SUMMARY
In the human fungal pathogen Cryptococcus neoformans, the SREBP ortholog Sre1 is important for adaptation and growth in nutrient-limiting host tissues. In this study, we characterize the C. neoformans serotype A Sre1 and its activating protease, Stp1. We demonstrate that Stp1 is a functionally conserved ortholog of the mammalian Site-2 protease and that Stp1 cleaves Sre1 within its predicted first transmembrane segment. Gene expression analysis revealed that Stp1 is required for both Sre1-dependent and Sre1-independent gene transcription, indicating that other substrates of Stp1 may exist. Using gas chromatography, we showed that Sre1 and Stp1 are required for both normoxic and hypoxic ergosterol biosynthesis, and therefore cells lacking SRE1 or STP1 are defective for growth in the presence of low levels of the ergosterol biosynthesis inhibitors, itraconazole and 25-thialanosterol. Importantly, our studies demonstrated fungicidal effects of itraconazole and 25-thialanosterol toward sre1Δ and stp1Δ cells, demonstrating that the Sre1 pathway is required for both growth and survival in the presence of sterol biosynthesis-inhibiting antifungal drugs. Given the need for fungicidal drugs, we propose that inhibitors of Stp1, Sre1, or other regulators of Sre1 function administered in combination with a sterol synthesis inhibitor could prove an effective anti-cryptococcal therapy.
Keywords: Cryptococcus neoformans, SREBP, hypoxia, sterol, ergosterol, azole, Sre1
INTRODUCTION
Cryptococcus neoformans is a basidiomycetous yeast that causes life-threatening meningoencephalitis primarily in immunocompromised patients, particularly individuals with HIV/AIDS (Kwon-Chung and Bennett, 1992; Mitchell and Perfect, 1995). Yeast cells or possibly spores are inhaled and disseminate to the brain where they establish growth and formation of cystic lesions. Several factors have been identified as being required to promote disease, including the ability of C. neoformans cells to grow at host body temperature, the production of melanin, and the production of a polysaccharide capsule surrounding the cell wall of C. neoformans cells (Kwon-Chung and Bennett, 1992). Despite the identification of these well-known virulence requirements, mechanisms underlying the adaptation of C. neoformans cells to the host environment remain poorly understood.
The C. neoformans sterol regulatory element-binding protein (SREBP) pathway was recently found to be required for host adaptation and virulence (Chang et al., 2007; Chun, Liu, and Madhani, 2007). C. neoformans SREBP, called Sre1, is a membrane-bound transcription factor that stimulates ergosterol production in response to sterol depletion, for example when oxygen-dependent ergosterol synthesis is limited by hypoxia. Our studies in the C. neoformans var. neoformans serotype D strain demonstrated that Sre1 is proteolytically activated under low oxygen conditions and that SRE1 is required for virulence in a mouse model of infection (Chang et al., 2007). Although sre1Δ cells enter the brain, they failed to cause lethal infection in mice, suggesting a role for Sre1 in growth or survival in the host tissue. The Cryptococcus neoformans species complex is classified into five serotypes (A, B, C, D and A/D) and serotypes A and D cause the majority of human disease (Bennett, Kwon-Chung, and Howard, 1977). A parallel study of Sre1 in C. neoformans serotype A found that Sre1 is also required for virulence in mice (Chun, Liu, and Madhani, 2007), however, while serotype D SRE1 was completely required to cause fatal infection in mice, serotype A SRE1 was found to be only partially required for virulence.
Most fungal infections are currently treated with drugs that inhibit ergosterol biosynthesis, including the commonly used azole class of drugs. Azoles have been shown to be effective against a wide-range of pathogenic fungi, including species of Cryptococcus, Candida, Aspergillus, Coccidioides, and Histoplasma (Sheehan, Hitchcock, and Sibley, 1999; Scheinfeld, 2007). Insomuch as C. neoformans Sre1 regulates genes encoding ergosterol biosynthetic enzymes, SRE1 was shown to be required for growth in the presence of low levels of azoles (Chang et al., 2007; Chun, Liu, and Madhani, 2007; Lee et al., 2007). Importantly, SREBP displays a similar requirement in Aspergillus fumigatus (Willger et al., 2008). These findings suggest that Sre1, or regulators of Sre1, may be promising targets for antifungal drug design due to the fact that Sre1 inhibitors may display synergistic effects when used in combination with current antifungals.
SREBP transcription factors were first characterized in mammalian cells as regulators of lipid homeostasis (Goldstein, DeBose-Boyd, and Brown, 2006). Mammalian SREBPs are synthesized as ER membrane-bound inactive precursors. SREBPs contain two transmembrane segments separated by a short lumenal loop and the N- and C-termini face the cytosol (Fig. 2A). When cellular cholesterol levels decrease, the SREBP cleavage activating protein (Scap) escorts SREBP to the Golgi apparatus where SREBP undergoes two sequential proteolytic cleavage events catalyzed by the Site-1 and Site-2 proteases. The Site-1 protease is a Golgi-localized serine protease that performs the first cleavage event in the lumenal loop of SREBP. Following Site-1 cleavage, the Site-2 zinc metalloprotease catalyzes the cleavage of SREBP within its first transmembrane segment. Site-2 protease cleavage liberates the N-terminal bHLH transcription factor domain of SREBP from the membrane, allowing translocation to the nucleus and target gene transcription.
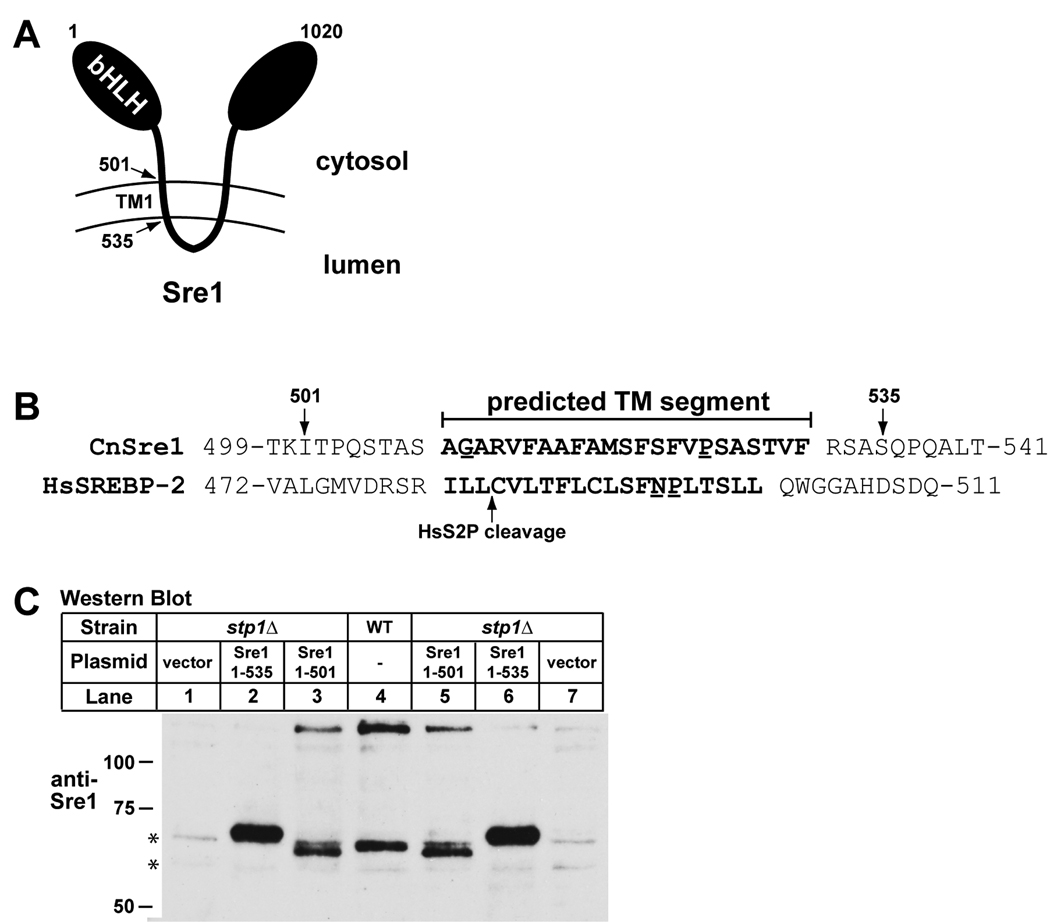
Figure 2. Sre1 cleavage occurs within the first transmembrane segment.
A. Diagram of Sre1 in the membrane. TM1 indicates predicted first transmembrane segment. Amino acid positions are given and bHLH denotes the N-terminal basic helix-loop-helix leucine zipper DNA binding domain. B. Protein sequence alignment of the predicted first transmembrane segments of C. neoformans and human SREBP-2. Putative helix-destabilizing residues are underlined. C. Western blot of phosphatase-treated whole cell extracts (40 µg) from wild-type and stp1Δ cells transformed with empty vector, or plasmids expressing the indicated Sre1 truncations. Lanes 1 and 7, 2 and 6, 3 and 5 are duplicates of stp1Δ cells expressing empty vector, Sre1 (aa1-535), and Sre1 (aa1-501), respectively. Cells were grown to exponential phase in YES medium at ambient oxygen concentrations. Asterisks indicate non-specific, cross-reacting proteins detected by anti-Sre1 antiserum.
Site-2 proteases have been identified in nearly all sequenced genomes, including prokaryotes (Rawson and Li, 2007; Makinoshima and Glickman, 2006). In bacteria which lack SREBPs and sterols, Site-2 proteases function in multiple pathways, including several types of stress responses (Rawson and Li, 2007; Makinoshima and Glickman, 2006). Interestingly, the Site-2 protease in the bacterial pathogen Mycobacterium tuberculosis plays a role in maintaining cell envelope lipid composition and is required for persistence of infection in mice (Makinoshima and Glickman, 2005). Despite their widespread conservation, most commonly studied fungi lack clear homologs of Site-2 proteases, particularly fungi belonging to the phylum Ascomycota. The basidiomycete C. neoformans contains an identifiable Site-2 protease homolog, named Site-2 Protease 1 (Stp1). Stp1 mutants display similar phenotypes to Sre1 mutant strains, suggesting a role for Stp1 in regulating Sre1 activity (Chun, Liu, and Madhani, 2007).
In this study, we performed a detailed characterization of Stp1. We provide evidence that Stp1 is a functional metalloprotease that cleaves Sre1 within its first transmembrane segment. Detailed gene expression analysis revealed that Stp1 is required for Sre1 transcriptional activity and that additional Stp1 substrates may exist. Finally, we further characterized the role of Sre1 and Stp1 in resistance to sterol biosynthesis inhibitors. Our results demonstrate that inhibition of the Sre1 pathway transforms these fungistatic inhibitors into fungicidal drugs. Based on our findings, we propose that regulators of the Sre1 pathway, including Stp1, are promising targets for the development of novel antifungal therapeutics.
RESULTS
STP1 encodes a protein required for Sre1 proteolysis
The serotype A reference strain C. neoformans H99 contains a single sequence homolog of the human Site-2 protease called STP1 that is required for hypoxic growth and virulence (Chun, Liu, and Madhani, 2007). STP1 codes for a 594 amino acid protein (Broad ID: CNAG_05742.2), and protein sequence alignment shows a low level of sequence identity to the human Site-2 protease (Fig. 1A). Similar to other Site-2 protease orthologs, Stp1 is predicted to be extremely hydrophobic. Hydropathy plots of the human Site-2 protease and Stp1 obtained using Phobius software (http://phobius.sbc.su.se/) show a similar number and position of transmembrane domains between the two proteins (data not shown). Importantly, the key catalytic residues for Site-2 protease function are conserved, including two histidines and a glutamate in the N-terminus and one aspartate near the C-terminus (Fig. 1A, underlined residues). These residues coordinate the catalytic zinc ion and water molecule in the enzyme active site (Feng et al., 2007).
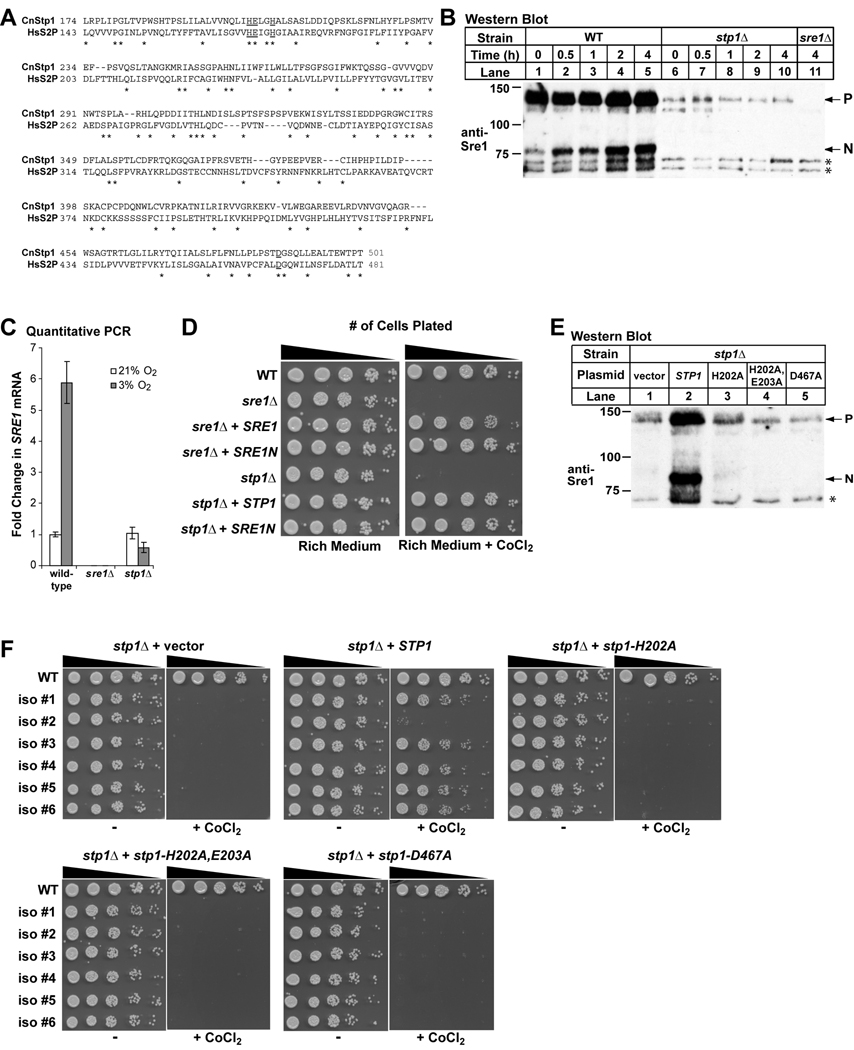
Figure 1. C. neoformans Stp1 is a conserved Site-2 protease required for Sre1 activation.
A. Protein alignment of human Site-2 protease (NCBI refseq ID: NP_056968) and C. neoformans Stp1 (BROAD ID: CNAG_05742.2) using SIM alignment tool (http://ca.expasy.org/tools/sim.html). Putative catalytic residues are underlined. B. C. neoformans serotype A wild-type and stp1Δ cells were shifted from 21% oxygen to 3% oxygen for indicated periods of time. Immunoblot analysis was performed on whole cell extracts (40 µg) using anti-Sre1 antiserum. P and N denote the precursor and nuclear forms, respectively. Asterisks indicate non-specific, cross-reacting proteins detected by anti-Sre1 antiserum. C. C. neoformans cells from the indicated strains were grown under ambient (21%) or 3% oxygen for 2 hours, and RNA was harvested. SRE1 transcript levels were quantified by real time RT-PCR and normalized to that in wild-type cells at 21% oxygen. Error bars represent standard deviation from three biological replicate experiments. D. Five-fold serial dilutions of C. neoformans cells from the indicated strains were spotted on rich medium and rich medium containing 0.3 mM CoCl2 and incubated at 30°C for 3 days. E. C. neoformans stp1Δ cells transformed with either empty vector, STP1, STP1 (H202A), STP1 (H202A, E203A) and STP1 (D467A) were grown for 2 hours at 3% oxygen. Immunoblot analysis was performed on whole cell extracts (40 µg) using anti-Sre1 antiserum. P and N denote the precursor and nuclear forms, respectively. Asterisk indicates non-specific, cross-reacting proteins detected by anti-Sre1 antiserum. F. C. neoformans stp1Δ cells were transformed with the indicated plasmids and five-fold serial dilutions of cells were spotted on rich medium and rich medium containing 0.3 mM CoCl2 and incubated at 30°C for 3 days. Six independent isolates were plated.
In mammals, Site-2 protease liberates the soluble N-terminal transcription factor domain of SREBP from the Golgi membrane. To determine whether cells lacking C. neoformans STP1 produce cleaved Sre1 N-terminus (Sre1N), we generated stp1Δ cells by homologous recombination. Using previously generated anti-Sre1 antiserum to detect both the full-length form and the cleaved N-terminal form of Sre1, we assayed Sre1 cleavage in stp1Δ cells by immunoblot analysis (Fig. 1B). Hypoxia strongly induces the production of Sre1N in wild-type cells (Chang et al., 2007). Wild-type and stp1Δ cells were grown at ambient oxygen and then switched to 3% oxygen for increasing amounts of time. Unlike wild-type cells where Sre1N (~75 kD) rapidly accumulates under low oxygen, stp1Δ cells failed to produce Sre1N (Fig. 1B). In addition, stp1Δ cells contained reduced amounts of Sre1 precursor. These data indicate that Stp1 is required for Sre1 proteolysis. STP1 is additionally required for Sre1 expression, possibly due to the fact that SREBP orthologs regulate their own synthesis or that Stp1 is somehow required for the stability of full-length Sre1 protein. To investigate this further, we measured levels of SRE1 mRNA using quantitative PCR (Fig. 1C). Compared to wild-type, SRE1 transcript levels in stp1Δ cells were similar under normoxic conditions but reduced under hypoxic conditions, suggesting that under Sre1-activating conditions STP1 is required for SRE1 synthesis.
Since STP1 is required to produce Sre1N, we tested whether stp1Δ cells display similar growth phenotypes as sre1Δ cells. Sre1 is required for normal growth under low oxygen conditions and in the presence of the hypoxia mimetic, cobalt chloride (CoCl2) (Chang et al., 2007; Lee et al., 2007). Serial dilutions of cells were plated on rich medium in the absence or presence of 0.3 mM CoCl2 and grown for 3 days at 30°C (Fig. 1D). In the absence of CoCl2, all strains grew similar to wild-type. However, both sre1Δ and stp1Δ cells failed to grow on the CoCl2-containing plates. Growth on CoCl2 was rescued by transforming sre1Δ and stp1Δ with wild-type copies of SRE1 and STP1, respectively. To test whether the growth defect of stp1Δ cells on CoCl2-containing medium is due to loss of Sre1 cleavage, sre1Δ and stp1Δ cells were generated that stably-express the soluble N-terminal domain of Sre1 (Sre1N). Sre1N rescued growth of sre1Δ cells on CoCl2, indicating that the gene truncation codes for a functional transcription factor. Interestingly, Sre1N also rescued growth of stp1Δ cells on CoCl2, demonstrating that the growth defect of stp1Δ cells in the presence of CoCl2 is due to the loss of Sre1 activity. Together, these data suggest that Sre1 proteolytic cleavage is required for adaptation to growth on CoCl2 conditions, and that Stp1 is required for Sre1 proteolysis.
C. neoformans Stp1 is a conserved Site-2 protease
Stp1 contains conserved residues required for metalloprotease activity of Site-2 protease orthologs (Fig. 1A) (Feng et al., 2007). To test whether these residues are required for cleavage of Sre1, we performed site-directed mutagenesis on STP1 changing codons coding for histidine 202, glutamate 203, and aspartate 467 to alanine. Plasmids containing wild-type and mutant STP1 were transformed into stp1Δ cells and maintained extra-chromosomally. To assay Sre1 cleavage, we grew cells for 2 hours at 3% oxygen and Sre1 protein was detected by immunoblot analysis. stp1Δ cells transformed with wild-type STP1 accumulated Sre1N under low oxygen compared to cells transformed with empty vector (Fig. 1E, lanes 1 and 2). In contrast, cells expressing mutant STP1 failed to induce Sre1N and precursor levels remained low (Fig. 1E, lanes 3–5). To test whether these conserved residues are required for Sre1-dependent growth on CoCl2, six independent isolates of stp1Δ strains expressing empty vector, wild-type STP1 and the STP1 mutants were plated in serial dilution on rich medium in the absence and presence of 0.3 mM CoCl2 and grown for 3 days at 30°C (Fig. 1F). Wild-type STP1 restored growth on CoCl2, but empty vector and the STP1 mutants did not rescue growth. These data suggest that these conserved residues are required for Stp1 function and that Stp1 is a functional Sre1 protease. We have been unable to detect Stp1 by immunoblot, possibly because Stp1 is extremely hydrophobic. While the analogous mutations in human Site-2 protease do not affect protein levels (Rawson et al., 1997), we cannot rule out the possibility that these mutations lead to reduced amounts of Stp1 protein.
All characterized Site-2 protease orthologs perform a unique proteolytic mechanism in which substrate cleavage occurs within the lipid bilayer (Urban and Shi, 2008). In the case of SREBP, cleavage occurs between a leucine and cysteine residue predicted to be within the first transmembrane segment (Duncan et al., 1998). However, these residues are not present within the first predicted transmembrane segment of C. neoformans Sre1 (determined using online Phobius software, http://phobius.sbc.su.se/). If Stp1 cleaves Sre1 at a similar position relative to the membrane, Sre1N should be 502 to 530 amino acids in length. To estimate the position of Stp1 cleavage, we generated stp1Δ strains expressing two different C-terminal truncations of Sre1 (Fig. 2A). These truncated genes encode proteins of 501 and 535 amino acids (Fig. 2B). Wild-type and stp1Δ cells expressing empty vector and the truncated SRE1 genes were grown in log-phase for 2 hours at ambient oxygen. Immunoblot analysis was performed to detect Sre1 protein (Fig. 2C). Since Sre1N is heavily phosphorylated, we treated cell lysates with phosphatase to collapse Sre1N to a single band. Phosphatase-treated Sre1N migrated ~60 kD, ~15 kD lower than non-phosphatase treated Sre1N (compare Fig. 2C, lane 4 and Fig. 1B, lane 5). Endogenous Sre1N migrated between the two truncated proteins (Fig. 2C, lanes 2–6), consistent with Sre1 being cleaved within the first predicted transmembrane segment.
STP1 is required for virulence in mice
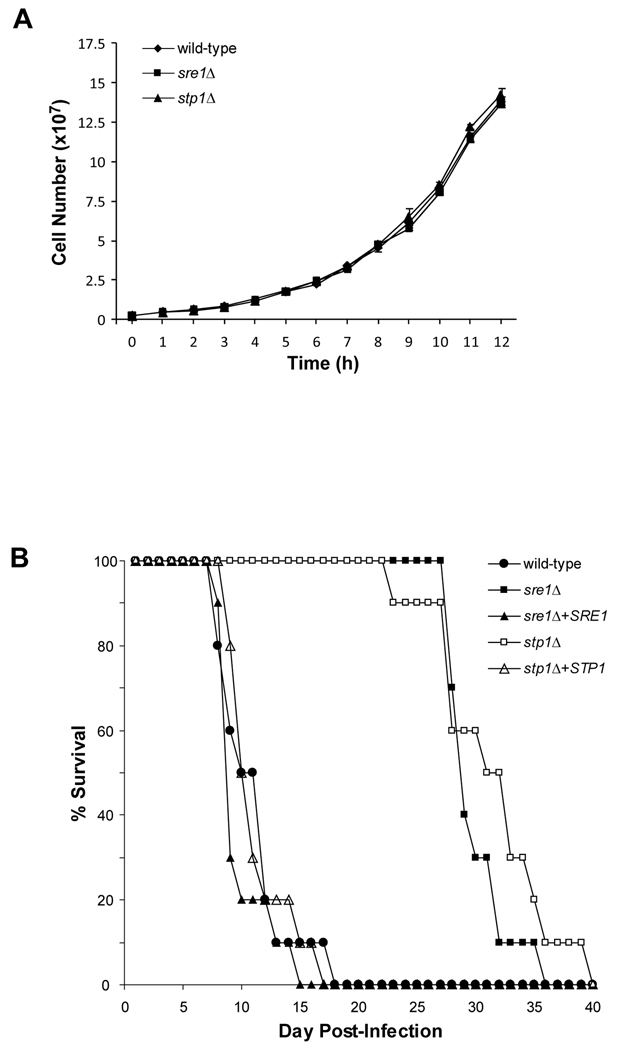
A previous study showed that C. neoformans serotype A (H99) SRE1 and STP1 are partially required for virulence in an A/J mouse model of infection (Chun, Liu, and Madhani, 2007). Interestingly, these results differed from our previous studies using the C. neoformans serotype D strain (B3501A) in BALB/c mice (Chang et al., 2007). In this experiment, SRE1 was essential for fatal infection. To test whether these different findings were due to C. neoformans serotype differences or mouse strain differences, we performed virulence studies using independently-generated deletion strains in BALB/c mice. First, we tested whether the mutant strains displayed growth defects under lab culture conditions. We monitored growth of wild-type, sre1Δ and stp1Δ strains in rich medium for 12 hours at 30°C and 37°C (Fig. 3A and data not shown). Under these conditions, we observed no significant difference in growth between the mutant and wild-type strains. In addition, we tested whether sre1Δ and stp1Δ cells display significant defects in other known virulence factors, including melanin production and polysaccharide capsule synthesis. Confirming previous studies, we observed a decrease in melanin production on norepinephrine-containing medium and no difference in capsule size (Fig. S1, S2). Next, we infected groups of 10 BALB/c mice with wild-type, sre1Δ, stp1Δ, sre1Δ + SRE1 and stp1Δ + STP1 strains via lateral tail-vein injection and monitored mouse survival over time (Fig. 3B). As expected, mice infected with wild-type serotype A cells succumbed to fatal infection from 7–18 days post-infection. However, cells lacking SRE1 or STP1 showed delayed lethality in mice after ~25–40 days post-infection, indicating that Sre1 activation is partially, but not completely, required for normal disease progression in this model of infection. Finally, complementing sre1Δ and stp1Δ cells with the corresponding wild-type genes restored virulence to these strains. Overall, this study highlights the differential requirement for the Sre1 pathway in the virulence of C. neoformans serotypes A and D.
Figure 3. STP1 is required for virulence.
A. C. neoformans serotype A wild-type and stp1Δ cells were grown in YES medium at 37°C. Cell numbers were determined at one hour intervals for 12 hours. Error bars represent one standard deviation from three biological replicates. B. Female Balb/c mice (n=10/strain) were infected via tail vein injection with the indicated C. neoformans strains and mouse survival was monitored.
STP1 regulates Sre1-dependent and Sre1-independent gene expression
In mammals, Site-2 protease cleaves substrates in addition to SREBP, including the stress-response transcription factor ATF6 (Ye et al., 2000b). To determine whether C. neoformans Stp1 regulates other proteins, we designed experiments to determine Stp1-dependent gene expression. In addition, our previous work defining Sre1-dependent gene expression in serotype D differs from reported serotype A Sre1 gene expression profiles (Chang et al., 2007; Chun, Liu, and Madhani, 2007). Therefore, we also evaluated Sre1-dependent gene expression changes in serotype A. Using gene lists provided from published C. neoformans H99 microarray studies, we designed whole genome microarrays containing probes to 6798 unique transcripts (Chun, Liu, and Madhani, 2007). Preliminary microarray studies comparing wild-type, sre1Δ and sre1Δ + SRE1 cells revealed several abnormalities in the sre1Δ strain background. In both the sre1Δ and sre1Δ + SRE1 strains, we noticed two large chromosomal regions that showed significantly higher gene expression than in wild-type cells (encompassing 23 genes on chromosome 10 and 20 genes on chromosome 14). Since this strain variation was independent of Sre1, we determined Sre1-dependent gene expression by comparing the sre1Δ strain to the sre1Δ + SRE1 strain. RNA was harvested from wild-type, stp1Δ, sre1Δ + SRE1 and sre1Δ strains grown for 2 hours at 3% oxygen at 30°C, the earliest time point at which Sre1 cleavage is maximally induced (Fig. 1B). We performed three different strain comparisons: 1) wild-type vs. stp1Δ, 2) sre1Δ + SRE1 vs. sre1Δ and 3) wild-type at 21% oxygen vs. 3% oxygen. All data can be found in supplemental Tables S4-S6.
Table 1 lists all statistically significant genes expressed at higher levels in wild-type or sre1Δ + SRE1 cells compared to cells lacking either STP1 or SRE1, respectively. Genes were categorized into functional groups. As observed previously, sre1Δ cells showed lower expression of genes involved in ergosterol biosynthesis (Table 1) (Chang et al., 2007; Chun, Liu, and Madhani, 2007). Consistent with the requirement of Stp1 for Sre1 activation, stp1Δ cells also showed reduced expression of these genes. In addition to the 12 genes regulated by both Sre1 and Stp1, 7 genes involved in sterol synthesis were found to be Stp1-dependent, but not Sre1-dependent, suggesting a possible role for Stp1 in controlling other regulators of sterol biosynthesis. In C. neoformans serotype D, Sre1 regulates genes required for metal homeostasis, including multiple pathways involved in high affinity transport of iron and copper (Chang et al., 2007). However in serotype A, these genes were largely not Sre1 transcriptional targets. In fact, only two genes from this category appeared on the list of statistically significant genes: CTR4, a copper transporter and CNBF4480, a poorly characterized protein also involved in copper transport (Table 1).
Table 1. Low oxygen gene expression analysis.
List of all statistically significant genes expressed 1.4-fold or higher in wild-type vs. stp1Δ or in sre1Δ + SRE1 vs. sre1Δ. Only annotated genes are listed. Full data set can be found in Table S4.
| Fold change | |||||
|---|---|---|---|---|---|
| BROAD ID (CNAG_#) |
S. cerevisiae homolog |
B3501A homolog | Description | wild-type vs. stp1 Δ 3% O2 |
sre1Δ+ SRE1 vs. sre1Δ 3% O2 |
| Ergosterol Biosynthesis | |||||
| 519 | ERG3 | CNBA4830 | Sterol-C5-desaturase | 6.7 | 6.5 |
| 4804 | SRE1 | CNBJ1170 | bHLH transcription factor | 4.2 | 17.0 |
| 1737 | ERG25 | CNBC4830 | C-4 methyl sterol oxidase | 4.0 | 3.5 |
| 40 | ERG11 | CNBA0300 | Lanosterol 14 alpha-demethylase | 3.6 | 2.8 |
| 3819 | ERG6 | CNBB2600 | Sterol methyltransferase | 3.6 | 2.7 |
| 6644 | ERG5 | CNBF1100 | C-22 sterol desaturase | 3.5 | 3.2 |
| 5742 | CNBF2350 | Metalloprotease | 3.5 | ||
| 3311 | ERG13 | CNBC2090 | Hydroxymethylglutaryl-CoA synthase | 3.4 | 3.5 |
| 2830 | ERG4 | CNBC2700 | Sterol C-24 reductase | 3.1 | 2.5 |
| 854 | ERG2 | CNBA8120 | C-8 sterol isomerase | 2.9 | 2.6 |
| 6829 | ERG1 | CNBD0290 | Squalene epoxidase | 2.8 | 2.5 |
| 2896 | ERG13 | CNBG2100 | Hydroxymethylglutaryl-CoA synthase | 2.7 | 2.1 |
| 2084 | ERG20 | CNBE4510 | Farnesyl diphosphate synthetase | 2.3 | |
| 2918 | ERG10 | CNBC1890 | Acetyl-CoA C-acetyltransferase | 2.1 | |
| 1129 | ERG7 | CNBD3820 | Lanosterol synthase | 2.0 | 2.2 |
| 6534 | HMG2 | CNBF0070 | Hydroxymethylglutaryl-CoA reductase | 1.8 | 1.7 |
| 6001 | ERG8 | CNBM0120 | Phosphomevalonate kinase | 1.6 | |
| 5125 | ERG19 | CNBI1880 | Diphosphomevalonate decarboxylase | 1.6 | |
| 4605 | ERG26 | CNBJ3060 | C-3 sterol dehydrogenase | 1.5 | |
| 117 | ERG24 | CNBA1040 | C-14 sterol reductase | 1.5 | |
| 1003 | NCP1 | CNBD4990 | Cytochrome P450 oxidoreductase | 1.5 | |
| Translation | |||||
| 640 | RPS4A | CNBA6010 | 40s ribosomal protein s4 | 1.9 | |
| 5976 | NOP58 | CNBF4620 | Involved in pre-rRNA processing | 1.8 | |
| 513 | YPL146C | CNBA4770 | Involved in 60S ribosome subunit biogenesis | 1.8 | |
| 4445 | RPS7B | CNBH1070 | 40S ribosomal protein S7 | 1.7 | |
| 4348 | SRP4004 | CNBH1900 | Involved in pre-rRNA processing | 1.7 | |
| 6126 | PXR1 | CNBM1170 | Involved rRNA maturation | 1.7 | |
| 2754 | RPS12 | CNBC3450 | 40S ribosomal protein S12 | 1.7 | |
| 3064 | NOP16 | CNBC6990 | Involved in 60S ribosome subunit biogenesis | 1.7 | |
| 4571 | DIM1 | CNBJ3380 | 16S rRNA demethylase | 1.7 | |
| 5755 | MAK16 | CNBF2480 | Involved in 25S and 5.8S rRNA maturation | 1.7 | |
| 3302 | LTV1 | CNBG2020 | Involved in ribosome nuclear export | 1.7 | |
| 3789 | MRT4 | CNBB2850 | Involved in mRNA turnover and ribosome assembly | 1.7 | |
| 5975 | TRZ1 | CNBF4610 | tRNA 3'-end processing endonuclease | 1.6 | |
| 7257 | NOG2 | CNBD0720 | GTPase associated with 60S pre-ribosomal subunits | 1.6 | |
| 3724 | NOP2 | CNBB3490 | Required for 27S pre-rRNA maturation | 1.6 | |
| 6468 | NOG1 | CNBN1850 | GTPase associated with 60S pre-ribosomal subunits | 1.6 | |
| 3127 | RPS23B | CNBG0330 | 40S ribosomal protein S23 | 1.6 | |
| 4799 | RPL14B | CNBJ1220 | Ribosomal protein L14 | 1.6 | |
| 3747 | RPL28 | CNBB3280 | 60S ribosomal protein L27a | 1.5 | |
| 3053 | RPL25 | CNBC6880 | Ribosomal protein L23a | 1.5 | |
| 3975 | ERB1 | CNBB1130 | Involved in 25S and 5.8S rRNA maturation | 1.5 | |
| 6007 | YOR004W | CNBM0170 | Involved in 40S ribosomal subunit biogenesis | 1.5 | |
| 4072 | NIP7 | CNBB0230 | Involved in 60S ribosome subunit biogenesis | 1.5 | |
| 358 | MOD5 | CNBA3320 | Involved in biosynthesis of modified tRNAs | 1.5 | |
| 7878 | NOC2 | CNBL1270 | Binds to 90S and 66S pre-ribosomes | 1.5 | |
| 5202 | SET7 | CNBI2600 | Methyltransferase involved in methylation of Rpl42 | 1.5 | |
| 3293 | TSR2 | CNBG1930 | Involved in pre-rRNA processing | 1.4 | |
| 5228 | LCP5 | CNBI2760 | Involved in maturation of 18S rRNA | 1.4 | |
| Carbohydrate metabolism and transport | |||||
| 6031 | SKN1 | CNBM0410 | Protein involved in sphingolipid biosynthesis | 2.2 | 3.2 |
| 5607 | YIR007W | CNBL2520 | C9 endoglycoceramidase | 2.0 | 2.4 |
| 5607 | YIR007W | CNBL2520 | C9 endoglycoceramidase | 1.8 | 1.8 |
| 2588 | CNBC4720 | Beta-glucosidase | 1.7 | 1.9 | |
| 4516 | CNBA4380 | Beta-1, 3 exoglucanase precursor | 1.5 | 1.6 | |
| 6832 | KRE6 | CNBD0270 | Beta-1,6-glucan synthetase | 1.5 | |
| 6323 | CNBN0500 | L-fucose permease | 1.8 | ||
| 6253 | CNBM2380 | Maltose permease | 1.6 | ||
| 4210 | MAL11 | CNBH3340 | Alpha-glucoside transport protein | 1.4 | |
| RNA helicases | |||||
| RNA Helicase | 1.8 | 2.0 | |||
| 764 | HCA4 | CNBA7250 | ATP-dependent RNA helicase | 1.8 | |
| 3205 | ROK1 | CNBG1110 | ATP-dependent RNA helicase | 1.6 | |
| 5757 | CNBF2500 | ATP-dependent RNA helicase | 1.6 | ||
| 2502 | DBP3 | CNBE0530 | ATP-dependent RNA helicase | 1.5 | |
| 1140 | DRS1 | CNBD3700 | ATP dependent RNA helicase | 1.5 | |
| 3603 | HAS1 | CNBB4650 | ATP-dependent RNA helicase | 1.5 | |
| 6741 | DBP10 | CNBB5090 | ATP-dependent RNA helicase | 1.5 | |
| 1831 | DBP6 | CNBC4030 | ATP-dependent RNA helicase | 1.4 | |
| 4580 | MSS116 | CNBJ3280 | RNA helicase | 1.4 | |
| Energy production | |||||
| 7316 | ADH4 | CNBA1780 | Iron-dependant alcohol dehydrogenase | 2.3 | 2.2 |
| 6699 | TDH1 | CNBF1640 | Glyceraldehyde-3-phosphate dehydrogenase | 1.8 | |
| 716 | CYC7 | CNBA6760 | Cytochrome c isoform 2 | 2.1 | 1.7 |
| 1078 | ALD5 | CNBD4300 | Aldehyde dehydrogenase | 1.6 | |
| 6663 | QCR6 | CNBF1240 | Ubiquinol-cytochrome c reductase component | 1.6 | |
| 4000 | COX11 | CNBB0900 | Cytochrome-c oxidase assembly protein | 1.5 | |
| 47 | YIL042C | CNBA0360 | Pyruvate dehydrogenase kinase isoform 2 | 1.5 | 1.8 |
| 6923 | CNBK3360 | Phosphoketolase | 1.4 | 2.0 | |
| 4659 | PDC1 | CNBJ2540 | Pyruvate decarboxylase | 1.5 | |
| Cell cycle and division | |||||
| 5968 | CDC42 | CNBF4550 | GTPase required for cell the maintenance of polarity | 2.4 | |
| 2712 | BUD32 | CNBK1570 | Kinase invovled in bud site selection | 1.8 | 1.5 |
| 5890 | STE18 | CNBL0880 | Involved in activating the mating signaling pathway | 1.7 | |
| 1907 | CDC5 | CNBK1050 | Kinase involved in mitosis and cytokinesis | 1.7 | |
| 7689 | CWC22 | CNBF4440 | Cell cycle control protein | 1.6 | |
| 1922 | PHO80 | CNBK0910 | Cyclin, negatively regulates phosphate metabolism | 1.5 | 1.5 |
| 3315 | RHO1 | CNBG2140 | GTP binding protein involved in regulation of cell polarity | 1.5 | |
| Response to oxidative stress | |||||
| 2849 | CNBC2520 | Glutaredoxin | 2.8 | 1.6 | |
| 3340 | CNBK0670 | Flavanoid synthase | 2.3 | 1.9 | |
| 6448 | CYS3 | CNBN1680 | Cystathionine gamma-lyase | 1.9 | 2.7 |
| 1464 | YHB1 | CNBC0030 | Flavohemoglobin | 1.8 | |
| Pantothenate uptake and acetyl-CoA production | |||||
| 2361 | Fen2 | CNBE1850 | Pantothenate transporter | 2.3 | 2.3 |
| 2361 | Fen2 | CNBE1850 | Pantothenate transporter | 2.2 | 2.3 |
| 2222 | YIL083C | CNBE3190 | Phosphopantothenoylcysteine synthetase | 1.6 | 1.7 |
| 5027 | FMS1 | CNBI0930 | Polyamine oxidase | 1.6 | 1.6 |
| Mating/hyphal growth | |||||
| 5890 | STE18 | CNBL0880 | Involved in activating the mating signaling pathway | 1.7 | |
| 6808 | STE3 | CNBD0590 | Phermone receptor | 1.5 | |
| 5970 | STE20 | CNBF4570 | Kinase in pheromone response and pseudohyphal growth | 1.5 | |
| 406 | STE20 | CNBA3750 | Kinase in pheromone response and pseudohyphal growth | 1.4 | |
| 4495 | MUC1 | CNBH0590 | Glycoprotein involved in pseudohyphal formation | 1.5 | |
| Phosphate metabolism | |||||
| 2944 | PHO5 | CNBC1640 | Acid phosphatase | 2.0 | |
| 2777 | PHO84 | CNBC3220 | High-affinity inorganic phosphate transporter | 1.9 | |
| 1922 | PHO80 | CNBK0910 | Cyclin, negatively regulates phosphate metabolism | 1.5 | 1.5 |
| Methionine metabolism | |||||
| 6448 | CYS3 | CNBN1680 | Cystathionine gamma-lyase | 1.9 | 2.7 |
| 418 | SAM1 | CNBA3860 | Methionine adenosyltransferase | 1.7 | |
| 5950 | MUP1 | CNBF4390 | High affinity methionine permease | 1.7 | |
| 886 | SAH1 | CNBD5970 | S-adenosylhomocysteine hydrolase | 1.5 | |
| Copper Transport | |||||
| 979 | CTR4 | CNBD5220 | High affinity copper transporter | 3.8 | 1.7 |
| 5959 | CNBF4480 | Copper transporter | 1.9 | ||
| NAD metabolism | |||||
| 5951 | NMA1 | CNBF4380 | Nicotinic acid mononucleotide adenylyltransferase | 1.7 | |
| 2868 | BNA2 | CNBC2350 | Indoleamine 2,3-dioxygenase | 1.4 | |
| 1865 | BNA3 | CNBK1490 | Arylformamidase | 1.5 | |
| Fatty acid beta-oxidation | |||||
| 5657 | CNBL3020 | Peroxisomal 2,4-dienoyl-CoA reductase-like protein | 1.6 | ||
| 1836 | PCS60 | CNBC3980 | Acyl-CoA synthetases | 1.6 | 1.6 |
| 2553 | SPS19 | CNBE0040 | Peroxisomal 2,4-dienoyl-CoA reductase | 1.5 | |
| 4531 | EHD3 | CNBH0240 | Enoyl-CoA hydratase | 1.8 | |
| 675 | CRC102 | CNBA6340 | Carnitine/acyl carnitine carrier | 1.4 | |
| Heme biosynthesis | |||||
| 3939 | HEM1 | CNBB1490 | Aminolevulinic acid synthetase | 1.8 | |
| 2460 | HEM13 | CNBE0930 | Coproporphyrinogen oxidase | 1.5 | |
| Other transporters | |||||
| 1946 | DAL5 | CNBK0660 | Allantoate transporter | 2.3 | 2.0 |
| 5952 | HNM1 | CNBF4370 | Amino acid permease | 2.1 | 1.5 |
| 5952 | HNM1 | CNBF4370 | Amino acid permease | 2.1 | 1.5 |
| 869 | PDR15 | CNBD6120 | ABC transporter | 2.0 | |
| 5952 | HNM1 | CNBF4370 | Amino acid permease | 2.0 | |
| 904 | YKR105C | CNBD5800 | AflT-like major facilitator superfamily protein | 1.8 | |
| 3474 | YOR378W | CNBG3620 | Permease of the major facilitator family | 1.7 | |
| 2561 | TPO2 | CNBK2950 | Polyamine transporter | 1.6 | |
| 2254 | HXT1 | CNBE2930 | Hexose transporter | 1.5 | |
| 1074 | AVT1 | CNBD4340 | Amino acid permease | 1.4 | 1.6 |
| 6503 | FUI1 | CNBN2320 | Uracil permease | 1.8 | |
| 5952 | HNM1 | CNBA5400 | Amino acid permease | 1.7 | |
In total, 87 genes were more highly expressed in sre1Δ + SRE1 versus sre1Δ cells under low oxygen, and 185 genes were more highly expressed in wild-type versus stp1Δ cells (Fig. 4A). Similarly, 98 genes were more highly expressed in sre1Δ than sre1Δ + SRE1 cells under low oxygen, and 176 genes were more highly expressed in stp1Δ than in the wild-type cells (Fig. 4B). The fact that more genes were Stp1-dependent than Sre1-dependent suggested a role in addition to Sre1 cleavage for Stp1 in regulating gene expression. To further classify the genes regulated by Sre1 and Stp1, we performed Gene Ontology (GO) analysis. GO term assignment for C. neoformans H99 genes was based on homology to S. cerevisiae genes. As expected, genes involved in lipid metabolism, particularly sterol metabolism, were significantly enriched in the list of genes regulated by both Sre1 and Stp1 (Table 2). In addition, genes involved in pantothenate and coenzyme-A metabolism were also enriched in this list, possibly due to the fact that these genes are required for the production of acetyl-CoA, the initial substrate for ergosterol biosynthesis. GO analysis of genes regulated by Stp1, but not Sre1, revealed a significant enrichment of genes involved in translation and ribosome biogenesis (Table 2). In total, 28 genes involved in ribosome biogenesis and protein translation were significantly higher in wild-type versus stp1Δ cells (Table 1).
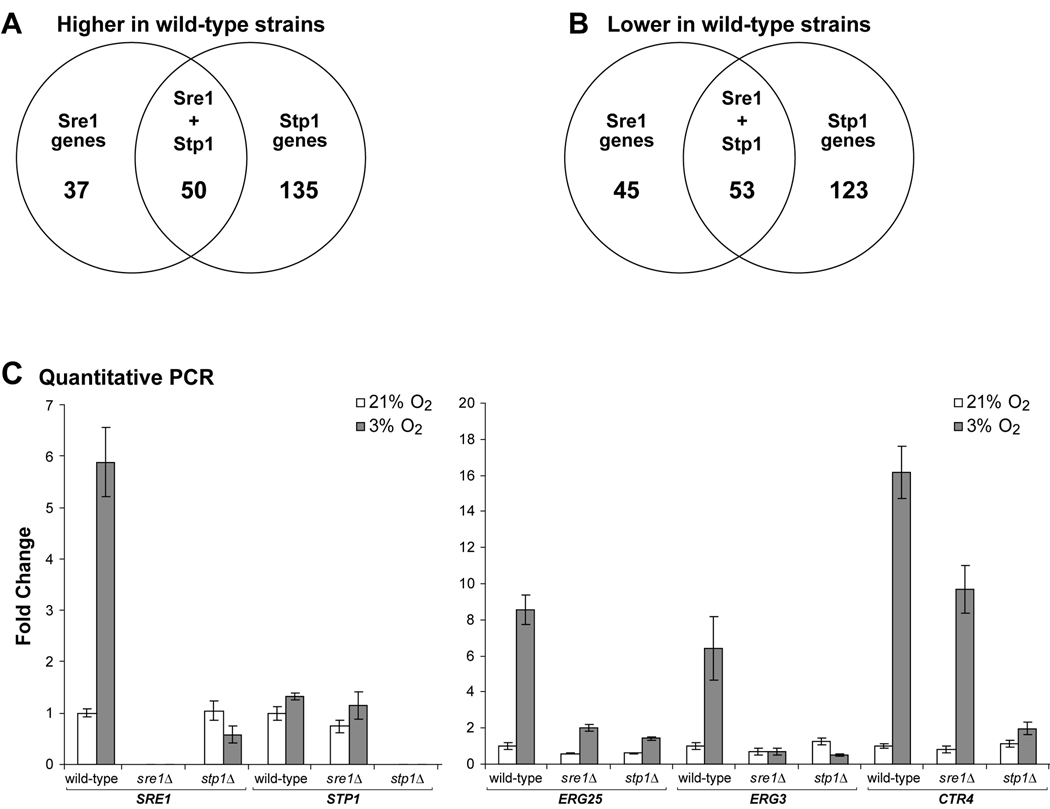
Figure 4. STP1 is required for both Sre1-dependent and Sre1-independent gene expression under low oxygen.
A. Venn diagram comparing statistically-significant genes more highly expressed in wild-type versus sre1Δ and stp1Δ cells under low oxygen. B. Venn diagram comparing statistically-significant genes with reduced expression in wild-type versus sre1Δ and stp1Δ cells under low oxygen. C. Quantitative PCR analysis on genes more highly expressed in wild-type versus sre1Δ or stp1Δ cells. Wild-type, sre1Δ, and stp1Δ cells were grown under ambient (21%) or 3% oxygen for 2 hours, and RNA was harvested. RNA level for the indicated genes was quantified by real time RT-PCR and normalized to that in wild-type cells at 21% oxygen. Error bars represent standard deviation from three biological replicate experiments.
Table 2. Gene ontology analysis for Sre1- and Stp1-dependent genes.
GO terms were assigned to C. neoformans genes based on homology to S. cerevisiae genes. P-value represents the probability that a particular GO term is enriched in the indicated microarray gene lists. The p-value cutoff was <0.05.
| GO group | GO subgroup | p-value |
|---|---|---|
| Up in wild-type vs. sre1Δ and stp1Δ | ||
| Cellular lipid metabolism | 4.5×10−9 | |
| Sterol biosynthesis | 1.7×10−12 | |
| Fatty acid metabolism | 1.5×10−3 | |
| Coenzyme metabolism | 6.1×10−3 | |
| Pantothenate metabolism | 0.031 | |
| Co-A metabolism | 0.040 | |
| Pantothenate transport | 0.023 | |
| Amino acid transport | 0.013 | |
| Up in wild-type vs. stp1Δ only | ||
| Ribosome biogenesis and assembly | 6.7×10−6 | |
| Ribosome bigenesis | 2.0×10−5 | |
| rRNA processing | 1.6×10−4 | |
| Ribosome export from the nucleus | 0.046 | |
| S-adenosyl methionine metabolism | 1.6×10−3 | |
| RNA processing | 5.5×10−3 | |
| Ergosterol biosynthesis | 0.014 | |
| Isoprenoid biosynthesis | 0.020 | |
| Signal transduction and during conjugation | 0.016 | |
| NAD metabolism | 0.034 | |
| Maintenance of actin cytoskeleton polarity | 0.046 | |
| Up in wild-type vs. sre1Δ only | ||
| Amino acid catabolism | 8.3×10−3 | |
| Branched chain amino acid catabolism | 1.1×10−4 | |
| Down in wild-type vs. sre1Δ and stp1Δ | ||
| Phosphoinositide-mediated signalling | 1.4×10−3 | |
| Down in wild-type vs. stp1Δ only | ||
| Disaccharide biosynthesis | 1.3×10−4 | |
| Glycerolipid biosynthesis | 2.5×10−4 | |
| Down in wild-type vs. sre1Δ only | ||
| Carbohydrate metabolism | 2.5×10−5 | |
| Hydrogen peroxide metabolism | 7.2×10−3 | |
Finally, independent experiments were performed to analyze gene expression changes in wild-type cells in response to hypoxia. Strikingly, 1929 significant gene expression changes occurred when cells were shifted from 21% oxygen to 3% oxygen for 2 hours at 30°C. This finding demonstrates the strong requirement for hypoxic gene expression regulators in C. neoformans. GO analysis of genes both up- and down-regulated under low oxygen conditions is summarized in Table 3. Under low oxygen, cells induced non-respiratory pathways for energy derivation, including fermentation. In addition, cells up-regulated oxygen-requiring synthetic pathways such as for ergosterol and sphingolipids. Cells also induced pathways for several types of stress responses, including chemical and oxidative stress. Genes repressed under low oxygen included those involved in protein synthesis, particularly genes involved in ribosome biogenesis and assembly (Table 3). In addition, genes that promote cell division were down-regulated, consistent with an observed decrease in growth rate under low oxygen (data not shown).
Table 3. Gene ontology (GO) term analysis for the C. neoformans hypoxic response.
GO terms were assigned to C. neoformans genes based on homology to S. cerevisiae genes. P-value represents the probability that a particular GO term is enriched in the indicated microarray gene lists. GO groups with p-values <0.005 and subgroups with p-values <0.05 are listed.
| GO group | GO subgroup | p-value |
|---|---|---|
| Up at 3% O2 vs. 21% O2 | ||
| Energy derivation by oxidation | 2.7×10−7 | |
| Gluconeogenesis | 2.3×10−4 | |
| Fermentation | 1.5×10−3 | |
| Glycolysis | 3.3×10−3 | |
| Pentose phosphate shunt | 4.0×10−3 | |
| Coenzyme metabolism | 1.2×10−6 | |
| Nicotinamide metabolism | 1.5×10−5 | |
| Response to abiotic stimulus | 2.9×10−4 | |
| Response to chemical stimulus | 1.3×10−3 | |
| Response to drug | 0.047 | |
| Response to oxidative stress | 0.043 | |
| Cellular lipid metabolism | 3.1×10−4 | |
| Triacylglycerol biosynthesis | 0.013 | |
| Phospholipid catabolism | 0.013 | |
| Ergosterol biosynthesis | 0.019 | |
| Sphingolipid metabolism | 0.035 | |
| Amine catabolism | 4.6×10−4 | |
| Amino acid catabolism | 1.9×10−3 | |
| Down at 3% O2 vs. 21% O2 | ||
| Organelle organization and biosynthesis | 1.1×10−22 | |
| Ribosome biogenesis and assembly | 7.7×10−44 | |
| Mitotic spindle organization and biogenesis | 1.4×10−5 | |
| Nucleosome assembly | 8.7×10−3 | |
| Chromosome condensation | 0.015 | |
| Protein biosynthesis | 7.4×10−15 | |
| Translation | 1.3×10−3 | |
| Translation initiation | 2.2×10−3 | |
| RNA metabolism | 1.5×10−13 | |
| rRNA metabolism | 4.4×10−24 | |
| RNA methylation | 1.3×10−7 | |
| tRNA modification | 1.1×10−6 | |
| pseudouridine synthesis | 1.8×10−4 | |
| snRNA modification | 2.7×10−4 | |
| mRNA stabilization | 0.037 | |
| Cellular biosynthesis | 3.4×10−9 | |
| Protein biosynthesis | 7.4×10−15 | |
| Glutamate biosynthesis | 2.5×10−4 | |
| Nucleoside monophosphate biosynthesis | 2.5×10−3 | |
| Mitotic cell cycle | 4.8×10−5 | |
| Mitotic spindle organization and biogenesis | 1.4×10−5 | |
| M phase of mitotic cell cycle | 3.8×10−5 | |
| G/M transition of mitotic cell cycle | 3.2×10−3 | |
| Mitotic chromosome condensation | 8.9×10−3 | |
To confirm the microarray results, SRE1, STP1, ERG25, ERG3 and CTR4 mRNA levels were measured in ambient and 3% oxygen conditions by quantitative PCR. In wild-type cells, expression of SRE1 was induced under low oxygen (Fig. 4C, left). This induction required Sre1 activation since stp1Δ cells failed to induce SRE1 transcript. STP1 transcript levels did not change significantly in either wild-type or sre1Δ cells in response to low oxygen (Fig. 4C, left). Interestingly in serotype D, Sre1 regulates the expression of the Stp1 homolog, CNF02350 (Chang et al., 2007). Consistent with Sre1 directly regulating genes involved in ergosterol biosynthesis, ERG25 and ERG3 were up-regulated under low oxygen in wild-type cells, and this required both SRE1 and STP1 (Fig. 4C, right). CTR4, coding for a high affinity copper transporter, was induced under low oxygen in wild-type and low oxygen expression was slightly reduced in sre1Δ cells. Strikingly, CTR4 was not induced under low oxygen in stp1Δ cells, indicating an independent requirement for STP1 in CTR4 expression. Collectively, these genome-wide gene expression studies identify roles for Sre1 and Stp1 in hypoxic gene expression and reveal functions for Stp1 in gene expression that are independent of Sre1 proteolytic activation.
STP1 is required for hypoxic and normoxic sterol synthesis
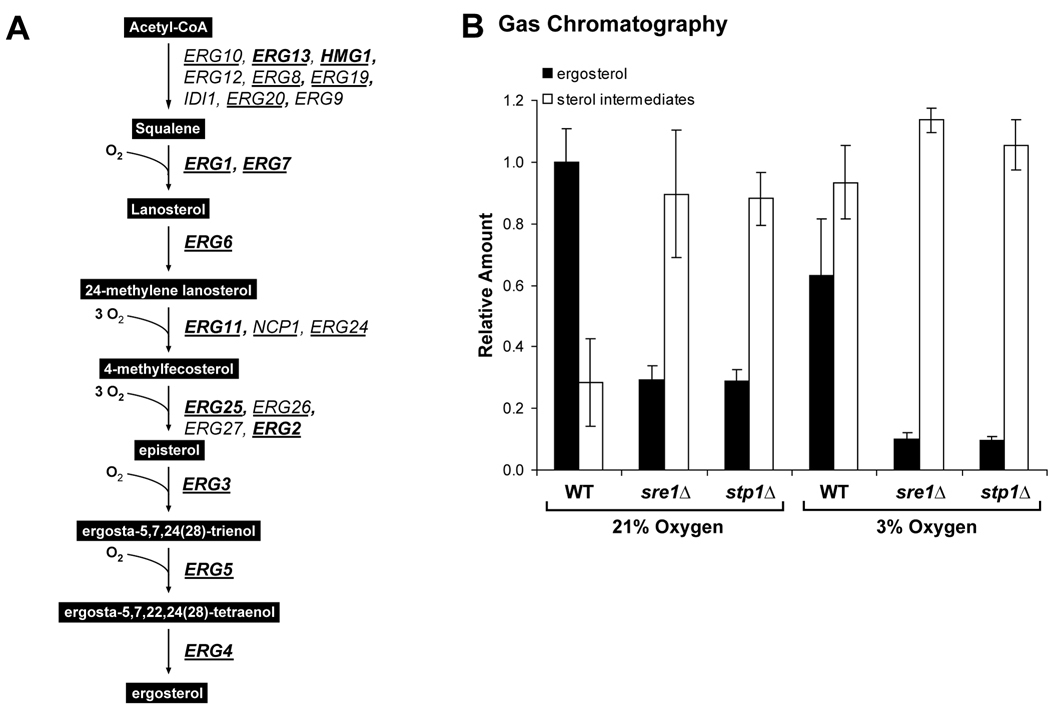
The gene expression studies demonstrated that C. neoformans serotype A Sre1 and Stp1 regulate genes involved in the ergosterol biosynthetic pathway (Table 1). Figure 5A outlines the sterol biosynthetic pathway in C. neoformans and indicates the Sre1-dependent genes (bold) and Stp1-dependent genes (underlined) (Lee et al., 2007). To determine the requirement of SRE1 and STP1 for sterol homeostasis, we used gas chromatography to analyze sterol content in wild-type, sre1Δ and stp1Δ cells. Total sterols were extracted from cells grown for 2 hours in rich medium at 30°C under ambient oxygen (21%) or 3% oxygen. Levels of ergosterol and all detectable sterol pathway intermediates were quantified and plotted (Fig. 5B). Under normoxic conditions, wild-type cells contained mostly ergosterol (gray bars) and a low level of sterol intermediates (white bars). When wild-type cells were grown at 3% oxygen, ergosterol production was reduced and pathway intermediates accumulated, consistent with the fact that ergosterol biosynthesis requires oxygen (Rosenfeld and Beauvoit, 2003). Interestingly, under normoxic conditions sre1Δ and stp1Δ cells showed decreased levels of ergosterol and increased levels of intermediates compared to the wild-type strain. This indicates a requirement for Sre1 activation even at ambient oxygen concentrations. However, when sre1Δ and stp1Δ cells were grown under hypoxic conditions, this defect was exaggerated. This experiment demonstrates that cells require Sre1 activation to control ergosterol homeostasis under both normoxic and hypoxic conditions.
Figure 5. STP1 is required for hypoxic and normoxic sterol homeostasis.
A. Outline of C. neoformans ergosterol biosynthetic pathway. Genes encoding Sre1-dependent (bold) and Stp1-dependent (underlined) enzymes are listed. B. Wild-type, stp1Δ and sre1Δ cells were grown for 2 hours at 21% or 3% O2. Total sterols were extracted and analyzed by gas chromatography. Ergosterol and sterol intermediates were quantified and normalized to wild-type 21% oxygen ergosterol levels and plotted. Error bars represent the standard deviation from three biological replicate experiments.
Azole drugs are fungicidal to sre1Δ and stp1Δ cells
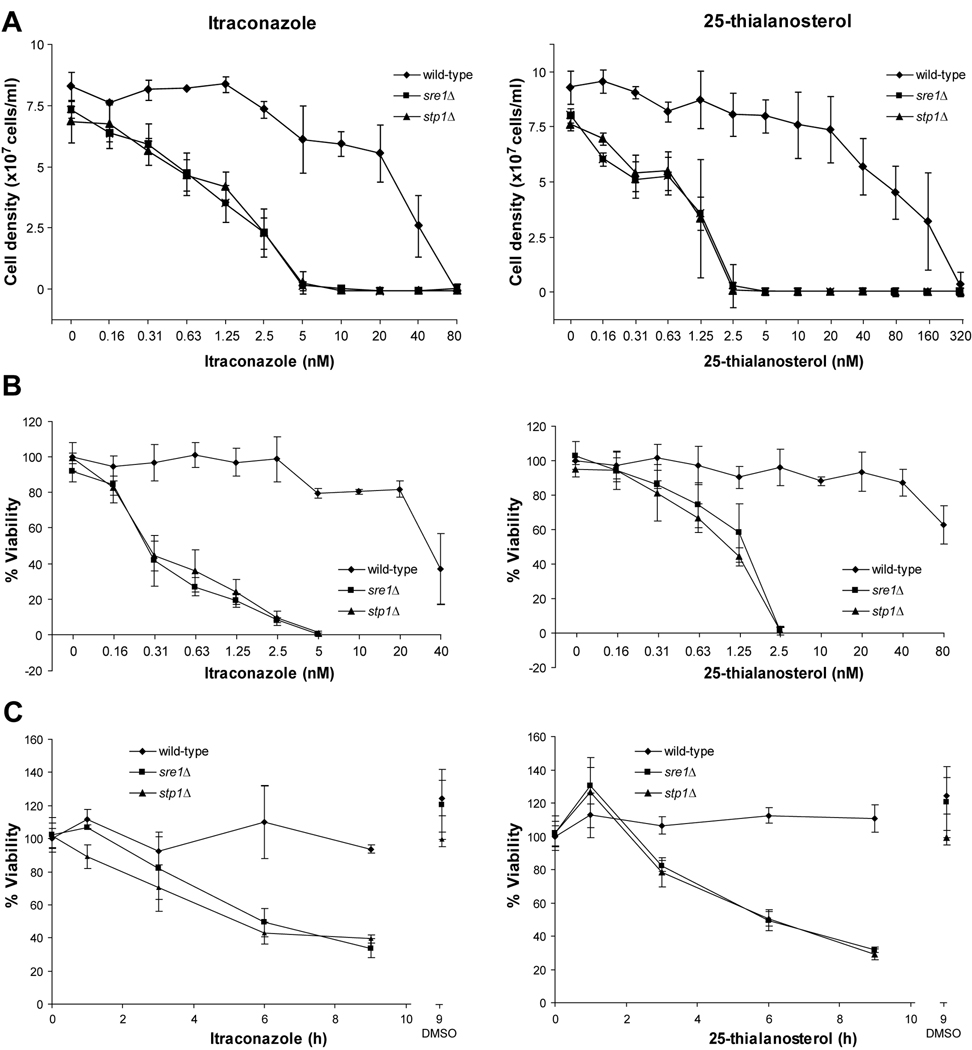
Ergosterol is an essential component of fungal cell membranes (Lees, Bard, and Kirsch, 1999). Consequently, antifungal therapeutics including the widely-used azole class of drugs target ergosterol biosynthesis. Previous studies demonstrated that SRE1 and STP1 are required for growth in the presence of the azole drugs, fluconazole, voriconazole and ketoconazole (Chang et al., 2007; Chun, Liu, and Madhani, 2007). To investigate this result further, we performed detailed studies analyzing both the fungistatic and fungicidal properties of ergosterol biosynthesis inhibitors on wild-type, sre1Δ and stp1Δ strains. We analyzed the effects of the antifungal azole drug, itraconazole, and a new sterol biosynthesis inhibitor, 25-thialanosterol. 25-thialanosterol is an inhibitor of the fungal-specific sterol biosynthetic enzyme, sterol 24-C-methyltransferase (Erg6) (Nes et al., 2009). Wild-type, sre1Δ and stp1Δ cells were grown for 48 hours in rich medium at 30°C in the presence of two-fold serial dilutions of itraconazole or 25-thialanosterol. As observed previously, low nanomolar concentrations of itraconazole inhibited growth of sre1Δ and stp1Δ cells, but not wild-type cells (Fig. 6A, left). Importantly, sre1Δ and stp1Δ strains also displayed growth sensitivity to 25-thialanosterol (Fig. 6A, right). Given that these two compounds inhibit different ergosterol biosynthetic enzymes, we conclude that the growth sensitivity phenotype of the two mutants is due to defects in ergosterol biosynthesis and not other non-sterol related effects of azole drugs.
Figure 6. Sre1 pathway is required for growth and viability in the presence of sterol synthesis inhibitors.
A. Wild-type, sre1Δ and stp1Δ cells were grown for 48 hours in liquid YES medium. Final cell density was determined using a spectrophotometer (1 OD600 = 2×107 cells) at the indicated concentrations of itraconazole (left panel) or 25-thialanosterol (right panel). Error bars represent standard deviation from three biological replicate experiments. B. Wild-type, sre1Δ and stp1Δ cells were grown for 48 hours in liquid YES medium. Equal numbers of cells were plated and grown on YES medium for 2 days. Colony-forming units were counted and percent viability calculated. C. Wild-type, sre1Δ and stp1Δ cells were grown for the indicated times in the presence of 2.5 nM itraconazole, 5 nM 25-thialanosterol or DMSO as a vehicle control. Equal numbers of cells were plated and grown on YES medium for 2 days. Error bars represent the standard deviation from three biological replicate experiments.
For the treatment of fungal infection, azole antifungals are considered fungistatic but not fungicidal, meaning that the drugs inhibit fungal cell growth but do not actually kill cells (Lewis and Graybill, 2008). Next, we examined whether Sre1 activation is required for cell viability under the same culture conditions. Interestingly, the viability of sre1Δ and stp1Δ strains was dramatically reduced compared to wild-type cells in the presence of both itraconazole and 25-thialanosterol (Fig. 6B). Treating cells with 2.5 nM itraconazole had no effect on wild-type cell viability, but reduced viability of sre1Δ and stp1Δ cells to 10%. Similarly, treating cells with 2.5 nM 25-thialanosterol had no effect on wild-type cells, but reduced viability of mutant cells to 2%. These data indicated that the decreased growth of sre1Δ and stp1Δ strains in the presence of these sterol synthesis inhibitors is due, at least in part, to cell-death.
To investigate the kinetics of this killing effect, we assayed viability of cells treated for short times with either 5 nM itraconazole or 2.5 nM 25-thialanosterol, conditions that killed >98% of sre1Δ and stp1Δ cells after a 48 hour treatment (Fig. 6A). Viability of wild-type cells was not significantly affected by either drug during the 9 hour experiment (Fig. 6C). However, treatment with either itraconazole or 25-thialanosterol rapidly decreased sre1Δ and stp1Δ cell viability to 37% and 30% after 9 hours for itraconazole and 25-thialanosterol, respectively. This rapid decrease in viability suggests that the Sre1 transcriptional response provides a strong resistance mechanism at early times following exposure to sterol synthesis-inhibiting drugs.
DISCUSSION
In this study, we investigated the mechanism for proteolytic processing of the Sre1 transcription factor in C. neoformans serotype A. Multiple lines of evidence suggest that Stp1 is a conserved Site-2 protease required for Sre1 activation. First, Stp1 shows ~14% identity and 22% similarity to the human Site-2 protease ortholog and predicted catalytic residues are conserved (Fig. 1A). Second, stp1Δ cells fail to accumulate the activated N-terminal transcription factor domain of Sre1 under low oxygen (Fig. 1B). Third, when conserved catalytic residues of Stp1 were mutated, Sre1 processing was defective and cells displayed growth phenotypes similar to sre1Δ and stp1Δ strains (Fig. 1E and 1F). Importantly, ectopic expression of the soluble N-terminus of Sre1 in stp1Δ cells rescued growth on CoCl2, demonstrating that Stp1 is required for Sre1 transcriptional activity (Fig. 1D). Finally, using Sre1 C-terminal truncations as size standards, we determined that Sre1N cleavage occurs between amino acids 501–535. Thus, proteolytic cleavage likely occurs within the first transmembrane segment of Sre1 at a position close to that determined for mammalian SREBP-2 (Fig. 2) (Duncan et al., 1998). The Site-2 protease cleavage site of mammalian SREBP is not present in the first transmembrane segment of C. neoformans Sre1. However, previous studies demonstrated that the mammalian Site-2 protease cleavage site in SREBP can be mutated without affecting cleavage (Ye et al., 2000a). Taken together, these data indicate that C. neoformans Stp1 is a functional Site-2 protease required for the proteolytic activation of Sre1. Notably, Schizosaccharomyces pombe and Aspergillus fumigatus have functional SREBP transcription factors, but lack an identifiable Stp1 homolog, suggesting alternate mechanisms for Sre1 cleavage in these fungi (Willger et al., 2008; Hughes, Todd, and Espenshade, 2005).
In addition to its role in Sre1 cleavage, Stp1 is required to maintain levels of the full-length precursor form of Sre1. While Sre1 precursor is present in stp1Δ cells, levels are greatly reduced under both normoxic and hypoxic conditions (Fig. 1B). Based on our results, the decrease is not due to changes in SRE1 transcription since SRE1 expression is unchanged in stp1Δ cells under normoxic conditions (Fig. 1C). Recent studies in the fission yeast Schizosaccharomyces pombe demonstrated that in the absence of the Sre1 binding partner Scp1, Sre1 precursor is degraded via the ER-associated degradation (ERAD) pathway, leading to reduced Sre1 precursor (Hughes, Nwosu, and Espenshade, 2009). While C. neoformans stp1Δ cells contain wild-type amounts of Scp1 (Fig. S3), Sre1 in the ER may not bind Scp1 due to mislocalization of Scp1. Preliminary studies to address this hypothesis employed the drug brefeldin A (BFA), which causes mixing of ER and Golgi compartments (Lippincott-Schwartz et al., 1989). Interestingly, treating stp1Δ cells with BFA restored Sre1 precursor to wild-type levels (Fig. S4). While further experimentation is required, these data are consistent with a model in which the localization of Scp1 is altered in stp1Δ cells. Consequently, free Sre1 in the ER may be degraded by ERAD. In addition, Sre1 nuclear form was decreased in wild-type cells treated with brefeldin A, suggesting that Stp1 is unable to cleave Sre1 under these conditions.
Cleavage of mammalian SREBP by the Site-2 protease requires prior cleavage in the lumenal loop of SREBP by the Site-1 protease, a subtilisin-like serine protease (Espenshade and Hughes, 2007). This intermediate cleavage product can be detected by SREBP immunoblot and accumulates in Site-2 protease deficient cells (Rawson et al., 1997). Despite the conservation of Stp1 function and cleavage position, we tested several candidate Site-1 protease homologs and have not identified a protein involved in Sre1 processing (data not shown). Furthermore, we have been unable to identify an intermediate form of Sre1 in stp1Δ cells by immunoblot, indicating that perhaps no Site-1 protease cleavage exists or that the intermediate cleavage product is unstable.
The Cryptococcus neoformans species complex is classified into 5 serotypes (A, B, C, D, A/D), each with distinct disease characteristics. Serotypes A and D cause the majority of human disease, primarily in immunocompromised patients (Mitchell and Perfect, 1995; Bennett, Kwon-Chung, and Howard, 1977). Our previous work showed that in C. neoformans serotype D cells SRE1 is essential for virulence in a mouse model of infection (Chang et al., 2007). Here, our virulence studies using C. neoformans serotype A strains confirmed previous results from Chun et al. and demonstrated that sre1Δ and stp1Δ cells show attenuated virulence, indicating that Sre1 activation is required for full virulence (Chang et al., 2007). Further experiments are required to determine whether the virulence phenotypes observed in the sre1Δ and stp1Δ strains are due to a decrease in melanin synthesis. However, Chun et al. argued that this is unlikely since sre1Δ and stp1Δ cells showed decreased colonization in both the lungs and brains of infected mice (Chun, Liu, and Madhani, 2007), while lac1 mutant cells, with no melanin synthesis, showed decreased colony formation only in the brain (Chun, Liu, and Madhani, 2007).
To investigate the differences in the virulence phenotypes of sre1Δ cells between serotype A and D strains, we determined Sre1-dependent gene expression in serotype A cells and compared these with our previous data for serotype D (Chang et al., 2007). Interestingly, serotype A Sre1 regulates similar but not identical genes as serotype D Sre1 (Table 1). Similar to serotype D Sre1, serotype A Sre1 is required for hypoxic expression of genes encoding ergosterol biosynthetic enzymes. However, serotype A Sre1 is largely not required for hypoxic expression of genes required for iron and copper acquisition. This is in contrast to serotype D Sre1, which regulates siderophore, iron and copper transporters (Chang et al., 2007). Given that iron acquisition in the host is a virulence requirement for many pathogens, serotype D cells lacking SRE1 may be limiting for iron, but not serotype A cells (Howard, 1999; Jung and Kronstad, 2008). Thus, differences in regulation of metal homeostasis between serotypes A and D may explain the differential requirement for Sre1 in virulence. Continued investigation and comparison of Sre1 in serotypes A and D may provide insight to the mechanism of C. neoformans adaptation to the host environment.
Site-2 proteases are highly conserved throughout evolution, and many substrates other than SREBPs exist, including the stress response transcription factor ATF6 (Rawson and Li, 2007; Makinoshima and Glickman, 2006; Ye et al., 2000b). While a clear C. neoformans ATF6 homolog has not been identified, we investigated whether Stp1 acts on other transcriptional regulators by comparing Stp1 and Sre1-dependent gene expression. In our analysis, we found 258 genes regulated by Stp1 and not Sre1 (Fig. 4A–B, Table 1). Stp1-dependent only genes included 28 genes involved in translation and ribosome biogenesis. Although the fold change in gene expression was significant, it was less than two-fold (Table 1). Overall, the genes with the highest fold difference between wild-type and stp1Δ strains were also dependent upon Sre1. These data indicate that the primary transcriptional regulator controlled by Stp1 under the conditions tested is Sre1. Importantly, Sre1 is the most important substrate of Stp1 for virulence, due to the fact that we observed similar virulence phenotypes in the sre1Δ and stp1Δ strains (Fig. 3B).
Sre1 is activated under low oxygen and functions as a hypoxic transcription factor in fungi (Chang et al., 2007; Chun, Liu, and Madhani, 2007; Willger et al., 2008; Hughes, Todd, and Espenshade, 2005). In fission yeast, Sre1 is a major regulator of hypoxic gene expression controlling two-thirds of genes highly regulated under low oxygen, and sre1Δ cells show a severe low oxygen growth defect (Hughes, Todd, and Espenshade, 2005; Todd et al., 2006). In contrast, sre1Δ strains from C. neoformans serotype A and D display mild growth defects under hypoxia (Chang et al., 2007; Chun, Liu, and Madhani, 2007). Our analysis of C. neoformans low oxygen gene expression in wild-type cells revealed that expression of 28% of genes (1929/6798) changed significantly after 2 hours at 3% oxygen. However, only a small subset (87/1929 or 4.5%) of these hypoxic genes required Sre1 for regulation under low oxygen. These data indicate that while activated under low oxygen, C. neoformans Sre1 is not the principal regulator of low oxygen gene expression and that hypoxic regulators remain unidentified.
Ergosterol is an essential component of fungal cell membranes and is not present in mammalian cells (Espenshade and Hughes, 2007). Consequently, many antifungal therapeutics target ergosterol biosynthesis including the widely-used azole class of drugs. However, issues of fungal resistance to azole drugs have become a concern, particularly in severely immunocompromised patients (Lewis and Graybill, 2008; Kanafani and Perfect, 2008). Due to this, azole drug regimens are frequently given in combination with other antifungals and are administered for long periods of time (Lewis and Graybill, 2008). One proposed reason for the appearance of azole resistance in immunocompromised hosts is that azole antifungals are generally thought to be fungistatic and not fungicidal, meaning that they inhibit fungal cell growth but do not lead to cell death (Lewis and Graybill, 2008; Kanafani and Perfect, 2008). Consequently, fungistatic drugs may only be completely effective in individuals with normal immune function that can rapidly clear non-dividing yeast.
Our gene expression and cellular sterol composition analyses demonstrate that Sre1 regulates ergosterol biosynthetic enzymes and that Sre1 activation is essential for sterol homeostasis (Fig. 5A). Excitingly, our studies demonstrated that itraconazole and 25-thialanosterol display fungicidal effects against sre1Δ and stp1Δ cells (Fig. 6B). Mutant cells lost viability rapidly, with significant effects as early as three hours post-treatment (Fig. 6C). Taken together, these results demonstrate that the Sre1 pathway is required for cellular growth and survival in the presence of sterol biosynthesis-inhibiting antifungal drugs. Given the need for fungicidal drugs, we propose that inhibitors of Stp1, Sre1, or other regulators of Sre1 function administered in combination with a sterol synthesis inhibitor could prove an effective strategy to treat cryptococcosis.
EXPERIMENTAL PROCEDURES
Materials and Culture Conditions
For all experiments, yeast cells were grown to exponential phase (~2×107 cells/ml) at 30°C in YES medium (0.5% [w/v] yeast extract plus 3% [w/v] glucose and supplements, 225 µg/ml each of uracil, adenine, leucine, histidine, and lysine). Low oxygen conditions were maintained using an Invivo2 400 workstation (Biotrace, Inc.) at 30°C. Itraconazole (Sigma) and 25-thialanosterol iodide salt (W. David Nes, Texas Tech University) were dissolved in DMSO and incorporated into media at indicated concentrations indicated. Brefeldin A (Sigma) was dissolved in 100% ethanol. Cobalt (II) chloride (Sigma) was dissolved in H2O and used at 0.3 mM in YES medium.
Strain Generation
Strains used were derived from Cryptococcus neoformans var. grubii serotype A strain (H99) (Perfect, Lang, and Durack, 1980). SRE1 and STP1 deletion strains were generated by biolistic transformation and homologous recombination using standard techniques (Toffaletti et al., 1993). The STP1 deletion strain was stably-complemented by pCB59-1 plasmid containing a wild-type copy of the STP1 expressed from the C. neoformans H99 ACT1 promoter using electroporation transformation techniques (Edman and Kwon-Chung, 1990). pCB59-1 was derived from the pYCC725 plasmid (Lee et al., 2007). Plasmids expressing truncations of SRE1 were generated using standard cloning techniques. Briefly, 1000 bp of SRE1 upstream sequence and truncated SRE1 coding sequence was amplified from C. neoformans H99 genomic DNA and digested with SfiI (New England Biolabs). PCR products were cloned into SfiI sites in pCB5-1 and pCB6-1 plasmids, which were derived from pYCC725 (containing NEO resistance gene) and pYCC726 (containing NAT resistance gene) respectively (Lee et al., 2007). Plasmids derived from pCB5-1 and pCB6-1 were transformed by electroporation into sre1Δ cells and stp1Δ cells respectively.
Site-Directed Mutagenesis
Point mutations were introduced in STP1 using Quickchange XLII PCR mutagenesis (Stratagene). pCB59-1 was used as the template for the PCR reactions. Mutagenized plasmids were sequenced and transformed into stp1Δ cells by electroporation.
Protein Extracts and Immunoblot Analysis
C. neoformans cell lysates were prepared as previously described (Chang et al., 2007). Briefly, cells were grown in YES medium under the indicated conditions. Yeast (4×107 cells) were washed in sterile H2O and lysed in a vortexer by glass bead beating (425–600 µm, Sigma) in 25 mM NaOH and 128 mM 2-mercaptoethanol for 5 min at 4°C. Proteins were precipitated with 6.4% (w/v) trichloroacetic acid followed by two washes with cold acetone. Protein was resuspended in SDS lysis buffer [1% (w/v) SDS, 10 mM Tris-HCl (pH 6.8), 1 mM EDTA, 1 mM EGTA] plus protease inhibitors (25 µg/ml ALLN, 10 µg/ml leupeptin, 2 µg/ml aprotinin, 5 µg/ml pepstatin A, 0.5 mM PMSF and 1 mM DTT). When needed, 8 µg protein was treated with alkaline phosphatase (Roche) for one hour at 30°C. SDS-PAGE and immunoblot analysis were performed as previously described (Chang et al., 2007). Immunoblots were performed using anti-Sre1 antiserum at a dilution of 1:1000 (Chang et al., 2007).
Virulence Studies
Female BALB/c mice (~8 weeks old) were injected via lateral tail vein with 2×106 yeast cells suspended in 0.1 ml saline. Mouse survival was monitored and mice displaying severe morbundity were sacrificed as described previously (Chang et al., 2007). Kaplan–Meier analysis of survival was performed using JMP software (SAS Institute, Cary, NC). Both mutant strains were found to be significantly different from wild-type (p<0.0001).
RNA Isolation and Quantitative PCR
RNA was prepared as described previously (Chang et al., 2007). Yeast (4×108 cells) grown under the indicated conditions were harvested and washed in sterile H2O. Cell pellets were frozen on dry ice, lyophilized for 15 hours and then lysed by glass bead beating. Total RNA was isolated using RNA STAT-60 reagent (Tel-Test) and chloroform followed by ethanol precipitation according to manufacturer’s instructions. RNA was treated with DNase I enzyme (Roche) to eliminate contaminating genomic DNA. cDNA was synthesized using SuperScript II kit (Invitrogen). cDNAs were quantified by Real Time PCR (Bio-Rad) using SyBr Green QPCR Master Mix (Stratagene). A list of primers used to amplify cDNAs is in Table S2.
Gene Expression Profiling
Whole genome Cryptococcus neoformans H99 microarrays were designed using Agilent eArray online software (https://earray.chem.agilent.com). Each microarray contains 6798 60-base probes printed in duplicate, with melting temperatures of ~80°C. Probes were designed using previously generated microarray probes (Chow et al., 2007; Chun, Liu, and Madhani, 2007), and cDNA sequences as templates. In general, probes were generated near the 3’ end of transcripts, while maximizing the number of unique transcript isoforms detected per gene. A list of probe sequences and ID numbers is in Supplemental Table S3. For microarray experiments, RNA was prepared as described above and treated with DNase I (Roche) to eliminate genomic DNA contamination. RNA was processed for microarray by the Johns Hopkins SKCCC DNA Microarray Core Facility (http://microarray.onc.jhmi.edu/). Experiments were performed using RNA harvested from 2 independent experiments, yielding four data points per gene. For data analysis, features were examined and flagged using Agilent Feature Extraction software. Per spot, per chip Lowess normalizations were performed using GenespringGX 7.3 software. Statistically significant genes were identified by Significance Analysis of Microarrays (SAM) using a 90th percentile false discovery rate (Tusher, Tibshirani, and Chu, 2001). The SAM method assigns a value to each gene by analyzing the change in gene expression relative to the standard deviation of replicate samples.
Gene Ontology Analysis
Gene ontology identifiers (GO terms) are assigned to help categorize a list of genes into funcational groups (Ashburner et al., 2000). To identify (GO terms) for C. neoformans H99 genes, we systematically performed homology searches against S. cerevisiae proteins using BLASTP at NCBI. C. neoformans proteins with homology to S. cerevisiae proteins (E value cutoff of <0.001) were assigned the corresponding S. cerevisiae GO terms.
Sterol Analysis
Sterols were extracted from 1×108 log-phase cells as described previously (Lee et al., 2007). Briefly, cells suspended in 9 ml methanol and 4.5 ml 60% (w/v) KOH and 5 µg cholesterol (an internal recovery standard). Sterols were saponified at 75°C for 2 hours, cooled to room temperature, and extracted with 4 ml petroleum ether. Sterols were dried by evaporating the petroleum ether under a stream of nitrogen gas and resuspended in 200 µl heptane. 2 µl were injected into an Agilent 6850 gas chromatograph with an HP-1 column and FID. Retention times for sterol intermediates were determined using standards.
Growth and Viability Assays
For drug growth and viability experiments, 500 cells were seeded in 200 µl YES medium in 96-well microtiter plates. Cells were grown for 48 hours at 30°C shaking. Optical density was measured at 600 nm and cell number was determined using the conversion: 1.0 OD600 = 2.5×107 cells. Control experiments were performed to ensure this conversion did not vary greatly between different C. neoformans strains and culture conditions. Cell cultures were diluted, and 1,000 cells were plated on rich medium and grown for 48 hours at 30°C. Colony-forming units were counted and percent viability relative to wild-type untreated cells was determined.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health HL077588 and AI72186 (to PJE), grants from the National Science Foundation MCB-0417436 and the Welch Foundation D-1276 (to WDN), and funds from the intramural program of the National Institutes of Allergy and Infectious Diseases, National Institutes of Health (to KJK-C). PJE is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease. We thank J. Burg for compiling and assigning GO terms for C. neoformans H99 genes, and S. Zhao for outstanding technical assistance.
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JE, Kwon-Chung KJ, Howard DH. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am.J.Epidemiol. 1977;105:582–586. doi: 10.1093/oxfordjournals.aje.a112423. [DOI] [PubMed] [Google Scholar]
- Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol.Microbiol. 2007;64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- Chow ED, Liu OW, O'Brien S, Madhani HD. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var grubii: nitric oxide stress and body temperature. Current Genetics. 2007;52:137–148. doi: 10.1007/s00294-007-0147-9. [DOI] [PubMed] [Google Scholar]
- Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS.Pathog. 2007;3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EA, Dave UP, Sakai J, Goldstein JL, Brown MS. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J.Biol.Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- Edman JC, Kwon-Chung KJ. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol.Cell Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu.Rev.Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Howard DH. Acquisition, transport, and storage of iron by pathogenic fungi. Clinical Microbiology Reviews. 1999;12:394–404. doi: 10.1128/cmr.12.3.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Hughes BT, Nwosu CC, Espenshade PJ. Degradation of SREBP precursor requires the ERAD components UBC7 and HRD1 in fission yeast. J.Biol.Chem. 2009;284:20512–20521. doi: 10.1074/jbc.M109.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Kronstad JW. Iron and fungal pathogenesis: a case study with Cryptococcus neoformans. Cellular Microbiology. 2008;10:277–284. doi: 10.1111/j.1462-5822.2007.01077.x. [DOI] [PubMed] [Google Scholar]
- Kanafani ZA, Perfect JR. Resistance to antifungal agents: Mechanisms and clinical impact. Clinical Infectious Diseases. 2008;46:120–128. doi: 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE. Medical Mycology. Philadelphia: Lea & Febiger; 1992. [Google Scholar]
- Lee H, Bien CM, Hughes AL, Espenshade PJ, Kwon-Chung KJ, Chang YC. Cobalt chloride, a hypoxia-mimicking agent, targets sterol synthesis in the pathogenic fungus Cryptococcus neoformans. Mol.Microbiol. 2007;65:1018–1033. doi: 10.1111/j.1365-2958.2007.05844.x. [DOI] [PubMed] [Google Scholar]
- Lees ND, Bard M, Kirsch DR. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit Rev.Biochem.Mol.Biol. 1999;34:33–47. [PubMed] [Google Scholar]
- Lewis JS, Graybill JR. Fungicidal versus fungistatic: what's in a word? Expert Opinion on Pharmacotherapy. 2008;9:927–935. doi: 10.1517/14656566.9.6.927. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinoshima H, Glickman MS. Regulation of Mycobacterium tuberculosis cell envelope composition and virulence by intramembrane proteolysis. Nature. 2005;436:406–409. doi: 10.1038/nature03713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinoshima H, Glickman MS. Site-2 proteases in prokaryotes: regulated intramembrane proteolysis expands to microbial pathogenesis. Microbes.Infect. 2006;8:1882–1888. doi: 10.1016/j.micinf.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS--100 years after the discovery of Cryptococcus neoformans. Clin.Microbiol.Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes WD, Zhou W, Ganapathy K, Liu J, Vatsyayan R, Chamala S, Hernandez K, Miranda M. Sterol 24-C-methyltransferase: an enzymatic target for the disruption of ergosterol biosynthesis and homeostasis in Cryptococcus neoformans. Arch.Biochem.Biophys. 2009;481:210–218. doi: 10.1016/j.abb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Perfect JR, Lang SDR, Durack DT. Chronic Cryptococcal Meningitis - A New Experimental-Model in Rabbits. American Journal of Pathology. 1980;101:177–193. [PMC free article] [PubMed] [Google Scholar]
- Rawson RB, Li W. Intramembrane-Cleaving Proteases (I-CLiPs) Springer; 2007. The Site-2 Protease at Ten; pp. 1–15. [Google Scholar]
- Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol.Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- Rosenfeld E, Beauvoit B. Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae. Yeast. 2003;20:1115–1144. doi: 10.1002/yea.1026. [DOI] [PubMed] [Google Scholar]
- Scheinfeld N. A review of the new antifungals: posaconazole, micafungin, and anidulafungin. Journal of Drugs in Dermatology. 2007;6:1249–1251. [PubMed] [Google Scholar]
- Sheehan DJ, Hitchcock CA, Sibley CM. Current and emerging azole antifungal agents. Clinical Microbiology Reviews. 1999;12:40–79. doi: 10.1128/cmr.12.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol.Cell Biol. 2006;26:2817–2831. doi: 10.1128/MCB.26.7.2817-2831.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene-Transfer in Cryptococcus neoformans by Use of Biolistic Delivery of DNA. Journal of Bacteriology. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc.Natl.Acad.Sci.U.S.A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA., Jr A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS.Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by Site-2 protease. Proc.Natl.Acad.Sci.U.S.A. 2000a;97:5123–5128. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol.Cell. 2000b;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.