Abstract
Reactive oxygen species (ROS) are produced in growth factor signaling pathways leading to cell proliferation, but the mechanisms leading to ROS generation and the targets of ROS signals are not well understood. Using a focused siRNA screen to identify redox-related proteins required for growth factor induced cell cycle entry, we show that two ROS generating proteins, the NADPH oxidases NOX4 and DUOX2, are required for platelet-derived growth factor (PDGF) induced retinoblastoma protein (Rb) phosphorylation in normal human fibroblasts. Unexpectedly, NOX4 and DUOX2 knockdown did not inhibit the early signaling pathways leading to cyclin D1 upregulation. However, hours after growth factor stimulation, NOX4 and DUOX2 knockdown reduced ERK1 phosphorylation and increased levels of the tumor suppressor protein p53 and a cell cycle inhibitor protein p21 (Waf1/Cip1) that is transcriptionally regulated by p53. Co-knockdown of NOX4 or DUOX2 with either p53 or with p21 overcame the inhibition of Rb phosphorylation that occurred with NOX4 or DUOX2 knockdown alone. Our results argue that rather than primarily affecting growth factor receptor signaling, NOX4 and DUOX2 regulate cell cycle entry as part of a p53-dependent checkpoint for proliferation.
Keywords: NADPH oxidases, redox signaling, p53, NOX4, DUOX2
Introduction
ROS are generated in response to growth factors in the signaling pathways that lead to cell proliferation (Bae et al 1997, Sundaresan et al 1995). Cancer cells produce elevated levels of ROS (Szatrowski and Nathan 1991, Trachootham et al 2009), and NADPH oxidases, proteins that are involved in growth factor induced ROS generation, have been found to be over-expressed in tumor cell lines (Laurent et al 2008, Yamaura et al 2009). While these and a number of other studies point towards an important role of ROS in cell proliferation, the components and mechanisms of the redox-signaling pathways involved in growth factor-induced ROS generation and the mechanisms by which growth-factor induced ROS generation affect cell cycle control are not well understood.
Current experimental evidence supports a model in which NADPH oxidases generate ROS in response to growth factors and the ROS in turn can affect signaling pathways through the reversible oxidation of susceptible amino acids that are critical for protein activity (typically low pKa, solvent-exposed cysteine residues) (Janssen-Heininger et al 2008, Lambeth 2004, Rhee et al 2000, Winterbourn 2008, Winterbourn and Hampton 2008). For example, the NADPH oxidase NOX1 has been implicated in ROS production in response to PDGF and EGF (Lassegue et al 2001, Park et al 2004) and NOX4 in ROS production in response to PDGF or TGF-β (Park et al 2005, Sturrock et al 2007). One of the main protein families that are regulated by growth factor induced ROS are the protein tyrosine phosphatases (PTPs). EGF stimulation of A431 cells and PDGF stimulation of Rat1 fibroblasts have been shown to lead to inhibition of PTPs and enhanced phosphorylation of the growth factor receptors (Lee et al 1998, Meng et al 2002). More recently, the NADPH oxidase NOX4 was shown to promote oxidation of PTP1B in response to insulin and EGF (Chen et al 2008, Mahadev et al 2004). Other signaling proteins such as transcription factors and protein kinases also have redox-sensitive cysteines and may be redox regulated within signaling pathways (reviewed in (Janssen-Heininger et al 2008, Rhee et al 2000)).
The production of ROS in early growth factor signaling pathways has led to the proposal that ROS regulate the G0 to G1 transition of the cell cycle by activating the signaling pathways that promote cyclin D expression (Burch and Heintz 2005, Burhans and Heintz 2009). Increases in CyclinD levels promote the activation of CyclinD/CDK4/6 complexes that phosphorylate retinoblastoma (Rb) protein, a key step in the initiation of cell cycle entry (Yao et al 2008, Zarkowska and Mittnacht 1997). CyclinD1 expression and cell proliferation are increased by overexpression of NOX1 (Ranjan et al 2006) and, in response to TGF-Beta, Rb phosphorylation and proliferation are inhibited by NOX4 knockdown (Sturrock et al 2007).
Here we show a different role for the NADPH oxidases NOX4 and DUOX2. Our data suggests that they primarily act by downregulating the p53-dependent signaling pathways that inhibit Rb phosphorylation. We identified NOX4 and DUOX2 in a focused siRNA screen in normal human fibroblasts aimed at identifying specific redox-associated proteins (primarily proteins involved in ROS generation and thiol or cysteine reduction) that affect PDGF-induced cell cycle entry. For the screen, we developed a high-throughput fluorescence microscopy assay to measure Rb-phosphorylation and DNA content at the single cell level directly from fluorescence microscopy images. Surprisingly, we did not observe an effect of NOX4 and DUOX2 knockdown on Akt or ERK phosphorylation minutes after growth factor stimuli or on CyclinD1 expression. However, NOX4 and DUOX2 siRNA knockdown caused a delayed reduction in ERK1 phosphorylation and an increase in p53 and p21 levels. These data support a model in which NOX4 and DUOX2 activity are required to inactivate the p53-dependent checkpoint machinery.
Materials and Methods
Sources of siRNA
Diced pools of siRNA were generated according to previously published methods (Liou et al 2005, Myers et al 2003) using the PCR primers and methods in the Supplementary Information and Supplementary Table 1. NOX4 and DUOX2 siGENOME siRNA and siCONTROL NON-Targeting siRNA Pool #2 (D-001206-14-05) were purchased from Dharmacon (Lafayette, CO, USA).
Cell Culture
HS68 cells from the American Tissue Culture Collection (ATCC, Manassas, VA, USA) were cultured in 10%CO2 in Dulbucco’s modified eagle medium (DMEM) (Invitrogen, Carlsbad, CA, USA) with 10% FBS (ATCC or Invitrogen) plus penicillin-streptomycin-glutamine (Invitrogen) between passage numbers 16 and 30.
Cell cycle entry assay
Co-star 96-well plates were coated with 0.1mg/ml poly-D-lysine (Sigma-Aldrich (USA)) for 10 minutes washed with water and dried. HS68 cells were reverse transfected with 10-40nM siRNA (final concentration in 150μl) using Lipofectamine 2000 (Invitrogen) following the manufacturers protocol except 3000 cells were plated per well. For the co-knockdown experiments, we transfected 20nM of each siRNA for a total of 40nM siRNA. The cells were incubated with the transfection reagents overnight (12–16 hours) and were then washed five times with 0.1% Bovine Serum Albumin (BSA, Sigma-Aldrich (USA)) plus DMEM. Forty six to fifty hours after serum removal, the cells were stimulated with 10ng/ml PDGF-BB (PeproTech, Inc. (Rocky Hill, NJ, USA) for twenty seven hours and were then fixed in 4% paraformaldehyde and washed 5X with phosphate buffered saline (PBS). The cells were permeabilized in 0.2% Triton X for 15-30 minutes and blocked in 3% BSA for one to two hours. Cells were incubated in pSer807/pSer811 Rb antibody (#9308, Cell Signaling Technologies (CST) (Danvers, MA, USA)) 1:500 in 3%BSA overnight at 4°C, washed 5X in PBS, then incubated in Alexa 514 goat anti-rabbit IgG antibodies (A-31558, Invitrogen) 1:1000 in 3% BSA for two hours at room temperature. The plates were washed 5X in PBS and then Hoechst 33342 (H3570, Invitrogen) was added to a final concentration of 0.1ng/ml. The plates were stored for a minimum of 12 hours at 4°C in Hoechst stain to ensure saturation of the Hoechst staining prior to imaging. The 96-well plates were imaged with an Image Xpress 5000A and analyzed using MATLAB for their DNA content and intensity of Rb staining (see Supplementary Information for details).
Nested PCR and quantitative RT-PCR
HS68 cells were plated, transfected, serum starved and PDGF stimulated (see figure legends for details) as described for the cell cycle entry assay except the cells were plated in a 12-well dish so the volumes and cell numbers were scaled by a factor of 12. Total RNA was isolated from HS68 cells using an RNAeasy kit (Qiagen, Valencia, CA, USA) and was then reverse transcribed using Superscript III (Invitrogen) following the manufacturer’s protocol with random hexamer primers. For the nested PCR, one μl of cDNA per sample was used as a template from a starting concentration of 26ng/μl total RNA. The nested PCR was carried out following the same protocol and using the same primers that were used to generate the diced siRNA library. For the quantitative RT-PCR, Taqman Gene Expression assays (NOX4 (Hs00418356_m1), DUOX2 (Hs00204187_m1), GAPDH (Hs99999905_m1) and cyclophilin (Hs99999904_m1) from Applied Biosystems (Foster City, CA, USA) were used following the manufacturers protocol with fifty ng of total RNA as starting material per reaction for the synthetic siRNA pools or 100,000 cells for the single synthetic siRNA. For the synthetic siRNA pools, cDNA was prepared as described above and IQsupermix (BioRad, (Hercules, CA, USA) was substituted for the master mix and the reactions were run on a BioRad icycler. For the single synthetic siRNA reactions, the TaqMan Gene Expression Cells-to-CT kit was used (Applied Biosystems/Ambion, Austin, Texas, USA) and reactions were run on an Applied Biosystems 7500 real-time PCR instrument.
Western blots
HS68 cells were plated in a 12-well dish, transfected and serum starved as described above for quantitative RT-PCR. Approximately forty eight hours after serum removal, cells were stimulated with PDGF for various amounts of time (see figure legends). Cells were washed with cold PBS and lysed in SDS lysis buffer containing 130mM Tris pH 8.0, 20% Glycerol, 4.6% sodium dodecyl sulfate, 2% dithiothreitol and .02% bromophenol blue with phosphatase inhibitor cocktail I and II (Sigma) and complete protease inhibitor (Roche). The lysates were boiled at 95°C for five minutes and collected by centrifugation. Gel electrophoresis and western blots were carried out following the Tris-Glycine or NuPAGE gel system protocols (Invitrogen).
Western blots were blocked and stained following the Licor-western blot protocol (Li-COR Biosciences, Lincoln, Nebraska, USA). The primary antibodies and dilutions were: pT202/pY204 ERK (#9106, CST) 1:2000, pT308 Akt (#9275, CST) 1:1000, total-ERK (#9102,CST) 1:500, total-Akt (#MAB2055, R&D systems (Minneapolis, MN, USA) 1:500, cyclin D1 antibody (#2926, CST) 1:1000 and GAPDH (ab8245, abcam, Cambridge, MA). All primary antibodies were diluted in Odyssey Blocking Buffer (Li-COR) and the incubation times were overnight at 4°C except GAPDH for which the incubation was one hour at room temperature. For blots co-stained with pERK/ERK or pAkt/AKT the blots were first incubated with the ERK and AKT phosphospecific antibodies and then the antibodies for total protein. Secondary antibodies were diluted 1:15,000 in Odyssey Blocking Buffer and 1% goat serum. The antibodies were Alexa Fluor 680 Goat anti-Rabbit IgG conjugated antibodies (A-21109) and IRDye 800 conjugated Anti-Mouse antibodies (610-132-121, Rockland Immunochemicals (Gilbertsville, PA, USA)).
The blots were scanned on a Li-COR infrared imager. Ratios of phosphorylated ERK and AKT to total ERK and AKT or total ERK and AKT to GAPDH were quantified in MATLAB using the methods described in the supplementary material.
Immunocytochemistry for p53 or p21
Cells were plated, transfected, PDGF stimulated, fixed after 18 hours of PDGF stimuli, blocked and permeabilized using the same methods described for the cell-cycle entry assay. The cells were transfected with the DUOX2 or NOX4 Dharmacon synthetic siRNA sequences listed in Figure legend 3 or Supplementary Figure 7. NOX4-1 exhibited a different effect on p21 levels from the five other NOX4-targeting siRNA and was therefore omitted from subsequent analysis. Cells were stained overnight with pRb antibody or with p53 antibody (#9282, CST) 1:500 then stained with Alexa-514 conjugated rabbit secondary antibody as described above. The cells were then washed 5X with PBS and stained overnight with 1:1000 p21 (#6246, Santa Cruz Biotechnology Inc (Santa Cruz, CA, USA). They were then washed with 5X PBS, stained for two hours with 1:1000 Alexa-568 conjugated mouse secondary antibody (Invitrogen) and washed again with PBS, Hoechst stained and imaged as described above.
Results
Optimization and validation of a cell cycle entry assay
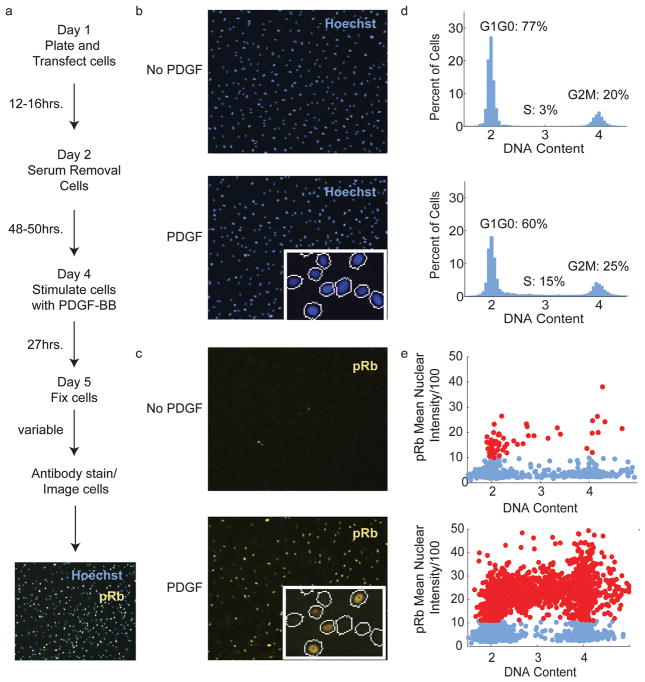
We developed an assay to monitor cell cycle entry using phosphorylated Rb as a readout for cyclin/CDK activity and Hoechst stain as a nuclear marker to segment the image and measure DNA content (Figure 1a, Supplementary Information and Supplementary Figures 1 and 2)). We chose HS68 normal human fibroblasts because they can be serum starved, are readily transfected with siRNA and are non-transformed (which avoids effects on redox-signaling that might arise due to transformation). Phosphorylation of Rb on Ser807/811 can be detected in HS68 cells with phospho-specific antibodies prior to the initiation of DNA synthesis starting at approximately 8 hours after growth factor stimuli and reaching maximum levels at approximately 27 hours (Figure 1b–e). We developed an analysis method to estimate the percentage of cells in G0/G1, S and G2/M phases directly from images of Hoechst stained cells (Figure 1d, Supplementary Information). The Rb-antibody staining intensities were also analyzed on a well-to-well basis to determine a threshold level to consider the cells Rb-positive (red circles Figure 1c and 1e). We validated that several control siRNAs inhibited or promoted cell cycle entry as expected (Supplementary Figure 2c).
Figure 1. Development of cell cycle entry assay.
a) Schematic of the cell cycle entry assay. b) Images of Hoechst stained cells that are serum starved (top) and PDGF stimulated (bottom). The lower right hand corner of the image of cells that have been stimulated by PDGF shows a zoomed region of the image with the mask that was used to calculate the Hoechst stain intensities. c) Images of Rb staining for the same cells shown in Figure 1b. The lower right hand corner of the image of cells that have been stimulated by PDGF shows a zoomed region with the mask that was determined form the Hoechst stain and was used to calculate the Rb staining intensities. Cells labeled with a red dot have an Rb staining intensity above the intensity limit that was set using the kmeans algorithm (see Supplementary Information) to consider cells as Rb positive. d) Histograms of the DNA content determined from the images shown in b). e) Scatter plots of the mean intensity of the phosphorylated Rb stain versus DNA content. Cells that fall above the Rb limit determined in the analysis by a kmeans algorithm are shown in red.
Screening the diced siRNA library identifies NOX4 and DUOX2 as necessary for the PDGF proliferative response
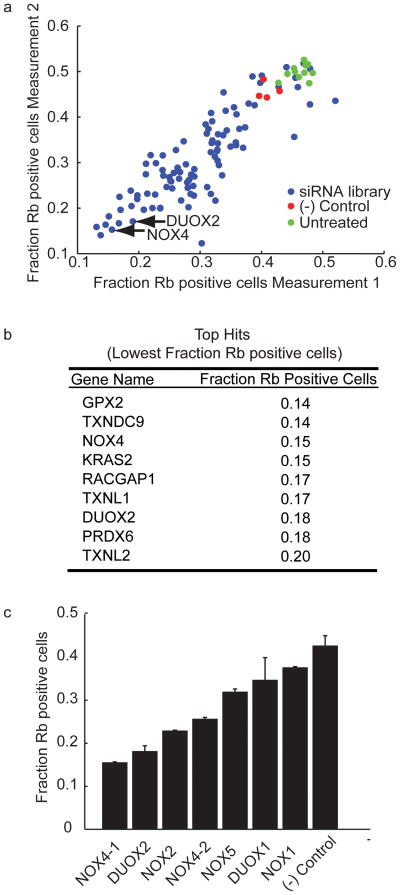
To investigate the role of ROS related signaling components in PDGF induced proliferation, we generated diced siRNA pools for a targeted set of 96 genes implicated in ROS production, in the regulation of ROS production or in cysteine/thiol reduction (Table 1 and Supplementary Table 1). For the screen, we transfected duplicate samples of HS68 cells with each of the siRNAs and measured the effect of each of the knockdowns in the cell cycle entry assay (Figure 2a). All of the siRNA pools either inhibited PDGF-induced cell cycle entry or did not have a significant effect compared to cells transfected with negative control siRNAs or to cells that were not transfected (Figure 2a). The 10% of siRNAs most strongly suppressing cell cycle entry are listed in Figure 2b. Of the ROS generating enzymes included in the siRNA library, the NADPH oxidases NOX4 and DUOX2 had the most significant effect on PDGF-induced cell cycle entry (Figure 2a–c). These two proteins were selected for follow-up studies.
Table 1.
Composition of the siRNA library.
| Category | Gene Names |
|---|---|
| NADPH OXIDASES | NOX1, CYBB (NOX2), NOX3, NOX4, NOX5, DUOX1, DUOX2, SYTL1 |
| NADPH OXIDASE Regulatory Subunits and Related Proteins | CYBA (p22 phox), NCF1 (p47phox), NOXO1, NOXA1, RUFY1 |
| Protein Kinase C Isoforms | PRKC1, PRKCQ, PRKD3, PRKCH, PRKCD, PKCB1, PRKCD, PRKCB1, PRKCABP, PRKCE PRKCA, PRKCDBP, PRKCG, PRKCZ |
| Small GTPases, GEFs, GAPs, GDIs | HRAS, KRAS, RAC2, CDC42, RACGAP1, ARHGEF7, ARHGAP30, RAP1GDS, ARHGDIA, ARHGDIB |
| Lipoxygenases | LOXHD1, ALOX5AP, ALOXE3, ALOX15B, ALOX5, ALOX12B, ALOX12 |
| Cycloxygenases | PTGS1 (Cox1), PTGS2 (Cox2) |
| Peroxiredoxins | PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, PRDX6 |
| Glutaredoxins | GLRX, GLRX2 |
| Glutathione Peroxidase | GPX1, GPX2, GPX3, GPX4, GPX5, , GPX6, GPX7, LOC389839 |
| Thioredoxins/Thioredoxin Reductases | TXN, TXN2, TXNRD1, TXNRD2, TXNRD3 |
| Thioredoxin like | TXNL1, TXNL2, TXNL4 |
| Thioredoxin Domain Containing Proteins | TXNDC, TXNDC2, TXNDC3, TXNDC9 |
| Catalase | CAT |
Figure 2. siRNA screening results.
a) Results from the siRNA screen comparing the duplicate measurements (one on each axis) for each gene and for the controls (GL-3 diced siRNA as the negative control and untransfected cells for comparison). b) List of the genes for which the corresponding siRNA resulted in the lowest Rb staining intensity (the list was arbitrarily cut off to include approximately the lowest 10%). c) Comparison of the fraction of Rb positive cells for six members of the NADPH oxidase family. Due to a separate listing for a NOX4 variant in an earlier version of the NCBI RefSeq database (subsequently removed), we generated two diced siRNA pools for different regions of NOX4 and found that NOX4-1 siRNA. The NOX4-1 diced siRNA pool also caused a more significant reduction in NOX4 mRNA levels when measured by quantitative RT-PCR (Supplementary Figure 3d). The NADPH oxidase NOX3 was excluded because the siRNA preparation was unsuccessful.
Verification of the role of NOX4 and DUOX2 in the PDGF proliferative response
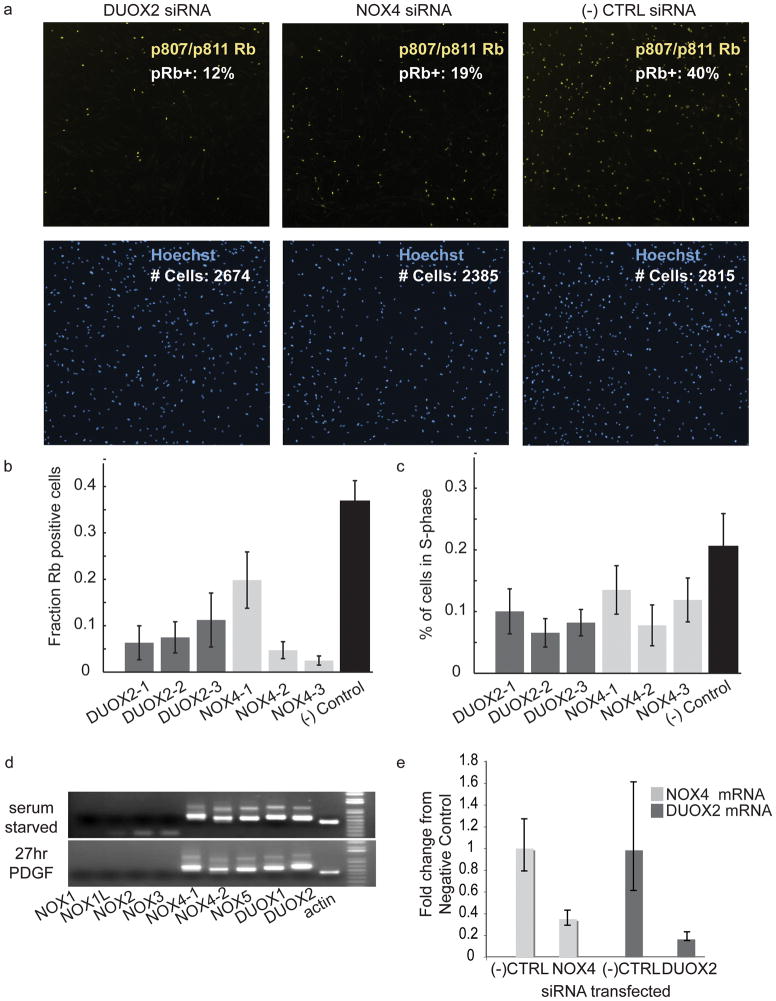
To verify that the effects of the siRNA against NOX4 and DUOX2 were not sequence- or preparation-specific, we repeated the cell cycle entry assay using synthetic siRNA from Dharmacon (Figure 3a–c). For both DUOX2 and NOX4, at least three independent synthetic siRNAs consistently caused a significant reduction in Rb phosphorylation compared to the negative control (Figure 3b and c). Knockdown of NOX4 and DUOX2 with Dharmacon synthetic pools also inhibited Rb-phosphorylation in the cell cycle entry analyzed by western blot (Supplementary Figure 3a–b). We also confirmed that transfecting cells with NOX4 and DUOX2 synthetic siRNA reduces fluorescence of the ROS-sensitive dye, 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA), which is consistent with a role for these two proteins in producing ROS (Supplementary Figure 3c). It is interesting that this reduction in ROS signaling occurs already in the absence of growth factor stimulation.
Figure 3. Verification that NOX4 and DUOX2 affect Rb phosphorylation in HS68 cells.
a) Images of cells transfected with negative control siRNA (10nM), NOX4 Dharmacon synthetic siRNA pool (10nM) and DUOX2 Dharmacon synthetic siRNA pool (10nM) stained with pSer807/pSer811 Rb antibodies (top) and stained with Hoechst stain (bottom). b) Combined results from three independent experiments (n=17 for each NOX4 and DUOX2 and n=13 for negative control) showing the effect of three individual siRNA for NOX4 (NOX4-1: CAGGAGGGCUGCUGAAGUA, NOX4-2: GGGCUAGGAUUGUGUCUAA, NOX4-3: GAUCACAGCCUCUACAUAU) and DUOX2 (DUOX2-1: GGAAUGGCCUCCCAGAUUU, DUOX2-2: GGAGUGAUCUCAACCCUAA and DUOX2-3: GAGGAUAAGUCCCGUCUAA) on the fraction of Rb positive cells. The final concentration of siRNA used was 10 or 20nM. The negative control was Dharmacon siGenome Non-Targeting pool number 2. c) Same as b) except figure shows quantification of fraction of cells in S-phase determined from measurements of the Hoechst stain intensity. d) Nested PCR from cDNA libraries generated from HS68 cells that were serum starved 48 hours (top) or serum starved for 48 hours then stimulated with PDGF for 27 hours (bottom). e) Quantitative RT-PCR from cDNA libraries generated from cells transfected with 20nM Dharmacon synthetic pools of NOX4 or DUOX2 siRNA or Dharmacaon negative control siRNA and serum starved 48 hours. Error bars represent the low and high fold change based on the standard deviation of the ΔΔCt values for three PCR reactions from the same cDNA library with the exception of detection of the DUOX2 transcript with DUOX2 siRNA which was only detectable in two of three PCR reactions.
To verify that NOX4 and DUOX2 are expressed in HS68 cells, we conducted RT-PCR (Figure 3d–e, Supplementary Figure 3d–f). The Ct values for DUOX2 determined by quantitative PCR were low (~ 35 for 50ng of starting total RNA). This could indicate that DUOX2 is a low abundance protein, that the synthesized protein is turned over slowly or that amplification by the Taqman primer set detects only a subset of the expressed splice variants. However, for both NOX4 and DUOX2, approximately a 70% knockdown was detected when we transfected siRNA pools against their respective transcripts (Figure 3e).
DUOX2 and NOX4 knockdown have a delayed effect on ERK1 phosphorylation
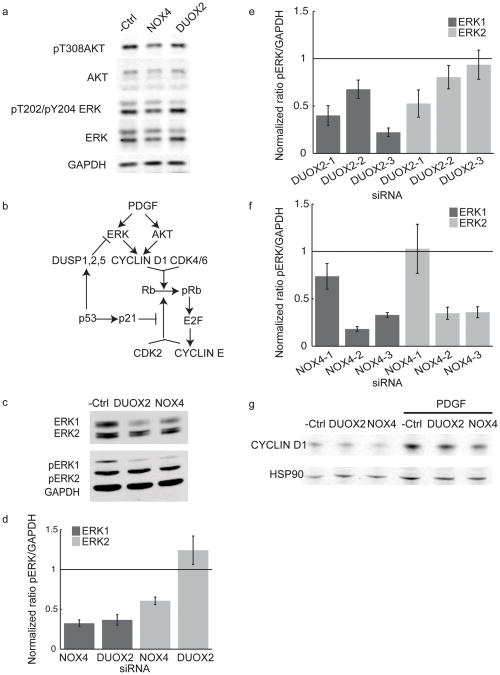
Our initial working hypothesis was that NOX4 and DUOX2 exert their effects on cell cycle entry via the production of ROS, and through inhibition of PTPs within minutes after the addition of growth factor stimuli. To test this hypothesis, we investigated the effects of NOX4 and DUOX2 knockdown on activation of the ERK and Akt signaling pathways which are rapidly activated in response to PDGF. After thirty minutes of PDGF stimulation, we were unable to observe a significant reduction in the ratio of pThr308 Akt to total Akt or the ratio of pThr202/pTyr204 ERK1/2 to total ERK in cells transfected with NOX4 or DUOX2 siRNA compared to those transfected with negative control siRNA (Figure 4a). These data suggest that NOX4 and DUOX2 knockdown do not inhibit the initial activation of the PDGF receptor kinase activity.
Figure 4. NOX4 and DUOX2 knockdown suppresses long term ERK1 phosphorylation but not short term ERK1 phosphorylation, Akt phosphorylation or Cyclin D1 expression.
a) Western blot showing ERK and Akt phosphorylation in response to PDGF after 30minutes for cells transfected with synthetic pools of NOX4, DUOX2 or non-targeting siRNA from Dharmacon. The complete western blot with molecular weight markers is shown in the supplementary information. b) Diagram of PDGF signaling pathways that were investigated to study the role of NOX4 and DUOX2 in proliferation. c) Western blot showing ERK phosphorylation in lysates from cells transfected with Dharmacon siGENOME synthetic pools of NOX4, DUOX2 or non-targeting siRNA after 9 hours of PDGF stimulation. The complete western blot with molecular weight markers is shown in the supplementary information. The bands were labeled as ERK1 and ERK2 based on their respective molecular weights, the identity of the two bands was also confirmed by transfecting cells with ERK1 and ERK2 specific siRNA. d) Quantification of ERK1 and ERK2 phosphorylation levels expressed as a ratio to levels of GAPDH from cell lysates transfected with synthetic pools of NOX4, DUOX2 or non-targeting siRNA (all Dharmacon siGENOME siRNA) after nine hours of PDGF stimuli. Error bars represent standard error of the mean, n=6. e) Quantification of ERK1 and ERK2 phosphorylation expressed as a ratio to GAPDH levels from cell lysates transfected with single synthetic siRNA targeting DUOX2 or non-targeting siRNA (all Dharmacon siGENOME siRNA) after nine hours of PDGF stimuli. Error bars represent standard error of the mean, n=6. f) Quantification of ERK1 and ERK2 phosphorylation expressed as a ratio to GAPDH levels from cell lysates transfected with single synthetic siRNA targeting NOX4 or non-targeting siRNA (all Dharmacon siGENOME siRNA) after nine hours of PDGF stimuli. Error bars represent standard error of the mean, n=6. g) Western blot showing CyclinD1 expression levels in response to PDGF with NOX4 and DUOX2 siRNA.
An alternative hypothesis is that NOX4 and DUOX2 do not significantly contribute to early growth factor receptor signaling but that they maintain long term ERK or Akt signaling or enhance a different proliferative signaling mechanism. Figure 4b shows some of the known alternative signaling events related to PDGF signaling and cell cycle entry. We tested for a delayed regulatory role of NOX4 and DUOX2 by measuring phosphorylation of ERK and Akt nine hours after PDGF stimulation. Akt Thr 308 phosphorylation was undetectable at this time point. However, nine hours after PDGF stimulation, NOX4 and DUOX2 knockdown with the siRNA pools reduced both pThr202/pTyr204 ERK1 and total ERK1 (Fig 4c–d, Supplementary Figure 4a, in comparison to GAPDH). The effect on the ERK1 levels is relatively small for several of the siRNAs (Supplementary Figure 4a–c), suggesting that the primary effect of NOX4 and DUOX2 knockdown on ERK signaling is at the level of ERK1 phosphorylation (Fig 4e–f). Since it has been shown that persistent ERK activation promotes the expression of cyclin D1 (Chambard et al 2007) and since an increase in cyclin D1 promotes Rb-phosphorylation and cell cycle entry, we tested for an effect of NOX4 and DUOX2 knockdown on PDGF triggered cyclin D1 expression. We did not find a significant effect of NOX4 or DUOX2 knockdown on cyclin D1 expression, indicating that these proteins regulate an alternative pathway to promote cell cycle entry (Figure 4g).
DUOX2 and NOX4 promote Rb phosphorylation through p21-p53 dependent pathways
We hypothesized that NOX4 and DUOX2 may promote cell cycle entry by downregulating the Cip/Kip (p21, p27 and p57) protein families that inhibit cyclin/cdk activity by binding to cyclin/cdk complexes (Figure 4b). Specifically, we focused on p21, since previous reports have shown that p53, a transcription factor that regulates the cyclin/cdk inhibitor p21, can be regulated by ROS (Hainaut and Mann 2001, Sun et al 2003, Velu et al 2007). We therefore decided to test if NOX4 and DUOX2 promote PDGF-induced Rb phosphorylation and cell cycle progression through regulation of p53 and/or p21.
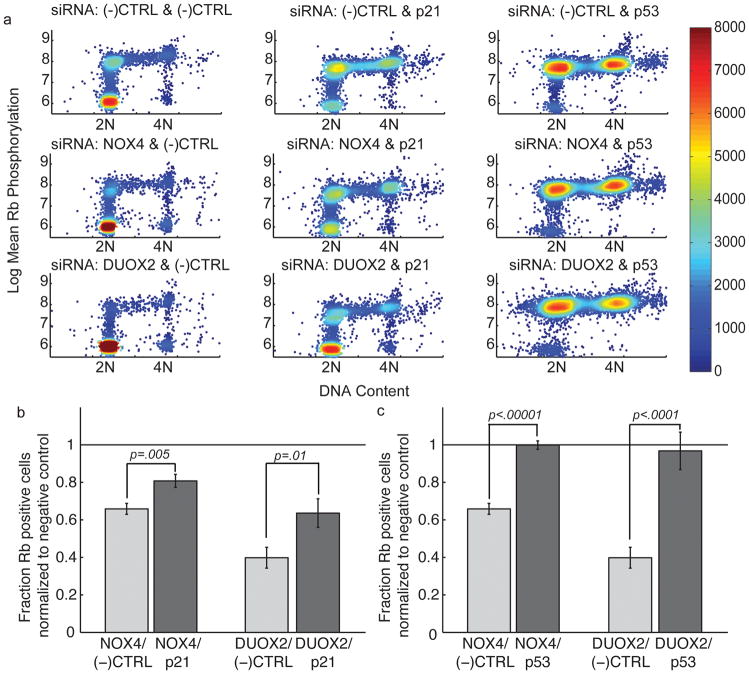
To test this, we first conducted co-knockdown experiments to see if simultaneously transfecting p21 or p53 siRNA with NOX4 or DUOX2 siRNA would reduce PDGF induced Rb phosphorylation. We show that the effects of NOX4 or DUOX2 knockdown could be partially compensated for by p21 knockdown (Figure 5a and b). In the context of a p53 knockdown, NOX4 and DUOX2 knockdown no longer had a strong effect on Rb-phosphorylation levels, indicating that NOX4 and DUOX2 only affect Rb-phosphorylation when p53 is present (Figure 5a and c). Importantly, p21 and p53 siRNA could not compensate for the inhibition of PDGF induced Rb phosphorylation caused by cyclin D1 knockdown, providing further support that NOX4 and DUOX2 exert their effects on PDGF signaling through mechanisms that are independent of the signaling pathways leading to cyclin D1 expression (Supplementary Figure 6a–b).
Figure 5. Co-knockdown of NOX4 or DUOX2 with p53 or p21 restores cell cycle entry.
a) DNA content scatter plots in which the color of the points represents the density of the points in a given region. The density is calculated by the reciprocal of the area of the voronoi region surrounding the centroid of a point. The two siRNA that were transfected are listed at the top of each plot. For all of the co-knockdown experiments the total siRNA concentration was kept constant by adding negative control siRNA to ensure that any effects we observed were not due to concentration differences. b) Results of the cell cycle entry assay for co-knockdown of synthetic pools of NOX4, DUOX2 or non-targeting siRNA (all Dharmacon siGENOME siRNA) with p21 or non-targeting negative control siRNA. The light grey bars were normalized to the fraction of Rb-positive cells in samples transfected with (−)CTRL/(−)CTRL siRNA. The dark grey bars were normalized to the fraction of Rb-positive cells for cells transfected with (−)CTRL/p21 siRNA. P-values were calculated using a t-test assuming a normal distribution. c) Results of the cell cycle entry assay for co-knockdown of synthetic pools of NOX4, DUOX2 or non-targeting siRNA (all Dharmacon siGENOME siRNA) with p53 or non-targeting negative control siRNA. The light grey bars were normalized to the fraction of Rb-positive cells in samples transfected with (−)CTRL/(−)CTRL siRNA. The dark grey bars were normalized to the fraction of Rb-positive cells for cells transfected with (−)CTRL/p53 siRNA.
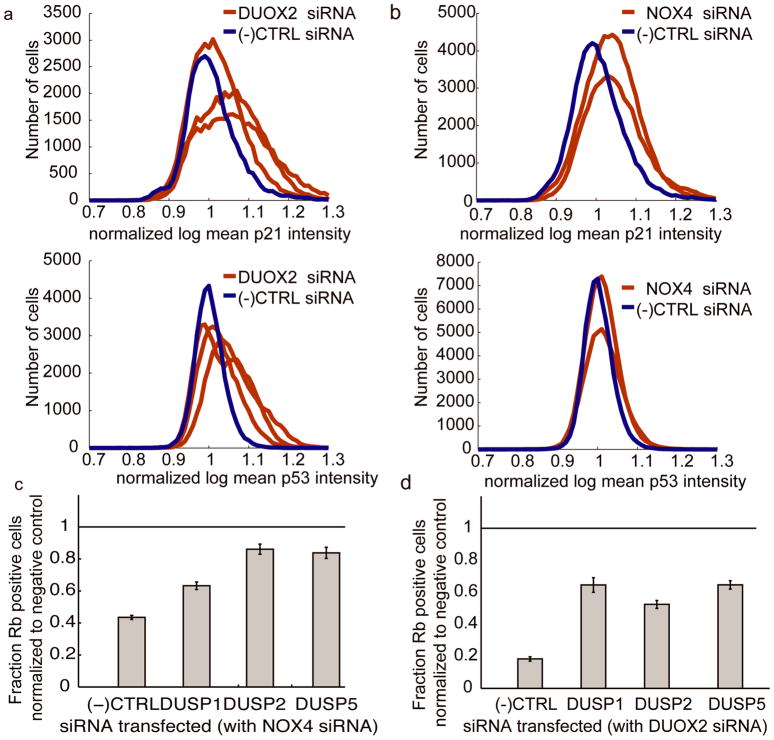
To demonstrate a more direct effect of DUOX2 and NOX4 knockdown on p53 or p21, we measured p53 and p21 immunofluorescence in the cell cycle entry assay in the presence of NOX4 or DUOX2 siRNA. We first validated with siRNA knockdown experiments that the p53 and p21 antibodies were effective (Supplementary Figure 5). All three DUOX2 synthetic siRNA sequences that inhibit Rb phosphorylation (Figure 3b) increased both p53 and p21 protein levels 18hours after PDGF stimulation (Figure 6a–b). The two NOX4 synthetic siRNA sequences that most strongly inhibited Rb phosphorylation (Figure 3b) as well as two additional NOX4-targeting siRNA sequences also increase p53 and p21 protein levels (Figure 6a–b, Supplementary Figure 7).
Figure 6. NOX4 and DUOX2 are required to suppress p53-dependent signaling pathways.
a) DUOX2 knockdown increases p21 and p53 levels. Intensity distributions of single cell immunofluorescence measurements of cells stimulated with PDGF for 18 hours and stained with p21 antibodies (top) or p53 antibodies (bottom). Cells were transfected with DUOX2-1, DUOX2-2 DUOX2-3 or negative control synthetic siRNA (N=12, for p21, p< .001 for all three DUOX2 siRNAs and, for p53, p<.05 for DUOX2-1 and DUOX2-2, see supplementary material). b) NOX4 knockdown increases p21 and p53 levels. Panels same as a) except that cells were transfected with NOX4-2, NOX4-3 or negative control siRNAs ( N=18, for p21, p<.001 for NOX4-2 and NOX4-3 siRNAs and, for p53, p<.05 for the same siRNAs). c) Compensation by dual specificity phosphatases. DUSP1, DUSP2 and DUSP5 phosphases were knocked down together with NOX4, showing partial compensation of Rb phosphorylation ( Dharmacon siGENOME siRNAs for NOX4 and dicer generated pools targeting the DUSPs). d) Same as c) except that DUOX2 was targeted instead of NOX4.
Since p53 compensated for NOX4 and DUOX2 knockdown more strongly than p21, it suggested that more than one pathway downstream of p53 may affect Rb phosphorylation. In addition to regulating p21 transcription, p53 has also been proposed to transcriptionally regulate the dual specificity phosphatases (DSPs) MKP-1/DUSP1, PAC1/DUSP2 and DUSP5 (Li et al 2003, Liu et al 2008, Wu 2004). These three nuclear localized dual specificity phosphatases can all dephosphorylate ERK (reviewed in (Keyse 2008)). We therefore tested if siRNA targeting MKP-1/DUSP1, PAC1/DUSP2 or DUSP5 could also compensate for NOX4 or DUOX2 knockdown. Using dicer generated siRNA, MKP-1/DUSP1, PAC1/DUSP2 and DUSP5 knockdown all partially compensated for NOX4 and DUOX2 knockdown (Figure 6c–d). These results are consistent with the hypothesis that NOX4 and DUOX2 target the MAPK pathway via the dual-specificity phosphatases and that inhibition of DUSP transcription may be an additional mechanism by which p53 knockdown compensates for NOX4 and DUOX2 knockdown.
Discussion
We used a targeted siRNA screen for redox related proteins and identified DUOX2 and NOX4 as regulators of PDGF induced cell cycle entry. Markedly, rather than acting on the early receptor signaling phosphorylation events that happen within minutes after growth factor stimulation (Bae et al 2000, Meng et al 2002, Park et al 2004) and lead to cyclin D expression (Ranjan et al 2006), we found a key role of NOX4 and DUOX2 in PDGF signaling after cyclin D1 accumulation through the down-regulation of p53 and p21 protein levels. The involvement of p53 and p21 in NOX4 and DUOX2 signaling downstream of PDGF is also supported by our co-knockdown compensation experiments in which we combined siRNAs against NOX4 or DUOX2 with siRNAs against p53 or p21 and show that p53 or p21 siRNA were able to wholly or partially revert the suppression of proliferation by NOX4 and DUOX2 knockdown. Together these findings provide evidence that NOX4 and DUOX2 play a role in a p53-dependent checkpoint mechanism for cell cycle entry.
Regulation of cell cycle entry by NOX4 and DUOX2
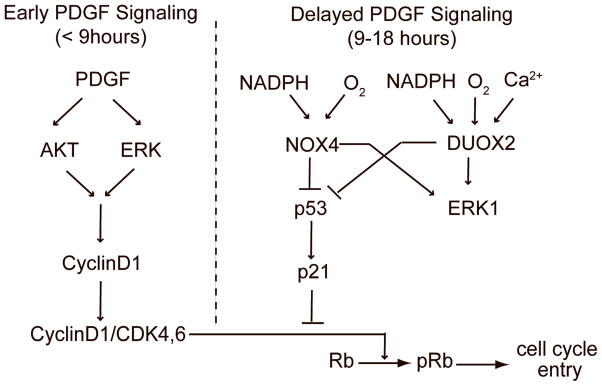
Previous studies on the causes and consequences of ROS generation in PDGF signaling have primarily focused on the signaling pathways immediately downstream of growth factor receptor stimulation (5–60 minutes) (Bae et al 2000, Meng et al 2002, Park et al 2004). Our study points towards a different role for NOX4 and DUOX2 in regulating pathways that can influence longer term signaling events (Figure 7). NOX4 and DUOX2 knockdown did not affect early signaling events including activation of the AKT and ERK pathways and induction of cyclin D1 expression. However, NOX4 and DUOX2 knockdown did induce a delayed effect on ERK1 phosphorylation nine hours after PDGF receptor stimulation. While this observation suggests a role for NOX4 and DUOX2 in MAPK signaling, the lack of a significant effect on cyclin D expression points to alternative roles of this phosphorylation. For example, ERK activity is also important for cell cycle progression by regulating CDK2 nuclear localization and by inhibiting the expression of anti-proliferative genes (reviewed in (Chambard et al 2007).
Figure 7.
Proposed model for the role of NOX4 and DUOX2 in promoting cell cycle entry.
Since the effect of NOX4 and DUOX2 knockdown on MAPK signaling does not explain how these proteins affect cell cycle entry, we investigated whether NOX4 or DUOX2 might target inhibitors of cyclin/cdk complexes. Surprisingly, we show that co-knockdown of NOX4 or DUOX2 together with p21, a cyclin/cdk complex inhibitor, or p53, which regulates p21 transcription, compensated for NOX4 or DUOX2 knockdown. The compensating effect of p53 was more consistent and significant than that of p21, suggesting that p53 affects Rb phosphorylation through more than one pathway. This also suggested that p53 is an important target for NOX4 and DUOX2. This was further supported by our finding that NOX4 or DUOX2 knockdown increased levels of both p53 and p21. Furthermore, knockdown of three dual specificity phosphatases that are transcriptionally regulated by p53 also partially compensated for NOX4 or DUOX2 knockdown. This was of particular interest since it could account for how an effect of NOX4 or DUOX2 on p53 signaling could be mediated by a delayed effect of NOX4 and DUOX2 knockdown on MAPK signaling via regulation of these phosphatases.
In addition to a regulation of the concentration of p53, it is also interesting to note that p53 has redox sensitive cysteine residues in the DNA binding domain and oxidation of these residues have been shown to inhibit p53 DNA binding activity, providing a second likely mode of regulation (Hainaut and Mann 2001, Sun et al 2003, Velu et al 2007). Together, this provides support for a model of a signaling pathway that links ROS production by NOX4 and DUOX2 to oxidation and inhibition of p53. Given the known complexity of p53 and p21 associated regulatory mechanisms, alternative or additional mechanisms cannot be completely excluded and it is for example conceivable that p53 may be targeted by the NADPH oxidases through ROS independent mechanisms. Further clarification of the detailed regulatory mechanisms of this NOX4 and DUOX2 pathway is an avenue for future studies that could be important for understanding the redox regulation of p53.
Our evidence that NOX4 acts through a p53 dependent pathway could reconcile several disparate previously proposed NOX4 functions. In addition to the role in PDGF signaling that we have demonstrated, NOX4 has been linked to endothelial cell proliferation (Petry et al 2006), TGF-Beta induced cell proliferation (Sturrock et al 2006, Sturrock et al 2007), and TGF-beta induced differentiation (Cucoranu et al 2005). TGF-beta activates the SMAD family of transcription factors. In vascular smooth muscle cells, NOX4 was shown to maintain the differentiated phenotype of cells by promoting SMAD phosphorylation and smooth muscle actin expression. Recently, several papers have revealed that p53 is an essential partner of SMADs in regulating the transcription of target genes (Wilkinson et al 2005, Wilkinson et al 2008). While further investigation is required, our data demonstrating that p53 is a target of NOX4 could explain how NOX4 is involved in both TGF-beta and PDGF signaling pathways.
NOX4 and DUOX2 as part of a p53-dependent NADPH sensing checkpoint
Our data raise the question of what function NOX4 and DUOX2 may have in regulating p53-p21 dependent signaling pathways. In addition to the effect of NOX4 and DUOX2 on PDGF induced cell cycle entry, we also observed that NOX4 and DUOX2 knockdown can affect basal ROS levels, suggesting that these proteins have a function that is at least partially independent of the PDGF receptor (Supplementary Figure 3c). This led us to consider the interesting hypothesis that NOX4 and DUOX2 may be acting in basal and stimulated cells as sensors for their two substrates, NADPH and oxygen. Both NADPH and oxygen are necessary for cell proliferation. Signaling from NOX4 and DUOX2 to p53 could then provide a mechanism for cells to sense if sufficient levels oxygen and NADPH are present to complete the cell division cycle. NADPH oxidases have previously been suggested to act as sensors for oxygen (reviewed in (Ward 2008). NADPH, which is utilized in proliferation in the synthesis of cholesterol, fatty acids and deoxyribonucleic acids could also affect the activity of the NADPH oxidases. P53 is a target for other enzymes that sense precursors necessary for cell proliferation. Low glucose levels and depletion of ribonucleotides can both lead to p53 activation (Jones et al 2005, Linke et al 1996). Based on our new data, we propose that NOX4 and DUOX2 may also act through p53 and function as joint sensors for oxygen and NADPH. This control mechanism could ensure that proliferation persists only when sufficient NADPH and oxygen are present.
In conclusion, we have demonstrated that knockdown of NOX4 and DUOX2 in human normal fibroblasts results in an inability to enter the cell cycle. Co-knockdown of these proteins with p53 restores normal cell cycle entry and knockdown of either of these two proteins alone leads to an increase in p53 and p21 levels. We propose that NADPH levels may be a limiting factor that is sensed in cells by the NADPH oxidases. Proper activation of NADPH production could then be sensed by p53, providing an additional checkpoint that is necessary for the initiation of cell cycle entry.
Supplementary Material
Acknowledgments
We thank Dr. Won Do Heo for his assistance with the cell cycle entry assay, Dr. Dan Kaplan, Dr. Sean Collins and Dr. Karlene Cimprich for their critical reading of the manuscript and members of the Meyer lab and Renee Paulsen for helpful discussions. This work was supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund (AS), by a Helen Hay Whitney Foundation/Paul Sigler Agouron Institute post-doctoral fellowship (AS) and by an NIH grant R33 CA 120732 (TM).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Bae YS, Kang SW, Seo MS, Baines IC, Tekle E, Chock PB, et al. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. The Journal of biological chemistry. 1997;272:217–221. [PubMed] [Google Scholar]
- Bae YS, Sung JY, Kim OS, Kim YJ, Hur KC, Kazlauskas A, et al. Platelet-derived growth factor-induced H(2)O(2) production requires the activation of phosphatidylinositol 3-kinase. The Journal of biological chemistry. 2000;275:10527–10531. doi: 10.1074/jbc.275.14.10527. [DOI] [PubMed] [Google Scholar]
- Burch PM, Heintz NH. Redox regulation of cell-cycle re-entry: cyclin D1 as a primary target for the mitogenic effects of reactive oxygen and nitrogen species. Antioxidants & redox signaling. 2005;7:741–751. doi: 10.1089/ars.2005.7.741. [DOI] [PubMed] [Google Scholar]
- Burhans WC, Heintz NH. The cell cycle is a redox cycle: Linking phase-specific targets to cell fate. Free radical biology & medicine. 2009 doi: 10.1016/j.freeradbiomed.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Chambard JC, Lefloch R, Pouyssegur J, Lenormand P. ERK implication in cell cycle regulation. Biochimica et biophysica acta. 2007;1773:1299–1310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Chen K, Kirber MT, Xiao H, Yang Y, Keaney JF., Jr Regulation of ROS signal transduction by NADPH oxidase 4 localization. The Journal of cell biology. 2008;181:1129–1139. doi: 10.1083/jcb.200709049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucoranu I, Clempus R, Dikalova A, Phelan PJ, Ariyan S, Dikalov S, et al. NAD(P)H oxidase 4 mediates transforming growth factor-beta1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circulation research. 2005;97:900–907. doi: 10.1161/01.RES.0000187457.24338.3D. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Mann K. Zinc binding and redox control of p53 structure and function. Antioxidants & redox signaling. 2001;3:611–623. doi: 10.1089/15230860152542961. [DOI] [PubMed] [Google Scholar]
- Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, et al. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free radical biology & medicine. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molecular cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer metastasis reviews. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nature reviews. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- Lassegue B, Sorescu D, Szocs K, Yin Q, Akers M, Zhang Y, et al. Novel gp91(phox) homologues in vascular smooth muscle cells : nox1 mediates angiotensin II-induced superoxide formation and redox-sensitive signaling pathways. Circulation research. 2001;88:888–894. doi: 10.1161/hh0901.090299. [DOI] [PubMed] [Google Scholar]
- Laurent E, McCoy JW, 3rd, Macina RA, Liu W, Cheng G, Robine S, et al. Nox1 is over-expressed in human colon cancers and correlates with activating mutations in K-Ras. International journal of cancer. 2008;123:100–107. doi: 10.1002/ijc.23423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Kwon KS, Kim SR, Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. The Journal of biological chemistry. 1998;273:15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- Li M, Zhou JY, Ge Y, Matherly LH, Wu GS. The phosphatase MKP1 is a transcriptional target of p53 involved in cell cycle regulation. The Journal of biological chemistry. 2003;278:41059–41068. doi: 10.1074/jbc.M307149200. [DOI] [PubMed] [Google Scholar]
- Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes & development. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YX, Wang J, Guo J, Wu J, Lieberman HB, Yin Y. DUSP1 is controlled by p53 during the cellular response to oxidative stress. Mol Cancer Res. 2008;6:624–633. doi: 10.1158/1541-7786.MCR-07-2019. [DOI] [PubMed] [Google Scholar]
- Mahadev K, Motoshima H, Wu X, Ruddy JM, Arnold RS, Cheng G, et al. The NAD(P)H oxidase homolog Nox4 modulates insulin-stimulated generation of H2O2 and plays an integral role in insulin signal transduction. Molecular and cellular biology. 2004;24:1844–1854. doi: 10.1128/MCB.24.5.1844-1854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng TC, Fukada T, Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Molecular cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- Myers JW, Jones JT, Meyer T, Ferrell JE., Jr Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nature biotechnology. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- Park HS, Lee SH, Park D, Lee JS, Ryu SH, Lee WJ, et al. Sequential activation of phosphatidylinositol 3-kinase, beta Pix, Rac1, and Nox1 in growth factor-induced production of H2O2. Molecular and cellular biology. 2004;24:4384–4394. doi: 10.1128/MCB.24.10.4384-4394.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Jin DK, Shin SM, Jang MK, Longo N, Park JW, et al. Impaired generation of reactive oxygen species in leprechaunism through downregulation of Nox4. Diabetes. 2005;54:3175–3181. doi: 10.2337/diabetes.54.11.3175. [DOI] [PubMed] [Google Scholar]
- Petry A, Djordjevic T, Weitnauer M, Kietzmann T, Hess J, Gorlach A. NOX2 and NOX4 mediate proliferative response in endothelial cells. Antioxidants & redox signaling. 2006;8:1473–1484. doi: 10.1089/ars.2006.8.1473. [DOI] [PubMed] [Google Scholar]
- Ranjan P, Anathy V, Burch PM, Weirather K, Lambeth JD, Heintz NH. Redox-dependent expression of cyclin D1 and cell proliferation by Nox1 in mouse lung epithelial cells. Antioxidants & redox signaling. 2006;8:1447–1459. doi: 10.1089/ars.2006.8.1447. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Bae YS, Lee SR, Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. 2000;2000:PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- Sturrock A, Cahill B, Norman K, Huecksteadt TP, Hill K, Sanders K, et al. Transforming growth factor-beta1 induces Nox4 NAD(P)H oxidase and reactive oxygen species-dependent proliferation in human pulmonary artery smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2006;290:L661–L673. doi: 10.1152/ajplung.00269.2005. [DOI] [PubMed] [Google Scholar]
- Sturrock A, Huecksteadt TP, Norman K, Sanders K, Murphy TM, Chitano P, et al. Nox4 mediates TGF-beta1-induced retinoblastoma protein phosphorylation, proliferation, and hypertrophy in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1543–1555. doi: 10.1152/ajplung.00430.2006. [DOI] [PubMed] [Google Scholar]
- Sun XZ, Vinci C, Makmura L, Han S, Tran D, Nguyen J, et al. Formation of disulfide bond in p53 correlates with inhibition of DNA binding and tetramerization. Antioxidants & redox signaling. 2003;5:655–665. doi: 10.1089/152308603770310338. [DOI] [PubMed] [Google Scholar]
- Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science (New York, NY) 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer research. 1991;51:794–798. [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry. 2007;46:7765–7780. doi: 10.1021/bi700425y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JP. Oxygen sensors in context. Biochimica et biophysica acta. 2008;1777:1–14. doi: 10.1016/j.bbabio.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Wilkinson DS, Ogden SK, Stratton SA, Piechan JL, Nguyen TT, Smulian GA, et al. A direct intersection between p53 and transforming growth factor beta pathways targets chromatin modification and transcription repression of the alpha-fetoprotein gene. Molecular and cellular biology. 2005;25:1200–1212. doi: 10.1128/MCB.25.3.1200-1212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DS, Tsai WW, Schumacher MA, Barton MC. Chromatin-bound p53 anchors activated Smads and the mSin3A corepressor to confer transforming-growth-factor-beta-mediated transcription repression. Molecular and cellular biology. 2008;28:1988–1998. doi: 10.1128/MCB.01442-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nature chemical biology. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free radical biology & medicine. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Wu GS. The functional interactions between the p53 and MAPK signaling pathways. Cancer biology & therapy. 2004;3:156–161. doi: 10.4161/cbt.3.2.614. [DOI] [PubMed] [Google Scholar]
- Yamaura M, Mitsushita J, Furuta S, Kiniwa Y, Ashida A, Goto Y, et al. NADPH oxidase 4 contributes to transformation phenotype of melanoma cells by regulating G2-M cell cycle progression. Cancer research. 2009;69:2647–2654. doi: 10.1158/0008-5472.CAN-08-3745. [DOI] [PubMed] [Google Scholar]
- Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nature cell biology. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. The Journal of biological chemistry. 1997;272:12738–12746. doi: 10.1074/jbc.272.19.12738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.