV-based vectors, suggesting that YY1 facilitates in vivo integration events. Similar results were obtained by nested PCR 24 h postinfection (Fig. 5E). This result is in contrast to the viral cDNA synthesis, which increased following siRNA treatment (Fig. 5B). As the nuclear entry of the MoMLV PIC is dependent on the disappearance of the nuclear envelope during cell growth (31, 42) and is required for successive integration events, growth retardation following siRNA treatment was evaluated. We found that cell growth was not significantly affected by siRNA, as assessed by direct counting of cell numbers in parallel runs. We observed that the increase in cell number in the siRNA-treated dishes was 60 to 80% of that observed in the control culture during the first 24 h after virus infection. Afterward, obvious retardation of cell growth was not observed (data not shown). We hypothesized that this level of growth retardation did not profoundly influence nuclear entry of PIC and its subsequent integration, although we could not rule out a limited effect on integration. We also studied the expression of virally encoded GFP. As demonstrated by the results in Fig. 5F, the mean intensity of GFP fluorescence in the knockdown cells was increased 1.4-fold at 48 h postinfection, although no increase was observed at 24 h. This suggests that the expression of GFP was partly repressed by YY1. Taking these results together, siRNA appears to facilitate viral cDNA synthesis while abrogating integration. Thus, we speculated that YY1 activates integration events in vivo, even though the reverse transcriptase reaction was partly inhibited.
DISCUSSION
The present study demonstrated that YY1 is able to bind to MoMLV, HIV-1, and ASV INs. This interaction was found to occur at the catalytic core and C-terminal domains of the MoMLV and HIV-1 INs. IN proteins can be divided into three functional domains. The N-terminal domain contains a conserved His-His/Cys-Cys (HHCC) that is involved in protein multimerization (65, 70), the catalytic core domain contains a conserved DDE motif that is essential for catalytic activity (11, 12, 26), and the less conserved C-terminal domain is involved in nonspecific DNA-binding activity (13, 59). The amino acid sequence of MoMLV IN has extra sequences at both the C and N termini and an additional 36-amino-acid sequence in the C-terminal region that is close to the boundary of the catalytic core domain. In addition, various amino acids differ in the common region from those of HIV-1 and ASV INs (23, 41). Despite the diversity in their amino acid sequences, MoMLV and HIV-1 INs share a similar array of helix and β-strand structures (41), suggesting that interaction with YY1 may rely on their secondary rather than primary structure.
YY1 could not be detected in MoMLV virions, although IN was detected (A. Mizutani, unpublished data). The immunoprecipitation experiments whose results are shown in Fig. 3 demonstrated that YY1 associates with viral cDNA in the cytoplasm of infected cells. These results suggest that YY1 may associate with PIC in the cytoplasm. Since viral cDNA synthesis was partly hampered by YY1, it is possible that YY1 may interact with IN before PIC formation. Extensive analysis is necessary to clarify this.
We also found that YY1 stimulated in vitro integration reactions of MoMLV, HIV-1, and ASV INs. Maximum activation was observed for the IN/YY1 molar ratio of 1:2 with MoMLV and a higher ratio with HIV-1 IN. Thus, an intriguing question is how YY1 facilitates the integration reaction in vitro. As YY1 showed an apparently positive effect on the in vitro activity of three different INs, we assumed that YY1 might have a common and fundamental function for basic IN catalytic mechanisms. In the present study, it was demonstrated that the distribution of host target duplication of two-end concerted integration was not affected by YY1. And the MoMLV IN C209A mutant, which shows a higher oligomerization state, was also activated by YY1, suggesting that the activation may not result from the changes in the oligomerization state of IN. However, we have not yet obtained any direct evidence to demonstrate the molecular basis of this activation. In this relationship, it is interesting that YY1, together with the INO80 chromatin-remodeling complex, is essential for mammalian homologous recombination and was shown to bind to a recombination intermediate termed the holiday junction in vitro (60). Based on the results of this study, an attractive hypothesis would be that YY1 might facilitate catalytic reactions by stabilizing specific reaction intermediates. Further analyses on this point will provide us with a better understanding of the mechanisms underlying the enzymatic reaction of IN. We also demonstrated that YY1 facilitated the IN-mediated integration events in vivo, even though viral cDNA synthesis was impaired. This positive effect on in vivo integration may in part mirror the activation of in vitro IN activity by YY1.
Retroviruses display preferences for target DNA sequences in the immediate vicinity of the integration site which appear to be virus specific, and IN may select specific sequences (10, 20). In addition to this nucleotide-scale selection of target sites, previous studies have demonstrated that viruses have favored and disfavored chromosomal regions for integration. For instance, chromosomal regions rich in expressed genes are favored for HIV-1 integration (10, 30, 37, 48). MoMLV prefers integration near the start sites of transcription and CpG islands that are genomic regions enriched in the rare CpG dinucleotide and commonly associated with the TATA box-less transcription start site (10, 37, 54, 61). IN is supposed to be the principal viral determinant of the integration specificity, probably through tethering to cellular proteins bound near preferred genomic regions (30). LEDGF/p75 is considered to act as a tethering factor for HIV-1 PIC (6, 36, 48). YY1-binding sites have been shown to reside in promoter and translation start sites with a certain frequency (more than 3% of human promoters contain the binding site) (63). Moreover, YY1 interacts with basal transcription machinery, such as TBP and TFIIB (1), and forms physical complexes, suggesting that YY1 may modulate the access of MoMLV PIC to chromosomes. This might cause a bias in integration site selection to a certain extent. However, as of now there is no clear evidence that MoMLV integration sites are enriched in the close vicinity of YY1-binding sites.
In Drosophila, the gene known as Pho, an ortholog of YY1, binds to the Polycomb-responsive elements of the Hox and Engrailed genes and recruits the Polycomb-repressive complex to the genes. A similar function in mammals has not been reported for YY1 (reviewed in reference 49). Instead, YY1 was found to interact with the Polycomb protein Ezh2 that shows histone lysine methyltransferase activity (4) and is a member of Polycomb-repressive complex 2 with EED. In Xenopus, YY1 interacts with EED (43), and EED physically interacts with HIV-1 IN (56). Thus, it is possible that these repressive proteins may interact with PIC, as is the case for HDAC (19), and may be eventually delivered to integration sites and repress the integrated proviral DNA under some physiological conditions. In the present case, YY1 seemed to partially repress GFP expression, but the molecular basis of its action and its relationship to other repressive proteins remains unclear.
Although no direct evidence for the physiological functions of the IN-YY1 interaction has been found except for stimulation of the integration reaction in vitro and in vivo, we hypothesized that YY1 plays an important role in integration and related events that occur after integration. Detailed analyses are required to clarify the precise biological function of the IN-YY1 interaction.
Acknowledgments
This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and by the Bio-oriented Technology Research Advancement Institution (BRAIN).
Footnotes
Published ahead of print on 2 June 2010.
REFERENCES
- 1.Austen, M., B. Lüscher, and J. M. Lüscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. J. Biol. Chem. 272:1709-1717. [DOI] [PubMed] [Google Scholar]
- 2.Belak, Z. R., and N. Ovsenek. 2007. Assembly of the Yin Yang 1 transcription factor into messenger ribonucleoprotein particles requires direct RNA binding activity. J. Biol. Chem. 282:37913-37920. [DOI] [PubMed] [Google Scholar]
- 3.Busschots, K., A. Voet, M. De Maeyer, J.-C. Rain, S. Emiliani, R. Benarous, L. Desender, Z. Debyser, and F. Christ. 2007. Identification of the LEDGF/p75 binding site in HIV-1 integrase. J. Mol. Biol. 365:1480-1492. [DOI] [PubMed] [Google Scholar]
- 4.Caretti, G., M. Di Padova, B. Micales, G. E. Lyons, and V. Sartorelli. 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18:2627-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 6.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. [DOI] [PubMed] [Google Scholar]
- 7.Coull, J. J., F. Romerio, J.-M. Sun, J. L. Volker, K. M. Galvin, J. R. Davie, Y. Shi, U. Hansen, and D. M. Margolis. 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74:6790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dar, M. J., B. Monel, L. Krishnan, M.-C. Shun, F. Di Nunzio, D. E. Helland, and A. Engelman. 2009. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology 6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLorbe, W. J., P. A. Luciw, H. M. Goodman, H. E. Varmus, and J. M. Bishop. 1980. Molecular cloning and characterization of avian sarcoma virus circular DNA molecules. J. Virol. 36:50-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derse, D., B. Crise, Y. Li, G. Princler, N. Lum, C. Stewart, C. F. McGrath, S. H. Hughes, D. J. Munroe, and X. Wu. 2007. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J. Virol. 81:6731-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drelich, M., R. Wilhelm, and J. Mous. 1992. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology 188:459-468. [DOI] [PubMed] [Google Scholar]
- 12.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman, A., A. B. Hickman, and R. Craigie. 1994. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman, A., I. Oztop, N. Vandegraaff, and N. K. Raghavendra. 2009. Quantitative analysis of HIV-1 preintegration complexes. Methods 47:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flanagan, J. R., K. G. Becker, D. L. Ennist, S. L. Gleason, P. H. Driggers, B.-Z. Levi, E. Appella, and K. Ozato. 1992. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol. Cell. Biol. 12:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García, E., C. Marcos-Gutiérrez, M. del Mar Lorente, J. C. Moreno, and M. Vidal. 1999. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 18:3404-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon, S., G. Akopyan, H. Garban, and B. Bonavida. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25:1125-1142. [DOI] [PubMed] [Google Scholar]
- 18.Greger, J. G., R. A. Katz, A. M. Ishov, G. G. Maul, and A. M. Skalka. 2005. The cellular protein Daxx interacts with avian sarcoma virus integrase and viral DNA to repress viral transcription. J. Virol. 79:4610-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, G., and D. M. Margolis. 2002. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol. Cell. Biol. 22:2965-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holman, A. G., and J. M. Coffin. 2005. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. U. S. A. 102:6103-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hotta, A., Y. Saito, K. Kyogoku, Y. Kawabe, K. Nishijima, M. Kamihira, and S. Iijima. 2006. Characterization of transient expression system for retroviral vector production. J. Biosci. Bioeng. 101:361-368. [DOI] [PubMed] [Google Scholar]
- 22.Iba, H., T. Mizutani, and T. Ito. 2003. SWI/SNF chromatin remodelling complex and retroviral gene silencing. Rev. Med. Virol. 13:99-110. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, M. S., M. A. McClure, D.-F. Feng, J. Gray, and R. F. Doolittle. 1986. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc. Natl. Acad. Sci. U. S. A. 83:7648-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 25.Kamihira, M., K. Ono, K. Esaka, K. Nishijima, R. Kigaku, H. Komatsu, T. Yamashita, K. Kyogoku, and S. Iijima. 2005. High-level expression of single-chain Fv-Fc fusion protein in serum and egg white of genetically manipulated chickens by using a retroviral vector. J. Virol. 79:10864-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulkosky, J., K. S. Jones, R. A. Katz, J. P. G. Mack, and A. M. Skalka. 1992. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 12:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai, L., H. Liu, X. Wu, and J. C. Kappes. 2001. Moloney murine leukemia virus integrase protein augments viral DNA synthesis in infected cells. J. Virol. 75:11365-11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J.-S., K. M. Galvin, R. H. See, R. Eckner, D. Livingston, E. Moran, and Y. Shi. 1995. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 9:1188-1198. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J.-S., K. M. Galvin, and Y. Shi. 1993. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc. Natl. Acad. Sci. U. S. A. 90:6145-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewinski, M. K., M. Yamashita, M. Emerman, A. Ciuffi, H. Marshall, G. Crawford, F. Collins, P. Shinn, J. Leipzig, S. Hannenhalli, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2006. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, M., and R. Craigie. 2005. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 280:29334-29339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 35.Mansharamani, M., D. R. M. Graham, D. Monie, K. K. Lee, J. E. K. Hildreth, R. F. Siliciano, and K. L. Wilson. 2003. Barrier-to-autointegration factor BAF binds p55 Gag and matrix and is a host component of human immunodeficiency virus type 1 virions. J. Virol. 77:13084-13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall, H. M., K. Ronen, C. Berry, M. Llano, H. Sutherland, D. Saenz, W. Bickmore, E. Poeschla, and F. D. Bushman. 2007. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2:e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell, R. S., B. F. Beitzel, A. R. W. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono, K., M. Kamihira, Y. Kuga, H. Matsumoto, A. Hotta, T. Itoh, K. Nishijima, N. Nakamura, H. Matsuda, and S. Iijima. 2003. Production of anti-prion scFv-Fc fusion proteins by recombinant animal cells. J. Biosci. Bioeng. 95:231-238. [PubMed] [Google Scholar]
- 39.Palko, L., H. W. Bass, M. J. Beyrouthy, and M. M. Hurt. 2004. The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J. Cell Sci. 117:465-476. [DOI] [PubMed] [Google Scholar]
- 40.Petkova, V., M. J. Romanowski, I. Sulijoadikusumo, D. Rohne, P. Kang, T. Shenk, and A. Usheva. 2001. Interaction between YY1 and the retinoblastoma protein. J. Biol. Chem. 276:7932-7936. [DOI] [PubMed] [Google Scholar]
- 41.Puglia, J., T. Wang, C. Smith-Snyder, M. Cote, M. Scher, J. N. Pelletier, S. John, C. B. Jonsson, and M. J. Roth. 2006. Revealing domain structure through linker-scanning analysis of the murine leukemia virus (MuLV) RNase H and MuLV and human immunodeficiency virus type 1 integrase proteins. J. Virol. 80:9497-9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satijn, D. P. E., K. M. Hamer, J. den Blaauwen, and A. P. Otte. 2001. The Polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 21:1360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segura-Totten, M., and K. L. Wilson. 2004. BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 14:261-266. [DOI] [PubMed] [Google Scholar]
- 45.Serhan, F., M. Penaud, C. Petit, T. Leste-Lasserre, S. Trajcevski, D. Klatzmann, G. Duisit, P. Sonigo, and P. Moullier. 2004. Early detection of a two-long-terminal-repeat junction molecule in the cytoplasm of recombinant murine leukemia virus-infected cells. J. Virol. 78:6190-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, Y., J.-S. Lee, and K. M. Galvin. 1997. Everything you have ever wanted to know about Yin Yang 1… . Biochim. Biophys. Acta 1332:F49-F66. [DOI] [PubMed] [Google Scholar]
- 47.Shields, A., O. N. Witte, E. Rothenberg, and D. Baltimore. 1978. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell 14:601-609. [DOI] [PubMed] [Google Scholar]
- 48.Shun, M.-C., N. K. Raghavendra, N. Vandergraaff, J. E. Daigle, S. Hughes, P. Kellam, P. Cherepanov, and A. Engelman. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon, J. A., and R. E. Kingston. 2009. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10:697-708. [DOI] [PubMed] [Google Scholar]
- 50.Studamire, B., and S. P. Goff. 2008. Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology 5:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki, Y., and R. Craigie. 2007. The road to chromatin-nuclear entry of retroviruses. Nat. Rev. Microbiol. 5:187-196. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki, Y., H. Yang, and R. Craigie. 2004. LAP2α and BAF collaborate to organize the Moloney murine leukemia virus preintegration complex. EMBO J. 23:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas, M. J., and E. Seto. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197-208. [DOI] [PubMed] [Google Scholar]
- 54.Tsukahara, T., H. Agawa, S. Matsumoto, M. Matsuda, S. Ueno, Y. Yamashita, K. Yamada, N. Tanaka, K. Kojima, and T. Takeshita. 2006. Murine leukemia virus vector integration favors promoter regions and regional hot spots in a human T-cell line. Biochem. Biophys. Res. Commun. 345:1099-1107. [DOI] [PubMed] [Google Scholar]
- 55.Villanueva, R. A., C. B. Jonsson, J. Jones, M. M. Georgiadis, and M. J. Roth. 2003. Differential multimerization of Moloney murine leukemia virus integrase purified under nondenaturing conditions. Virology 316:146-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Violot, S., S. S. Hong, D. Rakotobe, C. Petit, B. Gay, K. Moreau, G. Billaud, S. Priet, J. Sire, O. Schwartz, J.-F. Mouscadet, and P. Boulanger. 2003. The human Polycomb group EED protein interacts with the integrase of human immunodeficiency virus type 1. J. Virol. 77:12507-12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, C.-Y., Y.-J. Liang, Y.-S. Lin, H.-M. Shih, Y.-S. Jou, and W. C. Y. Yu. 2004. YY1AP, a novel co-activator of YY1. J. Biol. Chem. 279:17750-17755. [DOI] [PubMed] [Google Scholar]
- 58.Wilkinson, T. A., K. Januszyk, M. L. Phillips, S. S. Tekeste, M. Zhang, J. T. Miller, S. F. J. Le Grice, R. T. Clubb, and S. A. Chow. 2009. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J. Biol. Chem. 284:7931-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woerner, A. M., and C. J. Marcus-Sekura. 1993. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 21:3507-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, S., Y. Shi, P. Mulligan, F. Gay, J. Landry, H. Liu, J. Lu, H. H. Qi, W. Wang, J. A. Nickoloff, C. Wu, and Y. Shi. 2007. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Mol. Biol. 14:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 62.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xi, H., Y. Yu, Y. Fu, J. Foley, A. Halees, and Z. Weng. 2007. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res. 17:798-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan, N., P. Cherepanov, J. E. Daigle, A. Engelman, and J. Lieberman. 2009. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 5:e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang, F., O. Leon, N. J. Greenfield, and M. J. Roth. 1999. Functional interactions of the HHCC domain of Moloney murine leukemia virus integrase revealed by nonoverlapping complementation and zinc-dependent dimerization. J. Virol. 73:1809-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, W.-M., C. Inouye, Y. Zeng, D. Bearss, and E. Seto. 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. U. S. A. 93:12845-12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, W.-M., Y.-L. Yao, J.-M. Sun, J. R. Davie, and E. Seto. 1997. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 272:28001-28007. [DOI] [PubMed] [Google Scholar]
- 68.Yao, Y-L., W.-M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yung, E., M. Sorin, E.-J. Wang, S. Perumal, D. Ott, and G. V. Kalpana. 2004. Specificity of interaction of INI1/hSNF5 with retroviral integrases and its functional significance. J. Virol. 78:2222-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng, R., T. M. Jenkins, and R. Craigie. 1996. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. U. S. A. 93:13659-13664. [DOI] [PMC free article] [PubMed] [Google Scholar]