Abstract
The non-LTR retrotransposon LINE-1 (L1) comprises ~17% of the human genome, and the L1-encoded proteins can function in trans to mediate the retrotransposition of non-autonomous retrotransposons (i.e., Alu and probably SVA elements) and cellular mRNAs to generate processed pseudogenes. Here, we have examined the effect of APOBEC3G and APOBEC3F, cytidine deaminases that inhibit Vif-deficient HIV-1 replication, on Alu retrotransposition and other L1-mediated retrotransposition processes. We demonstrate that APOBEC3G selectively inhibits Alu retrotransposition in an ORF1p-independent manner. An active cytidine deaminase site is not required for inhibition of Alu retrotransposition and the resultant integration events lack G to A or C to T hypermutation. These data demonstrate a differential restriction of L1 and Alu retrotransposition by APOBEC3G, and suggest that the Alu ribonucleoprotein complex may be targeted by APOBEC3G.
Keywords: LINE-1, Retrotransposon, APOBEC, Transposable element
1. Introduction
The non-LTR retrotransposon LINE-1 (L1) comprises ~17% of the human genome (Lander et al., 2001). Although most L1s are immobile, it is estimated that the average human genome contains approximately 60–100 retrotransposition-competent L1s (RC-L1s) and their mobility can cause human disease (Sassaman et al., 1997, Brouha et al., 2003, Hulme et al., 2006, Kazazian et al., 1988). RC-L1s are composed of a 5’ UTR that harbors an internal promoter, two non-overlapping open reading frames (ORF1 and ORF2), and a 3’ UTR that ends in a poly (A) tail (Swergold, 1990, Scott et al., 1987, Dombroski et al., 1991, Minakami et al., 1992). The ORF2 encoded protein (ORF2p) contains both endonuclease and reverse transcriptase activities, which are required for retrotransposition (Mathias et al., 1991, Feng et al., 1996, Moran et al., 1996). The ORF1 encoded protein (ORF1p) is a nucleic acid binding protein that also is required for retrotransposition (Moran et al., 1996, Holmes et al., 1992, Hohjoh and Singer, 1996). ORF1p and ORF2p exhibit cis-preference, such that after translation they bind back to the transcript from which they were derived to form a L1 ribonucleoprotein particle, which is a proposed intermediate (Kulpa and Moran, 2005, Kulpa and Moran, 2006, Hohjoh and Singer, 1997, Wei et al., 2001).
The L1 encoded proteins also can function in trans to mobilize other RNAs. ORF2p is required for retrotransposition of the non-autonomous retrotransposon Alu, which comprises ~11% of human DNA (Lander et al., 2001, Dewannieux et al., 2003). ORF1p and ORF2p seemingly are required to mobilize cellular mRNAs, leading to the formation of processed pseudogenes (Esnault et al., 2000). Therefore, L1s are responsible for creating at least 30% of human DNA. L1-mediated retrotransposition events also can cause human disease and sometimes can lead to genomic instability (Gilbert et al., 2002, Symer et al., 2002, Hulme et al., 2006, Callinan et al., 2005, Han et al., 2005, Gilbert et al., 2005). Thus, we hypothesize that the cell may have evolved proteins that can inhibit L1 retrotransposition.
Recently, the cellular protein APOBEC3G (A3G) has been identified as part of the intrinsic cellular defense against Vif-deficient HIV-1 infection. Virions produced in cells expressing A3G are less effective at infecting a target cell (Sheehy et al., 2002, Cullen, 2006). A3G is a cytidine deaminase (CDA) that is packaged into the viral particle, where it acts on the nascent minus strand of the viral cDNA to mutate dC residues to dU. This process results in extensive G to A editing of the proviral plus strand DNA and, through the action of DNA repair proteins, may degrade the cDNA and abolish viral replication (Sheehy et al., 2002, Mangeat et al., 2003, Zhang et al., 2003, Harris et al., 2003). A3G contains two CDA sites, one in each half of the protein, of which only the C-terminal site is enzymatically active (Newman et al., 2005, Navarro et al., 2005, Hache et al., 2005, Jarmuz et al., 2002). However, expression of only the C terminal half of A3G (C-A3G) is not sufficient to inhibit Vif-deficient HIV-1 replication (Bogerd et al., 2006b).
A3G is a member of the human APOBEC3 protein family, which has at least 5 members (Cullen, 2006, Jarmuz et al., 2002). APOBEC3B (A3B) and APOBEC3F (A3F) can inhibit Vif-deficient HIV-1 replication, while APOBEC3C (A3C) is only weakly active and APOBEC3A (A3A) has no effect (Cullen, 2006, Bishop et al., 2004, Yu et al., 2004). The APOBEC3 proteins, with the possible exception of A3A, have undergone multiple rounds of positive selection, suggesting that they have functions in addition to inhibition of HIV-1 replication (Sawyer et al., 2004, Zhang and Webb, 2004). Consistent with this notion, APOBEC3 proteins can inhibit retrotransposition of some LTR retrotransposons in mouse and yeast, and A3A and A3B have been shown to inhibit L1 retrotransposition and Alu retrotransposition (Bogerd et al., 2006b, Esnault et al., 2005, Schumacher et al., 2005, Bogerd et al., 2006a, Chen et al., 2006, Muckenfuss et al., 2006, Stenglein and Harris, 2006, Dutko et al., 2005) (Table 1). Here, we have examined the effect of A3G and A3F on Alu retrotransposition and other L1-mediated retrotransposition processes.
Table 1.
Ability of APOBEC3 proteins to inhibit HIV-1 replication and retrotransposition.
| Yeast | Mouse | Human | ||||
|---|---|---|---|---|---|---|
| APOBEC3 | Δvif HIV-1 | Ty1 | IAP | MusD | L1 | Alu |
| A3A | No1,4,5,11 | N.D. | Yes1,4,5 | Yes5 | Yes1,5,6 | Yes1 |
| A3B | Yes1,4,11 Modest7,12 |
N.D. | Yes1,4,5 | Yes5 No9 |
Yes1,5,6,7 | Yes1 |
| A3C | Modest1,4,11,12 | Yes10 | Yes5 Modest4 |
Yes5 No9 |
Yes5,6 Modest1 |
Modest1 |
| A3F | Yes1,4,7,11 | Yes3,10 | Yes5 Modest4 |
Yes5,9 | Yes5,6,7 No1 |
No |
| A3G | Yes1,4,5,7,8,11,12 | Yes3,10 | Yes2,4,5 Modest9 |
Yes2,9 No5 |
No1,2,5,6,7,8 | Yes |
| mA3 | Yes11 | Yes10 | Yes2,5 | Yes2,5,9 | No2,5 | N.D. |
Depicted is the ability of five human APOBEC3 proteins and mouse APOBEC3 (mA3) to inhibitvif-deficient (Δvif) HIV-1 replication, retrotransposition of LTR-retrotransposons (Ty1, IAP, MusD), and retrotransposition of non-LTR retrotransposons (L1, Alu). Species of the retrotransposon is indicated. Yes = inhibition. No = no inhibition. Modest = modest or weak inhibition. N.D. = not determined.
Publication reporting level of inhibition is indicated (1=Bogerd et al., 2006b; 2= Esnault et al., 2005; 3= Schumacher et al., 2005; 4= Bogerd et al., 2006a; 5= Chen et al., 2006; 6= Muckenfuss et al., 2006; 7= Stenglein and Harris, 2006; 8= Turelli et al., 2004; 9= Esnault et al., 2006; 10= Dutko et al., 2005; 11= Bishop et al., 2004; 12= Yu et al., 2004).
2. Methods
2.1 Plasmids
The pK/β-arr control plasmid and expression plasmids for A3G, N-A3G, C-A3G, and A3F have been described previously (Bogerd et al., 2006a, Bogerd et al., 2006b). The pA3GE259Q (A3Gm) expression plasmid was derived from pA3G by recombinant PCR-mediated mutagenesis. This mutation changes a critical active site glutamic acid to glutamine and has been shown to result in a stable A3G protein lacking detectable CDA acitivity (Newman et al., 2005). The following constructs used in the retrotransposition and trans-complementation assays have been described previously: pDK101, SP101/L1.3K7, JM101/L1.3Δneo, pCep5’UTRORF2Δneo, AluneoTet, and ORF1mneoI (Gilbert et al., 2005, Kulpa and Moran, 2005, Alisch et al., 2006, Dewannieux et al., 2003, Wei et al., 2001).
2.2 Retrotransposition and trans-complementation assays
The L1 and Alu retrotransposition assays and the trans-complementation assay have been described previously (Wei et al., 2001, Wei et al., 2000, Dewannieux et al., 2003). For the L1 retrotransposition assay and the trans-complementation assay, we used the HeLa cell line HeLa-JVM. For the Alu retrotransposition assay we used the HeLa cell line, HeLa-HA. Briefly, HeLa cells were plated (L1 retrotransposition assay- 2 × 103 cells/well and 2 × 104 cells/well, Alu retrotransposition assay- 5 × 105 cells/flask, trans-complementation assay- 2 × 106 cells/flask) and then transfected within 24 hours with the appropriate plasmids. Three days post-transfection, cells were selected with G418 for 10–12 days. Colonies were then fixed, stained with crystal violet, and counted. For the L1 retrotransposition assay, colonies were counted separately from 6 wells. For the Alu retrotransposition assay and trans-complementation assay, colonies were counted from two T75 flasks. In parallel, HeLa cells were transfected with the same plasmids and hrGFP. Three days post-transfection cells were subjected to flow cytometry and transfection efficiency was determined based on the number of GFP positive cells. Colony counts from replicate reactions within an experiment were averaged and adjusted for transfection efficiency. Retrotransposition efficiency was calculated by setting the control reaction (pK/β-arr) to 100%. Data is the result of at least two experiments with standard deviation indicated.
2.3 Sequence characterization of retrotransposed Alu elements
HeLa cells were plated at 2 × 105 cells per well and within 24 hours were co-transfected with AluneoTet, pDK101, and an APOBEC protein or control (pK/β-arr). Three days post-transfection, cells were selected with hygromycin for 4 days. DNA was harvested from two wells using the DNeasy tissue kit (Qiagen) to yield independent DNA samples. PCR was performed with primers 437S and 1808AS (Moran et al., 1996). This primer pair amplifies the neo gene across the intron to discriminate Alu sequences integrated into genomic DNA from those in the transfected plasmid. PCR products were purified, cloned, and sequenced. For each reaction, a total of 12–14 sequences were examined for base changes from two independent genomic DNA samples.
437S: 5’-GAGCCCCTGATGCTCTTCGTCC-3’
1808AS: 5’-CATTGAACAAGATGGATTGCACGC-3’
3. Results
3.1 A3G and A3F do not inhibit L1 retrotransposition
In order to test the ability of A3G and A3F to inhibit L1 retrotransposition we used a cultured cell retrotransposition assay, in which the 3’ UTR of a full length L1 is tagged with a retrotransposition indicator cassette (Moran et al., 1996, Wei et al., 2000). This cassette contains a copy of the neomycin phosphotransferase (neo) gene and is in the opposite transcriptional orientation relative to the L1. The neo gene is interrupted by a spliceable intron that is in the same transcriptional orientation as the L1. This arrangement ensures that G418 resistance results only when a transcript arising from the L1 promoter has undergone splicing, reverse transcription, and integration into the genomic DNA (Moran et al., 1996, Wei et al., 2000).
We used two L1 constructs in the cultured cell assay which were engineered from L1.3, a highly active L1 (Dombroski et al., 1993, Sassaman et al., 1997). The first (pDK101) is a wildtype L1 construct (Kulpa and Moran, 2005). The second (SP101/L1.3K7) is a wildtype L1 construct that contains the ColE1 origin of replication immediately 3’ of the neo cassette, resulting in ~900 bases of additional sequence that must be reverse transcribed before the neo gene during retrotransposition (Gilbert et al., 2005). As the mechanism by which APOBEC3 proteins affect retrotransposition is unknown, SP101/L1.3K7 provided a larger sequence window to see the effect of APOBEC3 proteins that may act during reverse transcription and/or integration of the newly retrotransposed L1.
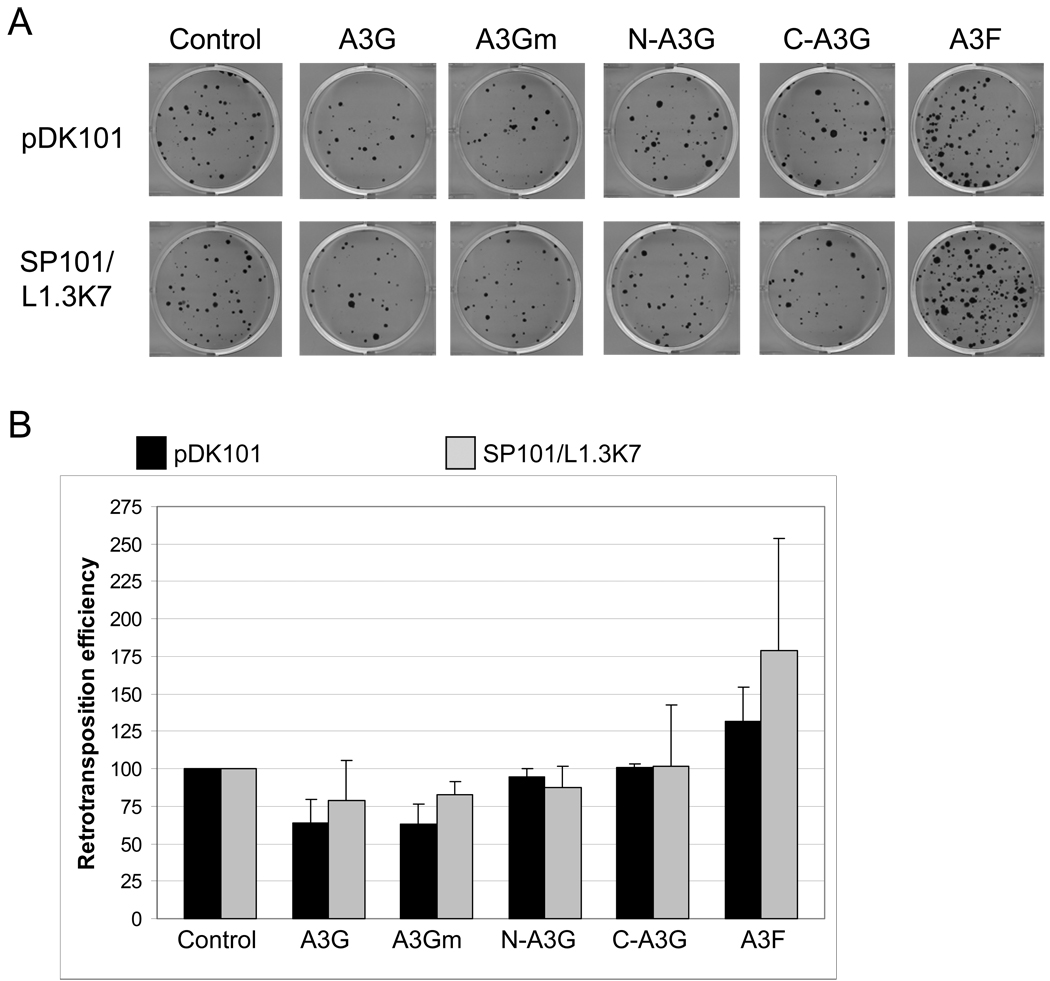
We co-transfected HeLa-JVM cells with a tagged L1 construct and either an APOBEC3 expression plasmid or a control plasmid expressing β-arrestin (pK/β-arr) that does not significantly affect L1 mediated retrotransposition (Bogerd et al., 2006b) (Fig. 1a). Neither A3G nor A3F affected the ability of either pDK101 or SP101/L1.3K7 to retrotranspose in cultured cells (Fig. 1b). A CDA active site mutant of A3G (A3GE259Q or A3Gm) which has been previously shown to produce a stable protein lacking CDA activity, and the N and C terminal halves of A3G (N-A3G and C-A3G) also did not affect L1 retrotransposition (Newman et al., 2005). Thus, consistent with previously published studies, we conclude that A3F and A3G do not significantly inhibit L1 retrotransposition (Turelli et al., 2004, Esnault et al., 2005, Bogerd et al., 2006b).
Fig. 1.
A3G and A3F do not inhibit L1 retrotransposition. HeLa-JVM cells were co-transfected with an L1 construct tagged with the retrotransposition indicator cassette and an APOBEC expression plasmid or control (pK/β-arr). Two different L1 constructs were used, a wildtype L1 (pDK101) and an L1 that contains the ColE1 origin of replication 3’ of the neomycin resistance cassette (SP101/L1.3K7). Three days post transfection cells were subjected to G418 for 12 days to select for retrotransposition events. (A) Representative results for the effect of A3G and A3F on L1 retrotransposition. Colonies were stained with crystal violet to visualize retrotransposition events. (B) For each experiment, colonies were counted from six wells and retrotransposition efficiency was determined relative to the control (pK/β-arr), which was set at 100%. Data are the average of at least two independent experiments, with standard deviation indicated. For A3G and A3F, similar results were observed in HeLa cell lines that stably expressed the A3G and A3F proteins (data not shown).
3.2 A3G inhibits Alu retrotransposition
A3A and A3B recently have been shown to inhibit both L1 and Alu retrotransposition, therefore we examined whether A3G and A3F would also affect Alu retrotransposition (Bogerd et al., 2006b, Chen et al., 2006, Muckenfuss et al., 2006, Stenglein and Harris, 2006). We co-transfected HeLa-HA cells with an Alu element tagged with the retrotransposition indicator cassette, an L1 expression construct, and either an APOBEC3 expression plasmid or a control plasmid (Fig. 2a). We used two L1 expression constructs, a wildtype L1 (JM101/L1.3Δneo) or an L1 construct that lacks the ORF1 coding sequence (ORF2Δneo).
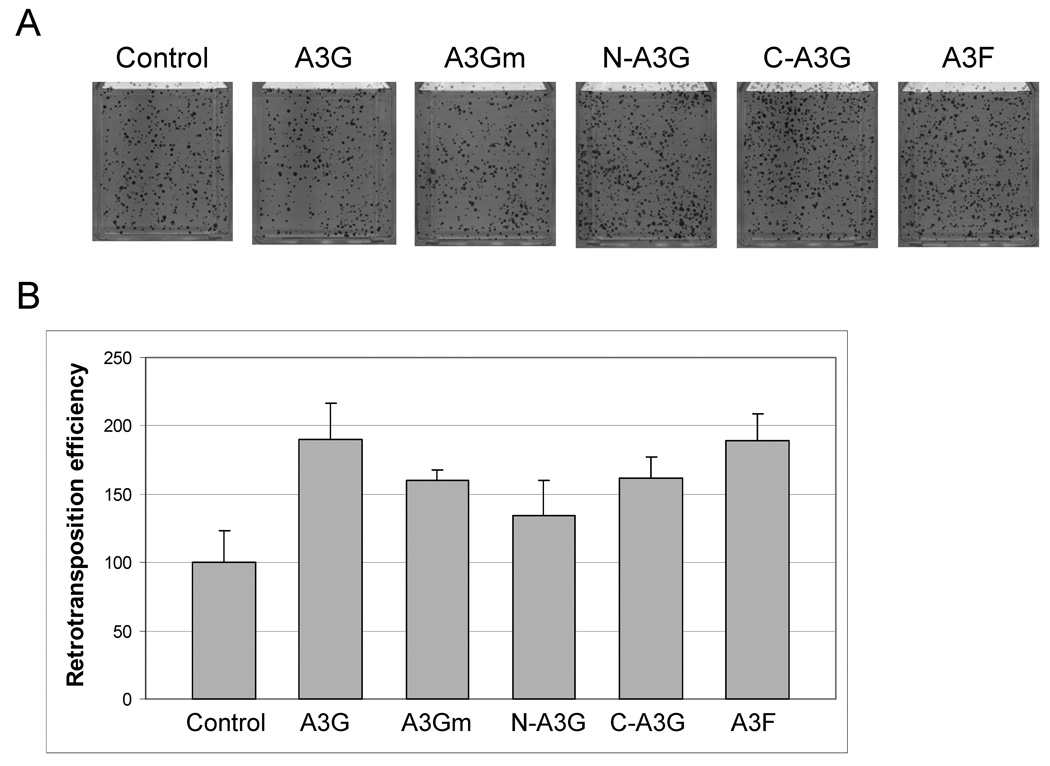
Fig. 2.
A3G inhibits Alu retrotransposition. HeLa-HA cells were co-transfected with an Alu construct tagged with the retrotransposition indicator cassette, an L1 construct, and an APOBEC protein or control (pK/β-arr). We used two different L1 constructs that lack the retrotransposition indicator cassette, a wildtype L1 (JM101/L1.3Δneo) and a L1 that lacks ORF1 coding sequence (ORF2Δneo). Three days post-transfection, cells were subjected to G418 for 12 days to select for retrotransposition events. (A) Representative results for the effect of A3G and A3F on Alu retrotransposition. (B) For each experiment, colonies were counted from two flasks and retrotransposition efficiency was determined relative to the control (pK/β-arr), which was set at 100%. Data are the average of at least two independent experiments, with standard deviation 20 indicated. For A3G, the retrotransposition frequency range was 5–23% (JM101/L1.3Δneo) in 5 independent experiments and 11–34% (ORF2Δneo) in 4 independent experiments as one experiment was removed due to low transfection efficiency. For A3Gm, the retrotransposition frequency range was 11–31% (JM101/L1.3Δneo) and 13–35% (ORF2Δneo) in 4 independent experiments.
A3G inhibited Alu retrotransposition to ~15% that of wildtype levels (Fig. 2b). C-A3G and N-A3G did not decrease Alu retrotransposition, so inhibition by A3G can not be attributed to only the N or C terminal halves of the protein. The failure of A3F and C-A3G to significantly inhibit Alu retrotransposition indicates that simply expressing an active CDA site is not sufficient to mediate the decrease in Alu retrotransposition. Consistent with this data, A3Gm showed a similar decrease in Alu retrotransposition to that seen with wildtype A3G. Finally, inhibition of Alu retrotransposition is ORF1-independent, as similar decreases in retrotransposition were observed with both the full length L1 and the ORF2 only construct (ORF2Δneo). Therefore, we conclude that A3G inhibits Alu retrotransposition in an ORF1p-independent manner and that an active CDA site is neither necessary nor sufficient for this inhibition.
3.3 A3G does not induce detectable hypermutation of retrotransposed Alu elements
We next examined the newly integrated Alu elements for hypermutation (Fig. 3). We did not observe editing of Alu elements that retrotransposed in the presence of A3G, A3Gm, or A3F. These results agree with previous experiments examining the effect of APOBEC3 proteins on L1 and Alu retrotransposition (Bogerd et al., 2006b, Turelli et al., 2004, Stenglein and Harris, 2006, Muckenfuss et al., 2006, Esnault et al., 2005). While it is possible that edited transcripts are rapidly degraded, the fact that an active CDA site is not required for restriction of retrotransposition (Fig. 2) implies that A3G inhibits Alu retrotransposition independent of hypermutation.
Fig. 3.
A3G, A3Gm, and A3F do not induce detectable hypermutation of retrotransposed Alu elements. HeLa-HA cells were transfected as described in Figure 2. As an additional control, cells were co-transfected with only an Alu construct and an L1 construct (No APOBEC). After four days of hygromycin selection, DNA was harvested from two wells to yield independent DNA samples. PCR was then performed to amplify the spliced neo gene from integrated Alu elements. PCR products were purified, cloned, and sequenced. For each reaction, a total of 12–14 sequences were examined for base changes from two independent genomic DNA samples. As seen previously, we sometimes detected the simultaneous mutation of three nucleotides (two C-to-T changes and one A-to-C change) surrounding the splice site present in the neo indicator cassette in controls or in cells transfected with an APOBEC3 protein (Bogerd et al., 2006b). These mutations have been removed from the data presented.
3.4 A3G does not inhibit ORF1p-mediated processed pseudogene formation
We have shown that A3G inhibits Alu retrotransposition, but not L1 retrotransposition. One of the differences between these two processes is that ORF1p is not required for Alu retrotransposition (Dewannieux et al., 2003). In order to test if simply providing ORF2p in trans was sufficient to mediate a decrease in retrotransposition, we conducted a trans-complementation assay (Wei et al., 2001, Alisch et al., 2006). In this assay, HeLa-JVM cells were transfected with two L1 constructs. The driver construct provides a source of ORF2p that can be used in trans to mediate retrotransposition of the reporter construct. As the driver, we used an L1 construct that lacks the ORF1 coding sequence and is not tagged with the retrotransposition indicator cassette (ORF2Δneo). For the reporter construct, we used an L1 construct that lacks any ORF2 coding sequence and contains the retrotransposition indicator cassette (ORF1mneoI). For this assay, we drove retrotransposition of ORF1mneoI with ORF2Δneo in the presence of either APOBEC3 proteins or a control protein.
Although A3G and A3Gm inhibit Alu retrotransposition (Fig. 2), they do not inhibit ORF1mneoI retrotransposition (Fig. 4). N-A3G, C-A3G, and A3F also did not affect ORF1mneoI retrotransposition, consistent with Alu and L1 retrotransposition data. These results indicate that A3G does not interfere with Alu retrotransposition by inhibiting ORF2p, and suggest that it may target a component of the Alu retrotransposition machinery that is not required for L1 retrotransposition.
Fig. 4.
A3G and A3F do not inhibit ORF1mneoI retrotransposition. HeLa-JVM cells were cotransfected with an L1 construct tagged with the retrotransposition indicator cassette that lacks the ORF2 coding sequence (ORF1mneoI), an L1 construct (pCep5’UTRORF2Δneo), and an APOBEC protein or control (pK/β-arr). Three days post-transfection, cells were subjected to G418 to select for retrotransposition events. (A) Representative results for the effect of A3G and A3F on ORF1mneoI retrotransposition. (B) For each experiment, colonies were counted from two flasks and retrotransposition efficiency was determined relative to the control (pK/β-arr), which was set at 100%. Data are the average of two independent experiments, with standard deviation indicated.
4. Discussion
We have presented data indicating that A3G selectively inhibits Alu retrotransposition, but not other L1-mediated retrotransposition processes. These data provide an example of a differential inhibition of retrotransposition, as A3A and A3B inhibit both Alu and L1 retrotransposition (Bogerd et al., 2006b, Stenglein and Harris, 2006, Muckenfuss et al., 2006, Chen et al., 2006). These results suggest that A3A, A3B, and A3G may inhibit Alu retrotransposition by different mechanisms or at different stages of the retrotransposition cycle. This hypothesis is supported by subcellular localization data, as both A3A and A3B localize to the nucleus where reverse transcription and integration occurs, while A3G is a cytoplasmic protein (Mangeat et al., 2003, Bogerd et al., 2006b, Stenglein and Harris, 2006, Muckenfuss et al., 2006, Chen et al., 2006).
Recently, three studies have shown that A3F can inhibit L1 retrotransposition to ~20% or ~35% of wildtype (Stenglein and Harris, 2006, Muckenfuss et al., 2006, Chen et al., 2006) (Table 1). Notably, the A3F protein used in those experiments differs at two amino acid residues (aa108 and aa231) from the A3F protein used here and in Bogerd et al., 2006b. While variations in the retrotransposition assay used in these studies could account for these differences, it also is possible that the amino acid changes are responsible for the differing action of A3F. Further experiments are required to determine if these residues do in fact affect the ability of A3F to inhibit L1 retrotransposition.
The data indicate that A3G-mediated inhibition of Alu retrotransposition does not require an active CDA site (Fig. 2), which is consistent with the lack of hypermutation observed in newly integrated Alu elements (Fig. 3). The data further suggest that A3G inhibits Alu retrotransposition in an ORF1p-independent manner, which is consistent with the finding that ORF1p is not required for Alu retrotransposition (Dewannieux et al., 2003). Finally, it has been speculated that ORF2p may directly interact with A3B and A3F to inhibit L1 retrotransposition (Stenglein and Harris, 2006). Our experiments on ORF1p-mediated processed pseudogene formation indicate that A3G does not interfere with Alu retrotransposition by inhibiting ORF2p (Fig. 4). Thus, components of the Alu RNP that are not required for L1 retrotransposition may be targeted by A3G.
The APOBEC3 family has expanded from one gene in the mouse to as many as seven genes in primates (Sawyer et al., 2004, Zhang and Webb, 2004). Alu elements demonstrated a burst of activity around 40 million years ago, followed by a steady decline such that the majority of elements had retrotransposed by 30 million years ago (Lander et al., 2001, Britten, 1994). Although the effect of A3G on Alu retrotransposition has not been assayed in a whole animal model, it remains possible that A3G may have evolved, in part, to curtail Alu retrotransposition.
Acknowledgements
We thank Michael Malim for reagents and Astrid Roy-Engel for the HeLa-HA cell line. We would also like to thank lab members for helpful discussion. This research was funded by National Institute of Health grants GM60518 (to J.V.M.) and AI65301 (to B.R.C.). A.E.H was supported in part by a Michigan Predoctoral Training Grant from the NIH (5T32GM07544).
Abbreviations
- A
adenosine
- APOBEC3
Apolipoprotein B mRNA catalytic editing peptide 3
- CDA
cytidine deaminase
- dC
deoxycytidine
- dU
deoxyuridine
- G
guanine
- G418
geneticin
- HIV-1
human immunodeficiency virus 1
- IAP
intracisternal A particle
- LINE-1 or L1
long interspersed element 1
- Neo
neomycin phosphotransferase
- non-LTR
non-long terminal repeat
- ORF
open reading frame
- RC-L1
retrotransposition competent L1
- RNP
ribonucleoprotein particle
- SVA
short interspersed element R-VNTR (variable nucleotide tandem repeat)-Alu
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006a;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006b;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ. Evidence that most human Alu sequences were inserted in a process that ceased about 30 million years ago. Proc Natl Acad Sci U S A. 1994;91:6148–6150. doi: 10.1073/pnas.91.13.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, Wang J, Herke SW, Garber RK, Liang P, Batzer MA. Alu retrotransposition-mediated deletion. J Mol Biol. 2005;348:791–800. doi: 10.1016/j.jmb.2005.02.043. [DOI] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J Virol. 2006;80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Scott AF, Kazazian HH., Jr Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc Natl Acad Sci U S A. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko JA, Schafer A, Kenny AE, Cullen BR, Curcio MJ. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr Biol. 2005;15:661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of l1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- Han K, Sen SK, Wang J, Callinan PA, Lee J, Cordaux R, Liang P, Batzer MA. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 2005;33:4040–4052. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. Embo J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J Mol Biol. 1997;271:7–12. doi: 10.1006/jmbi.1997.1159. [DOI] [PubMed] [Google Scholar]
- Holmes SE, Singer MF, Swergold GD. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem. 1992;267:19765–19768. [PubMed] [Google Scholar]
- Hulme AE, Kulpa DA, Garcia-Perez JL, Moran JV. The Impact of LINE-1 Retrotransposition on the Human Genome. In: Lupski J, Stankiewicz P, editors. Genomic Disorders: The Genomic Basis of Disease. Totowa, NJ: Humana Press; 2006. [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nature Structural and Molecular Biology. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Minakami R, Kurose K, Etoh K, Furuhata Y, Hattori M, Sakaki Y. Identification of an internal cis-element essential for the human L1 transcription and a nuclear factor(s) binding to the element. Nucleic Acids Res. 1992;20:3139–3145. doi: 10.1093/nar/20.12.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, Deberardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006 doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, Deberardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Nissley DV, Harris RS. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc Natl Acad Sci U S A. 2005;102:9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1:113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F Inhibit L1 Retrotransposition by a DNA Deamination-independent Mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- Turelli P, Vianin S, Trono D. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J Biol Chem. 2004;279:43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- Wei W, Morrish TA, Alisch RS, Moran JV. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1438. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum Mol Genet. 2004;13:1785–1791. doi: 10.1093/hmg/ddh183. [DOI] [PubMed] [Google Scholar]