Abstract
Nur77 is a nuclear orphan steroid receptor that has been implicated in negative selection. Expression of Nur77 in thymocytes and cell lines leads to apoptosis through a mechanism that remains unclear. In some cell lines, Nur77 was reported to act through a transcription-independent mechanism involving translocation to mitochondria, leading to cytochrome c release. However, we show here that Nur77-mediated apoptosis in thymocytes does not involve cytoplasmic cytochrome c release and cannot be rescued by Bcl-2. Microarray analysis shows that Nur77 induces many genes, including two novel genes (NDG1, NDG2) and known apoptotic genes FasL and TRAIL. Characterization of NDG1 and NDG2 indicates that NDG1 initiates a novel apoptotic pathway in a Bcl-2-independent manner. Thus Nur77-mediated apoptosis in T cells involves Bcl-2 independent transcriptional activation of several known and novel apoptotic pathways.
Keywords: Bcl-2/microarray/nuclear orphan steroid receptor/Nur77/TRAIL
Introduction
Central tolerance in the developing immune system of a mammalian organism is established through the removal of potentially autoreactive lymphocytes. For T cells, autoreactive cells are cleared by negative selection. The mechanism of apoptosis during negative selection is largely unknown, although molecules like PTEN, a tumor suppressor, and Bim, a BH3 pro-apoptotic Bcl-2 family member, have been implicated in playing a role in the process(Sohn et al., 2003). Nur77 (TR3, NGFI-B), a member of the orphan steroid receptor family which consists of Nor1 and Nurr1, has also been shown to play a role in negative selection (reviewed by Winoto and Littman, 2002; Sohn et al., 2003). Nur77, originally identified as an immediate-early gene in fibroblasts treated with serum is rapidly upregulated in response to T-cell receptor (TCR) and CD28 stimulation in CD4+CD8+ double-positive (DP) thymocytes. Constitutive expression of either Nur77 or Nor1 in thymocytes leads to massive apoptosis (Cheng et al., 1997). While mice lacking individual genes exhibit no obvious thymic defect (Lee et al., 1995; Ponnio et al., 2002), overexpression of a dominant-negative Nur77 protein, which blocks the activity of all family members, can inhibit apoptosis associated with negative selection.
Consistent with its apoptotic activity in thymocytes, overexpression of Nur77 in some cell lines leads to apoptosis. This apoptotic activity of Nur77 can be altered through phosphorylation by the kinase Akt (Masuyama et al., 2001). Akt has been shown to phosphorylate Ser-350 in the DNA binding domain of Nur77 and substantially diminishes its ability to bind DNA and apoptotic activity (Pekarsky et al., 2001). Thus the ability of Akt to inhibit Nur77-mediated apoptosis correlates with its ability to inhibit the transcriptional activation of Nur77. Studies performed with transgenic animals are also consistent with this idea, where an N-terminal truncation of the transcription activation domain abolishes Nur77-dependent apoptosis (Calnan et al., 1995). In contrast, studies in a human prostate cancer cell line (LNCaP) suggested that Nur77-mediated apoptosis involves transcription-independent mitochondrial targeting of Nur77 (Li et al., 2000). This mitochondrial targeting of Nur77 coincided with the release of cytochrome c from mitochondria and apoptosis. However, no such change in subcellular distribution of Nur77 has been found in thymocytes undergoing apoptosis. In T-cell hybridomas, Nur77 is mainly localized in the nucleus (A.Rajpal, unpublished data) and the inability of Bcl-2 to antagonize Nur77-mediated apoptosis in DP cells (see Figure 1) suggests that the canonical mitochondrial apoptosis pathway is not the major downstream pathway of Nur77 in these cells. The correlation between transcriptional function of Nur77 and its apoptotic activity is further illustrated by previous characterization of transgenic mice expressing C-terminal truncation variants of Nur77 in the thymus (Kuang et al., 1999). Deleting 65 amino acids from the C-terminus of Nur77 reduced its transcriptional ability by 81% and abolished its ability to initiate apoptosis in thymocytes. Surprisingly, removal of another 122 amino acids increased Nur77 transcriptional activity 4-fold over wild type. Expression of this truncated Nur77 protein in transgenic mice increased apoptosis and the severity of the thymic phenotype (Kuang et al., 1999). Therefore we believe that identification and characterization of downstream transcriptional targets of Nur77 would be instrumental in elucidating its apoptotic function.
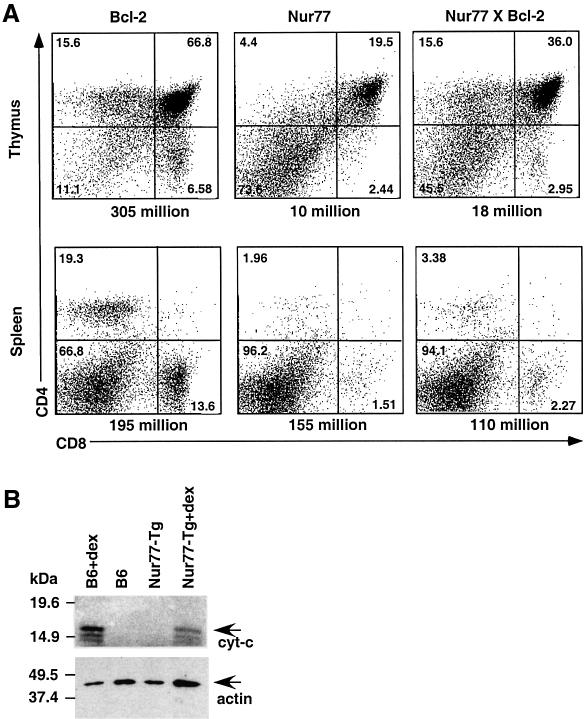
Fig. 1. Nur77-mediated apoptosis in T cells is mitochondria independent. (A) Representative flow cytometric analysis of thymocytes and splenocytes of 5-week-old Bcl-2, Nur77 and Nur77 × Bcl-2 transgenic mice. The cell numbers of the corresponding organs are indicated below each chart. (B) Cytoplasmic fractions of Nur77 transgenic (Nur77-Tg) and C57BL/6 (B6) non-transgenic thymocytes were isolated and blotted with anti-cytochrome c antibodies. Extracts from identical number of thymocytes were loaded. For positive controls, thymocytes were incubated with dexamethasone (dex) for 9 h. The cytochrome c reactive bands are denoted by cyt-c. This experiment was repeated several times with similar results.
Two possible downstream targets of Nur77 have been assessed in the past. The expression of both CD30 and FasL were shown to be elevated in Nur77 transgenic mice (Nur77 under expression of the T-cell-specific lck proximal promoter) (Weih et al., 1996; Zhang et al., 1999). However, Nur77-mediated apoptosis is intact in mice carrying the gld/gld FasL mutation or CD30 null mutation (Chan et al., 1998; Zhang et al., 1999). To examine globally the targets of Nur77 transcription that regulate apoptosis in thymocytes, we conducted differential gene expression experiments with RNA isolated from Nur77 transgenic and wild-type fetal thymi with cDNA microarrays. Several candidate genes were identified. These include known apoptotic genes, negative regulators of costimulation, transcription factors and two novel genes. These novel genes, which we termed NDG1 and NDG2 (for Nur77 Downstream Gene 1 and 2), were characterized in more detail by transgenic and overexpression studies. We show here that NDG1 encodes a novel protein possessing apoptotic activity. Thus Nur77 can activate transcription of several known and novel apoptotic pathways.
Results
The microarray experiments
To see if the mitochondria pathway is involved in Nur77-mediated apoptosis in thymocytes, we crossed the Bcl-2 transgenic mice (Linette et al., 1994) under the control of the lck proximal promoter with the Nur77 full-length transgenic mice (Calnan et al., 1995). Overexpression of Bcl-2 is known to inhibit cytochrome c release from mitochondria, and in T cells its overexpression leads to increased thymocyte cell number, accelerated positive selection and protection against spontaneous thymocyte apoptosis as well as apoptosis by a variety of agents (Linette et al., 1994). In contrast, constitutive Nur77 expression leads to massive cell death (Calnan et al., 1995). We analyzed the thymus and spleen cellularities as well as CD4 and CD8 profile of Nur77 and Nur77 × Bcl-2 mice. As reported previously (Calnan et al., 1995; Chan et al., 1998), constitutive expression of Nur77 leads to a dramatic decrease of their thymocyte cellularities (11.40 ± 0.98 × 106, n = 11; normal thymocyte cell number is ∼200 × 106) and a lower percentage of DP cells. Analysis of Bcl-2/Nur77 double transgenic thymocytes (n = 5) showed no significant increase in their thymocyte cellularities (21.75 ± 4.70 × 106) when compared with their Nur77 transgenic littermates. While flow cytometric analysis with CD4 and CD8 antibodies showed a modest increase in the percentages of DP and SP (single positive: CD4+CD8– and CD4–CD8+) thymocytes when Bcl-2 was present, the number of mature splenic T cells remained low (Figure 1A). To see if cytochrome c is released into cytoplasm during Nur77-mediated cell death, thymocytes from Nur77 transgenic mice were isolated and fractionated into cytoplasmic and mitochondrial fractions. Western blot analysis using cytochrome-c-specific antibodies was then carried out. As shown in Figure 1B, no cytoplasmic cytochrome c could be detected in apoptotic Nur77 transgenic thymocytes. In contrast, dexamethasone treatment of either wild-type or transgenic thymocytes led to cytochrome c release into the cytoplasm as expected (Figure 1B). We concluded that the major pathway of Nur77-mediated apoptosis in T cells is independent of the mitochondrial/Bcl-2 pathway and most likely involves transcriptional activation of downstream genes.
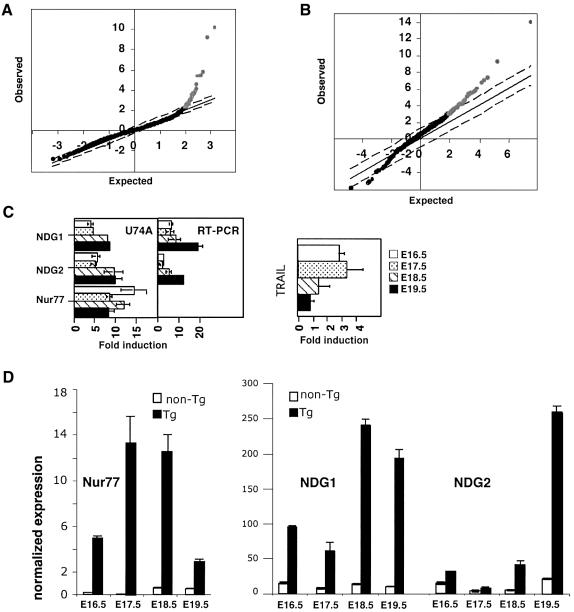
In order to determine potential downstream transcriptional targets of Nur77, we isolated total RNA from thymocytes of Nur77 transgenic and non-transgenic animals and investigated the differential expression of genes by microarray analysis. We used fetal instead of adult thymocytes, as thymocyte compositions are indistinguishable between the transgenic and wild-type mice at the fetal stage. αβ T-cell development starts in the thymus at E16.5 of gestation (Teh et al., 1990). In the past, we have shown that Nur77-mediated apoptosis in the transgenic thymocytes can be seen as early as E17.5 of gestation (Kuang et al., 1999) but apoptosis most likely starts 1 day earlier as the proximal lck promoter is already active at that time (Allen et al., 1992a). We isolated thymocytes from E16.5, E17.5, E18.5 and E19.5 embryos. Total RNA isolated from pools of transgenic and non-transgenic thymocytes was hybridized against the mouse U74A Affymetrix oligonucleotide or the custom UCB/UCSF cDNA microarrays. The experiments were conducted multiple times (Affymetrix eight times, twice for each fetal time point; UCB/UCSF four times, duplicates with dye swaps for E19.5) and the final datasets were analyzed by the statistics program SAM (Figure 2A and B) (Tusher et al., 2001). We found 38 significantly upregulated genes in the Nur77 transgenic thymocytes using the Affymetrix array, of which 20 were induced >2-fold. The custom microarray yielded 97 upregulated genes, of which 28 were induced >2-fold and four were repressed genes (for details see Supplementary figures 2 and 3 available at The EMBO Journal Online).
Fig. 2. Nur77 downstream genes. (A) A SAM plot of the differential expression data obtained from the U74A Affymetrix microarray hybridizations. The genes upregulated at significant levels are shown in gray and the remaining unperturbed genes are shown in black. (B) A SAM plot depicting the analysis of the data obtained with the UCB/UCSF cDNA microarray. (C) A comparison between the U74A hybridization and quantitative RT–PCR analysis of several differentially expressed genes, including quantitative RT–PCR analysis of TRAIL expression levels. Fold induction denotes the ratio of the expression levels between Nur77 transgenic and non-transgenic thymocytes. (D) Quantitative RT–PCR analysis of Nur77, NDG1 and NDG2 between Nur77 transgenic and non-transgenic thymocytes from different days of gestation. The expression is normalized to the actin controls.
A functional classification of some of the upregulated genes from both analyses are shown in Table I. As expected, Nur77 was among the most highly elevated genes and acted as a positive control in these experiments. As reported previously, FasL is also upregulated in thymocytes constitutively expressing Nur77. However, as discussed above, Nur77-mediated apoptosis is indistinguishable between gld/+ and gld/gld mice, suggesting that FasL is not the essential apoptotic gene downstream of Nur77 (Chan et al., 1998). The transcript level for another death receptor ligand, TRAIL, is moderately increased in Nur77 transgenic thymocytes. TGF-β3, which has been implicated in the modulation of cell death in the retina of developing chick embryos and murine intestinal mucosa (Dunker et al., 2001, 2002), was elevated 3.3-fold in the Nur77 transgenic thymocytes. However, there is no direct evidence to suggest that TGF-β3 is involved in apoptosis of developing thymocytes. Other known genes upregulated in Nur77 transgenic thymocytes include negative regulators of T-cell activation CTLA-4, PD-1, TGF-α, transcription factors and adaptor (Grb-2 like gene) molecules. Both CTLA-4 and PD-1 are important costimulatory molecules required for the generation of T-cell tolerance. Strikingly, two ESTs with unknown functions (AA636966 and AW125480, henceforth termed NDG1 and NDG2) are among the most highly elevated genes (5.7- and 7.7-fold). They are upregulated in transgenic fetal thymocytes from E16.5, E17.5, E18.5 and E19.5 of gestation (Figure 2C) and hence are good candidates for possible involvement in Nur77-dependent apoptosis. For that reason, we chose to characterize them further.
Table I. Genes upregulated by Nur77.
| Functional classification | Gene name | Fold inductiona |
|---|---|---|
| Apoptosis | Nur77 (control) | 9.2, 15.9 |
| EST AA636966 | 5.7 | |
| FasL | 3.0 | |
| TGF-β3 | 3.3 | |
| TRAIL | 1.9 | |
| Negative regulators ofT-cell activation | CTLA-4PD-1 | 2.62.6 |
| Growth and transcription | TGF-α | 26 |
| factors | Run×2 | 4.2 |
| IL-7R | 3.0, 2.3 | |
| Schlafen 3 | 3.0 | |
| TCF-4 | 3.3 | |
| COUP-TFII | 2.9 | |
| Thyroid hormone receptor | 2.3 | |
| Others | EST AW125480 | 7.7 |
| Grb2 like | 6.0 |
aValues in italics were obtained from the Affymetrix U74A hybridizations. The numbers represent fold induction of the corresponding gene from E16.5–19.5 thymocytes for the Affymetrix data or from E19.5 thymocytes for the custom microarray data.
Verification of microarray data by quantitative RT–PCR
Quantitative RT–PCR was performed to verify the differential levels of the two ESTs identified by the microarray analyses. To a first approximation, the RT–PCR data corroborate those obtained by the differential expression studies. The induction levels for NDG1 are already high at E16.5, while NDG2 is expressed highly only at E19.5 (Figure 2D). As the Nur77 level is already high at E16.5, this suggest a lag in the kinetics of Nur77-dependent expression of NDG2. Interestingly, the expression levels of TRAIL are high at days 16.5 and 17.5 but reduce over time (Figure 2C). Expression levels of PD-1 and NDG2 were also confirmed by northern blot analysis (data not shown).
Bioinformatics and gene cloning
The full-length cDNA sequences for the genes described by the ESTs were identified by aligning overlapping EST sequences and walking towards the 5′ and 3′ ends of the genes. This was repeated until no new EST could be obtained from the database. An open reading frame was found for each EST (166 amino acids for NDG1 and 138 amino acids for AW125480, henceforth termed NDG2). (The full-length DNA sequences of NDG1 and NDG2 have been submitted to DDBJ/EMBL/GenBank as AV238603 and AV238604, respectively.) The sequences obtained by this analysis were subsequently confirmed using the PHRAP program (Gordon et al., 1998) to assemble all the ESTs under the corresponding NCBI UniGene (AA636966 is UniGene Mm.26006 and AW125480 is UniGene Mm.30114). These gene sequences were then used to obtain IMAGE clones containing the coding sequences.
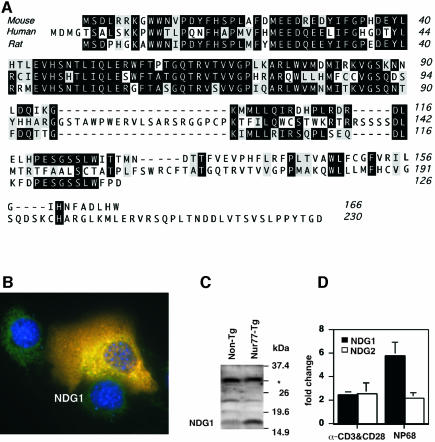
The amino acid sequence for the putative NDG1 protein was used to search protein databases, which revealed several murine and human isoforms. A ClustalW alignment of the polypeptide sequences for the murine protein used in this study, a human and a rat ortholog are shown in Figure 3A. Utilizing a Hidden Markov based PFAM (Bateman et al., 2002) database search with the polypeptide sequence, we were unable to detect any known protein domains in the NDG1 protein other than a putative transmembrane segment predicted by the program TMHMM (Krogh et al., 2001) between residues 136 and 158. However, while there is an approximately 40% similarity between the murine and human polypeptide sequences in this region, this domain is absent from the rat protein (see Figure 3A). Immunofluorescence analysis of NDG1-transfected NIH3T3 cells showed that NDG1 is a cytoplasmic protein and does not localize to the plasma membrane (Figure 3B).
Fig. 3. NDG1 amino acid alignments, protein expression levels and TCR-mediated induction. (A) A ClustalW alignment of the polypeptide sequences for murine, human and rat NDG1 orthologs. Identical residues are heavily shaded and similar residues are lightly shaded. (B) Subcellular localization of NDG1. NIH3T3 cells were transfected with NDG1 expression construct and stained with NDG1-specific antibodies (red). At the same time, DAPI staining for nuclear (blue) and HSP60 staining for mitochondria (green) in different colors were also performed. (C) Western blot depicting the NDG1 protein expression levels in Nur77 transgenic animals (Nur77-Tg) and littermate cohorts (Non-Tg). The asterisk indicates a non-specific band that serves as a loading control. (D) Upregulation of NDG1 and NDG2 transcript in wild-type thymocytes in response to stimulation by α-CD3 and α-CD28 antibodies in vitro and negatively selecting peptide NP68 injections into F5-TCR transgenic mice. The data were obtained by quantitative RT–PCR and normalized with levels of γ-actin in each sample and shown as fold over PBS control for each condition.
The NDG2 protein has human, yeast (Saccharomyces cerevisae), fly (Drosophila melanogaster) and worm (Caenorhabditis elegans) orthologs (Supplementary figure 4). Unlike NDG1, a PFAM database search using the NCBI conserved domains BLAST yielded a protein domain of unknown function (DUF657) that contains a central GXXXGH motif and four conserved C-terminal cysteines that may be involved in divalent metal binding. A search using MITOPROT (Scharfe et al., 1999) revealed that the N-terminus of the protein contains a consensus mitochondrial signal sequence with a putative cleavage site at position 89. Indeed, expression of the protein in NIH3T3 cells showed mitochondrial localization of the NDG2 molecule (Supplementary figure 5).
NDG1 and NDG2 expression is inducible by T-cell receptor activation
Western blotting of lysates obtained from thymocytes of Nur77 transgenic animals and littermate controls reveals a significantly higher amount of the NDG1 protein in the transgenic cells (Figure 3C). The 17 kDa size of the endogenous protein is in agreement with the computed molecular weight determined by its amino acid sequence. Quantitative RT–PCR analyses of RNA from neonatal thymocytes treated with α-CD3 and α-CD28 antibodies for 13 h in vitro indicate a slight but statistically significant increase of NDG1 and NDG2 transcripts. To assess the induction in vivo, F5-TCR transgenic animals (Mamalaki et al., 1992) were injected with either control buffer [phosphate-buffered saline (PBS)] or the F5 peptide NP68. NDG1 showed a 6-fold increase in NP68-stimulated thymocytes while NDG2 exhibited only a 2-fold upregulation (Figure 3D). The upregulation of NDG1 and NDG2 expression levels was detectable after 4 h of α-CD3 and α-CD28 stimulation (data not shown). This follows Nur77 expression, which is detected within an hour of TCR stimulation. The delay in kinetics of upregulation of the NDG1 and NDG2 transcript levels relative to those of Nur77 mRNA is consistent with the notion that they represent downstream targets of Nur77.
Generation and characterization of NDG1 and NDG2 transgenic mice
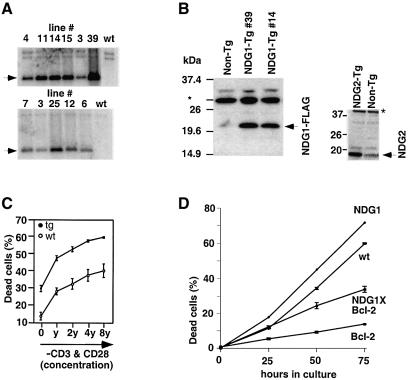
To assess the biological functions of the novel genes NDG1 and NDG2 we generated the corresponding transgenic mice using the lck proximal promoter (Allen et al., 1992b). Six founders for NDG1 and five founders for NDG2 (Figure 4A) were identified by Southern blot analysis, and the expression of the transgenes was determined by western blotting after lines were established from each founder (Figure 4B). Lines 14 and 39 (for NDG1) and 3 and 25 (for NDG2) represent the highest expressing lines and were chosen for further analysis. Judging from the intensity of the non-specific signals (indicated by asterisks in Figures 3C and 4B), expression of the NDG1 transgenic protein was not too dissmilar from that expressed in Nur77 transgenic thymocytes. Flow cytometric analyses with a variety of developmental markers showed no gross abnormalities in the composition of thymocyte subpopulations. Thymic cellularities of the transgenic animals were also normal (data not shown). The NDG2 transgenic mice displayed no detectable phenotypes and will not be discussed further (Supplementary figure 6). However, DP thymocytes from NDG1 transgenic mice of both lines consistently exhibit reduced viability in culture compared with their littermate cohorts (Figure 4C). Thymocytes from Nur77 transgenic mice also display reduced viability in culture (Chan et al., 1998). Similar to thymocytes from NDG1 transgenic mice, the Nur77 counterparts exhibit increased apoptosis after 18 h of culture although Nur77 transgenic thymocytes already have many apoptotic cells at time zero (Supplementary figure 6). The basal levels of death between the NDG1 transgenic and littermate control cells are similar, suggesting that either the dead cells in transgenic mice are cleared rapidly in vivo or there are compensatory signals that keep these cells alive in the thymus of the animals. The increased propensity for apoptosis in NDG1 transgenic thymocytes is most apparent when thymocytes from neonatal animals were used. The difference in cell viabilities between transgenic and non-transgenic DP cells is increased slightly when they are stimulated with α-CD3 and α-CD28 antibodies in vitro (Figure 4C).
Fig. 4. Characterization of transgenic NDG1 and NDG2 mice. (A) Southern blot analysis showing six founders for the NDG1 and five founders for the NDG2 transgene. The wild-type (wt) control shows the endogenous band that also serves as controls in other lanes. (B) The left panel shows western blot analysis of NDG1 transgenic thymocytes from two different founders (39 and 14) detected with NDG1-specific antibodies. The transgenic protein ran higher than the endogenous protein because it was tagged with the C-terminal FLAG tag. The right panelshows western blot analysis of thymocytes from NDG2 transgenic mice (NDG2-Tg) and their non-transgenic (non-Tg) littermates. NDG2 transgenic protein was tagged with C-terminal FLAG tag and was detected by α-FLAG (M2) antibody. the asterisk denotes non-specific binding of the antibodies that serves as a loading control. (C) Apoptosis of transgenic and littermate neonatal DP thymocytes in response to in vitro α-CD3 and α-CD28 stimulation. The antibodies were originally diluted to 1:1000 and 1:50, respectively, to yield the highest concentration point (8y) and serial diluted 2-fold three times to obtain the other points (4y, 2y and y). (D) Time course of spontaneous apoptosis of DP thymocytes from wild-type (wt), NDG1, Bcl-2 and NDG1 × Bcl-2 transgenic mice. All the experiments were done at least three times.
Spontaneous death of DP thymocytes in culture has been shown to be inhibitable by overexpressing either Bcl-2 or Bcl-xL (Chao and Korsmeyer, 1998). To address the role of the Bcl-2 pathway in NDG1-mediated death, double-transgenic NDG1 × Bcl-2 mice were generated. While the Bcl-2 single-transgenic DP thymocytes exhibit increased viability in culture as expected and can rescue the endogenous apoptosis, they failed to rescue apoptosis of NDG1 transgenic thymocytes completely (Figure 4D). Indeed, the difference in cell viability between NDG1/Bcl-2 and Bcl-2 transgenic DP thymocytes is even larger than those seen between NDG1 and wild-type DP thymocytes. This suggests that the mechanism of NDG1-mediated apoptosis is Bcl-2 independent and is consistent with our above finding that Bcl-2 failed to reverse the apoptotic fate of thymocytes in Nur77 × Bcl-2 double-transgenic mice.
NDG1-mediated apoptosis can be inhibited by CrmA
In order to evaluate the biochemical pathway of apoptosis regulated by NDG1, we utilized an in vitro system where transfection of NDG1 leads to apoptosis. Transient transfection of 293T cells with pCI-NDG1 expression construct led to significant death within 48 h post-transfection (Figures 5 and 6A). While the NDG1 polypeptide sequence does not contain any known domains, there is a significant conservation between the murine, human and rat sequences. Truncation studies suggest that both the N- and C-terminal regions are responsible for the NDG1 apoptotic activity (Figure 5). Consistent with previous data from NDG1/Bcl-2 transgenic mice, addition of a Bcl-2 expression construct, while protecting cells from staurosporine apoptosis, affords little protection from NDG1-dependent apoptosis (Figure 6A, left panel).
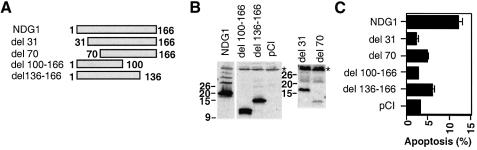
Fig. 5. Induction of apoptosis by N- and C-terminal truncation mutants of NDG1 in 293T cells. (A) A schematic diagram showing the regions of NDG1 that were truncated to generate a panel of NDG1 variants. An initiating Met codon was included at the N-terminus of the del 31 and 70 mutants. (B) Expression levels of the truncation mutants in transiently transfected 293T cells were analyzed by immunoblotting with anti-FLAG antibodies. (C) Induction of apoptosis by the truncation and full-length variants of NDG1. Apoptosis was scored by obtaining a ratio of punctated DAPI stained cells and the total number of GFP positive cells. For each time point, >200 cells were scored.
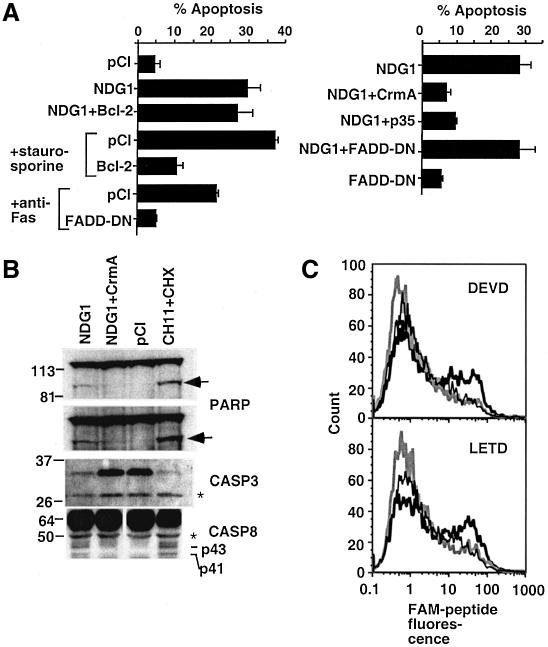
Fig. 6. The role of caspases in NDG1-mediated apoptosis. (A) Apoptosis of 293T cells transiently cotransfected with NDG1 and anti-apoptotic molecules was measured by the ratio of punctated DAPI stained cells to the total number of GFP-positive cells. For controls, staurosporine or anti-Fas antibody was added to the cells to initiate a Bcl-2- or FADD-dependent apoptosis. (B) Western blots indicating the activation of caspases and cleavage of downstream target PARP. Total lysates from transiently transfected 293T cells were analyzed by western blotting. Two exposures of the gel are shown and the arrow indicates the cleaved PARP product while the larger molecular weight band indicates the full-length species. Levels of the cotransfected full-length caspase-3 (CASP3) are greatly diminished in cells transfected with NDG1 and those treated with anti-Fas antibody and cycloheximide. Caspase-8 activation in NDG1-transfected and α-Fas-treated cells as depicted by the generation of p43 and p41 cleaved product of caspase-8. The asterisk indicates a non-specific band that serves as a loading control. (C) Activation of caspase-3 and caspase-8 was assessed by fluorescent labeling of 293T cells with FAM-labeled substrates. 293T cells transfected with NDG1 (bold black line) or cotransfected with NDG1 and CrmA (bold gray line) or CI (thin black line) were stained with FAM-labeled substrates for caspase-3 (DEVD) and caspase-8 (LETD) 48 h after transfection.
Activation of caspases is a common downstream event in apoptosis. To determine the specific role of caspases in NDG1-mediated death, we assessed the effects of expressing several caspase inhibitors in our system. As shown in Figure 6A (right panel), NDG1-induced apoptosis can be reversed by cotransfecting caspase inhibitors p35 or CrmA. Cotransfecting a dominant negative version of FADD (FADD-DN) fails to rescue NDG1-mediated apoptosis. Because FADD-DN can block apoptosis of all death receptors in the tumor necrosis factor receptor (TNF-R) family, which include Fas, TNF-RI, TRAIL-R, DR3 and DR6 (Yeh et al., 1998; Zhang et al., 1998; Kuang et al., 2000), these data suggest that NDG1-mediated apoptosis is independent of the death-receptor pathway. As a control, we showed that the same FADD-DN construct could protect cells from anti-Fas antibody induced apoptosis (Figure 6A).
CrmA binds preferentially to caspase-1 and caspase-8 (Zhou et al., 1997), but caspase-1 is not involved in apoptosis (Creagh et al., 2003). Thus we examined caspase-8 activation in our system, as evidenced by the generation of proteolytically processed versions of the protein (p41 and p43). Western blot analysis with caspase-8-specific antibodies revealed accumulation of p41 and p43 in NDG1 transfected cells and in Fas antibody (CH11) treated cells (Figure 6B). In contrast, they are not detectable in cells transfected with either the pCI control plasmid or with a combination of NDG1 and CrmA. We also tested activation of downstream caspase, caspase-3. Western blot analysis showed decreased levels of pro-caspase-3 in cells cotransfected with NDG1 and pro-caspase-3 (Figure 6B). Moreover, as expected for caspase-3-dependent cell death, PARP, a known substrate of caspase-3, is cleaved in NDG1 transfected cells and this proteolytic cleavage can be blocked by co-expressing CrmA (Figure 6B). The activation of caspase-8 and caspase-3 by NDG1 is further confirmed with LETD-FAM and DEVD-FAM peptides, respectively. These are cell-permeable fluorogenic substrates which fluoresce when the fluoromethylketone moiety on the substrate reacts irreversibly with the active site cysteine of caspases (Komoriya et al., 2000). The substrate specificities are defined by the tetrapeptide, originally determined by positional scanning of substrate libraries (Thornberry et al., 1997). As expected, fluorescence of these peptides can be detected in cells transfected with pCI-NDG1 but not in cells transfected with pCI expression plasmid or a combination of pCI-NDG1 and CrmA (Figure 6C). Together, these data suggest that NDG1-mediated apoptosis involves activation of caspase-3 and caspase-8 and is Bcl-2 independent.
Discussion
The role of Nur77 in negative selection and thymocyte apoptosis has been appreciated for some time, but the effector mechanisms of this process have not been addressed fully (Sohn et al., 2003). While data gathered from studies in cancer cell lines suggest that the mitochondrial translocation of Nur77 is involved in its apoptotic abilities (Li et al., 2000), we have not found such relocalization of the molecule in T-cell hybridomas or thymocytes. In addition, overexpression of Bcl-2, which is known to inhibit cytochrome c release from mitochondria (Chao and Korsmeyer, 1998), cannot rescue Nur77-mediated death in T cells and cytochrome c was not detected in the cytoplasm of apoptotic thymocytes from Nur77 transgenic mice. This apparent discrepancy is most likely due to cell type differences where Nur77 employs different strategies in initiating apoptosis. In this study, we identified several known apoptotic as well as two novel genes as transcriptional targets of Nur77 in thymocytes, further supporting the idea that Nur77 transcription is important for its apoptotic activity. While it remains unclear whether these genes are direct transcription targets of Nur77, their expression levels are significantly upregulated in thymocytes expressing a high level of Nur77. Transient transfection studies using reporter constructs containing upstream sequences of up to 3 kb of NDG1 or NDG2 gene with a Nur77 expression plasmid showed that these regions are not under direct regulation of Nur77 (unpublished data). However, there might be Nur77-regulated enhancers located in the introns, 3′ or further upstream of NDG1 or NDG2, that we have not identified. Two of the tumor necrosis factor family members, FasL and TRAIL, are upregulated in Nur77 transgenic thymocytes. Both FasL and TRAIL mediate apoptosis through caspase-8. We showed here that overexpression of one of the novel genes, NDG1, increases apoptosis of cultured thymocytes and 293T cell line through activation of caspase-8 and caspase-3. Thus activation of these downstream Nur77 target genes converges on the caspase-8 pathway. Interestingly, similar to Nur77-mediated cell death, apoptosis initiated by FasL (Strasser et al., 1995) and NDG1 in T cells is also Bcl-2 independent. NDG1 transcription is activated to a high level in Nur77 transgenic thymocytes. However, it is not yet clear whether NDG1 is the major effector molecule of Nur77-mediated apoptosis or just one of many genes that are activated by Nur77. Generation and characterization of mice with NDG1 loss of function might answer this question, but redundancy with TRAIL, FasL or other yet to be identified pro-apoptotic molecules downstream of Nur77 might also prevent a clear-cut phenotype in NDG1-deficient mice. Expression of NDG1 in T cells leads to increased propensity to die, but it does not lead to massive apoptosis akin to those observed in Nur77 transgenic mice, suggesting that activation of other genes also plays a role in Nur77-mediated apoptosis.
The role of caspases in negative selection is controversial. The ability of zVAD-FMK to halt negative selection (Clayton et al., 1997; Doerfler et al., 2000) and the impaired negative selection in transgenic mice expressing p35 baculoviral protein (Izquierdo et al., 1999) pointed to the involvement of caspases in this process. However, other groups have reported normal thymocyte apoptosis in p35 (Doerfler et al., 2000) or CrmA (Walsh et al., 1998) transgenic mice. While these studies might suggest that caspases are not absolutely required for negative selection, caspases are activated in NDG1-mediated apoptosis. Interestingly, NDG1-mediated death is independent of FADD. FADD, an adaptor molecule that facilitates the interaction between apical caspase-8 and the death-domain-containing cell-surface receptors, was originally considered to be required for caspase-8 activation. However. recent reports outline one possible mechanism of activation of caspase-8 that is FADD independent (Nguyen et al., 1998). Apoptotic signaling by oncogenic molecule E1A recruits the BAP31 complex protein in the endoplasmic reticulum to directly activate the longer isoform of procaspase-8 in FADD-deficient cells (Breckenridge et al., 2002). This novel mechanism for caspase-8 activation is inhibited by direct interaction of Bcl-2 with BAP31. However, NDG1-associated cell death cannot be blocked by anti-apoptotic molecules of the Bcl-2 family. Further studies are necessary to elucidate the exact molecular mechanisms of caspase-8 activation by NDG1 and the involvement of caspase-8 in negative selection.
Materials and methods
Antibodies and reagents
To generate NDG1 protein to immunize animals, the NDG1 coding sequence was subcloned into the pGEX-2T plasmid (Pharmacia) and transformed into Rosetta (Novagen) Escherichia coli cells. The cells were lysed by sonication in the presence of denaturing lysis buffer (PBS, 4 M urea, 1 mM dithiothreitol) on ice. The lysate was renatured by diluting with chilled PBS on ice over the span of a few hours to a final urea concentration of 0.5 M. This was then dialyzed against PBS overnight. The renatured lysate was then loaded on a glutathione–agarose column and used for antibody production. The α-caspase-8 antibodies were kindly provided by Dr Marcus Peter (University of Chicago) and were used at 1:70 dilution for immunoblotting.
RNA isolation and microarray hybridizations
Timed mating between Nur77 transgenic and C57BL/6 mice were conducted where the day after productive mating was considered E0.5. Embryos were harvested from pregnant females at E16.5–E19.5 (same as neonates at times) and their thymi were isolated. The genotype of the embryos was determined by PCR using primers hGH-5′ CAGGG TTTGG GGTTC TGAAT GTGAG and hGH-3′ TCTCC TCCTC TTATT TCCAG CGGG to identify Nur77 transgenic and non-transgenic embryos. Embryonic thymocytes were pooled according to their genotypes and total RNAs were isolated using the Qiagen RNeasy Mini Kit (Qiagen).
Total RNA from the above thymocytes was used for microarray screening of Murine Genome U74Av2 Arrays (MG-U74Av2) (Affymetrix) according to the manufacturer’s protocols. The data were analyzed using Microarray Suite (MAS) 4.0 and normalized to an average intensity value of 600. Fold changes were calculated after the baseline was adjusted to an intensity of 50. Qualitative analysis of the GeneChip screening included confirmation of complete cDNA synthesis of the housekeeping genes in the samples, positive detection of spike controls, and low and consistent background noise levels.
UCB/UCSF custom microarray
The UCB/UCSF custom microarray slides were generated by spotting the PCR products from around 2700 clones. The identities of the clones were confirmed or revealed by DNA sequencing. Data analysis of the clone sequences was done by several Perl scripts in MacOS X. Briefly, the text files of the DNA sequences were converted into FASTA files and combined into one file. They were then fed into the BLAST program to interrogate the mouse Expressed Sequence Tags database. The BLAST results were parsed using another Perl script to extract the clone names and the GenBank numbers of the corresponding top hits into a tab-delimited file. This was converted into a FileMaker Pro file and compared relationally with a FileMaker Pro file containing the latest mouse UniGene database (the FileMaker file of UniGene 116 was generated by parsing the UniGene text file download from the NCBI web site into a tab-delimited format. See supplementary figure 1 for the Perl scripts.)
Array fabrication and hybridization of the slides were performed according to standard protocol using http://www.microarray.org. Total RNA from Nur77 transgenic or non-transgenic fetal thymocytes were amplified using T7 polymerase after reverse transcription using an oligo(dT)-T7 primer (AAATT AATAC GACTC ACTAT AGGGA GACCA CATTT TTTTT TTTTT TTTTT TTT). Three micrograms of amplified RNA was labeled for each sample. The GenePix 4000A microarray scanner was used for acquisition of the fluorescent images, which were then analyzed by the GenePix Pro 3.01 software (Axon Instruments). The signals were normalized based on global hybridization signals and on the signals of several housekeeping genes (actin, tubulin, Sp1, Sp3, histone) spotted on the slides. Dye swap was also done for each experimental samples and the results were analyzed in Excel using the SAM statistical method in a Windows environment (Tusher et al., 2001).
Quantitative RT–PCR
Total RNA from thymocytes was reversely transcribed (RT) using Oligo(dT)16 or random hexamers using the Superscript II kit (Invitrogen). Primers for the PCR reaction were chosen using the Primer Express (Perkin Elmer) software. The sequences are as follows: AA-159F (TAGGT GTTCA TTTAA GTCCT TTACA GAGA) and AA-213R ( CAGCA AGAAT TCCTA GAAG ACAAC TTT) for NDG1; AW-97F (GATCA AGTTG GAGCT GAGTA AGAAG C) and AW-147R (TGAGC GAGCG GATGC TAC) for NDG2; TRAIL 983F (TGCCT GGAAA GCGAC TGAAC) and TRAIL 1033R (AGGTT TCTAC AGCCA GGCCA) for TRAIL; Nur77-1498F(GGTGT TGATG TTCCC GCCT) and Nur77-1550R(TCAGT GATGA GGACC AGAGC G) for Nur77; 5′ gamma actin (GCACC TAGCA CGATG AAGAT TAAG) and 3′ gamma actin (GCCAC CGATC CAGAC TGAGT) for γ-actin.
Quantitative PCR was conducted using the SYBR green PCR core reagents (PE Biosystems) and the Perkin Elmer GeneAmp 5700 system. Typically, 20 ng of the RT reaction was analyzed in triplicate PCR reactions. Standard curves were generated for individual primer sets with a predetermined amount of RT template. The amounts of template in unknown samples were determined by linear regression followed by normalization across samples using γ-actin expression levels as the internal control.
Generation of transgenic animals and genotyping
The NDG1 or NDG2 cDNA with C-terminal FLAG tag fusion was subcloned into the lck proximal promoter transgene expression vector p1017 (Abraham et al., 1991) in the BamHI site. Transgenic mice were genotyped by PCR using the following primers: AA(19–40) AAGGG ATGGT GGAAT GTGCC TG and hGH(571–548) CTTAC CTGTA GCCAT TGCAG CTAG for NDG1 lines; AW 116–137 (GTCTT ATGGC TCAGAT GGCAT C) and hGH for NDG2 lines; hgh 5′II (GACAC AAACT CACAC AACGA TGACG C) and hgh 3′II (ATGCC TGGAA CTCCA ACAAC TCGG) for lck12; bcl2tg-F (CATGT GTGTG GAGAG CGTCA AC) and bcl2tg-R (TAGCC ATTGC AGCTA GGTGA GC) for Bcl-2 transgenic mice (a kind gift from Stanley Korsmeyer). In the Bcl-2 × Nur77-Tg (lck12), the following Nur77 primers were used: MMN10R (CCAGA AGATG GACAG AGAAC TC) and bcl2-tgR (which corresponds to the human growth hormone sequence).
Thymocyte culture and apoptosis assay
For apoptosis, a million thymocytes were plated in each well of a 24-well tissue culture dish, which had been precoated with either α-CD3 (clone 500A2) and α-CD28 (clone 37N) antibodies or PBS overnight at 4°C. They were incubated at 37°C for varying periods of time and were stained with α-CD4-PE, α-CD8-FITC (Pharmingen) and 7-aminoactinomycin D (7AAD) (Sigma) for flow cytometric analysis. The CD4+CD8+7AAD+ population was scored as apoptotic/dead DP cells.
Transfection, cell death assay and labeling of active caspases
First, 0.8 × 105 293T cells were plated in each well of a 12-well dish and allowed to attach overnight. These were transfected with a total of 3.5 µg DNA, including 0.5 µg pcDNA–GFP (green fluorescent protein) for transfection efficiency normalizations, using 4 µl FUGENE 6 (Roche) according to the manufacturer’s protocol. Transfected cells were either visually scored by GFP fluorescence or quantified by flow cytometric analysis. The cells were detached from the plate by tryptic digest 24–48 h post-transfection and pooled with the floating cells, washed with cold PBS and stained with 4′,6-diamidino-2-phenyindole (DAPI). Cells displaying punctated DAPI staining were scored as dead, and this number was normalized against the number of GFP-positive GFP cells. Typically, 200 cells for each sample were counted to estimate the proportion of apoptotic cells.
For caspase activation, the floating and attached 293T cells were collected by centrifugation 48–72 h post-transfection and washed with cold PBS. They were resuspended in fresh media containing FAM-DEVD-FMK or FAM-LETD-FMK at 1:150 dilution (Cell Technology) and incubated for 1 h at 37°C. The cells were then washed and labeled with tricolor-conjugated α-CD8 antibody (pSV-CD8 was cotransfected to determine transfection efficiencies) and analyzed by flow cytometry.
Cell fractionation and cytochrome c release assay
Thymocytes were isolated from 4–6-week-old Nur77 transgenic or non-transgenic animals. Owing to their low cell number, the combined thymi of ≥15 Nur77 transgenic animals were used for each biochemical fractionation experiment. As a control for cytochrome c release, half of thymocytes were treated with1 µM dexamethasone for 9 h. The mitochondrial and cytoplasmic fractions were then separated according to the ApoAlert Cell Fractionation Kit (Clontech) protocol with minor modifications. Briefly, after washing, 2 ×107 thymocytes were resuspended in 0.25 ml fractionation buffer. They were incubated on ice for 10 min and homogenized (30 passes) in an ice-cold Dounce homogenizer. The homogenate was transferred to a fresh tube and centrifuged at 700g for 10 min. This was followed by a higher-speed centrifugation (10 000g) for 25 min. The resulting supernatant contained the cytosolic fraction. The mitochondria pellet was resuspended in 5 µl of 2× loading dye buffer and boiled before loading onto an SDS–PAGE gel. Cytochrome c was visualized using the antibody provided by the kit from Clontech. The β-actin control antibody was from Abcam.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Sue Sohn for critical reading of the manuscript, Zheng Xing for many of the reagents for apoptotic assays, Martin Guerbadot for excellent technical assistance, Marcus Peter for the caspase-8 antibodies, Vivian Peng and John Ngai for the use of their microarray scanner and advice, Patty Holman for the use of the CNR Genomics Facility arrayer, Chris Jamieson and Richard Locksley for exchanging the lymphochip clones to be spotted onto the custom microarray slides, Saira Mian for the initial analysis of the NDG1 and NDG2 sequences using the hidden Markov program, Phil Green for the PHRAP program, Stanley Korsmeyer for the Bcl-2 transgenic mice and Lianne Hsing for the initial compilation of the custom microarray clones. A.R. was supported by a Pfizer postdoctoral fellowship. Research described in this manuscript was supported by National Institutes of Health grant CA66236 (to A.W.).
References
- Abraham K.M., Levin,S.D., Marth,J.D., Forbush,K.A. and Perlmutter,R.M. (1991) Thymic tumorigenesis induced by overexpression of p56lck. Proc. Natl Acad. Sci. USA, 88, 3977–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.M., Forbush,K.A. and Perlmutter,R.M. (1992a) Functional dissection of the lck proximal promoter. Mol. Cell. Biol., 12, 2758–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.M., Forbush,K.A. and Perlmutter,R.M. (1992b) Functional dissection of the lck proximal promoter. Mol. Cell. Biol., 12, 2758–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. et al. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge D.G., Nguyen,M., Kuppig,S., Reth,M. and Shore,G.C. (2002) The procaspase-8 isoform, procaspase-8L, recruited to the BAP31 complex at the endoplasmic reticulum. Proc. Natl Acad. Sci. USA, 99, 4331–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calnan B., Szychowski,S., Chan,F.K.M., Cado,D. and Winoto,A. (1995) A role of the orphan steroid receptor Nur77 in antigen-induced negative selection. Immunity, 3, 273–282. [DOI] [PubMed] [Google Scholar]
- Chan F.K., Chen,A. and Winoto,A. (1998) Thymic expression of the transcription factor Nur77 rescues the T cell but not the B cell abnormality of gld/gld mice. J. Immunol., 161, 4252–4256. [PubMed] [Google Scholar]
- Chao D.T. and Korsmeyer,S.J. (1998) BCL-2 family: regulators of cell death. Annu. Rev. Immunol., 16, 395–419. [DOI] [PubMed] [Google Scholar]
- Cheng L.E., Chan,F.K., Cado,D. and Winoto,A. (1997) Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J., 16, 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton L.K., Ghendler,Y., Mizoguchi,E., Patch,R.J., Ocain,T.D., Orth,K., Bhan,A.K., Dixit,V.M. and Reinherz,E.L. (1997) T-cell receptor ligation by peptide/MHC induces activation of a caspase in immature thymocytes: the molecular basis of negative selection. EMBO J., 16, 2282–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creagh E.M., Conroy,H. and Martin,S.J. (2003) Caspase-activation pathways in apoptosis and immunity. Immunol. Rev., 193, 10–21. [DOI] [PubMed] [Google Scholar]
- Doerfler P., Forbush,K.A. and Perlmutter,R.M. (2000) Caspase enzyme activity is not essential for apoptosis during thymocyte development. J. Immunol., 164, 4071–4079. [DOI] [PubMed] [Google Scholar]
- Dunker N., Schuster,N. and Krieglstein,K. (2001) TGF-beta modulates programmed cell death in the retina of the developing chick embryo. Development, 128, 1933–1942. [DOI] [PubMed] [Google Scholar]
- Dunker N., Schmitt,K., Schuster,N. and Krieglstein,K. (2002) The role of transforming growth factor beta-2, beta-3 in mediating apoptosis in the murine intestinal mucosa. Gastroenterology, 122, 1364–1375. [DOI] [PubMed] [Google Scholar]
- Gordon D., Abajian,C. and Green,P. (1998) Consed: a graphical tool for sequence finishing. Genome Res., 8, 195–202. [DOI] [PubMed] [Google Scholar]
- Izquierdo M., Grandien,A., Criado,L.M., Robles,S., Leonardo,E., Albar,J.P., de Buitrago,G.G. and Martinez,A.C. (1999) Blocked negative selection of developing T cells in mice expressing the baculovirus p35 caspase inhibitor. EMBO J., 18, 156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoriya A., Packard,B.Z., Brown,M.J., Wu,M.L. and Henkart,P.A. (2000) Assessment of caspase activities in intact apoptotic thymocytes using cell-permeable fluorogenic caspase substrates. J. Exp. Med., 191, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kuang A.A., Cado,D. and Winoto,A. (1999) Nur77 transcription activity correlates with its apoptotic function in vivo. Eur. J. Immunol., 29, 3722–3728. [DOI] [PubMed] [Google Scholar]
- Kuang A.A., Diehl,G.E., Zhang,J. and Winoto,A. (2000) FADD is required for DR4- and DR5-mediated apoptosis: lack of TRAIL-induced apoptosis in FADD-deficient mouse embryonic fibroblasts. J. Biol. Chem., 275, 25065–25068. [DOI] [PubMed] [Google Scholar]
- Lee S.L., Wesselschmidt,R.L., Linette,G.P., Kanagawa,O., Russell,J.H. and Milbrandt,J. (1995) Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77). Science, 269, 532–535. [DOI] [PubMed] [Google Scholar]
- Li H., Kolluri,S.K., Gu,J., Dawson,M.I., Cao,X., Hobbs,P.D., Lin,B., Chen,G., Lu,J. et al. (2000) Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science, 289, 1159–1164. [DOI] [PubMed] [Google Scholar]
- Linette G.P., Grusby,M.J., Hedrick,S.M., Hansen,T.H., Glimcher,L.H. and Korsmeyer,S.J. (1994) Bcl-2 is upregulated at the CD4+ CD8+ stages during positive selection and promotes thymocyte differentiation at several control points. Immunity, 1, 197–205. [DOI] [PubMed] [Google Scholar]
- Mamalaki C., Norton,T., Tanaka,Y., Townsend,A.R., Chandler,P., Simpson,E. and Kioussis,D. (1992) Thymic depletion and peripheral activation of class I major histocompatibility complex-restricted T cells by soluble peptide in T-cell receptor transgenic mice. Proc. Natl Acad. Sci. USA, 89, 11342–11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama N., Oishi,K., Mori,Y., Ueno,T., Takahama,Y. and Gotoh,Y. (2001) Akt inhibits the orphan nuclear receptor Nur77 and T-cell apoptosis. J. Biol. Chem., 276, 32799–32805. [DOI] [PubMed] [Google Scholar]
- Nguyen M., Branton,P.E., Roy,S., Nicholson,D.W., Alnemri,E.S., Yeh,W.C., Mak,T.W. and Shore,G.C. (1998) E1A-induced processing of procaspase-8 can occur independently of FADD and is inhibited by Bcl-2. J. Biol. Chem., 273, 33099–33102. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y., Hallas,C., Palamarchuk,A., Koval,A., Bullrich,F., Hirata,Y., Bichi,R., Letofsky,J. and Croce,C.M. (2001) Akt phosphorylates and regulates the orphan nuclear receptor Nur77. Proc. Natl Acad. Sci. USA, 98, 3690–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnio T., Burton,Q., Pereira,F.A., Wu,D.K. and Conneely,O.M. (2002) The nuclear receptor Nor-1 is essential for proliferation of the semicircular canals of the mouse inner ear. Mol. Cell. Biol., 22, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfe C., Zaccaria,P., Hoertnagel,K., Jaksch,M., Klopstock,T., Lill,R., Prokisch,H., Gerbitz,K.D., Mewes,H.W. et al. (1999) MITOP: database for mitochondria-related proteins, genes and diseases. Nucleic Acids Res., 27, 153–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn S.J., Rajpal,A. and Winoto,A. (2003) Apoptosis during lymphoid development. Curr. Opin. Immunol., 15, 209–216. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris,A.W., Huang,D.C.S., Krammer,P.H. and Cory,S. (1995) Bcl-2 and Fas/APO-1 regulate distinct pathways to lymphocyte apoptosis. EMBO J., 14, 6136–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh H.S., Kishi,H., Scott,B., Borgulya,P.,H., v.B. and Kisielow,P. (1990) Early deletion and late positive selection of T cells expressing a male-specific receptor in T-cell receptor transgenic mice. Dev. Immunol., 1, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry N.A., Rano,T.A., Peterson,E.P., Rasper,D.M., Timkey,T., Garcia-Calvo,M., Houtzager,V.M., Nordstrom,P.A., Roy,S. et al. (1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem., 272, 17907–17911. [DOI] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani,R. and Chu,G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.M., Wen,B.G., Chinnaiyan,A.M., O’Rourke,K., Dixit,V.M. and Hedrick,S.M. (1998) A role for FADD in T cell activation and development. Immunity, 8, 439–449. [DOI] [PubMed] [Google Scholar]
- Weih F., Ryseck,R.P., Chen,L. and Bravo,R. (1996) Apoptosis of nur77/N10-transgenic thymocytes involves the Fas/Fas ligand pathway. Proc. Natl Acad. Sci. USA, 93, 5533–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winoto A. and Littman,D.R. (2002) Nuclear hormone receptors in T lymphocytes. Cell, 109, S57–S66. [DOI] [PubMed] [Google Scholar]
- Yeh W.-C., Pompa,J.L., McCurrach,M.E., Shu,H.-B., Elia,A.J., Shahinian,A., Ng,M., Wakeham,A., Khoo,W. et al. (1998) FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science, 279, 1954–1958. [DOI] [PubMed] [Google Scholar]
- Zhang J., Cado,D., Chen,A., Kabra,N.H. and Winoto,A. (1998) Absence of Fas-mediated apoptosis and T cell receptor-induced proliferation in FADD-deficient mice. Nature, 392, 296–300. [DOI] [PubMed] [Google Scholar]
- Zhang J., DeYoung,A., Kasler,H.G., Kabra,N.H., Kuang,A.A., Diehl,G., Sohn,S.J., Bishop,C. and Winoto,A. (1999) Receptor-mediated apoptosis in T lymphocytes. Cold Spring Harb. Symp. Quant. Biol., 64, 363–371. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Snipas,S., Orth,K., Muzio,M., Dixit,V.M. and Salvesen,G.S. (1997) Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J. Biol. Chem., 272, 7797–7800. [DOI] [PubMed] [Google Scholar]