Abstract
DNA replication results in interlinked (catenated) sister duplex molecules as a consequence of the intertwined helices that comprise duplex DNA. DNA topoisomerases play key roles in decatenation. We demonstrate a novel, efficient and directional decatenation process in vitro, which uses the combination of the Escherichia coli XerCD site-specific recombination system and a protein, FtsK, which facilitates simple synapsis of dif recombination sites during its translocation along DNA. We propose that the FtsK–XerCD recombination machinery, which converts chromosomal dimers to monomers, may also function in vivo in removing the final catenation links remaining upon completion of DNA replication.
Keywords: chromosome segregation/decatenation/FtsK/XerCD
Introduction
The structure determination of DNA 50 years ago immediately identified the topological problem of unlinking the two intertwined strands of the DNA double helix during its replication (Watson and Crick, 1953a, b). However, it was many years later that topoisomerases were shown to play key roles in facilitating the unlinking of DNA strands during DNA replication (Champoux, 2001; Postow et al., 2001; Wang, 2002). The separation of the parental DNA strands, which occurs as a replication fork progresses, results in an overwinding of the unreplicated duplex strands and a consequent accumulation of positive supercoiling ahead of the replication fork. For continued replication progression, this positive supercoiling must be removed by topoisomerase action on the supercoiling ahead of the fork, or by allowing the positive supercoiling to diffuse into newly replicated DNA, where it forms precatenanes, interlinks between the newly replicated sister duplex molecules. Precatenanes become catenanes at the completion of DNA replication.
Based on structure–mechanism, topoisomerases can be divided into two families. One contains the structurally related type II and type IA topoisomerases, which catalyse topological transitions by creating a double-stranded DNA-linked protein gate through which an intact duplex passes (type II), or a single-stranded gate (type IA) through which a single-stranded or double-stranded DNA segment passes. Both type II and type IA topoisomerases have been implicated in changing the supercoiling state of DNA and in decatenation, the process that separates interlinked DNA duplexes. In contrast, type IB topoisomerases, belonging to the second structurally unrelated family, use a catalytic mechanism in which strand cleavage is followed by rotation of one strand about the other prior to strand rejoining (Champoux, 2001; Wang, 2002), a mechanism well suited to changing the supercoiled state of DNA, but one not obviously applicable to decatenation. Type IB topoisomerases share structural features and biochemical mechanisms with the tyrosine recombinases (Cheng et al., 1998; Sherratt and Wigley, 1998).
In the 4.6 Mb circular Escherichia coli chromosome some 4.6 × 105 links must be removed during each round of replication at a rate of removal of ∼102 links/s, if the newly replicated sister chromosomes are to separate and be segregated to daughter cells. Failure to remove even a single link will result in the sister chromosomes being catenated and prevent sister chromosome separation. DNA gyrase, a type II topoisomerase, acts ahead of the progressing replication fork to remove positive supercoils as they accumulate ahead of the fork (Peebles et al., 1979; Adams et al., 1992; Zechiedrich and Cozzarelli, 1995; Postow et al., 2001). As the two converging replication forks approach each other at the termination of replication, removal of the final links by gyrase action ahead of the forks becomes compromised by the restricted accessibility to enzyme in the short unreplicated region. Diffusion of these links behind the replication fork, to produce precatenanes, allows completion of replication, although the consequent conversion of precatenanes to catenanes results in the sister molecules being topologically entangled (Ullsperger et al., 1995; Peter et al., 1998; Sogo et al., 1999). Topoisomerase IV (TopoIV), a second E.coli type II topoisomerase, efficiently removes precatenane and catenane links, and may act preferentially in the replication termination region (Peng and Marians, 1993; Ullsperger and Cozzarelli, 1996; Zechiedrich et al., 1997; Espeli et al., 2003). A type IA topoisomerase, TopoIII, can also remove precatenane links, presumably by acting at single-stranded gaps at the replication fork (Nurse et al., 2003).
In addition to the presence of catenation links, the separation and segregation of newly replicated circular chromosomes can also be prevented by the formation of circular chromosome dimers, which can arise during crossing over by homologous recombination (Blakely et al., 1991; Clerget, 1991; Kuempel et al., 1991). In E.coli, these dimers, which arise about once every six generations, are resolved to monomers by the action of the FtsK–XerCD–dif chromosome dimer resolution machinery (Steiner and Kuempel, 1998a, b; Recchia et al., 1999; Steiner et al., 1999). Two site-specific recombinases of the tyrosine recombinase family, XerCD, act at a 28 bp recombination site, dif, located in the replication terminus region of the E.coli chromosome to remove the crossover introduced by dimer formation, thereby converting dimers to monomers. A complete dimer resolution reaction during recombination at dif requires the action of the C-terminal domain of FtsK (FtsKC) (Steiner et al., 1999; Barre et al., 2000). FtsK is a multifunctional protein whose N-terminal domain acts in cell division, while the C-terminal domain functions in chromosome segregation (Liu et al., 1998; Wang and Lutkenhaus, 1998; Yu et al., 1998a, b). Therefore, FtsK is well suited to coordinate chromosome segregation and cell division. A purified protein, FtsK50C, containing a functional C-terminal domain, can translocate DNA in an ATP-dependent manner and activate Xer recombination at the recombination site dif, thereby reconstituting in vitro the expected in vivo activities of the C-terminal domain of the complete FtsK protein (Aussel et al., 2002).
Here, we demonstrate efficient and directional decatenation by FtsK50C-dependent XerCD recombination at dif in vitro. We show that the same molecular mechanism that converts dimeric chromosomes to monomers, can unlink catenated DNA monomers to free circles in reactions in which there is a sequential interconversion of catenated monomers and knotted dimers. Indeed, the FtsKC–XerCD–dif recombination machinery may not be able to discriminate between decatenation and dimer resolution; the DNA translocation activity of FtsKC and its ability to remodel the XerCD–dif recombination complex during each round of recombination are central to both of these activities. These findings make FtsKC–XerCD–dif recombination well suited to complete decatenation of chromosomes in vivo, and highlight a possible mechanistic link between decatenation, circular chromosome dimer resolution and cell division. Indeed, the action of the FtsKC–XerCD–dif machine may provide the final ‘fail-safe’ opportunity for ensuring that newly replicated chromosomes are decatenated, monomeric and moved away from the septal space prior to completion of cell division.
Results
Catenated DNA circles containing antiparallel dif recombination sites are unlinked by the action of FtsK and XerCD
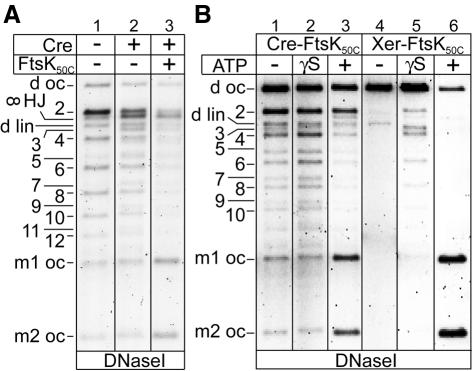
During dimer resolution, FtsKC-dependent XerCD site-specific recombination at directly repeated dif sites on supercoiled plasmids leads to the exclusive formation of free circles (Aussel et al., 2002). In contrast, the related recombinase Cre generates a spectrum of multiply linked catenated circles and free circle products when it recombines directly repeated loxP sites (Abremski et al., 1983). Differences in the topology of products between the Xer and Cre reactions are illustrated when a pseudo-dimeric 6 kb plasmid (d sc), which contains adjacent pairs of dif and loxP sites in direct repeat, is reacted with either FtsK50C–XerCD or Cre (Figure 1A, compare lanes 1 and 5).
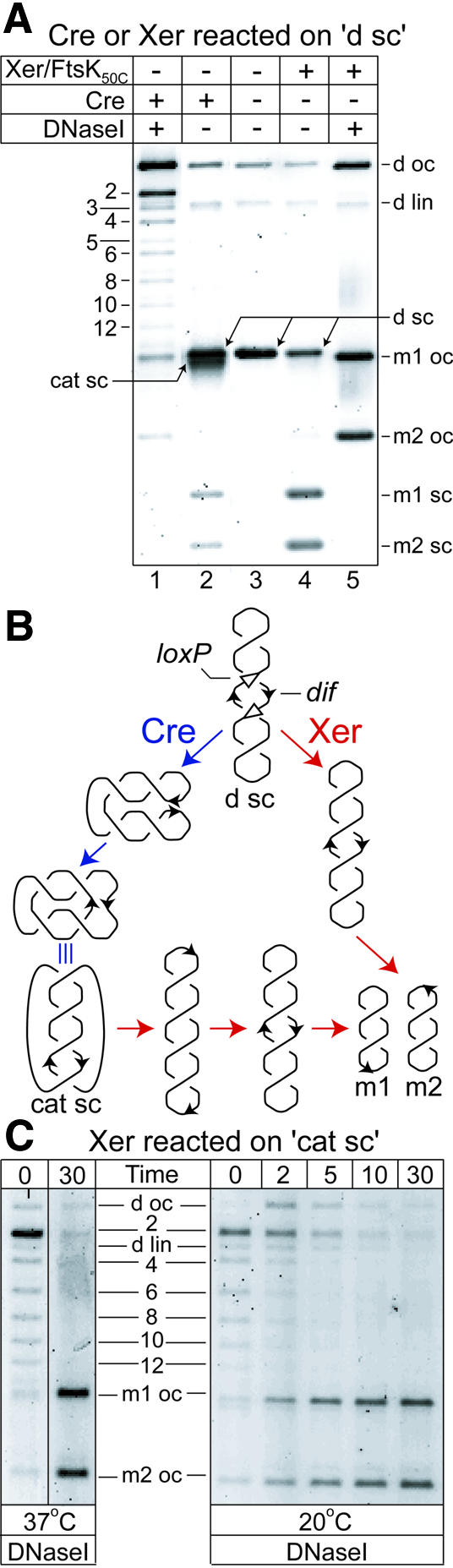
Fig. 1. FtsK50C–XerCD–dif recombination unlinks catenated circles containing antiparallel dif sites. (A) Topology of the products of Cre and FtsK50C–XerCD recombination on the 6 kb pseudo-dimeric deletion substrate, d sc (B), in which the pairs of adjacent dif and loxP sites are separated by 3.5 and 2.5 kb. The topological complexity of products is revealed after DNase I nicking (lanes 1 and 5). DNA bands are labelled: d sc, d oc and d lin, supercoiled, open circle and linear pseudo-dimeric plasmid, respectively; m1 oc and m2 oc, monomer open circles; m1 sc and m2 sc, monomer supercoils; cat sc, supercoiled catenanes; 2, 4, 6, 8, 10, 12, nicked catenanes with two, four, six, eight, 10 and 12 nodes, respectively; 3 and 5, nicked knots with three and five nodes. A small amount of HJ runs between 2-cat and 3-knot (see Figure 3A). (B) Schematic of Cre–loxP and FtsK50C–XerCD–dif recombination on the pseudo-dimeric supercoiled deletion substrate, d sc, and of FtsK50C–Xer recombination on supercoiled catenanes (cat sc). For simplicity, adjacent loxP and dif sites are shown as a single black arrow in all DNA molecules other than the initial d sc substrate. Cre–loxP and FtsK50C–XerCD–dif recombination pathways are delineated by blue and red arrows, respectively. (C) FtsK50C–Xer recombination on supercoiled catenanes containing antiparallel dif sites. Supercoiled catenanes (cat sc) were generated by Cre–loxP recombination on the supercoiled deletion substrate, d sc.
The exclusive formation of free circles (m1 and m2) by FtsK50C–XerCD recombination at dif is a direct consequence of Xer recombination occurring on simple synapses. In contrast, many of the products arising from Cre recombination at loxP are catenated circles of varying complexity. Catenanes arise from recombination of synapses with a variable number of supercoils trapped between the recombination sites, a characteristic of recombination systems that synapse recombination sites by random collision (Figure 1B, Cre pathway). The proportion of catenanes to free circles in Cre–loxP reactions is dependent on reaction conditions, but under all the conditions that we have used, mixtures of catenanes, with up to 12 catenation nodes, and free circles are detected. The exclusive free circle formation during FtsK50C-dependent Xer recombination at dif could be a consequence of FtsK50C either facilitating simple synapse formation, or only activating Xer recombination on simple synapses that form spontaneously.
The ability of FtsK50C–XerCD to recombine supercoiled DNA molecules with directly repeated dif sites exclusively to free circles, suggested to us that the same reaction might unlink catenated DNA molecules containing antiparallel dif sites, since the orientation of recombination sites on antiparallel catenanes is equivalent to the orientation of sites on a supercoiled substrate with directly repeated sites (Figure 1B). On a catenane with antiparallel dif sites, a single recombination event on simply synapsed sites would generate an unknotted dimer, irrespective of the number of catenation nodes in the substrate. This unknotted dimer has directly repeated dif sites, which can then be recombined, in a second event, to free circles, if the dif sites form a simple synapse.
In order to explore this possibility, we assayed for FtsK50C–XerCD–dif recombination on multiply linked supercoiled catenanes containing antiparallel dif sites, produced by Cre–loxP recombination (Figure 1A). Supercoiled catenanes, running slightly ahead of the substrate DNA (lane 2), were gel purified and reacted with FtsK50C–XerCD at 20°C or 37°C and analysed after DNase I nicking, which reveals product topology (Figure 1C). The catenanes were efficiently converted to free circles. Unknotted dimers appeared at early time points, but were quickly and efficiently converted to free circles, consistent with them being reaction intermediates. The reaction was >50% complete after 5 min incubation at 20°C.
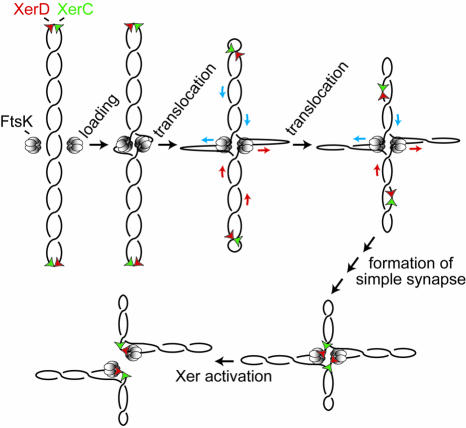
Therefore, FtsK-stimulated XerCD site-specific recombination at dif can efficiently decatenate linked DNA circles in a two-step reaction that converts a catenane to an unknotted dimer, which is then resolved to free circles. This reaction requires that recombination occurs only on simple synapses. Since the dimeric molecules formed in the first recombination reaction will have their dif sites distantly located in the supercoiled plectoneme (Figure 1B), the efficiency of the overall decatenation reaction suggests that the DNA translocation activity of FtsK50C might be used to disassemble the synaptic complex at the end of the first round of recombination and facilitate formation of a new simple synapse of dif sites in the dimer. Activation of Xer recombination by FtsK50C in the simple synapse then leads to free monomer circle formation. One way by which FtsK50C-mediated DNA translocation might synapse distant dif sites is shown in Figure 2 and in movie 1 (Supplementary data).
Fig. 2. One scheme by which FtsK50C (hexameric rings) might promote synapsis of directly repeated dif sites (red and green triangles) on a supercoiled DNA molecule, prior to activation of Xer recombination. Two units of FtsK50C form a double ring on DNA, by analogy with RuvB and other DNA translocases. Translocation by each ring reels in the distant dif sites and facilitates their synapsis and activation of recombination. We propose that once a XerCD-bound dif site interacts with a ring, its DNA translocation activity stalls. Models in which a single ring facilitates synapsis of dif sites are also possible, although the reaction is not as facile as that presented here. DNA translocation will generate separate positively and negatively supercoiled domains when two distant DNA segments interact simultaneously with a ring (Aussel et al., 2002). For simplicity, this differential supercoiling is not shown.
In order to address whether FtsK50C-mediated DNA translocation is responsible for the formation of the simple synaptic complexes that are essential for directional decatenation, we examined recombination by Cre of catenanes with antiparallel loxP sites in the absence and presence of FtsK50C. In the absence of FtsK50C, recombination by Cre of antiparallel loxP sites contained within catenanes generates predominantly twist knots with an odd number of nodes: the expected products of recombination between randomly synapsed sites (Figure 3A, lane 2; Bath et al., 1999; Crisona et al., 1999). Such twist knots cannot be untangled to free circles by further recombination events. In the presence of FtsK50C, more free circles and less knotted products were produced by Cre (lane 3). The enhanced levels of free circles produced by Cre suggests that the action of FtsK50C stimulated recombination on simply synapsed loxP sites. This result demonstrates that FtsK50C-mediated synapsis of recombination sites is not confined to Xer recombination at dif: FtsK50C can also use its DNA translocation activity to remove any topological nodes between loxP recombination sites even though Cre normally assembles its recombinational synapse by the random collision of sites and its recombination is not dependent on FtsKC. This result also explains why the ability of Cre–loxP to partly substitute for XerCD–dif in chromosome dimer resolution is dependent on FtsKC (Capiaux et al., 2002).
Fig. 3. FtsK50C promotes formation of free circles in recombination of plasmid dimers and catenated monomers by Cre–loxP as well as by XerCD–dif. (A) Effect of FtsK50C on Cre–loxP recombination of catenanes with antiparallel loxP sites. Samples were nicked with DNase I to reveal product topology. ∞ HJ, ‘figure 8’ nicked HJ intermediate; 2, 4, 6, 8, 10, 12, nicked catenanes with two, four, six, eight, 10 and 12 nodes, respectively; 3, 5, 7, 9 and 11, nicked knots with three, five, seven, nine and 11 nodes. (B) Influence of nucleotide on FtsK50C action during Cre–loxP and XerCD–dif recombination on the supercoiled deletion substrate, d sc (Figure 1B). γS is ATP-γS.
In an attempt to confirm rigorously that FtsK translocation activity on DNA is responsible for the formation of simple synaptic complexes, recombination of a supercoiled pseudo-dimeric plasmid at the respective dif and loxP sites was tested in the absence of ATP (Figure 3B, lane 4) or with the non-hydrolysable ATP analogue, AMPPNP (not shown). In both cases recombination was abolished. When the poorly hydrolysable nucleotide ATP-γS was used as a cofactor, recombination of supercoiled substrates with directly repeated dif sites was reduced in efficiency; however, products contained mainly catenanes (Figure 3B, lane 5). Similarly, twist knots, but no free circles, were obtained from antiparallel dif catenanes (data not shown). These findings reveal that FtsK50C can activate XerCD recombination independently of any role in defined synapse formation by DNA translocation. Nevertheless, it is the efficient formation of the simple synapse by the translocase activity of FtsK50C that ensures that XerCD normally converts dimers and catenated monomers to monomer free circles. This conclusion is reinforced by the observation that FtsK50C stimulates formation of free circles by Cre recombination of loxP-containing catenanes and pseudo-dimers.
These data suggest that the exclusive formation of free circles during FtsK50C-mediated XerCD recombination at dif in a pseudo-dimeric plasmid might arise from initial complex synapses as well as from simple synapses. Recombination on complex synapses would yield catenanes that are then efficiently converted to free circles via dimer intermediates. However, we have not seen the catenanes that would provide evidence for this pathway, even when we examined reaction products at very short reaction times.
In conclusion, the exclusive free circle formation during FtsK50C-dependent Xer recombination at dif is a consequence of FtsK50C mediating efficient simple synapse formation through its DNA translocation activity, rather than by acting only on simple synapses that form spontaneously. This mechanism is quite different from the one that determines selectivity for products of defined topology in other characterized site-specific recombination systems. These use wrapping of DNA around accessory proteins bound to accessory DNA sequences to trap supercoils, which are partitioned into defined yet topologically complex products (Stark and Boocock, 1995; Colloms et al., 1997).
Catenated circular DNA molecules equivalent to those produced by DNA replication are unlinked by the action of FtsK and XerCD
The ability of FtsK50C–XerCD to decatenate DNA molecules containing antiparallel dif sites suggested to us that it might also decatenate DNA molecules containing parallel dif sites, which could have important biological implications since replication of a dif-containing chromosome or plasmid produces such catenated molecules.
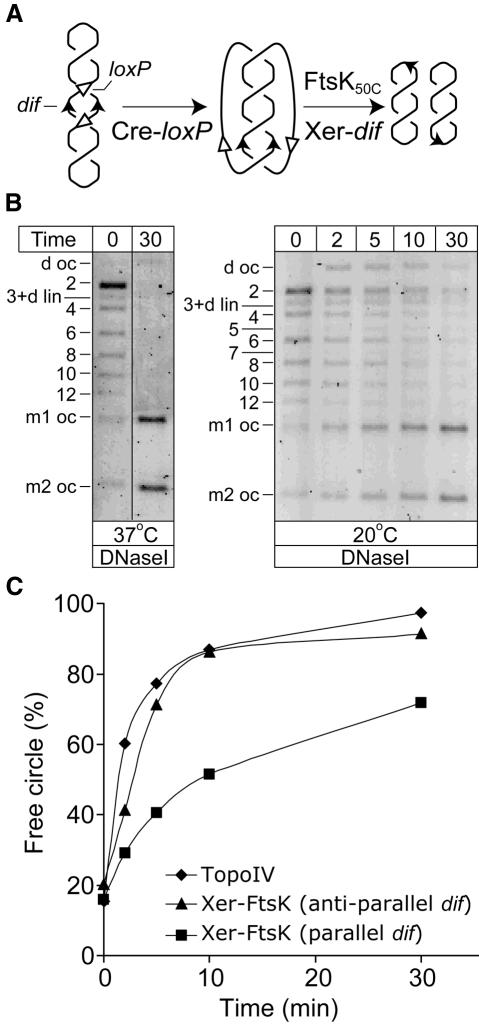
In order to produce catenanes with parallel dif sites, a plasmid with directly repeated loxP sites and adjacent dif sites that are inverted with respect to each other was constructed (Figure 4A). Cre–loxP recombination on this plasmid generates catenanes in which the dif sites are in a parallel orientation with respect to each other, and which therefore mimic replicative catenanes. When supercoiled catenanes with parallel dif sites were reacted with FtsK50C–XerCD, free monomer circles were again produced. The majority of catenanes had been converted to free circle products in a 30 min reaction at 37°C (Figure 4B). Consideration of possible reaction schemes led us to the view that the most probable decatenation mechanism would involve a sequential and stepwise loss of a catenation–knot linking node at each reaction step, with simple synapses being mandatory intermediate substrates throughout the reaction (Figure 5A). Such a gradual, stepwise disentanglement of parallel catenanes to free circles, via knotted intermediates of reducing complexity is consistent with the pattern of products observed when recombination was followed through a time course at 20°C (Figure 4B). Catenanes of low complexity were decatenated most efficiently and the level of unknotted pseudo-dimeric plasmids changed relatively slowly over time, as they are expected to be continuously produced and processed. Despite the stepwise nature of this reaction, the majority of catenation nodes were removed from catenanes having up to 12 catenation nodes after 30 min at 20°C. Again, this stepwise decatenation is dependent on FtsK50C and the efficiency was comparable to that of decatenation of the same substrate with TopoIV (Figure 4C).
Fig. 4. Decatenation of pseudo-replicative catenanes. (A) Schematic of catenanes with parallel dif sites formed by Cre recombination at directly repeated loxP sites, and their subsequent decatenation by FtsK50C–XerCD-mediated recombination at dif. The initial plasmid was constructed in a similar way to that described in Figure 1A, except that the dif sites were in inverted repeat with respect to each other. Therefore, the catenanes generated by Cre–loxP recombination have parallel dif sites and mimic replicative catenanes. (B) FtsK50C–XerCD recombination on catenanes containing parallel dif sites. The abbreviations are as in Figure 1A. (C) Comparison of decatenation by FtsK50C–XerCD and TopoIV at 20°C. Note that only a single FtsK50C–XerCD-dif decatenating unit can be active on each molecule, whereas multiple TopoIV molecules could be active.
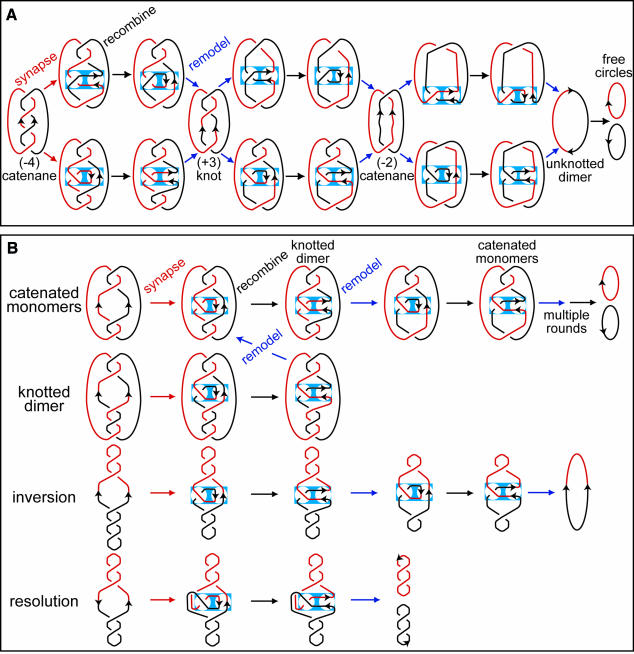
Fig. 5. Schematic of FtsK50C–XerCD-dif-mediated decatenation. (A) Comparison of alternative schemes for decatenation of pseudo-replicative catenanes by sequential rounds of Xer recombination. The consequences of the two schemes are identical, with a linking change (ΔLk) of +2 at each recombination round being accommodated through loss of one catenation–knot node and loss of one (–) supercoil. In the pathway shown at the top, a pre-existing (–) interdomainal catenation crossing is trapped in the recombination complex, and the recombination reaction generates an intradomainal (+) loop containing a single recombination site, which can cancel out a pre-existing (–) supercoil (not shown). In the pathway shown at the bottom, an intradomainal (–) supercoil loop containing a single recombination site is trapped into the recombination complex; this (–) loop can be derived from (–) plectonemic supercoiling elsewhere in the molecule (not shown). The blue rectangle represents the recombination complex formed by the XerCD heterotetramer on the synapsed dif sites. The recombination complex is portioned into two separate regions (circles). One contains the antiparallel- oriented dif sites and is changed by recombination (right-hand side), while the other is unchanged by recombination and contains the trapped (–) crossing (left-hand side). (B) Scheme showing recombination pathways in which an intradomainal supercoil is trapped into the recombination complex for a variety of recombination substrates. Decatenation, unknotting, inversion and deletion are compared. DNA contained within the recombination complex is the same in all cases. The two separate circles within the recombination complex correspond to the ‘parental (P) tangle’ (right-hand side) and ‘bound (Ob) tangle’ (left-hand side) of Crisona et al. (1999). In the configuration shown, tangle P = ∞ and recombinant tangle, R = 0; i.e the orientation of the synapsed dif sites is changed from vertical to horizontal by recombination. The (–) supercoil loop corresponding to Ob is assumed to be recruited from plectonemic supercoiling elsewhere in the molecule (only shown for the inversion substrate). Any other topological nodes are removed by FtsKC action as it facilitates synapsis. If any such nodes had been trapped, they would be incorporated into the product topology. The ∞ tangle for antiparallel recombination sites has been shown to be the most appropriate recombination site configuration for λ Int reactions, and the additional crossing bound by recombinase has been shown to be a (–) topological node for λ Int, Cre and Flp (Crisona et al., 1999; Grainge et al., 2000). The recombination sites are in exactly the same configuration before and after recombination during decatenation, unknotting, simple dimer resolution and inversion. Note that, in the plasmid deletion reaction, in addition to the (–) node trapped in the recombination complex, there is a compensatory (+) node outside the complex; this is required to maintain the correct connectivity and to ensure the products are free circles. The other way to ensure free circles during deletion is not to trap any crossings into the recombination complex. During decatenation–unknotting, each round of recombination leads to the loss of one catenation or knotting node and one (–) supercoil from within the catenane–knot, consistent with a ΔLk of +2, which has been shown experimentally for λ Int, Cre and Flp inversion.
The orientation of dif sites on parallel catenanes is equivalent to those present in a supercoiled molecule containing inverted dif sites (Figure 4B). Such a plasmid is a substrate for FtsK50C-dependent XerCD-mediated inversion with most (>80%) of the recombination product being unknotted circle (Aussel et al., 2002; data not shown), and therefore it is not surprising that decatenation of pseudo-replicative catenanes containing parallel dif sites was observed. Knotted products arise from recombination reactions in which the synapsis of recombination sites traps supercoils. Indeed, reaction of an inversion substrate with XerCD, FtsK50C and ATP-γS led to ∼70% of the products being knotted as a consequence of FtsK50C being compromised in its ability to remove randomly trapped supercoils. On a supercoiled plasmid containing inverted dif sites, the equivalent of the stepwise decatenation reaction described above leads to the stepwise loss of supercoiling, two supercoils being lost per recombination event (Figure 5B; Nash and Pollock, 1983; Abremski et al., 1986; Crisona et al., 1999; Grainge et al., 2000).
Discussion
Consideration of our data leads to a unifying model of FtsKC–XerCD action at synapsed dif sites contained within different substrates (Figure 5). This model explains how a FtsKC-mediated dimer resolution reaction can be equivalent to a decatenation reaction at the local level of the synaptic complex, and indicates why in principle the Xer recombination machinery can act on dimers, knotted dimers and catenated monomers, irrespective of the connectivity of the recombination sites. This model demands that the DNA translocation activity of FtsKC removes all non-essential supercoils from synapsed recombination sites in functional recombination complexes [Figure 2; movie 1 (Supplementary data)].
We assume a close to antiparallel arrangement of recombination sites, consistent with the Cre–loxP and Flp–frt structures (Guo et al., 1997, 1999; Gopaul et al., 1998; Chen et al., 2000) and a range of biochemical and topological data obtained using different tyrosine recombinases (Stark et al, 1989; Arciszewska et al., 1997; Crisona et al., 1999; Grainge et al., 2000; Lee et al., 2000). In a recombination reaction in which recombination sites are antiparallel-aligned and which proceeds through a HJ intermediate, there is no local linkage change (ΔLk) during the process of recombination (ΔLk = 0; Stark et al., 1989). The total linkage change during recombination is most conveniently measured in inversion reactions, and for λ Int, Flp and Cre, ΔLk = ±2 (Nash and Pollock, 1983; Abremski et al., 1986; Crisona et al., 1999; Grainge et al., 2000), a result most parsimoniously explained by a single crossing being trapped in the recombinational complex (Figure 5B). Recombination during inversion reverses the sign of that crossing, thereby leading to a ΔLk of 2, with the linkage change being preferentially +2 in a negatively supercoiled substrate. For supercoiled substrates containing the λ Int attL and attR sites, as well as for Cre and Flp, the (–) topological node trapped in the recombination complex presumably reflects the DNA–recombinase architecture in a recombining complex. This trapped (–) node can be derived from a pre-existing catenane, knot or supercoiled crossing involving distant DNA segments, with each segment containing a recombination site (interdomainal crossing) (Figure 5A, top). Alternatively, the (–) node can be a supercoil loop containing a single recombination site, which we define here as an intradomainal crossing. Such a (–) supercoil loop can be created without energetic cost from a pre-existing plectonemic supercoil (Figure 5A, bottom, and B). In the latter case, recombination generates an interdomainal node (bottom), while in the former case (top), recombination generates a (+) intradomainal node, which leads to the loss of a single (–) supercoil. The topological consequences of these alternative reaction schemes are the same; each round of recombination on a knotted or catenated substrate removes one node of catenation–knotting and one (–) supercoil from elsewhere in the molecule. The differences are whether the initial trapped crossing contains a single recombination site within a loop, or the two recombination sites are contained on different segments of the crossing. Since the action of FtsKC is to remove all topological complexity outside the synaptic complex, the schemes indicated sequentially simplify the topology as sequential recombination reactions proceed. FtsKC could also actively promote the formation of the (–1) node into the recombination complex, irrespective of whether it is intradomainal or interdomainal. Thus, the recombination mechanism mediated by a recombinase related to a type IB topoisomerase can remove a catenation or knot node per recombination event, while changing total linkage by 2.
We are trying to design experiments that distinguish between these alternative schemes. The scheme in which an intradomainal supercoil is initially trapped into the recombination complex and converted to an interdomainal crossing by strand exchange (Figure 5A, bottom), is in accord with biochemical data obtained for other tyrosine recombinases and the topological interpretation of these reactions in terms of ‘tangles’ (Figure 5 legend; Crisona et al., 1999). In Figure 5B, we show that recombination pathways using this scheme are essentially identical for substrates that are catenated monomers, knotted dimers, or inverted or direct repeats in a model plasmid substrate. The alternative scheme could equally well be drawn for the different substrates. A linkage change of ΔLk = +2 during each round of recombination on negatively supercoiled catenated or knotted DNA removes one catenation or knot node at each step along with one (–) supercoil. In a supercoiled inversion substrate the same reaction scheme would remove two (–) supercoils during each round of recombination, consistent with the published experimental data.
In either scheme, FtsKC plays two key roles. It ensures that productive recombination synapses do not trap non-essential supercoil, catenation or knot nodes. The translocation activity of FtsKC displaces such nodes, thereby preventing their incorporation into topologically complex products. Secondly, it ensures that recombination proceeds in a defined order with XerD initiating recombination to form HJs that are recombined by XerC to give complete recombinantion products (Aussel et al., 2002). Importantly, after each round of recombination, FtsKC mediates the necessary remodelling of the XerCD complex from a post-recombination conformation in which XerC is active (XerC having just catalysed the second pair of strand exchanges), to one in which XerD is primed to reinitiate recombination. This remodelling is not trivial, particularly if the XerCD recombinases adopt a cyclic configuration as for Cre; it requires either a change in pathway of the two DNA duplexes through a fixed arrangement of four recombinase molecules, or a breakage of all recombinase–recombinase interactions and reassembly in a different configuration (Bregu et al., 2002).
If after replication, two chromosomal dif sites are present in either a precatenane or a catenane, then the local organization of the dif sites is identical to that which would be found if homologous crossing over between newly replicated sisters was destining the sister chromosomes to be present in a single knotted dimer. The only obvious distinction is that in the former case the total number of precatenation or catenation nodes would be even, whereas in the latter case the number of pre-knot or knot nodes would be odd. This is true irrespective of whether the newly replicated dif sites are in precatenanes–preknots or catenanes–knots. Therefore, we expect the action of FtsK–XerCD at dif in the chromosome to be ‘blind’ to whether the sites are in (pre-)catenated monomers or (pre-)knotted dimers; the sequential action of the recombination machinery will both decatenate and convert dimers to monomers.
What are the relative roles of topoisomerases and FtsK–XerCD in facilitating the removal of interstrand links? Gyrase plays the major role in removing positive supercoiling as it accumulates ahead of replication forks (Peebles et al., 1979; Zechiedrich and Cozzarelli, 1995; Postow et al., 2001). TopoIV is essential and implicated in decatenation and precatenane removal, and may also act in positive supercoil removal (Adams et al., 1992; Peng and Marians, 1993; Zechiedrich et al., 2000). How frequently precatenanes arise during the main part of replication progression is unclear. Precatenane formation requires rotation of the replication fork along with any replication machinery assembled on the fork. If the two forks are coordinately replicated as part of a single replication factory, a barrier to such rotation would be expected. In vivo data suggest that the major role of TopoIV may be at the termination of replication (Espeli et al., 2003). Therefore, chromosomal precatenanes may form most frequently at the termination of replication when replication forks approach each other. Elsewhere, they may only form infrequently, perhaps when replication forks stall. The FtsK–XerCD decatenation reaction can only occur once the dif sites replicate late in the replication cycle. Whether the newly replicated dif sites immediately find themselves in catenane or knot interlinks or whether they inevitably start off in precatenane or preknot nodes remains to be determined. If the former is the case, then TopoIV may generally deal with precatenanes (and dimer preknots), while Xer may act preferentially once replication has completed and catenane and knot nodes are the recombination substrates.
In bacteria, chromosomes segregate as they replicate. At slow, or moderate growth rates, replication in E.coli occurs close to mid-cell and newly replicated DNA moves away from the division plane and condenses close to the cell one-quarter position. The replication terminus region remains close to mid-cell until late in the cell cycle, sometimes until cytokinesis is well advanced (Gordon et al, 1997; Niki et al., 2000; Li et al., 2002; Espeli et al., 2003; Lau et al., 2003). The localization of FtsK to the division septum means that it is ideally placed to act in the final events of chromosome separation and segregation, and to coordinate these with cytokinesis. TopoIV too appears to act preferentially at the later stages of the cell cycle, and at least under some conditions is associated with the replication machinery and with the dif-site region (Hojgaard et al., 1999; Espeli et al., 2003). The action of FtsK in dimer resolution has raised the question of how the FtsK–XerCD machinery discriminates between dimer resolution and formation by intermolecular reaction (Barre et al., 2000). The results here suggest that no such discrimination is necessary, since FtsKC-mediated XerCD recombination at dif can in principle remove catenation-, knotting- and dimer crossovers by the same recombination mechanism.
Materials and methods
Plasmids, DNA manipulations and proteins
All pseudo-dimeric reporter plasmids used in this work are derived from pMIN33, a pUC19-derived plasmid containing a single dif site (Blakely et al., 1991). Briefly, synthetic oligonucleotides were used to insert a loxP site 42 bp from (centre to centre) the dif site. A second copy of dif–loxP was added on a DNA fragment, generating plasmid d sc in which each pair of sites is in direct repeat with respect to each other. Recombination with either Cre or XerCD generates 2.5 kb and 3.5 kb circles (Figure 1A). In a related plasmid, the directly repeated loxP sites had the same spacing with respect to each other and the dif sites were inverted with respect to each other, thereby generating catenanes with parallel dif sites after Cre–loxP recombination (Figure 3A). Details of constructions are available on request. Plasmids were transformed into DS981(xerC), and supercoiled plasmid DNA was purified by QIAfilter Plasmid Kits (QIAGEN). The FtsK50C, XerC and XerD recombinases were purified as described previously (Colloms et al., 1996; Aussel et al., 2002).
Recombination assays
Recombination reactions containing 300 ng of supercoiled plasmid substrate or purified catenanes were normally performed at 37°C for 30 min in a buffer containing 10 mM Tris pH 7.9, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 100 µg/ml BSA and 2.5 mM ATP. To reveal the topology of products, DNase I nicking reactions were carried out for 15 min at 37°C in 10 mM Tris pH 7.9, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 100 µg/ml BSA, 0.3 mg/ml ethidium bromide and 1 µg/ml DNase I. Reactions were stopped by the addition of 0.6% SDS, 20 mM EDTA and 0.5 mg/ml proteinase K. Samples were then extracted with phenol and analysed on a 0.7% agarose gel in TAE. All gels were run at 3 V/cm for 18 h, stained with SyberGreen and visualized using a Fluorimager imaging system (Fuji).
Purification of catenanes
Cre–loxP was reacted with pseudo-dimeric plasmids at 37°C for 30 min in a buffer containing 50 mM Tris pH 7.5, 50 mM NaCl, 5 mM spermidine, 2 mM EDTA, 100 mg/ml BSA, 10% glycerol and 0.1 mM DTT. Reactions were stopped by the addition of 0.6% SDS, 20 mM EDTA and 0.5 mg/ml proteinase K. Samples were extracted with phenol and then analysed on a 0.7% agarose gel in TAE. Supercoiled catenanes, running ahead of the supercoiled substrate, were gel purified by QIAquick Gel Extraction Kit (Qiagen).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank B.Hallet and T.Massey for invaluable discussions. We thank G.Van Duyne for the gift of Cre and A.Bates for the gift of TopoIV. The research was supported by the Wellcome Trust and the Royal Society. S.C.Y.I. was supported by scholarships from the Croucher Foundation of Hong Kong and the Hong Kong Oxford Scholarship Fund, and F.-X.B. by an EMBO Fellowship.
References
- Abremski K., Hoess,R. and Sternberg,N. (1983) Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell, 32, 1301–1311. [DOI] [PubMed] [Google Scholar]
- Abremski K., Frommer,B. and Hoess,R.H. (1986) Linking-number changes in the DNA substrate during Cre-mediated loxP site-specific recombination. J. Mol. Biol., 192, 17–26. [DOI] [PubMed] [Google Scholar]
- Adams D.E., Shekhtman,E.M., Zechiedrich,E.L., Schmid,M.B. and Cozzarelli,N.R. (1992) The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell, 71, 277–288. [DOI] [PubMed] [Google Scholar]
- Arciszewska L.K., Grainge,I. and Sherratt,D.J. (1997) Action of site-specific recombinases XerC and XerD on tethered Holliday junctions. EMBO J., 16, 3731–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aussel L., Barre,F.X., Aroyo,M., Stasiak,A., Stasiak,A.Z. and Sherratt,D. (2002) FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell, 108, 195–205. [DOI] [PubMed] [Google Scholar]
- Barre F.-X., Aroyo,M., Colloms,S.D., Helfrich,A., Cornet,F. and Sherratt,D.J. (2000) FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev., 14, 2976–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J., Sherratt,D.J. and Colloms,S.D. (1999) Topology of Xer recombination on catenanes produced by λ integrase. J. Mol. Biol., 289, 873–883. [DOI] [PubMed] [Google Scholar]
- Blakely G., Colloms,S., May,G., Burke,M. and Sherratt,D. (1991) Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol., 3, 789–798. [PubMed] [Google Scholar]
- Bregu M., Sherratt,D.J. and Colloms,S.D. (2002) Accessory factors determine the order of strand exchange in Xer recombination at psi. EMBO J., 21, 3888–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capiaux H., Lesterlin,C., Perals,K., Louarn,J.M. and Cornet,F. (2002) A dual role for the FtsK protein in Escherichia coli chromosome segregation. EMBO Rep., 3, 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J.J. (2001) DNA topoisomerases: structure, function and mechanism. Annu. Rev. Biochem., 70, 369–413. [DOI] [PubMed] [Google Scholar]
- Chen Y., Narendra,U., Iype,L.E., Cox,M.M. and Rice,P.A. (2000) Crystal structure of a Flp recombinase–Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell, 6, 885–897. [PubMed] [Google Scholar]
- Cheng C., Kussie,P., Pavletich,N. and Shuman,S. (1998) Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell, 92, 841–850. [DOI] [PubMed] [Google Scholar]
- Clerget M. (1991) Site-specific recombination promoted by a short DNA segment of plasmid R1 and by a homologous segment in the terminus region of the Escherichia coli chromosome. New Biol., 3, 780–788. [PubMed] [Google Scholar]
- Colloms S.D., McCulloch,R., Grant,K., Neilson,L. and Sherratt,D.J. (1996) Xer-mediated site-specific recombination in vitro. EMBO J., 15, 1172–1181. [PMC free article] [PubMed] [Google Scholar]
- Colloms S.D., Bath,J. and Sherratt,D.J. (1997) Topological selectivity in Xer site-specific recombination. Cell, 88, 855–864. [DOI] [PubMed] [Google Scholar]
- Crisona N.J., Weinberg,R.L., Peter,B.J., Sumners,D.W. and Cozzarelli,N.R. (1999) The topological mechanism of phage lambda integrase. J. Mol. Biol., 289, 747–775. [DOI] [PubMed] [Google Scholar]
- Espeli O., Levine,C., Hassing,H. and Marians,K.J. (2003) Temporal regulation of topoisomerase IV activity in E. coli.Mol. Cell, 11, 189–201. [DOI] [PubMed] [Google Scholar]
- Gopaul D.N., Guo,F. and Van Duyne,G.D. (1998) Structure of the Holliday junction intermediate in Cre–loxP site-specific recombination. EMBO J., 17, 4175–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon G.S., Sitnikov,D., Webb,C.D., Teleman,A., Straight,A., Losick,R., Murray,A.W. and Wright,A. (1997) Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell, 90, 1113–1121. [DOI] [PubMed] [Google Scholar]
- Grainge I., Buck,D. and Jayaram,M. (2000) Geometry of site alignment during Int family recombination: antiparallel synapsis by the Flp recombinase. J. Mol. Biol., 298, 749–764. [DOI] [PubMed] [Google Scholar]
- Guo F., Gopaul,D.N. and Van Duyne,G.D. (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature, 389, 40–46. [DOI] [PubMed] [Google Scholar]
- Guo F., Gopaul,D.N. and Van Duyne,G.D. (1999) Asymmetric DNA bending in the Cre–loxP site-specific recombination synapse. Proc. Natl Acad. Sci. USA, 96, 7143–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojgaard A., Szerlong,H., Tabor,C. and Kuempel,P. (1999) Norfloxacin-induced DNA cleavage occurs at the dif resolvase locus in Escherichia coli and is the result of interaction with topoisomerase IV. Mol. Microbiol., 33, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Kuempel P.L., Henson,J.M., Dircks,L., Tecklenburg,M. and Lim,D.F. (1991) dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol., 3, 799–811. [PubMed] [Google Scholar]
- Lau I.F., Filipe,S.R., Soballe,B., Okstad,O.-A, Barre,F.X. and Sherratt,D.J. (2003) Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol., 49, 731–743. [DOI] [PubMed] [Google Scholar]
- Lee J., Tribble,G. and Jayaram,M. (2000) Resolution of tethered antiparallel and parallel Holliday junctions by the Flp site-specific recombinase. J. Mol. Biol., 296, 403–419. [DOI] [PubMed] [Google Scholar]
- Li Y., Sergueev,K. and Austin,S. (2002) The segregation of the Escherichia coli origin and terminus of replication. Mol. Microbiol., 46, 985–996. [DOI] [PubMed] [Google Scholar]
- Liu G., Draper,G.C. and Donachie,W.D. (1998) FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol. Microbiol., 29, 893–903. [DOI] [PubMed] [Google Scholar]
- Nash H.A. and Pollock,T.J. (1983) Site-specific recombination of bacteriophage λ. The change in topological linking number associated with exchange of DNA strands. J. Mol. Biol., 170, 19–38. [DOI] [PubMed] [Google Scholar]
- Niki H., Yamaichi,Y. and Hiraga,S. (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev., 14, 212–223. [PMC free article] [PubMed] [Google Scholar]
- Nurse P., Levine,C., Hassing,H. and Marians,K.J. (2003) Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J. Biol. Chem., 278, 8653–8660. [DOI] [PubMed] [Google Scholar]
- Peebles C.L., Higgins,N.P., Kreuzer,K.N., Morrison,A., Brown,P.O., Sugino,A. and Cozzarelli,N.R. (1979) Structure and activities of Escherichia coli DNA gyrase. Cold Spring Harbor Symp. Quant. Biol., 43, 41–52. [DOI] [PubMed] [Google Scholar]
- Peng H. and Marians,K. (1993) Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl Acad. Sci. USA, 90, 8571–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter B.J., Ullsperger,C., Hiasa,H., Marians,K.J. and Cozzarelli,N.R. (1998) The structure of supercoiled intermediates in DNA replication. Cell, 94, 819–827. [DOI] [PubMed] [Google Scholar]
- Postow L., Crisona,N.J., Peter,B.J., Hardy,C.D. and Cozzarelli,N.R. (2001) Topological challenges to DNA replication: conformations at the fork. Proc. Natl Acad. Sci. USA, 98, 8219–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia G.D., Aroyo,M., Wolf,D., Blakely,G. and Sherratt,D.J. (1999) FtsK-dependent and -independent pathways of Xer site-specific recombination. EMBO J., 18, 5724–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt D.J. and Wigley,D.B. (1998) Conserved themes but novel activities in recombinases and topoisomerases. Cell, 93, 149–152. [DOI] [PubMed] [Google Scholar]
- Sogo J.M., Stasiak,A., Martinez-Robles,M.L., Krimer,D.B., Hernandez,P. and Schvartzman,J.B. (1999) Formation of knots in partially replicated DNA molecules. J. Mol. Biol., 286, 637–643. [DOI] [PubMed] [Google Scholar]
- Stark W.M. and Boocock,M.R. (1995) Topological selectivity in site-specific recombination. In Sherratt,D.J. (ed.), Mobile Genetic Elements. IRL Press, Oxford, pp. 101–129. [Google Scholar]
- Stark W.M., Sherratt,D.J. and Boocock,M.R. (1989) Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell, 58, 779–790. [DOI] [PubMed] [Google Scholar]
- Steiner W.W. and Kuempel,P.L. (1998a) Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli. Mol. Microbiol., 27, 257–268. [DOI] [PubMed] [Google Scholar]
- Steiner W.W. and Kuempel,P.L. (1998b) Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J. Bacteriol., 180, 6269–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W., Liu,G., Donachie,W.D. and Kuempel,P. (1999) The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol. Microbiol., 31, 579–583. [DOI] [PubMed] [Google Scholar]
- Ullsperger C. and Cozzarelli,N.R. (1996) Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem., 271, 31549–31555. [DOI] [PubMed] [Google Scholar]
- Ullsperger C.J., Vologodskii,A.V. and Cozzarelli,N.R. (1995) In Lilley,D.M.J. and Eckstein,F. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, Berlin, pp. 115–142. [Google Scholar]
- Wang J.C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol., 3, 430–440. [DOI] [PubMed] [Google Scholar]
- Wang L. and Lutkenhaus,J. (1998) FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol., 29, 731–740. [DOI] [PubMed] [Google Scholar]
- Watson J.D. and Crick,F.H.C. (1953a) Genetical implications of the structure of deoxyribonucleic acid. Nature, 171, 964–967. [DOI] [PubMed] [Google Scholar]
- Watson J.D. and Crick,F.H.C. (1953b) Molecular structure of nucleic acids. Nature, 171, 737–738. [DOI] [PubMed] [Google Scholar]
- Yu X.-C., Tran,A.H., Sun,Q. and Margolin,W. (1998a) Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J. Bacteriol., 180, 1296–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.-C., Weihe,E.K. and Margolin,W. (1998b) Role of the C-terminus of FtsK in Escherichia coli chromosome segregation. J. Bacteriol., 180, 6424–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich E.L. and Cozzarelli,N.R. (1995) Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev., 9, 2859–2869. [DOI] [PubMed] [Google Scholar]
- Zechiedrich E.L., Khodursky,A.B. and Cozzarelli,N.R. (1997) Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev., 11, 2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich E.L., Khodursky,A.B., Bachellier,S., Schneider,R., Chen,D., Lilley,D.M.J. and Cozzarelli,N.R. (2000) Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem., 275, 8103–8113. [DOI] [PubMed] [Google Scholar]