Summary
Two age-1 nonsense mutants, truncating the class-I phosphatidylinositol 3-kinase catalytic subunit (PI3KCS) before its kinase domain, confer extraordinary longevity and stress-resistance to Caenorhabditis elegans. These traits, unique to second-generation homozygotes, are blunted at the first generation and are largely reversed by additional mutations to DAF-16/FOXO, a transcription factor downstream of AGE-1 in insulin-like signaling. The strong age-1 alleles (mg44, m333) were compared with the weaker hx546 allele on expression microarrays, testing four independent cohorts of each allele. Among 276 genes with significantly differential expression, 92% showed fewer transcripts in adults carrying strong age-1 alleles rather than hx546. This proportion is significantly greater than the slight bias observed when contrasting age-1 alleles to wild-type worms. Thus, transcriptional changes peculiar to nonsense alleles primarily involve either gene silencing or failure of transcriptional activation. A subset of genes responding preferentially to age-1-nonsense alleles was reassessed by real-time polymerase chain reaction, in worms bearing strong or weak age-1 alleles; nearly all of these were significantly more responsive to the age-1(mg44) allele than to age-1(hx546). Additional mutation of daf-16 reverted the majority of altered mg44-F2 expression levels to approximately wild-type values, although a substantial number of genes remained significantly distinct from wild-type, implying that age-1(mg44) modulates transcription through both DAF-16/FOXO-dependent and –independent channels. When age-1-inhibited genes were targeted by RNA interference (RNAi) in wild-type or age-1(hx546) adults, most conferred significant oxidative-stress protection. RNAi constructs targeting two of those genes were shown previously to extend life, and RNAi’s targeting five novel genes were found here to increase lifespan. PI3K-null mutants may thus implicate novel mechanisms of life extension.
Keywords: aging, Caenorhabditis elegans, dauer, gene expression, lifespan, life-span, longevity, stress resistance
Introduction
The age-1 gene of Caenorhabditis elegans encodes a class-I phosphatidylinositol 3-kinase catalytic subunit (PI3KCS) that is central to insulin/IGF-1 signaling (Friedman & Johnson, 1988a; Dorman et al., 1995; Guarente & Kenyon, 2000). Most age-1 mutants are temperature-sensitive and outlive wild-type controls by less than twofold (Friedman & Johnson, 1988a). Two stronger alleles of age-1 have been reported, nonsense-mutants that truncate the AGE-1 protein upstream of its kinase domain (Larsen et al., 1995; Morris et al., 1996; Tissenbaum & Ruvkun, 1998). These strong alleles are carried as heterozygotes, which segregate ~25% age-1−/− homozygous progeny that develop normally, are fertile and live 2.2–2.6 times as long as N2 controls (Larsen et al., 1995; Morris et al., 1996). Such first-generation homozygous age-1−/− worms (herein termed F1 worms) are thought to derive ‘maternal protection’ by oocyte transmission of age-1 transcripts, AGE-1 protein or PIP3 kinase products from their heterozygous parent. Their offspring, F2-generation age-1−/− homozygotes with little or no maternal protection, fail to mature at 25 °C (Larsen et al., 1995; Morris et al., 1996). At lower temperatures (15–20 °C), however, they slowly develop into adults sharing two remarkable phenotypes: (i) their mean and median lifespans are 8–11 times as long as the longest-lived cohort of near-isogenic N2DRM worms, and at least four times those of the next longest-lived insulin/IGF-1 signaling (IIS) mutant tested; and (ii) they are far more resistant to oxidative and electrophilic stresses than either wild-type or other age-1-mutant strains in an isogenic background (Ayyadevara et al., 2008). Both traits depend on the DAF-16/FOXO (ForkHead O) transcription factor, because they are largely or entirely reversed by mutations of the daf-16 gene (Ayyadevara et al., 2008; Tazearslan et al., 2009).
Expression microarrays have been used to discover transcriptional changes associated with daf-2 hypomorphic mutations, relative to either a wild-type strain (usually Bristol-N2) or a double mutant of daf-2 with daf-16 (Lee et al., 2003a; Murphy et al., 2003; Oh et al., 2005; Murphy, 2006). The daf-2 gene encodes an insulin/IGF-1-like membrane receptor, functionally upstream of age-1, whereas DAF-16 is a downstream FOXO transcription factor, through which flow most or all of the longevity and stress-resistance traits associated with IIS disruption (Ogg et al., 1997; Murphy, 2006).
In the present study, markers and mechanisms were sought that are associated specifically with the extreme traits unique to strong age-1 alleles: a four- to fivefold further increase in longevity, and comparable gains in stress resistance, relative to a weaker age-1 allele. We therefore screened for transcriptional changes that are more pronounced in worms bearing the longer-lived alleles, using genomewide expression arrays to compare eight independent biological cohorts of strong age-1 alleles (four each of mg44 and m333) to the weaker age-1(hx546) allele. These two strong alleles arose by chemical mutagenesis and thus, despite extensive outcrossing (including six generations in our laboratory), they might show transcript changes that derive from mutations to other genes tightly linked to age-1. Because such ‘background’ mutations would be distinctive to each strain, combining data from the two strains should markedly reduce significance for gene expression changes as a result of non-age-1 mutations.
A subset of genes emerging from the microarray screen was evaluated by real-time polymerase chain reaction (RT-qPCR) for multiple cohorts, to quantitatively address allelic differences in transcript levels, DAF-16-dependence and resemblance of the age-1(mg44) F2 transcriptional profile to those of their F1 parents, wild-type dauer larvae or daf-2 adults bearing a temperature-sensitive mutation in the insulin-like receptor upstream of AGE-1. Biological variance greatly exceeds technical variance in these experiments, so the use of independent cohorts is essential to ensure that biological variation is adequately represented.
We have included some data on age-dependent expression changes in early adulthood, demonstrating that for most genes implicated by microarray and RT-qPCR studies, genotype is a stronger determinant of expression than either chronological or biological age. The emphasis here, however, is on inter-strain differences observable in presenescent adults that might serve as predictive biomarkers of subsequent longevity, or that (more rarely) implicate new mechanisms that contribute to the substantial gene- and allele-specific differences in lifespan among IIS mutants.
Results
Microarray surveys reveal allele-specific expression changes in age-1 mutants
Three C. elegans strains, bearing distinct mutations of the age-1 gene, were outcrossed six generations into N2DRM, the longest-lived of six Bristol-N2 stocks tested (Gems & Riddle, 2000). The first mutant discovered at this locus, age-1(hx546) (Klass, 1983), is a temperature-sensitive missense allele (Ayyadevara et al., 2008) that allows worms to mature at 15 °C but then extends lifespan by 40–65% upon shifting adults to 20–25 °C (Friedman & Johnson, 1988b). Two unconditional alleles, age-1(mg44) (Morris et al., 1996) and age-1(m333) (Larsen et al., 1995), are nonsense mutations that truncate the AGE-1 protein upstream of its kinase domain (Morris et al., 1996). No deletion mutants of AGE-1 have been described.
To define gene expression changes peculiar to the extremelongevity phenotype, transcript profiles were assessed on fullgenome microarrays comprising > 22 000 synthetic oligomers (60-mers) printed on each epoxy slide (Genome Sequencing Center, Washington University, St Louis, MO, USA). Eight arrays compared eight biological expansions of very long-lived age-1 alleles mg44 and m333 (four arrays each) to four distinct expansions of the weaker age-1(hx546) allele, which in the N2DRM background confers approximately twofold life extension typical of long-lived IIS mutants. Another six arrays were used to compare the two age-1 nonsense alleles (three expansions each) to the wild-type N2DRM strain. All mutants utilized here had been outcrossed to N2DRM for at least six generations, to create a near-isogenic set of strains, and all cultures were maintained at 20 °C. Second-generation homozygotes of age-1(mg44) and age-1(m333) are infertile (Ayyadevara et al., 2008), in marked contrast to all other strains tested. Although ‘pregravid’ worms can be collected within the first 12 h after the L4/adult molt, this does not ensure complete exclusion of eggs and embryos; moreover, chemical or genetic methods to block reproduction may themselves perturb survival phenotypes and transcription profiles. We therefore harvested the fertile strains when postgravid (as assessed by microscopic examination of at least 30 worms) but prior to senescence (typically at 6 days of adult age, ~38% of the median adult lifespan), and F2 worms of the strong age-1 mutants at 9–12 days of adult age. With developmental time included, N2 worms were 8.5 days posteclosion, age-1(mg44) worms were at 18–21 days and age-1(m333) worms 27–30 days. Because strains could not be matched with respect to both chronological and physiological age, and in any case there is no consensus as to whether total or adult ages should be considered, we chose these conditions for initial comparisons and left relative-age issues to be addressed in subsequent experiments (see below).
Array data were combined from both very long-lived alleles, to reduce the likelihood of identifying gene expression changes caused by additional mutations at loci other than age-1, which would differ between any two strains arising from mutagenesis. Mean signal intensities of paired dyes Cy3 and Cy5 were within ±7%, as were the mean intensities for both strong alleles relative to the weaker hx546 allele. Although no technical repeats were performed, there was no discernable difference in spot ratios between arrays in which Cy3 or Cy5 was associated with the longer-lived alleles.
SAM Analysis
These two data sets [eight arrays using age-1(hx546) as the reference and six arrays with N2DRM as the reference] were analyzed separately by significance analysis of microarrays (SAM) [(Tusher et al., 2001) ver. 3.02, Stanford University], using the ‘two-class paired t-test’ procedure to identify genes preferentially or uniquely altered in the two longest-lived age-1 strains. Table S1 (Supporting information) lists 386 differentially expressed loci for the comparison between the very long-lived strains and age-1(hx546); 276 of these were significant at a nominal false-discovery rate (FDR or q) of < 5% (with true FDR estimated to be < 2.3%). Table S2 (Supporting information) is a parallel listing of 339 genes with significantly altered expression relative to N2DRM wild-type adults, at FDR < 5%. Each list comprises ~2% of all genes surveyed. These tables include brief descriptions of the encoded proteins, statistical parameters from SAM and notes of other relevant data [confirmation or contradiction by RT-qPCR assays, and life-span effects of RNA interference (RNAi)]. Numbers of significant genes found, and their FDR values determined empirically by permutation, differ somewhat (as expected) between the two comparisons.
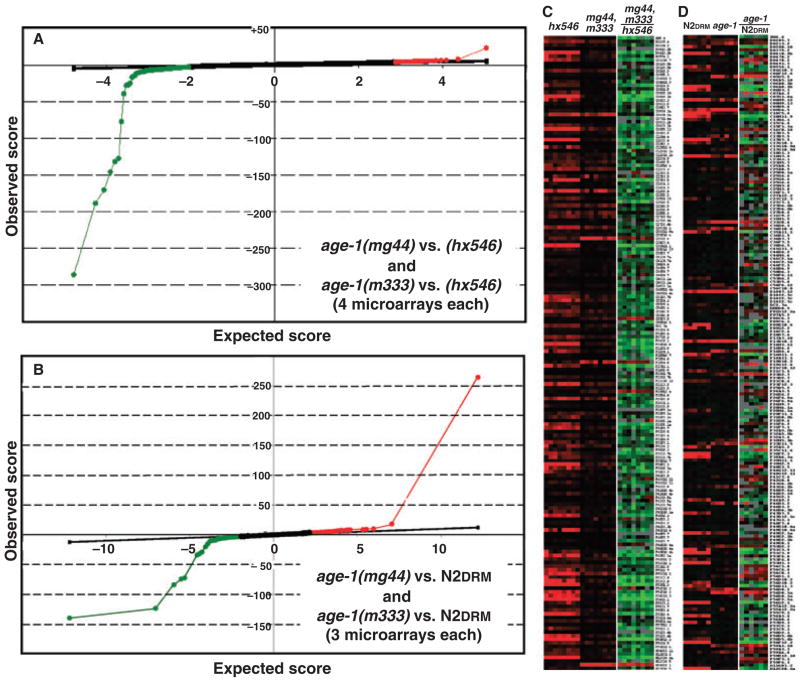
Significance analysis of microarray (SAM) ‘outlier plots’ for these two data analyses are shown in Fig. 1. The striking asymmetry of Fig. 1A reflects the greater number of differentially expressed genes that are downregulated, in strong age-1 mutants, than are upregulated. A ‘heat map’ of gene expression, by gene and array [Fig. 1C; Fig. S1 (Supporting information)], better illustrates this imbalance. Of genes significantly altered in transcript levels (at FDR < 5%), 92% were attenuated by the stronger age-1 alleles, while only 8% were upregulated (Fig. 1; Fig. S1; Table S1). It was this unexpected bias in significant transcript changes that prompted the second set of microarrays, in which the same strong age-1 mutants were contrasted to wild-type (N2DRM) near-isogenic controls. As depicted in Fig. 1B,D, a far less pronounced imbalance was seen in this comparison: at FDR< 5%, 39% of significant genes increased, while 61% decreased, in age-1 F2 adults (see also Table S2). The probability of such disparate proportions arising merely by chance, as in two random samplings from the same distribution of transcript levels, is P < 10−17 by chi-squared test.
Fig. 1.
Significance analysis of microarray (SAM) ‘outlier’ plots and heat maps from microarray analyses. (A) Four Caenorhabditis elegans full-genome microarrays were probed with cDNAs prepared from age-1(mg44)-F2 adults, mixed pairwise with cDNAs from adults bearing the age-1(hx546) allele (four independent cohorts of each strain), and four more arrays were probed with cDNAs from age-1(m333)-F2 adults, mixed pairwise with age-1(hx546) cDNAs (four cohorts of each strain). Data were combined from eight arrays (treating the two very long-lived strains as equivalent) and analyzed using SAM (Tusher et al., 2001). The SAM plot shows outliers in green if transcript levels are lower in the strong age-1 alleles (mg44 and m333) than in hx546, and in red if higher for the strong alleles, at a FDR of < 5%. (B) Three microarrays were probed with cDNAs from age-1(mg44)-F2 adults, mixed pairwise with cDNAs from N2DRM adults bearing the wild-type age-1 allele (three independent cohorts per strain), and three further arrays were probed with cDNAs from age-1(m333)-F2 adults, mixed pairwise with N2DRM cDNAs (three cohorts per strain). Data were combined from six arrays (treating the two very long-lived strains as equivalent) and analyzed by SAM as above. (C) Partial heat maps represent data from eight arrays probed with cDNA from age-1(hx546) adults (left), and four each probed with cDNAs from strong age-1 alleles mg44 and m333 (center). Expression ratios are shown for each array (right) as the log2(mg44/hx546) or (m333/hx546); red intensity indicates positive log-ratios (higher expression in mg44 or m333), whereas green implies negative log-ratios (lower expression in mg44 or m333). Full heat maps are presented in Fig. S1. (D) Partial heat maps represent data from six arrays probed with cDNA from N2DRM adults (left), and three each probed with cDNAs from age-1 alleles mg44 and m333 (center). Expression ratios were calculated for each array (right) as the log2(mg44/N2) or (m333/N2); red intensity indicates positive log-ratios (higher expression in mg44 or m333), whereas green implies negative log-ratios (lower expression in mg44 or m333). Full heat maps are presented in Fig. S2 (Supporting information).
The intersection of these two lists comprises genes that changed significantly in both comparisons: just 20 that increased and 104 that decreased, in strong age-1 mutants relative to each control. Together, these make up 37% of the genes that were differential with respect to N2DRM and 32% of genes in the age-1(hx546) comparison, whereas > 60% of differential genes were unique to each comparison.
The 23 genes significantly upregulated in strong age-1 alleles, vs. hx546 (Table S1a), include two of the three C. elegans catalase genes, ctl-2 and ctl-3; two genes involved in nucleoside diphosphate metabolism (mig-23, ndx-1); the ace-3 gene encoding an acetylcholinesterase; and two genes of lipid metabolism (dhs-3, lips-2). Among these 23 genes, RNAi had been reported to shorten lifespan only for the catalase gene ctl-2 (Murphy et al., 2003).
Of 253 genes significantly downregulated in strong age-1 mutants relative to the weaker age-1(hx546) allele (Table S1b), longevity has been reported as increased by RNAi targeting five genes, but reduced for two. The genes (function of encoded protein) for which RNAi extends life are Y71G12B.4 (a mono-oxygenase), akt-2 (part of the insulin/IGF-1 signaling pathway), erm-1 (a cytoskeletal linker protein), dyf-2 (involved in sensory cilia) and Y105C5B.12b (S-adenosylmethionine synthetase 2). A sixth gene reported to extend life by RNAi, coq-1 (required for ubiquinone/coQ9 biosynthesis), was underexpressed at marginal significance (FDR = 5.4%). Two further genes were reported to shorten life upon RNA interference: lin-40 (part of a complex mediating histone deacetylation and transcriptional repression) and gdi-1 (a Rab-GDP dissociation inhibitor). Taken together, six results agreed with expectation (lifespan up for five genes downregulated with increased lifespan, but down for one upregulated gene), based on allele-specific transcript levels. These data imply a modest 1.6- to 1.8-fold enrichment of RNAi life-span effects (exact Binomial P = 0.12–0.19 for X = 6, n = 276), over the 1.2–1.4% frequency found in unbiased screens for RNAi effects on nematode longevity (Lee et al., 2003b; Hamilton et al., 2005; Hansen et al., 2005).
Among the genes significantly downregulated by strong age-1 alleles relative to hx546 (exceptions are shown as [upregulated]), we find genes that encode 16 predicted ubiquitin-complex components (ubl-5, ufd-1, uba-2, ubc-22, nhl-3, tag-214, zer-1, rnf-113, C06C3.5, T20F5.6, T12E12.1, C44E4.7, C27A12.7b, F16B4.6, Y71F9AL.10, K09F6.7); 22 membrane receptors (aph-1, npr-9, sra-1, srab-21, srab-25, srbc-62, srh-140, srh-184, sri-43, srx-90, srz-28, [str-18], str-74, str-116, str-129, str-151, str-172, gar-2, C35A11.1, F21C10.12, F39B3.2, Y38F2AR.3); 11 transcription factors (hif-1, ngn-1, btf-1, athp-1, ztf-3, ztf-9, egl-5, C23H3.3, C27D6.4, Y43F8B.13, C55A6.9); two RNA polymerase subunits (Y105E8A.23, rbp-6); three protein/histone deacetylases (mys-3, mys-4, lin-40); four DNA repair enzymes (pms-2, him-6, rad54, Y43F8B.13); two cytochromes P450 (cyp-35A5, cyp-35C1); three K+-channel and one Na+-channel protein (twk-11, twk-21, C27F2.6, T28D9.7); eight other transporter proteins (chtl-1, dyf-2, cft-1, imb-2, cho-1, K08C7.1, F16H11.3, C13C4.5, H11E01.2); two FMRF-like peptides (flp-4, flp-11); six RNA splicing factors (lsm-1, prpf-4, rsr-1, [rsp-5], Y76B12C.4, F28C1.1); three chaperones (pfd-6, [uri-1], T24H7.2); four lipid dehydrogenases (sdz-1, alh-9, alh-11, [dhs-3]); seven other genes implicated in lipid metabolism (lact-3, W05E10.2, Y105C5B.12, C56E10.3, K02E10.5, C06A5.10, [lips-2]); three thioredoxin-related proteins (txl-1, ZK973.11, R06B9.1); seven F-box proteins (fbxa-24, -43, -73, -90, fbxb-60, C10E2.2, ZK262.8) and two UMP synthetases (ZK228.7, R12E2.11).
Over-representation of upstream motifs
The bias toward downregulation of genes in strong age-1 mutants suggested the possibility that these changes may be mediated by one or more transcription factors that are subject to transcriptional regulation by age-1, acting through DAF-16 and/or other factors affected post-translationally by PIP3 or a PIP3-dependant kinase. We screened for oligomeric sequences (six, seven or eight nucleotides) over-represented within 1 kilobase (kb) upstream of the 184 most significantly downregulated genes from the comparison of strong age-1 alleles to age-1(hx546) (at FDR< 0.7%, Table S1b). Oligo-analysis in Regulatory Sequence Analysis Tools (RSAT) (http://rsat.ulb.ac.be/rsat/) identified 12 octamers as the most significantly enriched upstream motifs. Overlap among these patterns implicated three highly over-represented consensus sequences of 9–11 nucleotides each: TTC(A/T)GAAAAT(T) (P < 10−21, binomial ‘E-value’ after Bonferroni adjustment); CTACAGTAA(C)(C) (P < 10−19); and AGAGA(A/C)G(A/C)AGA (P < 10−14). Within 1-kb downstream of the same genes, only one significant motif was found: CTGAAAAT(T/G) at P < 10−16, specifying a subset of TTC(A/T)GAAAAT(T). None of the above sequences is associated with specific binding of any known transcription factor. Near-identical results were obtained (with slightly reduced significance) on expanding upstream regions to 2 kb or on increasing the input gene set to include all significant genes. For purposes of comparison, we also analyzed 1-kb regions preceding all genes significantly downregulated in age-1 alleles relative to N2DRM controls. The same three upstream motifs defined in the first comparison were here observed at greatly reduced frequency and significance, contributing only six of the 16 octamers enriched at 10−7 < P < 0.01. These consensus motifs thus appear to be rather specific to genes whose expression is downregulated by strong but not weak age-1 alleles.
The same set of 184 upstream sequences, preceding genes downregulated in strong age-1 mutants (relative to the weaker hx546 allele), was also screened for transcription-factor binding sites from the Genomatix™ library. These consensus sites comprise over 750 nucleotide-frequency matrices, each summarizing many defined binding sites (on average, 36 per matrix) from diverse taxa. As a more conservative significance criterion than the P-values reported by Genomatix, we calculated the ‘P ratio’ for each site: the ratio of its binomial P-value from the set of 184 downregulated genes, to that obtained for 200 randomly selected upstream sequences. We observed 19 vertebrate sites that are significantly over-represented in 1-kb regions preceding downregulate genes, with P ratios ranging from 10−4 to 10−16. Twelve of these motifs are present upstream of 95–100% of the 184 most downregulated genes (Table 1).
Table 1.
Transcription-factor binding motifs (vertebrate consensus matrices) enriched in 1-kb regions immediately preceding start sites of 184 age-1-downregulated C. elegans genes. These sites are defined as nucleotide-frequency matrices describing vertebrate binding sites in the Genomatix™ library. P-values were calculated by Genomatix, as the fraction of surveyed genes with upstream motifs relative to the fraction expected in random-sequence DNA, without allowance for species differences in GC-content or nucleotide biases in regulatory regions upstream of coding sequences. To roughly correct for such biases, ‘Random P-values’ were calculated for 1-kb spans preceding 200 genes sampled at random from the C. elegans genome. The final column (P ratio) presents quotients obtained on dividing each ‘P-value’ by the corresponding ‘Random P ’. P ratios ≤ 0.01 are listed. Motif presence next to > 95% of genes, and P ratios ≤ 10−4, are shown in bold.
| Family | Transcription factor class | Genes with motifs (%) | P-value | Random P | P ratio |
|---|---|---|---|---|---|
| ATBF | AT-rich binding factor | 156/184 (85) | 3.08E-54 | 7.40E-47 | 4.2E-08 |
| BCL6 | POZ/Zn-finger protein, B cells | 141/184 (77) | 2.15E-20 | 8.80E-18 | 2.4E-03 |
| BRNF | Brn POU-domain factors | 182/184 (99) | 1.56E-28 | 2.70E-23 | 5.8E-06 |
| CART | Cartilage homeoprotein | 138/184 (75) | 7.58E-43 | 3.00E-36 | 2.5E-07 |
| CDXF | Caudal-related homeodomain | 158/184 (86) | 3.51E-44 | 5.40E-37 | 6.5E-08 |
| CHRF | Cell-cycle-gene homol: CDE, CHR | 162/184 (88) | 6.45E-55 | 1.60E-45 | 4.0E-10 |
| CLOX | Cut-like homeodomain: CLOX, CDP | 184/184 (100) | 6.04E-22 | 2.30E-17 | 2.6E-05 |
| COMP | Cooperating w. myogenic proteins | 115/184 (62) | 9.64E-18 | 4.60E-15 | 2.1E-03 |
| FKHD | ForkHead-domain factors (FoxO/A) | 183/184 (99.5) | 1.46E-19 | 2.30E-17 | 6.4E-03 |
| GATA | GATA-binding transcription factors | 182/184 (99) | 7.15E-22 | 1.30E-16 | 5.5E-06 |
| GFI1 | Growth fac. independent repressor | 138/184 (75) | 7.01E-27 | 2.40E-20 | 2.9E-07 |
| HNF6 | Onecut homeodomain factor | 178/184 (97) | 4.45E-57 | 3.30E-41 | 1.3E-16 |
| HOMF | Homeodomain binding factor | 182/184 (99) | 4.06E-35 | 7.80E-31 | 5.2E-05 |
| HOXF | Homeodomain binding factor | 184/184 (100) | 1.88E-13 | 8.30E-11 | 2.3E-03 |
| LHXF | Lim homeodomain binding factor | 176/184 (96) | 4.51E-54 | 1.20E-42 | 3.7E-12 |
| MYBL | Myb-like transcription factor | 172/184 (93) | 7.03E-17 | 7.00E-13 | 1.0E-04 |
| MYT1 | MyT1 C2HC Zn-finger protein | 181/184 (98) | 7.55E-25 | 1.00E-18 | 7.5E-07 |
| OCTP | Oct1 binding factor (POU-specific) | 142/184 (77) | 1.75E-31 | 5.60E-27 | 3.1E-05 |
| PARF | PAR/bZIP family, albumen D-box | 180/184 (98) | 1.69E-26 | 9.50E-20 | 1.8E-07 |
| PAX2 | PAX2 paired-domain protein | 144/184 (78) | 1.69E-32 | 4.30E-29 | 3.9E-04 |
| PDX1 | Pancreatic/intestinal homeoprotein | 157/184 (85) | 1.37E-28 | 3.10E-25 | 4.4E-04 |
| RUSH | RING or SWI/SNF helicases/ATPases | 170/184 (92) | 3.11E-22 | 3.10E-16 | 1.0E-06 |
| SATB | Special AT-rich binding factor | 136/184 (72) | 5.36E-46 | 1.60E-34 | 3.3E-12 |
| SORY | Sox/sRY sex/testis HMG-box factor | 184/184 (100) | 1.48E-22 | 7.80E-16 | 1.9E-07 |
| TBPF | TATA-binding transcription factor | 182/184 (99) | 3.07E-23 | 4.00E-19 | 7.7E-05 |
The consensus matrices were compiled from vertebrate databases, but are presumed to include related DNA-binding sites recognized by nematode proteins. A ForkHead binding motif, FKHD, might be expected to adjoin many of these genes, because most of them are regulated by DAF-16/FOXO. Although over 99% of the implicated genes (183/184, P < 7 × 10−3) are preceded within a kilobase by a FKHD site (actually an amalgamation of 16 distinct ForkHead binding sites defined by probability matrices), only 28% (51/184) have one or more exact matches with either the C. elegans consensus DAF-16 binding element (DBE), GTAAA(C/T)AA or a second DAF-16-associated element (DAE), CTTATCA (Kenyon & Murphy, 2006; Oh et al., 2006).
Why should there by less enrichment of two DAF-16-associated consensus sequences than of other regulatory motifs (either the 11-mers discussed above or transcription-factor binding sites)? This simply reflects the experimental design, in which strong age-1 mutants were contrasted to a weaker age-1 allele. Because these strains to some extent share the same DAF-16-mediated gene set, whereas the SAM gene-selection procedures are designed to detect strain differences, SAM results will necessarily tend to be depleted in DAF-16 consensus binding sites.
Gene ontology analysis
Gene sets implicated by microarray assays often show enrichment of genes with similar biological functions. Of the genes identified by SAM to have differential expression (FDR ≤ 0.1) between strong age-1 alleles (mg44 and m333) and the weaker hx546 allele, 382 are annotated in WormBase (http://www.wormbase.org). These genes were evaluated for enrichment of functional annotation terms, via penalty-modified Fisher’s exact tests (‘EASE scores’) as implemented in DAVID version 6 (2008; http://david.abcc.ncifcrf.gov) (Dennis et al., 2003; Huang et al., 2009).
Within this set of genes, 15 nonredundant gene ontology (GO) terms were significantly enriched, at < 5% FDR for this assessment (Table 2). These terms designate 30 metal-binding proteins, 25 of which bind zinc; 24 hydrolases; 21 ATP-binding proteins, including 14 kinases and seven Ser/Thr protein kinases; 17 genes involved in alternative splicing of RNA; 20 transferases; 20 nucleotide-binding proteins; 17 zinc finger proteins, of which 13 bind DNA; 19 transmembrane proteins; 17 nuclear proteins; 107 genes involved in cellular metabolic processes and 11 oxidoreductases. Each of these terms was chiefly enriched in the downregulated group, as indicated by arrows (↓) in the second column, with some further contribution from upregulated genes.
Table 2.
Analysis of enrichment of functional annotation terms. Over-representation of gene ontology (GO) terms was assessed among 382 annotated genes differentially expressed in F2 adults homozygous for strong age-1 alleles (mg44 and m333), relative to the weaker hx546 allele, and among 976 annotated genes differentially expressed in the same mutants relative to the N2DRM wild-type allele. ‘Functional Analysis Chart’ reports were generated by DAVID, ver. 6 (http://david.abcc.ncifcrf.gov), using default conditions and GO terms annotated in WormBase, for enrichment relative to a background set of all annotated C. elegans genes. Results are also shown for identical analyses performed on 514 annotated genes that were listed as differentially expressed in worms treated with RNAi to daf-2, relative to worms fed both daf-2 and daf-16 RNAi (Murphy et al., 2003), and for a set of ~2000 genes differentially expressed in N2 dauer larvae relative to postdauer worms 12 h after recovery, based on a reanalysis (McElwee et al., 2006) of primary data (Wang & Kim, 2003). Bold font indicates groups with Benjamini-Hochberg false-discovery rates (B-H FDR) < 0.05 (see Experimental procedures). For all microarray data generated in the present study and for data files provided to us from the dauer study (McElwee et al., 2006), the values shown indicate genes combined from up- and down-regulated sets. These analyses were also run, however, for those separate gene sets, and arrows in this table indicate whether enrichment occurred primarily in strong-age-1- or dauer-upregulated gene sets (↑), primarily in strong-age-1- or dauer-downregulated gene sets (↓), or was observed for both sets individually (↑↓). Blank spaces indicate that no enrichment for the indicated terms met the standard criteria (EASE score > 0.1 and n ≥ 2). It should be noted that the listing of significant terms is a complete one for the age-1 gene sets, but contains only the most significant terms for the daf-2 and dauer gene sets, which produced much larger lists of annotation terms. For every term listed in the first column, however, all reported results are presented from all four analyses.
| Comparison (n): |
mg44, m33 vs. hx546 (382) |
mg44, m33 vs. N2DRM (976) |
daf-2 vs. daf-16 + daf-2 RNAi (514) |
Dauer vs. postdauer (2000) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Term | * | No. | P-value | Fold | B-H FDR | * | No. | P-value | Fold | B-H FDR | No. | P-value | Fold | B-H FDR | † | No. | P-value | Fold | B-H FDR |
| Metal-binding | ↓ | 30 | 3.4E-11 | 4.3 | 8.6E-09 | ↑ ↓ | 46 | 2.6E-11 | 3.1 | 4.4E-09 | 29 | 1.6E-08 | 3.4 | 9.1E-07 | ↑ ↓ | 111 | 2.1E-19 | 2.5 | 9.1E-18 |
| Hydrolase | ↓ | 24 | 2.5E-11 | 5.7 | 1.3E-08 | ↓ | 32 | 2.0E-09 | 3.6 | 1.5E-07 | 29 | 1.5E-13 | 5.7 | 1.9E-11 | ↑ ↓ | 87 | 2.3E-22 | 3.3 | 1.7E-20 |
| Zinc | ↓ | 25 | 1.8E-09 | 4.4 | 3.1E-07 | ↑ ↓ | 36 | 2.1E-08 | 2.9 | 1.3E-06 | 13 | 4.3E-02 | 1.9 | 4.6E-01 | ↑ ↓ | 75 | 3.9E-09 | 2.1 | 6.4E-08 |
| Alternative splicing | ↓ | 17 | 1.1E-08 | 6.3 | 1.5E-06 | 8 | 4.1E-03 | 3.9 | 1.3E-01 | 11 | 1.6E-03 | 3.4 | 3.7E-02 | ↑ ↓ | 53 | 8.4E-13 | 3.1 | 2.4E-11 | |
| ATP-binding | ↓ | 21 | 2.7E-08 | 4.6 | 2.7E-06 | ↓ | 36 | 6.8E-11 | 3.7 | 5.8E-09 | ↑ ↓ | 56 | 4.6E-06 | 1.9 | 5.6E-05 | ||||
| Transferase | ↓ | 20 | 6.8E-08 | 4.6 | 5.8E-06 | ↑ ↓ | 31 | 2.1E-08 | 3.3 | 1.2E-06 | 16 | 2.7E-04 | 3.0 | 7.6E-03 | ↑ ↓ | 89 | 2.7E-22 | 3.2 | 1.7E-20 |
| Kinase | ↓ | 14 | 7.1E-07 | 5.9 | 5.2E-05 | ↓ | 9 | 4.0E-04 | 5.0 | 1.9E-02 | ↑ ↓ | 34 | 2.2E-05 | 2.3 | 2.3E-04 | ||||
| Zinc-finger | ↓ | 17 | 5.6E-06 | 4.0 | 3.2E-04 | ↑ ↓ | 28 | 5.7E-07 | 3.0 | 2.9E-05 | ↑ | 46 | 6.8E-04 | 1.7 | 5.5E-03 | ||||
| Nucleotide-binding | ↓ | 20 | 2.6E-06 | 3.6 | 1.7E-04 | ↑ ↓ | 40 | 6.7E-11 | 3.3 | 6.8E-09 | 12 | 7.3E-02 | 1.8 | 6.1E-01 | ↓ | 6 | 1.8E-02 | 3.9 | 9.1E-02 |
| Oxidoreductase | ↓ | 11 | 2.2E-04 | 4.3 | 1.1E-02 | ↑ ↓ | 18 | 3.4E-05 | 3.3 | 1.3E-03 | 34 | 7.6E-25 | 11.1 | 3.9E-22 | ↑ ↓ | 95 | 4.2E-46 | 5.9 | 1.1 E-43 |
| Transmembrane | ↓ | 19 | 3.0E-04 | 2.6 | 1.4E-02 | ↑ ↓ | 56 | 9.0E-18 | 3.6 | 5.7E-14 | 20 | 1.2E-03 | 2.3 | 2.9E-02 | ↑ ↓ | 121 | 9.4E-23 | 2.6 | 8.0E-21 |
| Nucleus | ↓ | 17 | 4.2E-04 | 2.8 | 1.8E-02 | 12 | 6.4E-03 | 2.6 | 1.6E-01 | ↑ ↓ | 79 | 4.7E-09 | 2.0 | 7.5E-08 | |||||
| Cellular metabolic process | ↓ | 107 | 1.2E-05 | 1.4 | 2.4E-02 | ||||||||||||||
| Serine/threonine-protein kinase | ↓ | 7 | 9.2E-04 | 6.2 | 3.6E-02 | 5 | 1.0E-02 | 5.9 | 2.2E-01 | 15 | 1.3E-02 | 2.1 | 6.8E-02 | ||||||
| DNA-binding | ↓ | 13 | 1.1E-03 | 3.0 | 4.1E-02 | ↑ | 48 | 1.8E-04 | 1.8 | 1.7E-03 | |||||||||
| Receptor | ↑ ↓ | 48 | 4.5E-11 | 2.9 | 5.8E-09 | ||||||||||||||
| Transport | 9 | 4.5E-02 | 2.3 | 6.9E-01 | ↑ | 25 | 5.1E-06 | 2.9 | 2.4E-04 | ||||||||||
| SH2 domain | 8 | 5.5E-05 | 8.1 | 2.0E-03 | |||||||||||||||
| WD repeat | 4 | 4.8E-02 | 4.9 | 7.0E-01 | ↓ | 10 | 6.8E-05 | 5.7 | 2.0E-03 | ||||||||||
| Ribonucleoprotein | 6 | 4.2E-02 | 3.1 | 4.2E-01 | ↓ | 20 | 3.4E-06 | 3.5 | 4.3E-05 | ||||||||||
| Membrane | ↓ | 14 | 1.6E-02 | 2.1 | 4.0E-01 | ↑ ↓ | 53 | 4.2E-16 | 3.7 | 1.1E-13 | 19 | 1.2E-03 | 2.3 | 2.8E-02 | ↑ ↓ | 114 | 4.3E-22 | 2.7 | 2.4E-20 |
| Glycoprotein | 6 | 8.5E-02 | 2.6 | 7.8E-01 | ↑ ↓ | 16 | 1.7E-04 | 3.2 | 4.9E-03 | 19 | 5.1E-10 | 6.7 | 4.3E-08 | ↑ ↓ | 56 | 2.9E-17 | 3.7 | 9.8E-16 | |
| Cytoplasm | 6 | 5.9E-04 | 8.6 | 4.9E-02 | 11 | 1.1E-04 | 4.8 | 3.5E-03 | ↓ | 256 | 1.0E-35 | 2.0 | 4.5E-33 | ||||||
| Determination of adult life span | 50 | 1.3E-37 | 10.8 | 1.3E-34 | 37 | 6.9E-03 | 1.5 | 2.7E-01 | |||||||||||
| CUB-like region | 21 | 9.0E-20 | 16.9 | 3.6E-16 | 16 | 4.1E-04 | 2.7 | 1.3E-01 | |||||||||||
| Signal | 12 | 1.9E-02 | 2.2 | 2.6E-01 | 28 | 2.5E-18 | 9.3 | 6.5E-16 | ↑ ↓ | 88 | 2.6E-40 | 5.5 | 4.4E-38 | ||||||
| Iron | 10 | 2.4E-03 | 3.5 | 5.1E-02 | 20 | 2.7E-15 | 12.4 | 4.5E-13 | ↑ ↓ | 35 | 3.7E-12 | 4.1 | 8.6E-11 | ||||||
| Secreted | 16 | 2.2E-12 | 12.7 | 2.2E-10 | ↑ ↓ | 41 | 9.8E-21 | 6.2 | 5.0E-19 | ||||||||||
| Monooxygenase | 11 | 2.1E-09 | 15.6 | 1.5E-07 | ↑ | 19 | 2.1E-08 | 5.1 | 3.0E-07 | ||||||||||
| Protease | 5 | 6.3E-02 | 3.3 | 7.6E-01 | 15 | 3.9E-09 | 8.3 | 2.5E-07 | ↑ ↓ | 45 | 8.7E-18 | 4.7 | 3.4E-16 | ||||||
| Heme | 11 | 3.7E-08 | 11.6 | 1.9E-06 | ↑ | 24 | 6.8E-10 | 4.8 | 1.2E-08 | ||||||||||
| Metalloprotein | 7 | 4.2E-03 | 4.6 | 7.9E-02 | 10 | 1.9E-07 | 11.7 | 8.8E-06 | ↑ | 20 | 9.2E-08 | 4.4 | 1.3E-06 | ||||||
| Storage protein | 4 | 1.1E-05 | 79.2 | 4.4E-04 | |||||||||||||||
| Lyase | 14 | 3.5E-05 | 4.1 | 1.1E-02 | ↑ ↓ | 21 | 1.1E-10 | 6.1 | 2.3E-09 | ||||||||||
| Signal peptide | 24 | 3.8E-06 | 2.8 | 1.5E-02 | ↓ | 70 | 2.6E-06 | 1.7 | 5.0E-03 | ||||||||||
| Carboxylic acid metabolic process | 18 | 6.8E-05 | 3.1 | 2.0E-02 | ↑ ↓ | 58 | 6.8E-07 | 1.9 | 1.4E-04 | ||||||||||
| Trehalose metabolic process | 5 | 5.2E-05 | 21.2 | 2.1E-02 | |||||||||||||||
| Neuropeptide | ↑ | 16 | 3.5E-12 | 10.6 | 8.6E-11 | ||||||||||||||
| Transthyretin-like | 7 | 1.7E-03 | 5.4 | 4.7E-01 | ↑ | 24 | 3.6E-09 | 3.9 | 3.6E-06 | ||||||||||
| Amidation | ↑ | 9 | 1.1E-06 | 10.2 | 1.4E-05 | ||||||||||||||
| Transcription | ↓ | 8 | 4.0E-02 | 2.5 | 6.7E-01 | ↑ | 34 | 4.1E-03 | 1.7 | 2.7E-02 | |||||||||
| Collagen | ↓ | 76 | 4.5E-67 | 13.1 | 2.3E-64 | ||||||||||||||
| Phosphate/anion transport | ↓ | 103 | 1.3E-47 | 4.6 | 2.6E-44 | ||||||||||||||
| Cell structure | ↓ | 71 | 2.3E-20 | 3.2 | 4.7E-17 | ||||||||||||||
| Muscle expressed genes | ↓ | 247 | 1.3E-18 | 1.7 | 2.6E-15 | ||||||||||||||
| Larval development | ↓ | 137 | 2.1E-18 | 2.1 | 4.3E-15 | ||||||||||||||
| Growth | ↓ | 117 | 2.1E-15 | 2.1 | 4.3E-12 | ||||||||||||||
| Locomotory behavior | ↓ | 118 | 4.4E-14 | 2.0 | 8.9E-11 | ||||||||||||||
| Proline-rich region | ↓ | 55 | 2.6E-11 | 2.7 | 5.4E-08 | ||||||||||||||
| Physiological process | ↓ | 73 | 2.1E-08 | 1.9 | 4.4E-05 | ||||||||||||||
| Embryonic development | ↓ | 149 | 6.7E-08 | 1.5 | 1.4E-04 | ||||||||||||||
| Biosynthesis | ↓ | 90 | 4.8E-07 | 1.7 | 9.8E-04 | ||||||||||||||
| RAS cluster C-variable | ↓ | 41 | 3.4E-06 | 2.1 | 6.9E-03 | ||||||||||||||
| Protein of unknown function, DUF141 | ↓ | 16 | 7.3E-06 | 3.9 | 1.5E-02 | ||||||||||||||
| Mitochondrion | ↓ | 44 | 3.1E-06 | 2.1 | 4.5E-04 | ||||||||||||||
| Amino acid metabolism | 8 | 5.2E-02 | 2.4 | 9.9E-01 | 10 | 1.0E-02 | 2.8 | 6.3E-01 | ↓ | 29 | 1.3E-05 | 2.4 | 2.7E-02 | ||||||
| Calcium | ↑ | 7 | 5.8E-06 | 3.9 | 6.7E-05 | ||||||||||||||
| Lipid metabolic process | ↑ ↓ | 65 | 6.3E-06 | 1.7 | 8.6E-04 | ||||||||||||||
Direction of change in age-1(mg44, m333) relative to hx546 or N2;
Direction of change in dauer larvae
We also analyzed GO terms for the second gene set, 976 annotated genes with expression altered in strong age-1 alleles relative to N2DRM controls (at FDR ≤ 0.1). This set duplicates only 28% of the genes in the first set, but shares ten of the 15 significant functional-annotation groups listed above and approaches significance for another three. A further eight GO terms distinguish the second set, being largely absent from the first: 48 receptor designations, 25 transport, eight SH2-domain terms, ten instances of WD repeats, six of ribonucleoprotein, 53 membrane (overlapping but not identical to the transmembrane group), 16 glycoprotein and six cytoplasm labels. As noted in the preceding section, the comparison of strong age-1 alleles to age-1(hx546) is expected to cancel out much of the effect on genes regulated via DAF-16 and that may account for the absence (or reduced presence) of these eight GO categories.
Conversely, several terms were more prominent when strong age-1 alleles were contrasted to hx546 than when they were compared with N2DRM. Of the 15 terms significantly enriched in the first comparison, ‘alternative splicing’ and ‘kinase’ were less enriched and less significant in the N2DRM comparison, while ‘cellular metabolic process’ and ‘DNA binding’ were absent from those results (i.e., they did not meet the criteria of an EASE score of ≥ 0.1 and 2 or more genes per term), despite the inclusion of over 2.5 times as many genes in the latter case. Such a shift implies the loss of positive transcriptional regulators in the strong age-1 mutants or the recruitment of negative regulators that are not invoked by the weaker allele. This is entirely consistent with the conclusions from the preceding section, from analyses of upstream regulatory motifs.
These age-1 GO patterns differ considerably from those reported for dauer larvae or long-lived daf-2 adults (Murphy et al., 2003; McElwee et al., 2004, 2006; Murphy, 2006) or for normal aging (Lund et al., 2002; Kim, 2007). As an example, 246 genes identified in an aging-profile study (Lund et al., 2002) included 12 insulin-related genes and six nuclear hormone receptors, but relatively sparse representation of the genes most enriched here, e.g. those encoding hydrolases, zinc-finger proteins or other metal-binding proteins. To provide a more direct comparison, DAVID was used to assess the annotation of 514 genes differentially expressed between worms defective in daf-2 expression (chiefly through RNAi) and controls (exposed to RNAi targeting both daf-2 and daf-16) in a quite extensive microarray study (Murphy et al., 2003), and to ~2000 dauer-specific genes specified by a careful reanalysis (McElwee et al., 2004, 2006) of microarray raw data generated previously (Wang & Kim, 2003), comparing N2DRM as dauer larvae to 12 h postrecovery. As noted above, arrows are used in Table 2 to indicate enrichment primarily in strong-age-1- or dauer-upregulated gene sets (↑), enrichment in strong-age-1- or dauer-downregulated gene sets (↓) or both (↑↓). The daf-2 gene sets were not separated by direction of change.
These results, summarized in Table 2, illustrate both similarities and differences in expression profiles among these groups. Genes annotated as impacting adult lifespan are especially prominent in the daf-2 set [in some measure as a result of the prior study (Murphy et al., 2003)], but failed to attain significance in age-1 and dauer comparisons. Nine significant terms are shared between the daf-2 and age-1 studies; of these, metal binding, hydrolase and transmembrane are comparably enriched, whereas zinc, alternative splicing, transferase and nucleotide binding are more enriched within one or both age-1 gene sets, and 12 additional terms are enriched only in those sets but not in the daf-2 comparison. Several terms (e.g. storage protein and trehalose metabolism), are unique to the daf-2 study, but many more are shared only with dauer larvae.
Pearson correlation coefficients were calculated for the ‘fold-enrichment’ factors reported in these four DAVID analyses. Significant correlations were seen between the two age-1 gene-annotation enrichments (R = 0.59, n = 19, P < 0.02), and between the daf-2 and dauer enrichments (R = 0.47, n = 23, P< 0.04). However, enrichments in the dauer gene list have essentially no correlation to those for strong-age-1 alleles vs. hx546, and an inverse correlation (R = −0.32) to those for strong-age-1 alleles vs. N2 [see Table S3 (Supporting information)].
This side-by-side comparison is intended primarily to make the point that the present transcript analysis is not redundant with previous studies of disruptions to the same pathway. Specific changes must be interpreted with caution, however, because enrichment of gene-ontology categories can depend strongly on the choice of control strains and on many subtler differences between the experimental protocols employed.
Expanded studies of allele-specific expression changes, by real-time PCR
We selected genes from the microarray surveys for more precise and sensitive quantitative analyses by RT-qPCR, allowing us to compare a variety of strains, to include larger numbers of biological replicates per strain, and to test two strains at multiple ages. The data thus generated also provide a ‘quality control’ to test for differential expression as detected in microarray surveys. Twenty-three genes were drawn at random (i.e. without regard to function, so as to avoid bias) from among those reported in SAM analyses of microarray data (q ≤ 5.5%). Sixteen of these genes appeared in gene lists for both sets of arrays, whereas four were found in only the hx546 comparison and two were unique to the N2DRM comparison. RNA levels were quantified by RT-qPCR in multiple independent cohorts of each of five strains (Table 3), for these 22 genes and three additional genes used as negative controls in RNAi experiments (described below), plus three further genes that varied little among the strains, used only for normalization (Table 3 legend).
Table 3.
Quantitation and functional assay of gene-expression changes implicated in microarray studies as specific to strong age-1 alleles. Twenty-two genes, initially implicated by microarray analysis of transcripts as more affected by strong age-1 alleles (mg44 and m333) than by a weak allele (hx546), were tested for transcript abundance by real-time polymerase chain reaction (RT-qPCR) for 3–8 biological replicates per group. Transcript levels, measured on a log2 scale, were normalized to the mean of three internal control genes that did not alter significantly among the strains (β-actin, T08G5.3 and Y71D11.3). Each is expressed as a ratio to the value for N2DRM, identically normalized, and is presented as the log2 of that ratio. Of these 22 genes, 21 (95%) differ significantly in transcript level between age-1(mg44)-F2 adults and wild-type N2DRM adults, at P < 0.05 (at which criterion, one false-positive would be expected), and 17 (77%) differ significantly at P < 0.001 (i.e. below the 0.0025 threshold after full Bonferroni correction for multiple assessments). In contrast, just five of 22 genes showed significant (P < 0.05) changes for the weaker age-1(hx546) allele and only two attained P < 0.001. Just one gene, evl-14, appeared to be less affected by the stronger allele, while 17 were significantly more affected in age-1(mg44)-F2 than in (hx546), at P < 0.01. The table reiterates three genes from Table 4, which were employed as negative controls for RNAi testing. Double-stranded-RNA producing bacterial clones were selected from the Ahringer RNAi library (Kamath & Ahringer, 2003) to target these controls, plus 10 genes strongly downregulated in age-1(mg44) F2 adults (by nine- to 32-fold, each P < 10−6) and one gene that declined 3.3-fold. Each RNAi clone was fed to N2DRM or age-1(hx546) worms, followed by tests of survival in 5-mM hydrogen peroxide. All positive RNAi results were confirmed in two independent experiments using different nematode expansions for testing.
| Gene (protein function) | Upstream motifs, ≤1 kb | Clone | Microarray vs. |
Log2 (expression) relative to N2DRM |
RNAi: H2O2 resistance in: |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| N2DRM | hx546 | age-1 (mg44)-F2 | daf-16(m26); age-1(mg44) | age-1 (hx546) | daf-16(m26); age-1(hx546) | N2DRM | age-1(hx546) | |||
| (predicted E3 ubiquitin ligase)† | M3 | T12E12.1 | ↓ | ↓ | −5.1***** | 0.0 | −1.0 | −0.7 | 1.39***** | 1.00 |
| dkf-2 (protein kinase D-3) | A, M1 | T25E12.4 | ↓ | −4.3***** | +0.7 | 0.0 | +1.0 | 1.06 | 1.02 | |
| (homologous to nucleoside transporter)† | A | F16H11.3 | ↓ | ↓ | −4.1***** | −0.4 | −0.5 | −0.1 | 1.36***** | 1.05 |
| cyp-35C1 (cytochr. P450, CYP2 subfam.) | A | C06B3.3 | ↓ | ↓ | −4.1***** | +0.4 | +1.9 | +0.8 | 1.15# | 2.92****** |
| F53H1.3 (unknown, highly conserved)† | F53H1.3 | ↓ | ↓ | −4.1***** | 0.0 | −1.0# | +1.0 | 1.53***** | 2.51****** | |
| (ubiquitin ligase homol. to Rb-BP-6) | B, B, M3 | F36F2.3 | ↓ | ↓ | −3.8***** | 0.0 | −0.5 | −0.7 | 1.33**** | 2.68****** |
| (ATP-dep’t RNA helicase, DEAD family) | R05D11.4 | ↓ | ↓ | −3.6***** | 0.0 | −0.3 | −0.3 | 1.03 | – | |
| ubl-5 (ubiquitin-like cytoplasmic protein) | F46F11.4 | ↓ | ↓ | −3.3***** | +0.3 | −0.5 | +0.1 | 1.10 | 2.62****** | |
| dna-2 (replication helicase, endonuclease) | A | F43G6.1 | ↑ | −3.3***** | −1.3* | −1.6** | −1.1 | 2.10**** | – | |
| let-502 (myotonic dystrophy S/T kinase) | A | C10H11.9 | ↓ | ↓ | −3.2***** | −1.0* | −1.3 | −0.7 | 1.02 | 1.76***** |
| pms-2 (DNA mismatch repair protein) | A, B | H12C20.2 | ↓ | ↓ | −2.8**** | −0.5 | −0.4 | −0.1 | – | – |
| (MTHFR, methylenetetrahydrofolate reductase) | A, A | C06A8.1 | ↓ | ↓ | −2.0***** | −0.4 | −1.0 | −0.3 | – | – |
| (aminoacylase ACY1, metallopeptidases) | A, A, B | C10C5.4 | ↓ | ↓ | −2.0** | −0.1 | +0.1 | −0.1 | – | – |
| (prob. ubiquitin conjugating enzyme E2)† | B | C03H5.3 | ↓ | ↓ | −1.8* | +0.3 | −0.3 | −0.5 | 1.31**** | 1.10 |
| uba-2 (ubiquitin-like activating enzyme 2) | W02A11.4b | ↓ | −1.7* | −2.4* | −0.4 | −0.1 | – | – | ||
| ife-1 (translational initiation factor eIF-4E) | F53A2.6 | ↓ | ↓ | −1.4# | 0.0 | +0.1 | +0.1 | – | – | |
| htp-2 (HORMA-domain protein) | M2 | Y73B6BL.2 | ↓ | ↓ | −1.3** | +0.2 | −0.8# | 0.0 | – | – |
| cah-5 (carbonic anhydrase family) | A | R173.1 | ↓ | ↓ | −1.3# | −0.7 | −0.3 | +0.3 | – | – |
| lact-3 (beta-lactamase domain protein) | B, M3 | M28.6 | ↓ | ↓ | −1.2**** | −0.7** | −0.6 | −0.9 | – | – |
| evl-14 (homol. yeast cohesion prot. Pds5p) | A, A | H38K22.1 | ↑ | ↑‡ | −0.2 | −0.3 | −2.4**** | +0.1 | – | – |
| (orthol. to human disease gene, Fukutin) | A | T07D3.4 | ↓ | ↓ | +1.1** | −0.1 | −0.5 | −0.7 | – | – |
| (probable G-protein coupled receptor) | C35A11.1 | ↓ | ↓ | +2.4***** | +0.8* | +0.8* | +0.4 | – | – | |
| Negative controls for RNAi assays (from Table 4) | ||||||||||
| ketn-1 (kettin, actin-binding titin paralog) | F54E2.3 | −0.6 | −0.5 | +0.5 | −0.1 | 1.04 | – | |||
| grd-14 (hedgehog-like signaling protein) | T01B10.2 | +0.8 | +0.1 | +0.4 | +0.6 | 0.96 | – | |||
| (VAP-like; phospholipid and inositol metab.) | C10H11.7 | +4.4***** | +0.9* | +1.6* | +1.3 | 0.98 | – | |||
Significance of difference from N2DRM (expression), or from empty-vector control (RNAi), by 2-tailed t-test:
P < 0.05;
P < 0.01;
P < 0.001;
P < 1E–4;
P < 1E–5;
P < 1E–6;
P < 1E–10.
–, not assessed; N.S., not significant; ↓, lower in age-1(mg44)-F2 adults than in N2DRM or age-1(hx546); ↑, higher in age-1(mg44)-F2 adults than in N2DRM or age-1(hx546); A, ‘DAE’, CTTATCA; B, ‘DBE’, GTAAAYAA; M1, TTCWGAAAAT; M2, CTACAGTAA; M3, AGAGAMGMAGA.
RNAi shown here to extend lifespan.
Note that all RT-qPCR results were calculated relative to N2DRM. Thus, the microarray and RT-qPCR results for evl-14 are consistent with regard to age-1 alleles mg44 and/or m333 vs. hx546, because −0.2 > −2.4.
Of 20 genes which microarrays had indicated to be differentially expressed between strong and weak age-1 alleles, 18 (90%) were confirmed by RT-qPCR. A lower confirmation rate (14/18 or 78%) was seen for genes significantly modulated by strong age-1 mutations relative to wild-type N2DRM adults. Two genes (T07D3.4 and C35A11.1) were discordant in both comparisons, showing downregulation by strong age-1 mutants in microarray assays, but upregulation when assessed by RT-qPCR. Two further discrepancies were seen only in comparisons with N2DRM: dna-2 and evl-14 appeared to be upregulated by strong age-1 mutations when assessed by microarrays, whereas RT-qPCR indicated tenfold downregulation of dna-2 and no change to evl-14. Discordant findings for these genes, highly significant in each analysis, could reflect differential splicing given that RT-qPCR and microarrays did not target the same exons.
Quantitative RT-PCR was also used to assess 37 genes that had not been significantly implicated by SAM analyses, but that appeared to differentially express low-abundance transcripts in the strongest age-1 alleles. The criteria for inclusion in this set were (i) differential expression among N2, age-1(hx546) and age-1(mg44)/(m333) adults in at least four microarrays; and (ii) low signal (< 3 × background) in the remaining microarrays. Heat maps for these 37 genes (some of them represented by more than one spot) are shown in Fig. S3 (Supporting information). Table 4 summarizes data for seven genes shown to be downregulated in age-1(mg44) F2 adults, of 18 tested, and for three of the 11 genes that did not change; data are also presented for seven genes found to be upregulated in age-1(mg44) F2 adults, of 18 tested. Confirmation rates are lower than these numbers suggest, because the direction of change was not always consistent between assays: only six genes appeared to be downregulated in strong age-1 alleles both by microarrays and by RT-qPCR (33%), while just five increased in both assays (28%). The highest expression ratio encountered, > 1000-fold higher transcript levels in age-1(mg44)-F2 than in N2DRM adults, was seen for one of these genes: clec-60, encoding an intestinally expressed C-type lectin.
Table 4.
Quantitation and functional assay of expression changes in genes weakly implicated on microarrays. Thirty-eight genes were initially suggested to be more affected by strong age-1 alleles (mg44 and m333) than by a weak (hx546) or wild-type (N2DRM) age-1 allele, based on at least four microarrays. When transcript abundances were assessed by real-time polymerase chain reaction (RT-qPCR), 14 of these 38 genes (37%) differed by over fourfold in transcript level between age-1(mg44)-F2 adults and wild-type N2DRM adults, at P < 10−5 (well below the 0.0025 threshold after full Bonferroni correction for multiple assessments). In contrast, only seven of 38 genes (18%) showed significant changes (P < 0.01) for the weaker age-1(hx546) allele, and just two of those altered by ≥4-fold. Two genes (T10B11.7 and lbp-6) appeared to be less affected by the stronger allele, while 13 genes were significantly more affected in age-1(mg44)-F2 than in (hx546), at P < 0.01. Double-stranded RNAi clones (Kamath & Ahringer, 2003) were selected to target seven genes strongly downregulated in age-1(mg44) F2 adults (by 4- to 20-fold, each P < 10−5), and one gene (lbp-6) downregulated primarily in age-1(hx546) (P < 0.01), as well as three control genes that were unchanged or upregulated in age-1(mg44).
| Gene (protein function) | Upstream motifs, ≤1 kb | Clone | Log2 (expression) relative to N2DRM |
RNAi: H2O2 resistance |
||||
|---|---|---|---|---|---|---|---|---|
| age-1 (mg44)-F2 | daf-16(m26); age-1(mg44) | age-1 (hx546) | daf-16(m26); age-1(hx546) | N2DRM | age-1(hx546) | |||
| Decreased in age-1(mg44) F2 adults (7 confirmed genes, 35% of 20 tested) | ||||||||
| (unknown; homol. to helicase, translation IF) | M2 | T10B11.7 | −4.3***** | −1.0* | −5.1*** | −1.0 | 1.05 | – |
| cpr-1 (cys-protease, sim. to cathepsin B)†,‡ | A, A | C52E4.1 | −4.3***** | −0.7# | −0.5 | +0.3 | 3.14***** | – |
| aka-1 (A-kinase anchor protein; FYVE Zn-finger) | D1022.7 | −4.1**** | −1.0# | −1.5 | 0.0 | 1.53***** | 1.50****** | |
| ung-1 (uracil N-glycosylase 1) | Y56A3A.29 | −3.8***** | −0.1 | −0.7 | −0.5 | 2.75*** | – | |
| sna-1 (snRNP-binding protein 1) | W02F12.6 | −3.2***** | −0.6# | −0.9* | −0.6 | 1.6** | – | |
| cdc-48.1 (ATPase, proteasomal sorting) | B | C06A1.1 | −2.8***** | 0.0 | −1.0* | −0.5 | 1.0 | – |
| rab-10 (intestinal RAS-like GTPase)‡ | A | T23H2.5 | −2.0**** | −0.4 | −0.8* | −0.1 | 1.82*** | – |
| ketn-1 (kettin, actin-binding titin paralog) | F54E2.3 | −0.6 | −0.5 | +0.5 | −0.1 | 1.03 | 1.01 | |
| lbp-6 (lipocalin-like fatty acid-binding protein)† | W02D3.5 | −0.5 | +0.1 | −1.4* | −0.8 | 1.37***** | 2.51***** | |
| grd-14 (hedgehog-like signaling protein) | T01B10.2 | +0.8 | +0.1 | +0.4 | +0.6 | 1.00 | – | |
| Increased in age-1(mg44) F2 adults (7 confirmed genes, 39% of 18 tested) | ||||||||
| (homol. to cruciform DNA binding protein) | Y75B7B.1 | +2.7**** | −0.3 | +0.1 | −0.5 | – | – | |
| hsp-12.3 (small heat-shock protein) | B, B | F38E11.1 | +3.7***** | −0.7* | 0.0 | −0.1 | – | – |
| (VAP-like; phospholipid, inositol metab.) | A, B | ZC196.2 | +4.2***** | −0.1 | −1.0 | −1.3 | – | – |
| (VAP-like; phospholipid, inositol metab.) | C10H11.7 | +4.4***** | +0.9* | +1.6* | +1.3 | 0.98 | N.S. | |
| nspd-2 (protein homologous to prions, RNA-BP) | K07F5.5 | +6.3***** | +0.9 | +4.9** | +1.1 | – | – | |
| cyc-2.2 (cytochrome C, of two in C. elegans) | M2 | ZC116.2 | +6.5***** | −2.0* | −0.7 | −0.5 | – | – |
| clec-60 (C-type lectin, expressed in intestine) | A | ZK666.6 | +10.1***** | −1.3* | ±0.8 | −1.0 | – | – |
Significance of difference from N2DRM (expression), or from empty-vector control (RNAi), by 2-tailed t-test:
P< 0.05;
P< 0.01;
P< 0.001;
P< 1E–4;
P< 1E–5;
P< 1E–6;
P < 1E-10.
–, not assessed; N.S., not significant; motifs seen within 1 kb upstream of each gene: A, ‘DAE’; B, ‘DBE’; M2, CTACAGTAA (see Table 3 key).
Genes shown here to extend life when inhibited by RNAi.
RNAi has been reported to extend lifespan when directed against cpr-1 (Murphy et al., 2003) or rab-10 (Hansen et al., 2005).
As expected, genes selected by SAM as highly significant in microarrays have a higher confirmation rate by RT-qPCR than genes with weaker support. However, 137 of the 185 most significant genes (i.e. 74% of genes with q < 0.7%) were also in the top quintile for total signal across all arrays, indicating a substantial skewing of significant results in favor of highly expressed genes. This well-established bias of microarray data sets (Miller et al., 2001) motivated our inclusion, for RT-qPCR and subsequent analyses, of lower-expression genes as well as those attaining the highest significance.
Five near-isogenic strains were compared here, as infertile or postgravid adults at the same ages as analyzed by microarrays: (i) N2DRM (wild-type), (ii) age-1(mg44) F2 homozygotes (very long-lived), (iii) daf-16(mu86); age-1(mg44) double mutants (largely reverted to wild-type longevity and stress resistance), (iv) age-1(hx546) (a weaker allele with ~2× normal longevity and intermediate stress resistance) and (v) daf-16(mu86); age-1(hx546) (fully reverted to wild-type longevity and stress resistance). Tables 3 and 4 together provide a quantitative comparison of transcript levels assessed by RT-qPCR across independent biological replicates of the five strains, for a total of 34 genes that are differentially regulated (25 down and nine up, at P < 0.01) in very long-lived age-1(mg44)-F2 adults, as well as two genes (lbp-6 and evl-14) that altered primarily in hx546. These results are discussed in greater detail in subsequent sections.
DNA sequences adjacent to the 34 genes differentially expressed in strong age-1 mutants at P < 0.01 were searched (using RSAT’s ‘dna-pattern’ module) for the known DAF-16-binding/associated elements DBE and DAE, and the three novel motifs discovered by RSAT oligo-analysis (see above). Within 1 kb of upstream sequence, on either strand, there are ten DBEs preceding eight genes (vs. 6.4 expected DBEs; N.S.); 18 DAEs preceding 14 genes (vs. 7.4 expected; exact Binomial P < 3E–4); one ‘M1’ motif, TTCWGAAAAT (0.5 expected, N.S.); two ‘M2’ motifs, CTACAGTAA (0.4 expected, P < 0.07); and three ‘M3’ motifs, AGAGAMGMAGA (0.06 expected; P< 5E–5). These upstream motif sites are indicated in Tables 3 and 4.
Transcript profiles differ between age-1(mg44) F2 adults and less long-lived worms
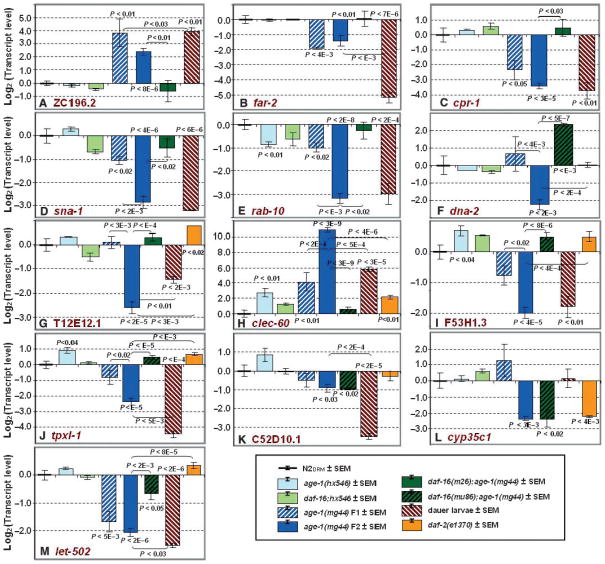
Of the 36 genes confirmed to differ significantly between either age-1 allele and N2DRM (Tables 3 and 4) at P < 0.01, 34 (94%) showed a more extreme shift in expression in mg44, while only two were more affected in hx546. These results indicate that mg44 is the stronger allele with regard to transcriptional effects, just as it is when assessing life-history traits such as dauer formation or survival (Ayyadevara et al., 2008).
For a subset of genes showing transcript modulation in age-1 mutants, we expanded the test panel by adding wild-type (N2DRM) dauer larvae, the first (F1) generation of age-1(mg44)−/− adults, and the extensively studied long-lived mutant daf-2(e1370) (Kenyon et al., 1993; Larsen et al., 1995). We also increased the number of independent worm expansions (the biological N) to as many as 14 for some groups, because biological variance for gene expression – among independently expanded cohorts – greatly exceeds variance observed for technical replicates. These RT-qPCR results (Fig. 2) documented seven genes for which age-1(mg44) F2 adults differ significantly from dauer larvae in transcript abundances and six genes for which they are quite similar. It thus appears that age-1 extreme longevity departs from the dauer strategies of gene expression about as often as it resembles it. As may be anticipated from the disparity in maternal protection (Ayyadevara et al., 2008), F2 homozygotes surpassed the parental F1 generation with respect to transcript changes in 11 of 13 genes, seven of which were significant at P < 0.02 (Fig. 2D–J). Unexpectedly, however, F1 adults also departed significantly from the expression phenotype of the weaker age-1(hx546) allele, which they resemble in terms of lifespan and stress resistance, for eight of 13 genes.
Fig. 2.
The age-1(mg44)-F2 transcript profile differs from those of F1 adults or N2 dauer larvae. We retested 13 age-1-regulated genes, in an expanded strain panel. Strains are as described in the legend to Table 3, with the addition of age-1(mg44) F1 (fertile first-generation homozygotes), daf-2(e1370), a long-lived mutant of the insulin/IGF-1 receptor (G–M), and daf-16(mu86); age-1(mg44), in which a daf-16 deletion allele, mu86, replaces the m26 allele used in Tables 3 and 4. Double mutants with daf-16(mu86) were assessed only for those genes that had appeared to be incompletely (or negligibly) reverted by daf-16(m26) (F, H–M; indicated by black striping on green bars). Orange bars [at the right of (G–M)] represent N2DRM dauer larvae, induced by crowding on starved plates and confirmed by > 95% resistance to 1% sodium dodecyl sulfate for samples of 500 worms. Sample numbers (n = the number of independent biological preparations) were as follows: 8–11 groups of postgravid N2DRM adults, 4–5 each of age-1(hx546) and daf-16(m26); age-1(hx546), 3 of age-1(mg44) F1, 10–14 of age-1(mg44)-F2 adults, 5–6 each of daf-16(m26); age-1(mg44) and daf-16(mu86); age-1(mg44), 3–4 of N2DRM dauers, and 3–4 of daf-2(e1370).
DAF-16 (FOXO) mediates most age-1 allele-specific changes in gene expression
Because the longevity and stress-resistance phenotypes of the age-1 mutants were largely or entirely reversed by a daf-16 (m26) mutation at the locus encoding DAF-16, a FOXO transcription factor (Ayyadevara et al., 2008; Tazearslan et al., 2009), the double mutants provide important controls in which proximal effectors of life extension are expected to also be nullified at the transcript level in the absence of active DAF-16. A substantial minority of genes violated this expectation, implying transcriptional effects that transpire via a DAF-16-independent route. Of 34 genes that altered expression in age-1(mg44)-F2 adults, with P < 0.01 by RT-qPCR, 23 (68%) were reverted to within 70–140% of wild-type by addition of the daf-16(m26) mutation, while 11 were not. Moreover, seven genes (21%) were expressed in daf-16(m26); age-1(mg44) double mutants at levels differing from N2DRM controls (each significant at P < 0.01; Tables 3 and 4) in the same direction as the age-1 mutant alone, indicating reversion that is incomplete at best. Those genes are dna-2, let-502, uba-2, lact-3, C35A11.1, C10H11.7, and ZC116.2. Three further genes (hsp-12.3, cyc-2.2, and clec-60) were shifted by daf-16 mutation in the opposite direction to the age-1-induced change.
Incomplete or negligible reversion may reflect residual FOXO activity in the daf-16(m26) allele we employed, although the same allele reversed the longevity phenotypes of both age-1(hx546) and age-1(mg44) worms (Ayyadevara et al., 2008). Nearly identical results were obtained; however, with a double mutant we constructed to combine age-1(mg44) with daf-16(mu86), a large-deletion allele. Among 26 genes tested, six (23%) showed no more than partial reversion (differing from N2DRM at P ≤ 0.01). Examples are shown in panels K–M of Fig. 2. These data support the existence of a DAF-16-independent route to transcriptional effects downstream of age-1, although this would account for only a small proportion of mg44-specific changes.
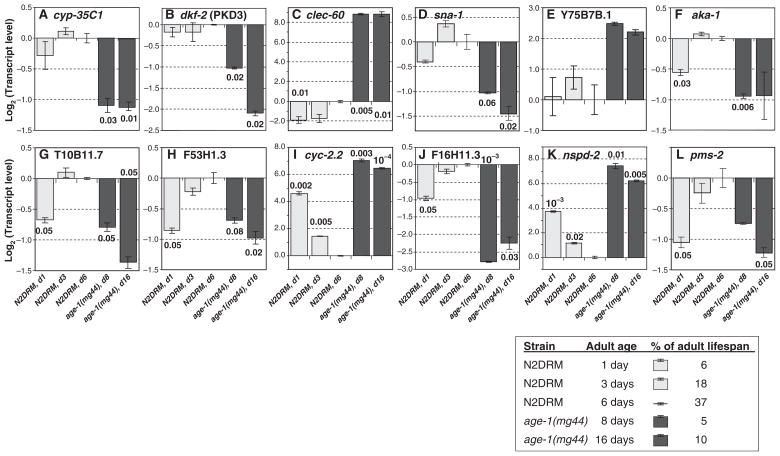
Expression changes in age-1(mg44) F2 adults reflect genotype rather than age
In the experiments described thus far, worms were approximately matched at an adult chronological age (days since the L4/adult molt) of at least 6 days. All fertile strains were post-gravid at this point, ensuring that egg transcripts could not account for any differential expression observed. Among strains differing markedly in longevity, however, alignment by chronological age creates considerable disparity in physiological age – which could itself underlie transcript-abundance changes in some cases (see, e.g. Helfand et al., 1995; Tazearslan et al., 2009). To compare the contributions of chronological vs. physiological age, we selected 12 genes that were strongly modulated in age-1(mg44) F2 adults – eight that decreased and four that increased. These genes were reassessed in wild-type N2DRM at three adult ages and in age-1(mg44) F2 adults at two ages. The resulting time series (Fig. 3) demonstrate that the expression changes seen in very long-lived age-1 worms cannot be interpreted as secondary consequences of their youthful physiological state relative to N2DRM. In particular, expression in age-1(mg44) F2 worms at 8 and 16 days of adult age (5 and 10% of their median adult lifespans) did not overlap the levels seen in N2DRM worms at 1, 3 or 6 days adult age (6, 18 or 37% of adult lifespan), for nine of the 12 genes examined (cyp-35C1, dkf-2, clec-60, sna-1, Y75B7B.1, aka-1, cyc-2.2, F16H11.3 and nspd-2). The remaining three genes shared a somewhat paradoxical pattern of expression, in which N2DRM expression was lowest at adult day 1, rising over time to the level observed at day 6 (to which other groups were normalized). Although in each case age-1(mg44) F2 adults at day 8 or 16 showed transcript levels somewhat close to those of day-1 wild-type adults, the age-1 mutant worms actually trended in the opposite direction, with expression declining over time. Thus, none of these gene expression patterns could be said to scale with lifespan. Transcript levels appeared independent of age in N2DRM for five of 12 genes (A–E), and in age-1(mg44) for 4–7 genes (A, C, E, F and probably I–K), but the genes that were stable with respect to age were not the same in the two strains.
Fig. 3.
Transcriptional changes in age-1(mg44) F2 adults are relatively stable with respect to age, and cannot be explained as a result of their physiological youthfulness. Transcript levels were assayed by real-time reverse-transcription polymerase chain reaction (RT-qPCR) as described in the legends to Table 3 and Fig. 2. Within each histogram, mean ± SEM are shown on a log2 scale for steady-state transcript level, comparing wild-type N2DRM adults at three ages to age-1(mg44) F2 homozygotes at two adult ages. All transcript values were first normalized to β-actin mRNA (to adjust for variation in RNA inputs), and then each biological group was normalized to the mean value for N2DRM day-6 adults (to correct for run-to-run variation in Ct values). Ages are measured from the L4/adult molt; a table (inset) indicates the percent of adult lifespan represented by each age. To monitor longitudinal changes, two biological preparations of each group were assessed (fewer than used for Fig. 2 or Table 3). Significance of differences was ascertained by two-tailed Behrens-Fisher t-tests, appropriate to samples of unequal or unknown variance. The transcript levels observed in this experiment differ from the means seen in previous experiments (e.g. Tables 3 and 4), but those differences are not significant.
Functional assessment of H2O2-resistance and longevity after RNAi knock-down
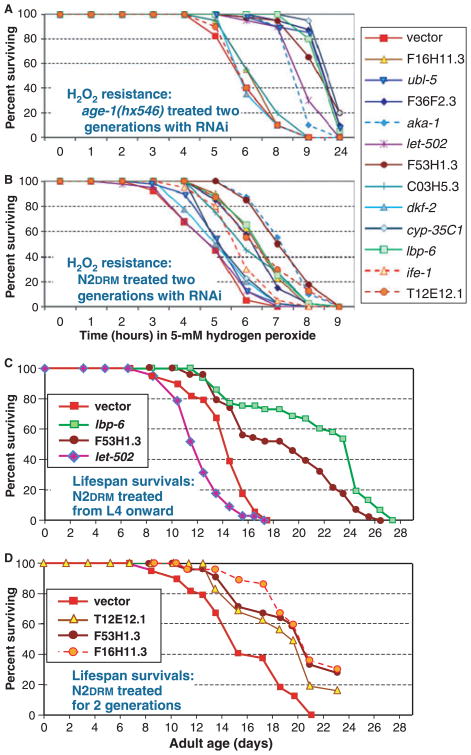
Alterations in gene expression, such as those described above, might contribute to the exceptional survival phenotypes of strong age-1 mutants or may be incidental side-effects. To distinguish between these possibilities, bacteria were retrieved from the Ahringer library (Kamath & Ahringer, 2003), which inducibly express double-stranded RNAs (dsRNA) corresponding to 18 of the 21 genes shown by RT-qPCR to be at least 3.3-fold downregulated by age-1(mg44) (each P < 10−5; see Tables 3 and 4), plus lbp-6, a gene of lesser and primarily hx546-specific effect, and three negative controls (with unchanged or increased expression in age-1 mutants). We then tested the effects of RNA interference in wild-type and age-1(hx546) worms, on survival in the presence of hydrogen peroxide – because resistance to this oxidative stress was shown previously to track with extreme longevity in comparisons of wild-type to five age-1 strains (Ayyadevara et al., 2008; Tazearslan et al., 2009).
RNAi knockdown, of genes showing lower expression in adults bearing strong age-1 alleles, significantly improved the hydrogen-peroxide resistance of N2DRM adults in 11 of 18 instances (61%; each P < 10−3). Targeting any of three control genes, however, produced no benefit relative to empty-vector controls (Tables 3–5). Feeding the same dsRNA-expressing bacteria to age-1(hx546) adults conferred peroxide protection for six of ten genes (60%). Despite this numerical equivalence, the benefit of RNAi was quite different for the two strains (see Fig. 4 and Table 5). RNAi targeting 6 genes (ubl-5, F36F2.3, let-502, F53H1.3, cyp-35C1 and lbp-6) conferred substantially more peroxide protection in age-1(hx546) adults (survival increases of 1.7- to threefold over feeding-vector controls) than in N2DRM, suggesting effects synergistic with a mild age-1 deficit. Conversely, three dsRNA clones (targeting F16H11.3, C03H5.3 and T12E12.1) had a greater impact on N2DRM, suggesting that their RNAi effects are partially redundant with the age-1(hx546) phenotype. Concordance between the two strains was seen for genes F36F2.3, aka-1, F53H1.3, lbp-6 and cyp-35C1 (dsRNA’s conferred significant protection in both strains), and dkf-2 (no protection in either strain). Two further genes (ubl-5, C03H5.3) elicited small RNAi effects that fell short of significance (0.05 < P < 0.07) in one of the strains.
Table 5.
Peroxide-resistance survival statistics for C. elegans strains N2DRM and age-1(hx546) fed dsRNA-expressing bacteria for two generations
| Gene targeted by RNAi | Mean survival (h), ±SD Experiment 1 |
% gain | Mean Survival (h), ±SD Experiment 2 |
% gain | Significance of difference vs. control* |
|---|---|---|---|---|---|
| N2DRM treated two generations | |||||
| None (vector) | 4.7 ± 1.0 | – | 4.6 ± 1.1 | – | – |
| F16H11.3 | 6.3 ± 1.0 | 34 | 6.3 ± 0.9 | 37 | < 10−6 |
| ubl-5 | 5.0 ± 0.8 | 6 | 5.2 ± 1.1 | 14 | 0.07 |
| F36F2.3 | 5.9 ± 1.0 | 27 | 6.3 ± 0.9 | 38 | < 10−6 |
| aka-1 | 7.1 ± 0.8 | 53 | 7.0 ± 0.9 | 53 | < 10−7 |
| let-502 | 4.8 ± 1.0 | 3 | 4.6 ± 1.4 | 0 | N.S. |
| F53H1.3 | 7.0 ± 1.1 | 51 | 7.1 ± 0.9 | 55 | < 10−7 |
| C03H5.3 | 5.8 ± 1.3 | 24 | 6.3 ± 1.1 | 37 | < 10−5 |
| dkf-2 | 5.2 ± 0.9 | 12 | 4.8 ± 1.3 | 5 | N.S. |
| cyp-35C1 | 5.2 ± 0.8 | 12 | 5.4 ± 0.9 | 18 | < 0.02 |
| lbp-6 | 6.2 ± 1.1 | 32 | 6.5 ± 0.9 | 42 | < 10−6 |
| ife-1 | 5.5 ± 0.9 | 18 | 5.7 ± 0.9 | 25 | < 10−4 |
| T12E12.1 | 6.4 ± 1.3 | 37 | 6.4 ± 1.2 | 40 | < 10−6 |
| age-1(hx546) treated two generations | |||||
| None (vector) | 6.1 ± 1.2 | – | 6.1 ± 1.2 | – | – |
| F16H11.3 | 6.3 ± 1.2 | 3 | 6.5 ± 1.0 | 7 | N.S. |
| ubl-5 | 15.5 ± 5.0 | 155 | 16.4 ± 4.1 | 169 | < 10−12 |
| F36F2.3 | 15.2 ± 5.0 | 150 | 17.4 ± 3.2 | 185 | < 10−12 |
| aka-1 | 9.0 ± 2.6 | 48 | 9.3 ± 2.4 | 52 | < 10−10 |
| let-502 | 10.6 ± 4.1 | 74 | 10.8 ± 3.8 | 77 | < 10−8 |
| F53H1.3 | 15.2 ± 6.3 | 150 | 15.3 ± 6.1 | 151 | < 10−12 |
| C03H5.3 | 6.5 ± 1.3 | 7 | 6.6 ± 1.2 | 8 | 0.07 |
| dkf-2 | 6.2 ± 0.9 | 1 | 6.2 ± 1.0 | 2 | N.S. |
| cyp-35C1 | 17.4 ± 4.6 | 185 | 18.2 ± 3.4 | 198 | < 10−12 |
| lbp-6 | 15.3 ± 4.0 | 151 | N.D. | – | < 10−8 |
| T12E12.1 | 6.2 ± 1.1 | 1 | N.D. | – | N.S. |
For each group, n = 20.
N.S., not significant.
Gehans-Wilcoxon log-rank test for experiments 1 and 2 combined.
Fig. 4.
RNAi knock-down of genes downregulated in age-1(mg44) confers peroxide resistance and can extend life. Caenorhabditis elegans bearing (A) age-1(hx546) or (B) the wild-type age-1 allele were fed bacteria expressing the indicated dsRNA. RNAi treatment in (A), (B) and (D) commenced during early larval development of the parental generation, so that the tested generation underwent knockdown throughout life. Worms in (C) were fed dsRNA-expressing bacteria continuously from the L4/adult molt onward. Survival statistics are summarized in Tables 5 and 6.
In these studies, feeding of dsRNA-producing bacteria began with the parents of the tested worms, so that the targeted RNAs would be depleted throughout their lives. In other experiments (not shown), similar results were observed for most genes when treatment commenced at the L4/adult molt, but a few genes (F36F2.3, ubl-5 and aka-1) were less influential after adult-only treatment, possibly because of incomplete inhibition.
Genes with low-level expression were also considered (Table 4), because such genes are generally under-represented among those identified on microarrays (Miller et al., 2001), whereas they are particularly good candidates for efficient inhibition by RNAi (Fire et al., 1998). The proportion of RNAi treatments conferring peroxide resistance was indeed a little higher in this group (5/7, 71%) than for genes implicated by SAM (6/11, 55%; Table 3), among genes comparably downregulated in age-1(mg44) adults – but this difference is not significant.
Twelve peroxide-protective RNAi clones (ten of which conferred significant protection in N2DRM, but two primarily in age-1(hx546)) were then tested for extension of N2DRM adult lifespan, with RNAi treatment beginning in the parental generation (Table 6, section A). Five RNAi treatments indeed improved the median lifespan of adults (lbp-6, F53H1.3, T12E12.1, F16H11.3 and C03H5.3), while RNAi clones targeting seven genes either had no significant effect or reduced longevity. Life extension was confirmed for F53H1.3 and lbp-6 in two other sets of assays (Table 6B, C), wherein RNAi treatment began only at the L4/adult molt. Examples of longevity survivals are shown in Fig. 4C,D.
Table 6.
Lifespan survival statistics for N2DRM worms fed dsRNA-expressing bacteria
| Gene targeted by RNAi | n | Median survival (d) | Mean survival (d) | Standard deviation (d) [SEM] | Significance of difference from control* |
|---|---|---|---|---|---|
| (A) RNAi treatment for 2 generations | |||||
| None (empty vector) | 32 | 15.4 | 16.7 | 2.4 [0.4] | – |
| F53H1.3 | 34 | 21.0 | 20.3 | 4.7 [0.8] | < 3 × 10−5 |
| T12E12.1 | 31 | 21.0 | 20.9 | 3.6 [0.6] | < 1 × 10−5 |
| F16H11.3 | 30 | 20.2 | 19.7 | 4.8 [0.9] | < 0.003 |
| C03H5.3 | 35 | 19.5 | 19.0 | 3.1 [0.5] | < 0.002 |
| cyp-35C1 | 37 | 14.4 | 14.6 | 2.8 [0.5] | N.S. |
| (B) RNAi treatment from late L4 | |||||
| None (empty vector) | 51 | 16.6 | 16.6 | 4.2 [0.6] | – |
| lbp-6 | 58 | 19.6 | 18.8 | 2.4 [0.3] | < 0.01 |
| F53H1.3 | 47 | 17.7 | 17.7 | 3.6 [0.5] | < 0.05 |
| (C) RNAi treatment from late L4 | |||||
| None (empty vector) | 35 | 14.6 | 14.1 | 2.3 [0.4] | – |
| let-502 | 37 | 11.5 | 12.2 | 1.9 [0.3] | < 10−3 |
| lbp-6 | 47 | 24.5 | 21.4 | 5.0 [0.7] | < 10−7 |
| F53H1.3 | 47 | 19.5 | 18.7 | 4.5 [0.7] | < 10−7 |
N.S., not significant.
Log-rank test, each experiment treated separately.
Discussion
The steps of selecting genes downregulated in age-1 mutants, and then narrowing that group to genes that confer oxidative-stress resistance when expression is blocked by RNAi in adults, produce successively greater enrichment of genes for which RNAi also extends lifespan. Thus, we were able to test functional effects of RNAi for 16 genes confirmed to be downregulated at least fourfold in age-1(mg44) worms (Tables 3 and 4), and 11 of these (69%) conferred hydrogen-peroxide resistance to wild-type worms – considerably more than the ~10% of unselected RNAi clones that increased resistance to another oxidative stressor, paraquat (Kim & Sun, 2007) (chi-squared and exact binomial P < 10−8), or to hydrogen peroxide (unpublished data). We were surprised by the high frequency at which RNAi, directed to genes selected for age-1-attenuated expression, resulted in hydrogen peroxide resistance. Although false-positives are possible, each survival was confirmed with an independent expansion of N2DRM worms. We know that the RNAi response itself does not confer protection against peroxide stress, because in many instances worms remained sensitive to H2O2 despite effective RNAi knock-down, as indicated by RT-qPCR.
Twelve peroxide-protective dsRNA clones were fed to wild-type worms to assess whether RNA inhibition extends lifespan. Five of the treatments significantly increased longevity in our studies; they targeted genes encoding an uncharacterized but highly conserved protein (F53H1.3), a lipid-binding protein (lbp-6), a putative E2 ubiquitin-conjugating enzyme (C03H5.3), a predicted E3 ubiquitin ligase (T12E12.1) and a possible nucleoside transporter (F16H11.3). Two of these were confirmed to extend lifespan in separate experiments, in which dsRNA feeding began only at maturity.
Among the 11 RNAi treatments that conferred substantial protection against hydrogen peroxide and were chosen to emulate under-expression in age-1(mg44) F2 adults (i.e. excluding lbp-6 because it is downregulated chiefly in hx546), those targeting rab-10 and cpr-1 had been reported previously to confer extended lifespan (Murphy et al., 2003; Hansen et al., 2005) and another four were first shown to do so in the present study (Table 6 and Fig. 4C,D). Our ‘rediscovery’ of two previously known longevity-control genes out of 11 (18%) is a 13-fold enrichment (exact Binomial P < 0.01) over the 1.2–1.4% frequencies of life extension reported in previous RNAi screens (Hamilton et al., 2005; Hansen et al., 2005; Smith et al., 2007). The probability of finding four novel longevity genes out of any 11 genes taken at random for survival testing (again omitting lbp-6) is P < 2 × 10−5, whereas to find all six longevity-implicated genes by chance, both previously reported and novel, P < 4 × 10−9.
Genes tested for RNAi enhancement of peroxide survival also included six genes for which transcripts were significantly reduced (at P < 0.01) by the weaker age-1(hx546) allele. Four of these were found to confer significant peroxide resistance, including one (rab-10) reported previously to extend lifespan through RNAi (Hansen et al., 2005) and another (lbp-6, encoding a lipid-binding protein) discovered here to do so (Fig. 4C). The probability of finding two life-extending RNAi targets among any six genes taken at random is < 0.003.
Conclusions
Based on the observations above, we conclude that (i) extreme age-1 deficiency invokes multiple transcriptional changes, many of which differ from those described previously; (ii) those changes are predominantly inhibitory and predominantly (but not entirely) mediated by DAF-16/FOXO; (iii) three novel upstream motifs are implicated, in addition to 19 previously established transcription-factor binding sites; (iv) a surprisingly large fraction of the genes significantly downregulated in age-1(mg44) F2 adults can confer resistance to oxidative stress when knocked down in wild-type worms; and (v) these genes are, in turn, highly and significantly enriched for RNAi targets that extend lifespan when suppressed.
We infer from these results that the extraordinary longevity and stress resistance of the age-1(mg44) strain is achieved, at least in part, by a concerted modulation of the gene-expression profile. This strongly implies a program, evolved in response to recurring selective pressures, which favors survival in the presence of oxidative and/or electrophilic stresses. That program is capable of producing far greater longevity and stress resistance than observed previously for IIS-pathway mutants, but presumably functions as an acute response to environmental signals, and would not be maintained over the full life-span potential because this precludes reproduction, and hence establishment through natural selection. Of genes primarily or exclusively inhibited or activated by the very-long-lived PI3KCS-null alleles of age-1, the great majority operate largely or entirely under the transcriptional control of DAF-16, and yet very few had been previously implicated in known lifespan-modulating pathways.
Of particular interest are the uncharacterized gene F53H1.3; the lbp-6 lipid-binding protein; 30 zinc- or other metal-binding proteins, including 17 zinc-finger proteins; 11 other transcription factors and 17 RNA splicing factors; 11 lipid-metabolism genes; 21 ATP-binding and 13 DNA-binding proteins; 11 oxidoreductases; 20 transferases; and 24 hydrolases. We also identified 16 genes that encode components of ubiquitin–ligase complexes, two of which were shown here to extend lifespan when targeted by RNAi; these are related to, but distinct from, genes that had been implicated previously in lifespan regulation (Ghazi et al., 2007; Li et al., 2007; Carrano et al., 2009). Such genes reflect expression patterns peculiar to (or exaggerated in) very long-lived mutant worms, which is consistent with the marked enrichment of potential transcription-factor-binding motifs, immediately upstream of affected genes. At least some of these genetic mechanisms of life extension may have eluded prior discovery because, when fully implemented, they interfere with normal development and propagation.
Experimental procedures
Strains
Nematode strains, supplied by the Caenorhabditis Genetics Center (CGC, Minneapolis, MN, USA) or derived in our laboratory from CGC strains, were maintained at 20 °C throughout their lives and for all experimental procedures. Cohorts were synchronized by alkaline-hypochlorite lysis of parents (sparing eggs/larvae), and propagated on 0.6% peptone NGM-agar plates seeded with E. coli strain OP50, as described (Ebert et al., 1993; Ayyadevara et al., 2001; Shmookler Reis et al., 2007).
Determination of lifespan
Nematodes, grown on NGM-agar plates containing 0.6% peptone (Sulston & Hodgkin, 1988; Ebert et al., 1993; Ayyadevara et al., 2003), were harvested by rinsing off each plate with S buffer (0.1 M NaCl, 0.05 M potassium phosphate, pH 6.0) (Sulston & Hodgkin, 1988; Ebert et al., 1993; Ayyadevara et al., 2003). Adults, recovered by differential settling (Ayyadevara et al., 2008), were then resuspended in alkaline hypochlorite (0.5 N NaOH, 1.05% hypochlorite; 5 min at 20 °C). The recovered eggs (containing uneclosed larvae) were rinsed in S Buffer and placed on fresh agar plates seeded with E. coli strain OP50. Survival cultures were established on 60-mm agar plates 1-day after the L4/adult molt; 25–50 adults were transferred to 60-mm dishes containing NGM agar and a central lawn of E. coli bacteria (strain OP50). Worms were maintained at 20 °C and live worms counted during daily transfer to fresh dishes, as described above. Worms were considered dead if they did not move either spontaneously or in response to touch. Those lost (stranded on dish walls or beneath the agar) or killed by internal hatching of progeny were censored at the midpoint of the time interval in which this occurred; worms inadvertently killed were censored at the time of the event.
Functional assessment of genes by peroxide survival after RNAi treatment
Adult worms (N2DRM or age-1(hx546), as indicated) were synchronized by alkaline hypochlorite lysis of hermaphrodites; surviving embryos were transferred to fresh NGM-agar plates and allowed to mature. On reaching the L4 stage, worms were placed on fresh NGM-agar plates seeded with bacteria expressing double-stranded RNAs from the Ahringer library (Kamath & Ahringer, 2003). After 48 h, they were transferred to 24-well plates (20–25 worms per well) containing S Medium (S buffer plus 0.5% cholesterol) and 5-mM hydrogen peroxide (Sigma, St Louis, MO, USA) at 20 °C, as previously described (Ayyadevara et al., 2008). Survival was scored as above, initially at 1-h intervals, until no worms remained alive. Assays were performed at least twice on each strain.
Microarray analysis of gene expression
Microarrays, 60-mer oligonucleotides printed on epoxy-coated slides, were obtained from The Genome Sequencing Center of Washington University (St Louis). Each array consists of > 22 000 spots representing nearly all predicted and confirmed genes of C. elegans. Nematode cultures were synchronized by alkaline-hypochlorite lysis of gravid hermaphrodites, followed by rinsing and propagation of unlysed eggs or embryos (see Ebert et al., 1993). Cultures were again synchronized by manual selection at the L4 stage, and full-sized adults were separated from runts at the time of harvest, by differential settling (Ayyadevara et al., 2008). Total RNA was prepared (RNEasy; Qiagen, Valencia, CA, USA) from fertile strains at 6–8 days of adult age, or from the F2 generation of strong age-1 alleles at 9–12 adult days, and reverse transcribed into cDNA using Superscript III (Invitrogen, Carlsbad, CA, USA) and oligo-dT26 primer. The cDNAs, labeled indirectly with Cy3 or Cy5 fluors using the Genisphere 900HS dendromer system, were combined in pairs (comprising one Cy3- and one Cy5-labeled cDNA) and hybridized to microarray slides in a HybAid annealing station (The Gel Company, San Francisco, CA, USA). After careful rinsing, slides were read using a ScanArray 5000 laser fluorescence scanner (GSI Lumonics, Bedford, MA, USA). Data pretreatment (spot recognition and background subtraction) utilized GeneSpring (Silicon Genetics/Agilent Technologies, Santa Clara, CA, USA), followed by statistical analysis with SAM [(Tusher et al., 2001), ver. 3.02; Stanford University]. Pathway analysis was conducted in Pathway Studio (ARIADNE, Rockville, MD, USA). Gene ontology analysis employed functional annotation chart and clustering reports, for which WormBase annotations (http://www.wormbase.org) of SAM significant-gene lists were compared by DAVID (Dennis et al., 2003) (ver. 4, 2008; http://david.abcc.ncifcrf.gov), to the full C. elegans gene set. Word patterns enriched in sequences adjoining identified genes were assessed by RSAT (J. van Helden, http://rsat.ulb.ac.be/rsat/), with background defined by an input-based 2nd order Markov model, and by Genomatix software and matrices for established consensus binding motifs (http://www.genomatix.de).
Transcript quantitation by RT-qPCR
Expression of selected genes was assessed by real-time polymerase chain reaction after reverse transcription. Total RNA was purified from each strain cohort (RNeasy; Qiagen), and cDNAs reverse-transcribed (SuperScript III; Invitrogen), followed by RT-qPCR (Opticon2, MJ Research, using SYBR Green, Roche).
Statistical analyses
False-discovery rate was estimated by the ‘q-value’ method of Storey & Tibshirani (2003) within SAM, and by the method of Hochberg and Benjamini (Hochberg & Benjamini, 1990; Klipper- Aurbach et al., 1995) within DAVID. Survivals were compared by log-rank tests (Lee, 1992) using NCSS™ software. Transcript levels were compared by two-tailed, heteroscedastic t-tests (i.e. for unpaired samples of unknown or unequal variance), conducted within Microsoft Excel 2003. The values compared were control-subtracted threshold cycle numbers, across biological replicates (independently generated cohorts), addressing purely inter-strain variation relative to variance among biological replicates. It should be noted that directly comparing values of Ct, the number of cycles required to reach a threshold yield of amplimers, is equivalent to working with the negatives of log-transformed RNA levels (−log2 [RNA]). Actual P-values are given in each case, prior to any Bonferroni adjustment.
Supplementary Material
Acknowledgments
We thank Rajani Ayyadevara for expert technical assistance, and David Gems and Eugene Schuster (University College, London) for kindly providing their gene lists and for helpful suggestions. This work was supported by a grant (to R.J.S.R.) from the U.S. National Institutes of Health, and by a Research Career Scientist Award (R.J.S.R.) from the U.S. Department of Veterans Affairs.
References
- Ayyadevara S, Ayyadevera R, Hou S, Thaden JJ, Shmookler Reis RJ. Genetic mapping of quantitative trait loci governing longevity of Caenorhabditis elegans in recombinant-inbred progeny of a Bergerac-BO x RC301 interstrain cross. Genetics. 2001;157:655–666. doi: 10.1093/genetics/157.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyadevara S, Ayyadevera R, Vertino A, Galecki A, Thaden JJ, Shmookler Reis RJ. Genetic loci modulating fitness and life span in Caenorhabditis elegans: categorical trait interval mapping in CL2a×Bergerac-BO recombinant-inbred worms. Genetics. 2003;163:557–570. doi: 10.1093/genetics/163.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- Carrano AC, Liu Z, Dillin A, Hunter T. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature. 2009;460:396–399. doi: 10.1038/nature08130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert RH, Cherkasova VA, Dennis RA, Wu JH, Ruggles S, Perrin TE, Shmookler Reis RJ. Longevity-determining genes in Caenorhabditis elegans: chromosomal mapping of multiple noninteractive loci. Genetics. 1993;135:1003–1010. doi: 10.1093/genetics/135.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988a;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE. Three mutants that extend both mean and maximum life span of the nematode, Caenorhabditis elegans, define the age-1 gene. J Gerontol. 1988b;43:B102–B109. doi: 10.1093/geronj/43.4.b102. [DOI] [PubMed] [Google Scholar]
- Gems D, Riddle DL. Defining wild-type life span in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2000;55:B215–B219. doi: 10.1093/gerona/55.5.b215. [DOI] [PubMed] [Google Scholar]
- Ghazi A, Henis-Korenblit S, Kenyon C. Regulation of Caenorhabditis elegans lifespan by a proteasomal E3 ligase complex. Proc Natl Acad Sci USA. 2007;104:5947–5952. doi: 10.1073/pnas.0700638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand SL, Blake KJ, Rogina B, Stracks MD, Centurion A, Naprta B. Temporal patterns of gene expression in the antenna of the adult Drosophila melanogaster. Genetics. 1995;140:549–555. doi: 10.1093/genetics/140.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Murphy CT. Enrichment of regulatory motifs upstream of predicted DAF-16 targets. Nat Genet. 2006;38:397–398. doi: 10.1038/ng0406-397. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kim SK. Common aging pathways in worms, flies, mice and humans. J Exp Biol. 2007;210:1607–1612. doi: 10.1242/jeb.004887. [DOI] [PubMed] [Google Scholar]
- Kim Y, Sun H. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell. 2007;6:489–503. doi: 10.1111/j.1474-9726.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- Klass MR. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech Ageing Dev. 1983;22:279–286. doi: 10.1016/0047-6374(83)90082-9. [DOI] [PubMed] [Google Scholar]
- Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M, Benjamini Y, Hochberg Y, Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ET. Statistical Methods for Survival Data Analysis. New York: J. Wiley & Sons Inc; 1992. [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003a;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003b;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Li W, Gao B, Lee SM, Bennett K, Fang D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev Cell. 2007;12:235–246. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thornton J, Gems D. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mech Ageing Dev. 2006;127:922–936. doi: 10.1016/j.mad.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Miller RA, Galecki A, Shmookler Reis RJ. Interpretation, design, and analysis of gene array expression experiments. J Gerontol A Biol Sci Med Sci. 2001;56:B52–B57. doi: 10.1093/gerona/56.2.b52. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Murphy CT. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp Gerontol. 2006;41:910–921. doi: 10.1016/j.exger.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci USA. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Shmookler Reis RJ, Kang P, Ayyadevara S. Quantitative trait loci define genes and pathways underlying genetic variation in longevity. Exp Gerontol. 2007;41:1046–1054. doi: 10.1016/j.exger.2006.06.047. [DOI] [PubMed] [Google Scholar]
- Smith ED, Kennedy BK, Kaeberlein M. Genome-wide identification of conserved longevity genes in yeast and worms. Mech Ageing Dev. 2007;128:106–111. doi: 10.1016/j.mad.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- Tazearslan C, Ayyadevara S, Bharill P, Shmookler Reis RJ. Positive feedback between transcriptional and kinase suppression confers extraordinary longevity and stress resistance. PLoS Genet. 2009;5:e1000452. doi: 10.1371/journal.pgen.1000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Ruvkun G. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics. 1998;148:703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.