Abstract
The incidence rate difference (IRD) is a parameter of interest in many medical studies. For example, in vaccine studies, it is interpreted as the vaccine-attributable reduction in disease incidence. This is an important parameter, because it shows the public health impact of an intervention. The IRD is difficult to estimate for various reasons, especially when there are quantitative covariates or the duration of follow-up is variable. In this paper, the authors propose an approach based on weighted least-squares regression for estimating the IRD. It is very easy to implement because it boils down to performing ordinary least-squares regression analysis of transformed variables. Furthermore, if the outcome events are repeatable, the authors propose that data on all events be analyzed instead of first events only. Four versions of the Huber-White robust standard error are considered for statistical inference. Simulation studies are used to examine the performance of the proposed method. In a variety of scenarios simulated, the method provides an unbiased estimate for the IRD, and the empirical coverage proportion of the 95% confidence interval is very close to the nominal level. The method is illustrated with data from a vaccine trial carried out in the Gambia in 2001–2004.
Keywords: incidence rate, least-squares analysis, recurrent events, standard error
The relative merits of the odds ratio, risk ratio, and risk difference and procedures for estimating them have been discussed by many epidemiologists and statisticians (1–3). The usage and relative merits of the incidence rate ratio (IRR) and the incidence rate difference (IRD) have received much less attention. Both IRR and IRD are commonly used in reporting results from vaccine trials. In vaccine trials, vaccine efficacy is estimated by 1 – (incidence rate in vaccine group/incidence rate in control group), or 1 – IRR, and vaccine-attributable reduction in incidence is estimated by (incidence rate in control group – incidence rate in vaccine group), or IRD (4, 5). Vaccine-attributable reduction represents the reduction in disease burden and is a useful measure of the public health importance of a vaccine. Furthermore, the inverse of vaccine-attributable reduction (or IRD) has the useful interpretation of “number needed to treat” in order to prevent 1 episode of disease per person-year (6). The impact of vaccines may also be studied in observational (nonrandomized) studies in which the evaluation can be subject to confounding, thus requiring statistical adjustment. The same concepts apply to the evaluation of other interventions or exposures. Hence, in this article, the generic term incidence rate difference is used instead of vaccine-attributable reduction, and it refers to the incidence rate in the control (unexposed) group minus that in the intervention (exposed) group.
There are 3 issues to consider when estimating IRD and IRR. Firstly, if the outcome events are repeatable, should one use data on time to first events only or all events? In the context of vaccine research, this issue has been the topic of recent debate (7, 8). A meeting convened by the World Health Organization in 2008 recommended that data on all events should be included in the evaluation of vaccine efficacy for malaria vaccines (8). It appears that the bias in estimating IRD by using first events only has been somewhat neglected (see Web Appendix 1, available on the Journal’s Web site (http://aje.oxfordjournals.org/), for an illustration).
Secondly, statistical estimation of IRD is more difficult than that of IRR. For a simple 2-group comparison without adjustment for covariates, there are established methods for estimating IRD on the basis of the Poisson distribution (first event) and the negative binomial distribution (all events) (9, 10). For analysis of a single quantitative exposure variable or multiple exposure variables, generalized linear models readily deal with estimation of the IRR by using the log of follow-up time as an “offset” term in the log(incidence) equation. However, unless follow-up time is equal for every participant, which is not true in many studies, it is not clear how well the generalized linear models estimate the IRD. Even in the simple case of equal follow-up time, the iterations for Poisson and negative binomial regression models with a quantitative exposure variable may not always converge (3). One possible option for multivariable analysis of IRD is standardization (11, 12), but this cannot accommodate quantitative exposure variables, and it becomes difficult in practice as the number of (categorical) exposure variables increases.
Thirdly, valid statistical inference for data on repeatable (or recurrent) events needs to avoid underestimation of variance and inflation of type 1 error rates due to correlated events within the same person.
In this article, we aim to provide a simple method based on a weighted least-squares regression approach and a robust standard error estimator for estimating IRD. The proposed method easily controls for unequal follow-up time and quantitative or multiple covariates. The method is general in that it is applicable to analysis of first events and all events. When there is only 1 binary exposure variable, the method will reduce to the one proposed by Stukel et al. (11) and Glynn and Buring (10) under mild conditions.
METHODS
Notations and model
Suppose there are n subjects in a study. For subject i (i = 1, …, n), for the analysis of recurrent-event data, Yi is the total number of events recorded and Zi is the total length of follow-up time, referring to the time from recruitment into the cohort to either loss to follow-up, which is random, or study closure, which is determined by the investigators. Moreover, Xi = (Xi0, Xi1, …, Xik) is the covariate vector for subject i. β = (β0, β1, …, βk) is the unknown regression coefficient vector associated with Xi. Let Xi0 = 1 be the intercept of the design matrix, Xi1 be an indicator of the exposure status, and Xi2–Xik be the other covariates. The estimate of the incidence rate for subject i is Yi/Zi. The observed incidence rate using data on all events in the unexposed (Xi1 = 0) group is
 |
whereas the observed IRD between groups is
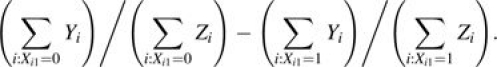 |
The gist of our proposed method, which will be elaborated below, is to generate new variables 
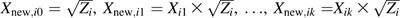 and to perform ordinary least-squares regression without an intercept for the regression equation
and to perform ordinary least-squares regression without an intercept for the regression equation
| (1) |
Note that IRD = −β1 by definition, and we will show below that the estimator 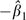 is unbiased. If a less conventional data coding scheme is used—for example, if the exposed (unexposed) group is indicated by Xi1 = 0 (Xi1 = 1)—then IRD = β1.
is unbiased. If a less conventional data coding scheme is used—for example, if the exposed (unexposed) group is indicated by Xi1 = 0 (Xi1 = 1)—then IRD = β1.
To accommodate possible unobserved heterogeneity in the event rates among subjects, a frailty term υi is introduced. Therefore, the incidence rate for subject i can be modeled as
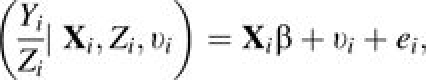 |
(2) |
where the ei’s are the independent individual residuals, with E(ei) = 0, Var(ei) = σ2/Zi, and Cov(ei, ej) = 0 if i ≠ j. υi is uncorrelated with Xi, Zi, and ei, and follows an arbitrary distribution with E(υi) = ν and Var(υi) = ϱ.
Estimation
Following model 2 (equation 2), the conditional mean and variance for the incidence rate of subject i are
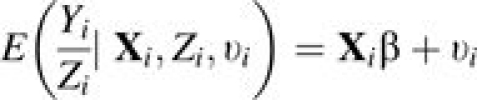 |
(3) |
and
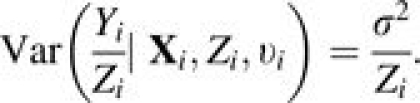 |
(4) |
Consequently, for any distribution of Yi, the marginal mean and variance of Yi/Zi are
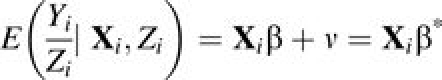 |
(5) |
and
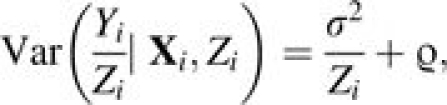 |
(6) |
respectively. β* = (β0 + ν, β1, …, βk), where the mean of the possible unobserved heterogeneity becomes part of the intercept. Since the primary interest is to estimate β from β1 (or equivalently, –IRD) to βk and subsequently make inference about β1,… βk, no further attention is paid to β0*.
Throughout this article, models and expectations are conditional on υi or marginal with respect to υi, while for each situation they are always conditional on Xi and Zi.
Note that equations 2–6 apply to recurrent-events data. When analysis is limited to the first events, Zi is subject to being censored by the first event. Moreover, the frailty υi is not required, and model 2 reduces to the linear probability model with a binary response variable. Nonetheless, the following estimation procedure still applies, but it is unbiased only if there is no heterogeneity (see Web Appendix 1 for an illustration).
We propose to estimate β* using the following estimation equation:
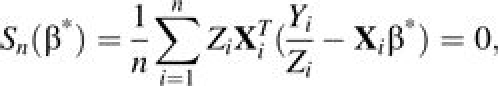 |
(7) |
which is a special case of generalized estimating equations (13). Using the conditional variance in equation 4 instead of the marginal variance in equation 6 in the estimating equation gives an explicit solution that allows one to incorporate some existing estimation methods. A robust variance estimator is used to correct for the misspecification of the working variance. By the general theory of generalized estimating equations (13), the resulting estimator of β* is consistent and asymptotically normal as long as equation 5 holds true.
For notational simplicity, let Y = (Y1, …, Yn)T, Z = (Z1, …, Zn)T, X = (X1T,…,XnT)T, and W = diag(Z1, …, Zn). Following equation 7,
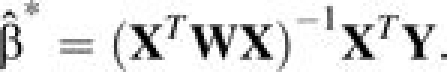 |
(8) |
Moreover, it can be shown, in the case of time-varying regression coefficients, that 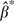 is estimating the average of the covariates’ effects over the total number of person-years studied using data on all of the events (the mathematical proof and simulation evidence are part of our ongoing work).
is estimating the average of the covariates’ effects over the total number of person-years studied using data on all of the events (the mathematical proof and simulation evidence are part of our ongoing work).
In the simplest case in which Xi = (1, Xi1), where Xi1 = 0 or 1 and β* = (β0*,β1)T, equation 8 yields
 |
and
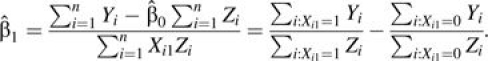 |
Therefore, 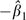 is the observed IRD in the sample.
is the observed IRD in the sample.
In the presence of L strata in the sample defined by 1 or more categorical confounders, using equation 8, 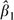 reduces to
reduces to
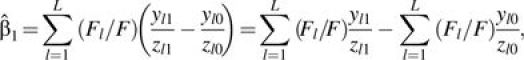 |
where ylj and zlj are the total number of events and the total follow-up time, respectively, for all subjects in the lth (l = 1, …, L) stratum in the unexposed group (j = 0) and the exposed group (j = 1). Fl = 1/(1/zl0 + 1/zl1) and F = ∑l = 1LFl. This agrees with the estimator proposed by Stukel et al. (11), which is a weighted average of the IRD between the exposed (j = 1) and the unexposed (j = 0) across the L strata.
Note that equation 8 can be equivalently estimated via an ordinary least-squares regression on the transformed variables Y and X—that is, Ynew = W–1/2Y and Xnew = W1/2X, as introduced above. Therefore, the robust variance estimator (14) for  is
is
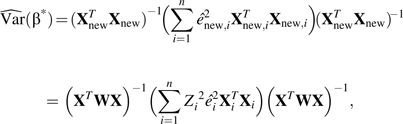 |
(9) |
where 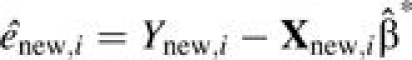 and
and 
For the simplest case in which Xi = (1, Xi1) and Xi1 = 0 or 1, the robust variance estimator for 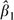 can be simplified as
can be simplified as
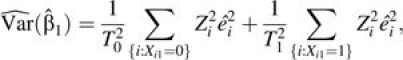 |
(10) |
where Tg = ∑{i:Xi1 = g}Zi and g = 0 or 1. It can be shown that the variance estimator in equation 10 is consistent with that given by Stukel et al. (11), provided that the residuals are independently and identically distributed within each group defined by Xi1 (see Web Appendix 2).
Small-sample adjustment
Equation 9 gives the asymptotic, or large-sample, version of the robust variance, which we name HCr. Three versions of adjustment for a small sample size have been proposed (15), conventionally named HC1, HC2, and HC3. The small-sample adjustment involves multiplying the ith summand in the middle term of HCr in equation 9 by a correction factor n/(n – k), 1/(1 – hi), or 1/(1 – hi)2 to obtain HC1, HC2, or HC3, respectively, where hi is the ith diagonal element in the matrix H:
The adjustment is smallest for HC1 and largest for HC3, and the 4 variance estimators are asymptotically the same.
SIMULATION STUDIES
The proposed method is general, but this work is motivated by our research in pediatric infectious diseases. The simulation scenarios will follow realistic situations seen in the studies of acute otitis media and radiologically confirmed pneumonia, representing diseases with relatively high and low incidence rates in young children (16, 17).
Data on first events and on all events will be used to estimate β*, and the differences in the estimates will be examined. The performance of the proposed estimator and robust variance estimator will be evaluated in various scenarios, using the average of parameter estimates, the average of the robust standard error estimates, the empirical standard deviation, and the 95% coverage proportion based on the robust standard error estimates. The variance estimate proposed by Stukel et al. (11) will also be calculated in the scenarios with only 1 binary exposure variable for comparison.
Simulation parameter configurations and data generation processes are described in detail in Web Appendix 3. For each scenario, 10,000 replications were simulated.
Table 1 shows the simulation results with n = 1,000 in the absence of confounding. It can be seen that the average of parameter estimates using all events was close to the true parameter value (β1 = −0.5 in high-incidence scenarios and β1 = −0.025 in low-incidence scenarios), regardless of whether the binary exposure variable Xi1 (Xi1 ∼ Bernoulli(p)) was evenly (p = 0.5) or unevenly (p = 0.7) distributed and regardless of heterogeneity (νi). However, the presence of heterogeneity resulted in biased estimates for β1 using first events for high-incidence scenarios (the averages of parameter estimates were –0.5151 and –0.5073 under 2 forms of heterogeneity, respectively), though little bias was observed for low-incidence scenarios (the averages of parameter estimates were very close to –0.025). Secondly, the 95% coverage proportions based on the robust standard error estimates were very close to their nominal level. When using data on all events, the inferences derived using robust standard errors were practically identical to those of the Stukel et al. (11) approach in terms of the average of parameter estimates and the 95% coverage proportion based on the robust standard error estimates. Moreover, regardless of whether data on first events or all events were used, the average of the robust standard error estimates and the empirical standard deviation agreed very well. Since this is also true in other simulation scenarios, we suppressed the display of empirical standard deviations in subsequent tables in the interest of space. Similar findings were observed for the cases with n = 200 (Web Table 1).
Table 1.
Simulation Results on the Estimates for β1 in the Absence of Confounding (n = 1,000)
| p and β1 | υi | First Events |
All Events |
||||||||
| Proposed Method |
Average Estimatea | ESD | Proposed Method |
Stukel et al. (11) |
|||||||
| Average Estimatea | ESD | Average SErb | CPrc | Average SErb | CPrc | Average SEd | CPe | ||||
| p = 0.5 | |||||||||||
| β1 = −0.5 | 0 | −0.5011 | 0.0537 | 0.0539 | 95.1 | −0.4999 | 0.0348 | 0.0347 | 95.0 | 0.0347 | 95.0 |
| ±0.25 | −0.5151 | 0.0520 | 0.0525 | 94.7 | −0.5001 | 0.0380 | 0.0381 | 95.2 | 0.0381 | 95.2 | |
| γ | −0.5073 | 0.0605 | 0.0599 | 94.6 | −0.5001 | 0.0408 | 0.0406 | 94.8 | 0.0406 | 94.8 | |
| β1 = −0.025 | 0 | −0.0250 | 0.0080 | 0.0080 | 94.9 | −0.0250 | 0.0077 | 0.0077 | 95.0 | 0.0077 | 95.0 |
| ±0.025 | −0.0251 | 0.0078 | 0.0079 | 95.3 | −0.0250 | 0.0077 | 0.0078 | 95.4 | 0.0078 | 95.3 | |
| γ | −0.0250 | 0.0086 | 0.0086 | 95.0 | −0.0249 | 0.0083 | 0.0084 | 94.9 | 0.0084 | 94.9 | |
| p = 0.7 | |||||||||||
| β1 = −0.5 | 0 | −0.5030 | 0.0648 | 0.0646 | 94.9 | −0.5005 | 0.0399 | 0.0402 | 94.9 | 0.0402 | 95.0 |
| ±0.25 | −0.5158 | 0.0689 | 0.0698 | 95.4 | −0.5004 | 0.0455 | 0.0454 | 95.0 | 0.0454 | 94.9 | |
| γ | −0.5074 | 0.0702 | 0.0703 | 95.0 | −0.4998 | 0.0459 | 0.0455 | 94.9 | 0.0455 | 94.9 | |
| β1 = −0.025 | 0 | −0.0250 | 0.0093 | 0.0093 | 94.7 | −0.0249 | 0.0091 | 0.0090 | 94.5 | 0.0090 | 94.6 |
| ±0.025 | −0.0251 | 0.0098 | 0.0098 | 94.9 | −0.0250 | 0.0096 | 0.0095 | 94.7 | 0.0095 | 94.7 | |
| γ | −0.0250 | 0.0097 | 0.0098 | 95.0 | −0.0249 | 0.0094 | 0.0095 | 94.9 | 0.0095 | 95.0 | |
Abbreviations: CP, coverage proportion; ESD, empirical standard deviation; SE, standard error.
Average of the parameter estimates.
Average of the robust standard error estimates.
95% coverage proportion based on the robust standard error estimates.
Average of the standard error estimates using the method proposed by Stukel et al. (11).
95% coverage proportion based on the standard error estimates using the method proposed by Stukel et al. (11).
Table 2 shows the simulation results when the intervention effect was confounded by a quantitative variable Xi2 which was slightly skewed. The analysis simultaneously included Xi1 and Xi2 in the regression model. The proposed method using all events performed very well for high-incidence scenarios, that is, (β1, β2) = (–0.5, 0.05). Regardless of the distribution of Xi1 (p = 0.5 or 0.7), the degree of collinearity, or the presence of heterogeneity, the mean estimates for β1 and β2 were always close to the targeted values. The 95% coverage proportions based on the robust standard error estimates were also very close to the nominal level. In the low-incidence scenarios with (β1, β2) = (–0.025, 0.005), the proposed method gave a mean estimate for β2 somewhat different from the true value. Nevertheless, the 95% coverage proportion based on the robust standard error estimates was still close to the nominal level. In only 1 case did the 95% coverage proportion based on the robust standard error estimates deviate from 95% by more than 0.5% (94.3% at p = 0.7, β1 = –0.025, υi = 0, and moderate collinearity). Results were very similar when n = 200 (Web Table 2).
Table 2.
Simulation Results on the Estimates for (β1,β2) With n = 1,000 and X2 Slightly Skewed
| p and β | υi | Moderate Collinearity | Strong Collinearity | ||||||||||
| First Events | All Events | First Events | All Events | ||||||||||
| Average Estimatea | Average SErb | CPrc | Average Estimate | Average SEr | CPr | Average Estimate | Average SEr | CPr | Average Estimate | Average SEr | CPr | ||
| p = 0.5 | |||||||||||||
| β1 = −0.5 | 0 | −0.5000 | 0.0637 | 94.7 | −0.4994 | 0.0393 | 94.9 | −0.5019 | 0.0727 | 94.8 | −0.5004 | 0.0450 | 94.7 |
| ±0.25 | −0.5124 | 0.0623 | 95.1 | −0.4993 | 0.0426 | 95.0 | −0.5145 | 0.0712 | 94.3 | −0.5003 | 0.0487 | 94.9 | |
| γ | −0.5058 | 0.0699 | 95.3 | −0.4993 | 0.0451 | 95.0 | −0.5077 | 0.0804 | 94.7 | −0.5001 | 0.0515 | 94.9 | |
| β2 = 0.05 | 0 | 0.0501 | 0.0404 | 94.8 | 0.0499 | 0.0265 | 95.1 | 0.0505 | 0.0449 | 94.8 | 0.0507 | 0.0288 | 94.9 |
| ±0.25 | 0.0506 | 0.0394 | 94.3 | 0.0496 | 0.0287 | 94.9 | 0.0519 | 0.0439 | 94.7 | 0.0505 | 0.0311 | 94.8 | |
| γ | 0.0503 | 0.0449 | 95.2 | 0.0499 | 0.0303 | 95.0 | 0.0511 | 0.0497 | 94.8 | 0.0499 | 0.0328 | 94.8 | |
| β1 = −0.025 | 0 | −0.0251 | 0.0084 | 94.9 | −0.0251 | 0.0081 | 94.8 | −0.0250 | 0.0095 | 94.8 | −0.0249 | 0.0093 | 94.6 |
| ±0.025 | −0.0251 | 0.0084 | 94.8 | −0.0251 | 0.0082 | 94.9 | −0.0251 | 0.0095 | 95.0 | −0.0251 | 0.0093 | 94.8 | |
| γ | −0.0249 | 0.0090 | 95.1 | −0.0249 | 0.0088 | 95.0 | −0.0251 | 0.0103 | 95.1 | −0.0251 | 0.0100 | 95.0 | |
| β2 = 0.005 | 0 | 0.0057 | 0.1129 | 95.3 | 0.0056 | 0.1098 | 95.1 | 0.0042 | 0.1231 | 95.2 | 0.0039 | 0.1196 | 95.0 |
| ±0.025 | 0.0043 | 0.1126 | 94.8 | 0.0044 | 0.1104 | 94.8 | 0.0056 | 0.1227 | 95.5 | 0.0059 | 0.1200 | 95.1 | |
| γ | 0.0043 | 0.1218 | 95.2 | 0.0049 | 0.1188 | 94.9 | 0.0048 | 0.1323 | 95.3 | 0.0046 | 0.1288 | 95.1 | |
| p = 0.7 | |||||||||||||
| β1 = −0.5 | 0 | −0.5041 | 0.0744 | 95.0 | −0.5001 | 0.0446 | 94.9 | −0.5023 | 0.0816 | 95.2 | −0.4995 | 0.0497 | 94.8 |
| ±0.25 | −0.5160 | 0.0800 | 94.8 | −0.4997 | 0.0496 | 94.9 | −0.5159 | 0.0878 | 94.5 | −0.4998 | 0.0553 | 94.8 | |
| γ | −0.5086 | 0.0807 | 94.9 | −0.5000 | 0.0497 | 95.1 | −0.5093 | 0.0885 | 95.1 | −0.5002 | 0.0555 | 94.9 | |
| β2 = 0.05 | 0 | 0.0503 | 0.0374 | 94.8 | 0.0505 | 0.0254 | 94.8 | 0.0498 | 0.0429 | 95.1 | 0.0498 | 0.0287 | 95.1 |
| ±0.25 | 0.0514 | 0.0407 | 94.9 | 0.0500 | 0.0289 | 95.1 | 0.0506 | 0.0466 | 95.1 | 0.0498 | 0.0323 | 94.7 | |
| γ | 0.0508 | 0.0411 | 94.9 | 0.0502 | 0.0289 | 95.2 | 0.0513 | 0.0470 | 94.7 | 0.0507 | 0.0324 | 94.9 | |
| β1 = −0.025 | 0 | −0.0250 | 0.0096 | 94.3 | −0.0249 | 0.0093 | 94.3 | −0.0253 | 0.0106 | 94.8 | −0.0251 | 0.0103 | 94.6 |
| ±0.025 | −0.0251 | 0.0102 | 94.5 | −0.0251 | 0.0099 | 94.7 | −0.0251 | 0.0113 | 95.0 | −0.0250 | 0.0109 | 94.7 | |
| γ | −0.0251 | 0.0103 | 94.9 | −0.0250 | 0.0099 | 94.6 | −0.0248 | 0.0113 | 94.8 | −0.0248 | 0.0109 | 94.5 | |
| β2 = 0.005 | 0 | 0.0068 | 0.1070 | 95.2 | 0.0071 | 0.1043 | 95.0 | 0.0067 | 0.1206 | 95.1 | 0.0065 | 0.1174 | 95.0 |
| ±0.025 | 0.0049 | 0.1149 | 95.3 | 0.0051 | 0.1124 | 95.1 | 0.0046 | 0.1290 | 95.3 | 0.0049 | 0.1259 | 95.0 | |
| γ | 0.0045 | 0.1151 | 95.2 | 0.0041 | 0.1126 | 94.9 | 0.0048 | 0.1293 | 95.1 | 0.0051 | 0.1263 | 95.0 | |
Abbreviations: CP, coverage proportion; SE, standard error.
Average of the parameter estimates.
Average of the robust standard error estimates.
95% coverage proportion based on the robust standard error estimates.
Table 3 shows the simulation results obtained when the intervention effect was confounded by a quantitative variable Xi2 which was highly skewed. The results were very similar to those reported for the slightly skewed data series in Table 2. Again, results were very similar when n = 200 (Web Table 3).
Table 3.
Simulation Results on the Estimates for (β1, β2) With n = 1,000 and X2 Highly Skewed
| p and β | υi | Moderate Collinearity | Strong Collinearity | ||||||||||
| First Events | All Events | First Events | All Events | ||||||||||
| Average Estimatea | Average SErb | CPrc | Average Estimate | Average SEr | CPr | Average Estimate | Average SEr | CPr | Average Estimate | Average SEr | CPr | ||
| p = 0.5 | |||||||||||||
| β1 = −0.5 | 0 | −0.5006 | 0.0678 | 95.0 | −0.4993 | 0.0410 | 94.9 | −0.5024 | 0.0767 | 95.1 | −0.5004 | 0.0459 | 95.0 |
| ±0.25 | −0.5148 | 0.0665 | 94.6 | −0.5006 | 0.0443 | 94.8 | −0.5139 | 0.0754 | 94.4 | −0.5002 | 0.0494 | 95.2 | |
| γ | −0.5078 | 0.0745 | 94.6 | −0.5006 | 0.0468 | 94.7 | −0.5069 | 0.0841 | 94.4 | −0.5003 | 0.0520 | 95.0 | |
| β2 = 0.05 | 0 | 0.0499 | 0.0308 | 95.3 | 0.0500 | 0.0199 | 95.2 | 0.0499 | 0.0367 | 94.9 | 0.0501 | 0.0227 | 94.7 |
| ±0.25 | 0.0509 | 0.0303 | 94.8 | 0.0497 | 0.0210 | 94.9 | 0.0504 | 0.0363 | 94.2 | 0.0501 | 0.0239 | 94.9 | |
| γ | 0.0501 | 0.0341 | 94.9 | 0.0497 | 0.0222 | 95.0 | 0.0501 | 0.0404 | 95.0 | 0.0501 | 0.0251 | 94.9 | |
| β1 = −0.025 | 0 | −0.0250 | 0.0085 | 94.7 | −0.0250 | 0.0083 | 94.8 | −0.0251 | 0.0095 | 94.5 | −0.0250 | 0.0092 | 94.6 |
| ±0.025 | −0.0250 | 0.0085 | 94.8 | −0.0250 | 0.0083 | 94.8 | −0.0250 | 0.0095 | 94.9 | −0.0249 | 0.0093 | 94.7 | |
| γ | −0.0250 | 0.0092 | 94.9 | −0.0250 | 0.0090 | 95.0 | −0.0250 | 0.0102 | 94.9 | −0.0249 | 0.0100 | 94.6 | |
| β2 = 0.005 | 0 | 0.0050 | 0.0836 | 95.1 | 0.0055 | 0.0812 | 94.7 | 0.0048 | 0.0964 | 94.9 | 0.0050 | 0.0931 | 94.8 |
| ±0.025 | 0.0044 | 0.0835 | 94.8 | 0.0049 | 0.0815 | 94.6 | 0.0037 | 0.0961 | 95.0 | 0.0040 | 0.0934 | 94.8 | |
| γ | 0.0033 | 0.0898 | 95.0 | 0.0035 | 0.0873 | 94.9 | 0.0042 | 0.1028 | 95.0 | 0.0042 | 0.0997 | 94.9 | |
| p = 0.7 | |||||||||||||
| β1 = −0.5 | 0 | −0.5024 | 0.0785 | 94.9 | −0.5001 | 0.0464 | 95.1 | −0.5026 | 0.0880 | 94.9 | −0.4996 | 0.0521 | 94.7 |
| ±0.25 | −0.5155 | 0.0844 | 94.7 | −0.5003 | 0.0514 | 95.2 | −0.5154 | 0.0946 | 94.8 | −0.5003 | 0.0576 | 94.9 | |
| γ | −0.5095 | 0.0848 | 94.7 | −0.5006 | 0.0516 | 94.8 | −0.5080 | 0.0950 | 94.5 | −0.4994 | 0.0577 | 94.8 | |
| β2 = 0.05 | 0 | 0.0497 | 0.0303 | 94.7 | 0.0498 | 0.0200 | 95.1 | 0.0490 | 0.0397 | 94.8 | 0.0500 | 0.0249 | 95.2 |
| ±0.25 | 0.0509 | 0.0328 | 94.4 | 0.0499 | 0.0225 | 94.8 | 0.0494 | 0.0429 | 94.7 | 0.0496 | 0.0278 | 95.0 | |
| γ | 0.0498 | 0.0331 | 95.3 | 0.0501 | 0.0225 | 95.0 | 0.0500 | 0.0431 | 94.7 | 0.0503 | 0.0278 | 94.7 | |
| β1 = −0.025 | 0 | −0.0252 | 0.0098 | 95.1 | −0.0251 | 0.0095 | 94.9 | −0.0252 | 0.0109 | 94.9 | −0.0251 | 0.0106 | 94.7 |
| ±0.25 | −0.0252 | 0.0104 | 94.8 | −0.0251 | 0.0101 | 94.6 | −0.0249 | 0.0115 | 95.0 | −0.0248 | 0.0112 | 94.7 | |
| γ | −0.0252 | 0.0104 | 94.7 | −0.0251 | 0.0101 | 94.7 | −0.0252 | 0.0116 | 94.5 | −0.0251 | 0.0112 | 94.2 | |
| β2 = 0.005 | 0 | 0.0065 | 0.0842 | 95.3 | 0.0066 | 0.0819 | 95.0 | 0.0033 | 0.1065 | 95.0 | 0.0039 | 0.1030 | 94.9 |
| ±0.025 | 0.0043 | 0.0899 | 95.2 | 0.0049 | 0.0877 | 94.9 | 0.0026 | 0.1129 | 94.8 | 0.0031 | 0.1094 | 94.3 | |
| γ | 0.0048 | 0.0902 | 94.6 | 0.0051 | 0.0880 | 94.5 | 0.0038 | 0.1131 | 95.0 | 0.0040 | 0.1096 | 94.8 | |
Abbreviations: CP, coverage proportion; SE, standard error.
Average of the parameter estimates.
Average of the robust standard error estimates.
95% coverage proportion based on the robust standard error estimates.
Furthermore, under all of the simulation scenarios, the variances were underestimated if the naive variance estimator was used, and consequently the coverage proportions were smaller than expected (details not shown).
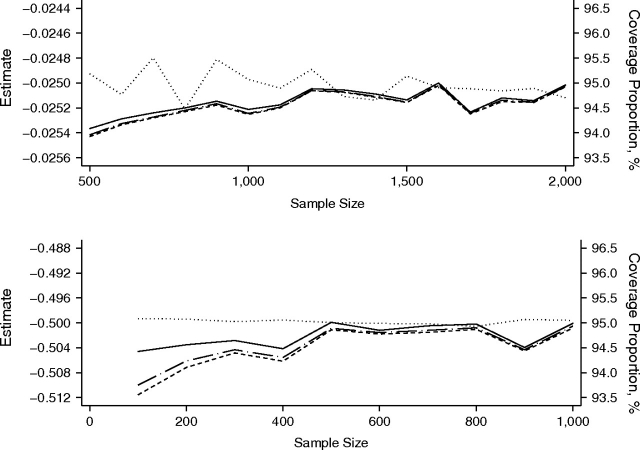
Figure 1 focuses on the performance of the asymptotic and small-sample versions of the robust variance estimator for the regression estimates using all events. Data were simulated under the unfavorable setup of unbalanced group size (p = 0.7), with a quantitative confounder Xi2 following a highly skewed distribution and being strongly correlated with the intervention status as previously described. The simulation replications for each scenario were 20,000. Because of the similarities between the 95% coverage proportions obtained using HCr and those obtained using HC1 as n/(n – k) ≈ 1, only the 95% coverage proportions obtained using HCr, HC2, and HC3 are shown in Figure 1. Mean estimates for β1 are also presented. For the low-incidence scenario with β1 = –0.025, the mean estimate for β1 fluctuated within 0.8% of the true value −0.025, and the 95% coverage proportion obtained using HCr was, at most, 1% different from the intended 95% coverage proportion when n was greater than 500. For the high-incidence scenario with β1 = –0.5, the estimate for β1 fluctuated within 0.1% of the true value –0.5, and the difference between the 95% coverage proportion obtained using HCr and the intended 95% coverage proportion was less than 1% when n was greater than 200. The 95% coverage proportion obtained using HC3 was always closer to the intended 95% level than the 95% coverage proportions obtained using HCr and HC2, but the differences were important only if n was less than or equal to 500.
Figure 1.
Average estimates and coverage proportions of 3 variance estimators for β1 using all events data in the scenario of γ random effects. X1 ∼ Bernoulli(0.7). X2 is highly skewed and highly correlated with X1. Dotted line, estimate; dashed line, 95% coverage proportion using robust variance estimator HCr; long-dashed-and-dotted line, 95% coverage proportion using HC2; solid line, 95% coverage proportion using HC3. There were 20,000 replications. Upper panel: β1 = –0.025; lower panel: β1 = –0.5.
CASE STUDY
Data from a randomized, double-blinded, placebo-controlled trial of a 9-valent pneumococcal conjugate vaccine conducted in the Gambia (17) were reanalyzed. Our purpose was 2-fold: firstly, to illustrate the proposed method and compare the analysis of first events with the analysis of all events in estimating the IRD; and secondly, to examine whether there was any ethnic difference in disease incidence. Details on the trial and the vaccine efficacy estimated using only first events can be found in the paper by Cutts et al. (17). Briefly, approximately 17,000 children aged 6–51 weeks were randomly allocated to one of the 2 regimens: receipt of either 3 doses of the vaccine or 3 doses of the placebo. Radiologically confirmed pneumonia was the primary endpoint. A disease episode was considered to be new only if at least 30 days had elapsed since the child's previous episode (18), and the 30 days were not counted in the person-time exposed. Following the method of Cutts et al. (17), we performed a per-protocol analysis and used time from 14 days after the third dose or placebo as the time scale. Permission to use the data for the present study was given by the Medical Research Council–Gambian Government Joint Ethics Committee.
Covariates included in the analyses were district (Bansang, Basse, or other), age at enrollment, gender, and ethnicity (Mandinka, Fula, Serahule, Wolof, or others). The placebo and vaccine groups were well balanced in terms of baseline covariates. In the study area, each of the 3 major ethnic groups (Mandinka, Fula, and Serahule) comprised approximately 30% of the local population. Moreover, across the 5 different ethnic groups, it was noted that: 1) the proportions of males were comparable; 2) the median ages were also comparable, but there were some differences at the upper percentiles (e.g., the 90th percentiles ranged from 0.44 to 0.50); and 3) the geographic distributions of ethnic groups varied considerably. For example, 27.5% of the Wolof participants lived in Bansang, where the disease incidence was the highest (see Table 4). This percentage was notably higher than percentages in the other ethnic groups (Mandinka, 5.0%; Fula, 11.3%; Serahule, 0.1%; and others, 14.1%). On the other hand, 3.9% of the Wolof participants lived in Basse, as compared with 36.0%, 26.9%, 31.8%, and 46.1% of the Mandinka, Fula, Serahule, and others, respectively.
Table 4.
Incidence Rate of Radiologically Confirmed Pneumonia in the Placebo Group and Incidence Rate Difference per Child-Year, by Ethnicity and District, the Gambia, 2001–2004
| First Events | All Events | |||||
| Variable | Placeboa | Incidence Rate Differenceb | Placebo | Incidence Rate Differenceb | ||
| Estimate | 95% CI | Estimate | 95% CI | |||
| Ethnicity | ||||||
| Mandinka | 0.0512 | −0.0239 | −0.0329, −0.0149 | 0.0549 | −0.0253 | −0.0352, −0.0155 |
| Fulla | 0.0391 | −0.0154 | −0.0233, −0.0076 | 0.0416 | −0.0163 | −0.0248, −0.0078 |
| Serahule | 0.0329 | −0.0076 | −0.0156, 0.0005 | 0.0358 | −0.0098 | −0.0185, −0.0012 |
| Wolof | 0.0425 | −0.0108 | −0.0262, 0.0046 | 0.0453 | −0.0109 | −0.0279, 0.0061 |
| Others | 0.0216 | 0.0026 | −0.0249, 0.0301 | 0.0211 | 0.0105 | −0.0220, 0.0430 |
| District | ||||||
| Bansang | 0.0951 | −0.0410 | −0.0663, −0.0156 | 0.1027 | −0.0451 | −0.0724, −0.0178 |
| Basse | 0.0459 | −0.0141 | −0.0230, −0.0052 | 0.0506 | −0.0176 | −0.0274, −0.0078 |
| Others | 0.0324 | −0.0125 | −0.0176, −0.0074 | 0.0338 | −0.0122 | −0.0177, −0.0067 |
Abbreviation: CI, confidence interval.
Incidence rate in the placebo group.
Incidence rate in the vaccine group minus incidence rate in the control group.
Of the 929 radiologically confirmed pneumonia episodes detected in the 16,340 children during the trial period, 567 were from the placebo group and 362 were from the vaccine group. The total numbers of child-years were 12,914 and 13,070 in the placebo and vaccine groups, respectively. Approximately 95% of the children had no episodes of pneumonia detected throughout this period, while 772 children had 1 episode, 65 children had 2, and 9 children had 3; 846 (91.1%) of the episodes were first episodes.
Without consideration of potential confounders, the IRD 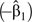 attributable to the vaccine was 0.0150 per child-year (95% confidence interval: 0.0104, 0.0195) using first episodes only and 0.0162 per child-year (95% confidence interval: 0.0113, 0.0211) using all episodes. Analysis of first events and all events did not make a big difference, as was seen in the simulation studies for low-incidence scenarios. The estimates for the incidence rate in the placebo group and the IRD in different ethnic groups and districts are shown in Table 4. Results suggested that regardless of whether first episodes only or all episodes were used, among the 5 ethnic groups, 1) persons of Mandinka ethnicity had the highest placebo-group incidence rate and Wolof the second-highest, and 2) the IRDs between the vaccine and placebo groups were higher in Mandinka and Fula than in the other ethnic groups, but their 95% confidence intervals mostly overlapped. When the analysis was based on first episodes only, the vaccine was found to have a significant protective effect only in Mandinka and Fula. When all of the episodes were used, the protective effect was also found to be significant among persons of Serahule ethnicity.
attributable to the vaccine was 0.0150 per child-year (95% confidence interval: 0.0104, 0.0195) using first episodes only and 0.0162 per child-year (95% confidence interval: 0.0113, 0.0211) using all episodes. Analysis of first events and all events did not make a big difference, as was seen in the simulation studies for low-incidence scenarios. The estimates for the incidence rate in the placebo group and the IRD in different ethnic groups and districts are shown in Table 4. Results suggested that regardless of whether first episodes only or all episodes were used, among the 5 ethnic groups, 1) persons of Mandinka ethnicity had the highest placebo-group incidence rate and Wolof the second-highest, and 2) the IRDs between the vaccine and placebo groups were higher in Mandinka and Fula than in the other ethnic groups, but their 95% confidence intervals mostly overlapped. When the analysis was based on first episodes only, the vaccine was found to have a significant protective effect only in Mandinka and Fula. When all of the episodes were used, the protective effect was also found to be significant among persons of Serahule ethnicity.
Table 5 shows the results of the multivariable analyses (STATA codes (Stata Corporation, College Station, Texas) are available in Web Appendix 4). In correspondence with the results in Table 4, this multivariable analysis suggested that Mandinka had a higher incidence rate than all of the other ethnic groups, although the difference with Wolof was not statistically significant. The covariate-adjusted difference between Wolof and Mandinka was similar to the unadjusted difference, showing that there was no major confounding by district. Moreover, a joint test of the 4 ethnic contrasts showed no difference among Fula, Serahule, Wolof, and others.
Table 5.
Multivariable Regression Analysis of the Incidence of Radiologically Confirmed Pneumonia per Child-Year, by Ethnicity and District, the Gambia, 2001–2004
| Variable | First Events | All Events | ||||
| Estimate | Robust SE | 95% CI | Estimate | Robust SE | 95% CI | |
| Ethnicity (referent: Mandinka) | ||||||
| Fulla | −0.0085 | 0.0030 | −0.0145, −0.0026 | −0.0097 | 0.0033 | −0.0161, −0.0032 |
| Serahule | −0.0072 | 0.0031 | −0.0132, −0.0011 | −0.0083 | 0.0034 | −0.0149, −0.0017 |
| Wolof | −0.0078 | 0.0045 | −0.0167, 0.0011 | −0.0089 | 0.0049 | −0.0186, 0.0007 |
| Others | −0.0207 | 0.0075 | −0.0353, −0.0061 | −0.0205 | 0.0089 | −0.0378, −0.0031 |
| Vaccine | −0.0151 | 0.0023 | −0.0196, −0.0105 | −0.0163 | 0.0025 | −0.0212, −0.0113 |
| District (referent: others) | ||||||
| Bansang | 0.0477 | 0.0066 | 0.0348, 0.0606 | 0.0518 | 0.0071 | 0.0379, 0.0656 |
| Basse | 0.0105 | 0.0027 | 0.0052, 0.0158 | 0.0116 | 0.0029 | 0.0059, 0.0174 |
| Age, years | −0.0398 | 0.0072 | −0.0540, −0.0257 | −0.0442 | 0.0076 | −0.0590, −0.0294 |
| Male gender | 0.0037 | 0.0023 | −0.0009, 0.0082 | 0.0036 | 0.0025 | −0.0013, 0.0085 |
| Intercept | 0.0484 | 0.0036 | 0.0413, 0.0556 | 0.0525 | 0.0039 | 0.0449, 0.0602 |
Abbreviations: CI, confidence interval; SE, standard error.
DISCUSSION
Statistical methods and software for estimation of the IRR are widely available. Much less attention has been given to the estimation of IRD. The IRD is an important parameter in medical research. It shows the public health impact of an intervention. We demonstrated here, using a hypothetical example (Web Appendix 1) and by simulation, that in the presence of unobserved heterogeneity, limiting the analysis of incidence rates for repeatable disease episodes to the first events results in bias. The severity of this bias depends on the disease incidence rate and on the degree and distribution of heterogeneity, which is usually unknown. The analysis of multiple events per person is more difficult than analysis of single events. The negative binomial regression model is an intuitive alternative for consideration. However, even in the simplest case in which the negative binomial model is fitted with an intercept only for a single group together with an “offset” term of log(follow-up time) in the log(incidence) equation, the point estimate does not necessarily agree with the observed disease incidence rate, such as when adopting the commonly used mean-variance relation that variance = mean + mean2 × dispersion parameter, which is usually called the “NB2” parameterization (19). This is counterintuitive. For example, applying the NB2 approach to all of the radiologically confirmed pneumonia episodes as described above in the Case Study section, the parameter estimates on the logarithmic scale for the intercepts are –3.1020 and –3.5636 for the placebo and vaccine groups, respectively. The corresponding estimates for the incidence rates in the placebo and vaccine groups were 0.0450 and 0.0283, respectively, for an IRD estimate of 0.0167. However, the observed incidence rates in the placebo and vaccine groups were 0.0439 and 0.0277, respectively, and the observed IRD was 0.0162. It is not clear how a negative binomial model can be parameterized to produce an IRD estimate that agrees with the observed IRD.
Furthermore, similar to the regression analysis of risk difference, the iterations for the Poisson and negative binomial regression models with an identity link function do not always converge (3) because of their implicit positiveness constraints on the value of the link function. In contrast, the proposed least-squares method has an analytic solution, but it may predict negative incidence rates for some persons. We agree with Spiegelman and Hertzmark (20) that in epidemiology and public health, it is usually more important to estimate a parameter of interest than to fit the data. In our case, the parameters of interest are at the group level instead of at the individual level, so we do not see a problem in the application. If the research purpose were to develop a prognostic model for application to individuals, then the present method would be undesirable, because it can give implausible individual-level parameter values.
We have proposed a simple yet flexible approach to estimating the IRD for analysis of first events or all events. The proposed method has several merits. Firstly, it boils down to ordinary least-squares regression of transformed variables, together with a robust variance estimator for inference. It can easily handle quantitative covariates and has an explicit solution for the parameter estimates. Many popular statistical software packages, such as STATA (21), can perform the proposed analysis without additional programming. Secondly, the proposed estimator is unbiased. Thirdly, the proposed method includes other existing methods as special cases, such as that of Stukel et al. (11) and Glynn and Buring (10). As with these methods, when comparing 2 groups without covariate adjustment, our estimate for IRD has the desirable property that it agrees with the observed IRD. Moreover, it can be shown (our ongoing work) that in the case of time-varying covariates’ effects, the estimates are measuring the average of the covariates’ effects over the total number of person-years using data on all of the events. A limitation of the present proposal is that the method does not model the time-varying IRD, which may be needed in some research situations. Further methodological development is needed.
We have also compared the asymptotic and small-sample versions of the robust sandwich estimator. The differences among them are fairly minor, especially when the sample size is larger than 500. However, there is no harm in always using the HC3 estimator. Interestingly, even HC3 tends to have a coverage proportion slightly lower than the nominal level, although the undercoverage is small in many realistic situations. This issue has been raised by other researchers in the context of analysis of recurrent events, but there remains no explanation (22).
Supplementary Material
Acknowledgments
Author affiliations: Department of Biostatistics, Singapore Clinical Research Institute, Singapore (Ying Xu, Y. B. Cheung); Office of Clinical Sciences, Duke-NUS Graduate Medical School, Singapore (Y. B. Cheung); Department of Statistics and Actuarial Science, Faculty of Science, University of Hong Kong, Hong Kong (K. F. Lam); Division of Clinical Trials and Epidemiological Sciences, National Cancer Center, Singapore (S. H. Tan); and Department of Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, University of London, London, United Kingdom (Paul Milligan).
The work of Y. B. C. and Y. X. was funded by the National Medical Research Council of Singapore (grant NMRC/1182/2008). The pneumococcal vaccine trial used for illustration was funded by grants from the US National Institute of Allergy and Infectious Diseases through contract N0-AI-25477; by the World Health Organization through contract V23/181/127; by the Children's Vaccine Program at PATH; and by the US Agency for International Development, with vaccine being kindly donated by Wyeth Vaccines.
Conflict of interest: none declared.
Glossary
Abbreviations
- IRD
incidence rate difference
- IRR
incidence rate ratio
References
- 1.Zou G. A modified Poisson regression approach to prospective studies with binary data. Am J Epidemiol. 2004;159(7):702–706. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
- 2.Spiegelman D, Hertzmark E. Easy SAS calculations for risk or prevalence ratios and differences. Am J Epidemiol. 2005;162(3):199–200. doi: 10.1093/aje/kwi188. [DOI] [PubMed] [Google Scholar]
- 3.Cheung YB. A modified least-squares regression approach to the estimation of risk difference. Am J Epidemiol. 2007;166(11):1337–1344. doi: 10.1093/aje/kwm223. [DOI] [PubMed] [Google Scholar]
- 4.Greenwood B. Interpreting vaccine efficacy. Clin Infect Dis. 2005;40(10):1519–1520. doi: 10.1086/429833. [DOI] [PubMed] [Google Scholar]
- 5.Cheung YB, Zaman SM, Ruopuro ML, et al. C-reactive protein and procalcitonin in the evaluation of the efficacy of a pneumococcal conjugate vaccine in Gambian children. Trop Med Int Health. 2008;13(5):603–611. doi: 10.1111/j.1365-3156.2008.02050.x. [DOI] [PubMed] [Google Scholar]
- 6.Cook RJ, Sackett DL. The number needed to treat: a clinically useful measure of treatment effect. BMJ. 1995;310(6977):452–454. doi: 10.1136/bmj.310.6977.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung YB, Xu Y, Tan SH, et al. Estimation of intervention effects using first or multiple episodes in clinical trials: the Andersen-Gill model re-examined. Stat Med. 2010;29(3):328–336. doi: 10.1002/sim.3783. [DOI] [PubMed] [Google Scholar]
- 8.Moorthy VS, Reed Z, Smith PG. MALVAC 2008: measures of efficacy of malaria vaccines in phase 2b and phase 3 trials—scientific, regulatory and public health perspectives. Vaccine. 2009;27(5):624–628. doi: 10.1016/j.vaccine.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 9.Rothman KJ. Modern Epidemiology. Boston, MA: Little, Brown and Company; 1986. [Google Scholar]
- 10.Glynn RJ, Buring JE. Ways of measuring rates of recurrent events. BMJ. 1996;312(7027):364–367. doi: 10.1136/bmj.312.7027.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stukel TA, Glynn RJ, Fisher ES, et al. Standardized rates of recurrent outcomes. Stat Med. 1994;13(17):1781–1791. doi: 10.1002/sim.4780131709. [DOI] [PubMed] [Google Scholar]
- 12.Greenland S. Model-based estimation of relative risks and other epidemiologic measures in studies of common outcomes and in case-control studies. Am J Epidemiol. 2004;160(4):301–305. doi: 10.1093/aje/kwh221. [DOI] [PubMed] [Google Scholar]
- 13.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73(1):13–22. [Google Scholar]
- 14.White H. A heteroskedasticity-consistent covariance matrix estimator and a direct test for heteroskedasticity. Econometrica. 1980;48(4):817–830. [Google Scholar]
- 15.Long JS, Ervin LH. Using heteroscedasticity consistent standard errors in the linear regression model. Am Stat. 2000;54(3):217–224. [Google Scholar]
- 16.Eskola J, Kilpi T, Palmu A, et al. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N Engl J Med. 2001;344(6):403–409. doi: 10.1056/NEJM200102083440602. [DOI] [PubMed] [Google Scholar]
- 17.Cutts FT, Zaman SM, Enwere G, et al. Efficacy of nine-valent pneumococcal conjugate vaccine against pneumonia and invasive pneumococcal disease in the Gambia: randomized, double-blinded, placebo-controlled trial. Lancet. 2005;365(9465):1139–1146. doi: 10.1016/S0140-6736(05)71876-6. [DOI] [PubMed] [Google Scholar]
- 18.Enwere G, Cheung YB, Zaman SM, et al. Epidemiology and clinical features of pneumonia according to radiographic findings in Gambian children. Trop Med Int Health. 2007;12(11):1377–1385. doi: 10.1111/j.1365-3156.2007.01922.x. [DOI] [PubMed] [Google Scholar]
- 19.Cameron AC, Trivedi PK. Regression Analysis of Count Data. Cambridge, United Kingdom: Cambridge University Press; 1998. [Google Scholar]
- 20.Spiegelman D, Hertzmark E. The authors reply [re: “easy SAS calculations for risk or prevalence ratios and differences”] [letter] Am J Epidemiol. 2006;163(12):1159–1161. [Google Scholar]
- 21.Stata Corporation. Stata Statistical Software, Release 9. College Station, TX: Stata Corporation; 2005. [Google Scholar]
- 22.Kelly PJ, Lim LL. Survival analysis for recurrent event data: an application to childhood infectious diseases. Stat Med. 2000;9(1):13–33. doi: 10.1002/(sici)1097-0258(20000115)19:1<13::aid-sim279>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.