Abstract
Fission yeast Spc1 (Sty1), a stress-activated mitogen-activated protein kinase (MAPK) homologous to human p38, orchestrates global changes in gene expression in response to diverse forms of cytotoxic stress. This control is partly mediated through Atf1, a transcription factor homologous to human ATF2. How Spc1 controls Atf1, and how the cells tailor gene expression patterns to different forms of stress, are unknown. Here we describe Csx1, a novel protein crucial for survival of oxidative but not osmotic stress. Csx1 associates with and stabilizes atf1+ mRNA in response to oxidative stress. Csx1 controls expression of the majority of the genes induced by oxidative stress, including most of the genes regulated by Spc1 and Atf1. These studies reveal a novel mechanism controlling MAPK-regulated transcription factors and suggest how gene expression patterns can be customized to specific forms of stress. Csx1-like proteins in humans may perform similar tasks.
Keywords: microarray/oxidative stress/post-transcriptional control of gene expression/RNA-binding protein/Schizosaccharomyces pombe
Introduction
Oxidative stress caused by reactive oxygen species (ROS), such as hydroxyl radicals (OH–), superoxide anions (O2–) and hydrogen peroxide (H2O2), can be highly toxic, causing damage to proteins, lipids and nucleic acids. ROS are formed as normal by-products of aerobic metabolism or can be derived from exogenous sources. Damage caused by ROS can be mitigated by DNA repair enzymes, lipases, proteases and other enzymes. These enzymes are supported by anti-oxidant defense mechanisms that include non-enzymatic molecules such as glutathione and several vitamins, as well as ROS scavenger enzymes such as superoxide dismutase (SOD), catalase and glutathione peroxidase. Failure to keep ROS under control can have severe consequences; indeed, damage from ROS is thought to play a major role in many of the most common and devastating diseases that afflict the human species (Finkel and Holbrook, 2000; Martindale and Holbrook, 2002).
Oxidative stress elicits a complex gene expression response that is orchestrated in large part by mitogen-activated protein kinase (MAPK) cascades (Robinson and Cobb, 1997; Chang and Karin, 2001; Kyriakis and Avruch, 2001). MAPKs have a variety of substrates ranging from effector kinases to transcription factors and translation factors. JNK1, p38 and ERK5, the three MAPKs activated by oxidative stress in mammalian cells, are controlled by specific MAPK/ERK kinases (MEKs) that are in turn controlled by MEK kinases (MKKs). Stress-activated MEKs activate MAPKs by phosphorylating specific tyrosine and threonine residues. MEK activity is counterbalanced by MAPK-directed tyrosine and threonine phosphatases. The existence of multiple MAPK cascades controlled by various combinations of upstream activators and inhibitors is thought to make these pathways extremely versatile and responsive to diverse forms of cytotoxic stress (Widmann et al., 1999).
The fission yeast Schizosaccharomyces pombe has a stress-activated MAPK module that is functionally and structurally similar to the p38 pathway in mammals. This pathway is defined by the MAPK Spc1 (Sty1, Phh1) (Millar et al., 1995; Shiozaki and Russell, 1995; Kato et al., 1996). Spc1 is responsible for the cellular response to many types of stress such as heat, hyperosmotic media, UV light, oxidation and nutrient deprivation (Millar et al., 1995; Shiozaki and Russell, 1995, 1996; Degols et al., 1996; Degols and Russell, 1997). Like its mammalian counterparts, Spc1 is regulated by a specific MEK, Wis1 (Millar et al., 1995; Shiozaki and Russell, 1995), several MKKs (Samejima et al., 1997; Shieh et al., 1998; Buck et al., 2001; Quinn et al., 2002) and tyrosine phosphatases (Pyp1 and Pyp2) (Millar et al., 1995; Shiozaki and Russell, 1995; Samejima et al., 1997; Shieh et al., 1997, 1998; Shiozaki et al., 1997, 1998). A two-component phosphorelay system acts upstream of the MKKs (Nguyen et al., 2000; Buck et al., 2001). Activation of the pathway leads to increased nuclear localization of Spc1 and elevated expression of a wide range of stress response genes (Gaits et al., 1998; Gaits and Russell, 1999). Spc1 has been shown to control two transcription factors: Atf1 and Pap1 (Toda et al., 1991; Takeda et al., 1995; Shiozaki and Russell, 1996; Wilkinson et al., 1996; Gaits et al., 1998; Toone et al., 1998). Atf1 is required to control gene expression in response to a broad variety of insults, including oxidative stress. On the other hand, Pap1 is especially important in the response to low levels of H2O2 and several other ROS-inducing agents (Toone et al., 1998; Quinn et al., 2002). Combined activities of these transcription factors create a highly tuned response to stress.
Atf1 is related to the human transcription factor ATF2, a substrate of p38 and other stress-activated MAPKs. ATF2 functions as a homodimer or a heterodimer when paired with c-Jun. Increases in ATF2’s transcriptional activity, its acetyltransferase activity and its cellular stability are associated with the dual phosphorylation on Thr69 and Thr71. Exactly how this phosphorylation regulates ATF2, and whether ATF2 is regulated by other means, is unknown (van Dam et al., 1995; Fuchs et al., 2000; Kawasaki et al., 2000). Similarly, Atf1 appears to function as a homodimer or a heterodimer with the c-Jun-related protein Pcr1. Spc1 regulates Atf1 phosphorylation, but the mechanism by which Spc1 controls Atf1 activity is unknown (Shiozaki and Russell, 1996; Wilkinson et al., 1996).
Regulation of mRNA stability is an important mechanism of modulating gene expression (Mitchell and Tollervey, 2000; Guhaniyogi and Brewer, 2001; Fan et al, 2002; Wang et al, 2002). The steady-state level of any mRNA is regulated not only by its transcription rate, but also by its rate of degradation. The best characterized mRNA cis-regulatory elements are the adenosine/uridine-rich elements (AREs) present in the 3′-untranslated region (UTR) of many mRNAs that encode proto-oncogenes, growth factors and cytokines. These AU-rich elements direct the deadenylation and consequent degradation of mRNAs (Chen and Shyu, 1995; Wilusz et al., 2001). AREs interact with trans-acting factors such as AUF1 (Zhang et al., 1993; DeMaria and Brewer, 1996), HuR (Brennan and Steitz, 2001), tristetraprolin (TTP) (Carballo et al., 1998) and TIA-1/TIAR (Gueydan et al., 1999; Piecyk et al., 2000). Some of these AU-rich element-binding proteins (AUBPs) target ARE-containing mRNAs for degradation by the exosome (Chen et al., 2001; Mukherjee et al., 2002). Interestingly, degradation of several ARE-containing mRNAs is regulated by p38 through its downstream effector, the kinase MAPKAPK2 (Winzen et al., 1999; Lasa et al., 2000). The obvious potential targets for this regulatory pathway are the AUBPs (Carballo et al., 2001; Kontoyiannis et al., 2001; Mahtani et al., 2001; Ming et al., 2001; Frevel et al., 2003).
Recent studies demonstrated ARE-targeted degradation of a specific mRNA species in the budding yeast Saccharomyces cerevisiae, indicating broad evolutionary conservation of this mechanism of controlling gene expression. Turnover of TIF51A mRNA, which encodes translation initiation factor eIF5A, and reporter transcripts containing mammalian AREs, was controlled by the RNA-binding protein Pub1 (Vasudevan and Peltz, 2001). Interestingly, the budding yeast p38 homolog Hog1 was shown to be essential for stabilization of these ARE-containing mRNAs (Vasudevan and Peltz, 2001). Thus, MAPKs also appear to regulate mRNA stability in lower eukaryotes.
The Spc1 pathway in fission yeast and the p38 pathway in mammals respond to diverse forms of stress by generating gene expression patterns that are tailored to each type of stress. For example, osmotic and oxidative stress responses are both transmitted by Spc1 and yet lead to different patterns of gene expression. How this specificity is achieved by a single pathway is unknown. Here we report the outcome of a genetic screen designed to discover mutants that are specifically involved in the response to oxidative stress. We describe an RNA-binding protein Csx1, that regulates global gene expression after oxidative stress and is essential for the oxidative stress response but dispensable for the osmotic stress response. Csx1 stabilizes atf1+ mRNA after oxidative stress, and thus reveals a novel mechanism of controlling the activity of transcription factors that are regulated by stress-activated MAPK cascades.
Results
Csx1 is required for survival of oxidative stress
An insertional mutagenesis screen using the ura4+ marker was carried out to identify mutants that were sensitive to oxidative stress but not osmotic stress. This approach yielded ∼500 stable uracil prototroph transformants, of which four were profoundly sensitive to oxidative stress caused by H2O2. Three of these four strains (1F7, 8F7 and 26F1) were also sensitive to osmotic stress caused by KCl, similar to the phenotype of an spc1Δ MAPK mutant (Figure 1A and data not shown). Strains 8F7 and 26F1 were refractory to the inverse PCR method required to identify integration sites, but we were able to show that 1F7 contained an ura4+ interruption of the wis1+ gene.
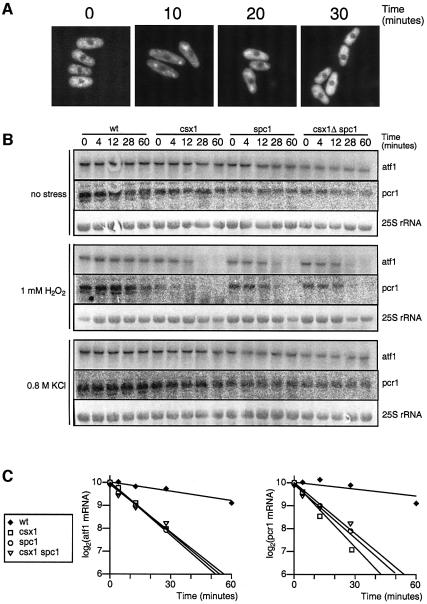
Fig. 1. Characterization of Csx1 as an oxidation-specific protein. (A) Serial dilutions (104–100) of wild-type, spc1Δ, 1F7 mutant and 9B8 mutant cultures were plated in rich medium (YES), rich medium with 0.8 M KCl (osmotic stress) and rich medium with 0.4 mM H2O2 (oxidative stress). Pictures were taken after incubation of the plates for 3–5 days at 30°C. (B) RNA recognition motif (RRM) conservation among Csx1, mammalian HuR and mammalian TIA-1. The numbers indicate the level of similarity. (C) Survival of wild-type, csx1Δ and spc1Δ under oxidative (1 mM H2O2) or osmotic (0.8 M KCl) stress. Cultures were incubated for the indicated times in the presence of stress and cells plated in rich medium (non-stress conditions). Colonies were counted after 3–5 days incubation at 30°C.
The fourth mutant, 9B8, was insensitive to osmotic stress (Figure 1A). DNA sequencing of inverse PCR products showed that 9B8 contained an ura4+ interruption of the csx1+ gene. This gene was identified previously as a multicopy suppressor of the lethality of cut6 temperature-sensitive mutants that are defective for a putative acetyl-CoA carboxylase (Saitoh et al., 1996). Csx1 is a protein of 632 amino acids, with three RNA recognition motifs (RRMs). This domain organization (2–4 RRMs) is found in many proteins involved in various steps of RNA processing. Two of them, human HuR and TIA-1, have been reported to be involved in the response to different types of stress (Gallouzi et al., 2000; Kedersha et al., 2000; Wang et al., 2000) (Figure 1B). Replacement of the entire open reading frame (ORF) of csx1+ with the kanMX6 gene reproduced the oxidation-sensitive phenotype of the 9B8 mutant, confirming that the 9B8 phenotype was caused by inactivation of csx1+ (Figure 1C). We confirmed the stress specificity of Csx1 by comparing the sensitivity of csx1Δ and spc1Δ mutants to oxidative and osmotic stress. While spc1Δ mutants were sensitive to osmotic and oxidative stress, csx1Δ mutants showed sensitivity uniquely to oxidative stress (Figure 1C).
Csx1 controls expression of pyp2 mRNA during oxidative stress
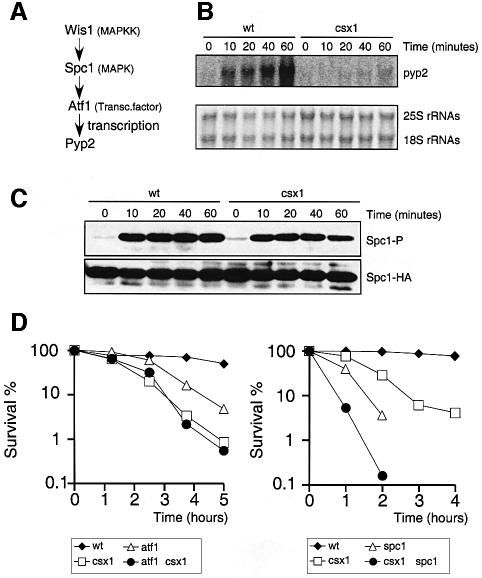
The pyp2 gene encodes a protein tyrosine phosphatase that dephosphorylates Spc1 as part of the stress adaptation response. Expression of pyp2 mRNA is induced by various forms of stress via an Spc1–Atf1-dependent pathway (Figure 2A) (Millar et al., 1995; Degols et al., 1996). Therefore, we measured the expression of pyp2 mRNA in csx1Δ. As shown in Figure 2B, H2O2 induction of pyp2 mRNA was abolished in csx1Δ cells.
Fig. 2. Interaction of Csx1 with the Spc1–Atf1 pathway. (A) Scheme of the Spc1–Atf1 pathway and pyp2 transcription up-regulation. (B) Northern blot analysis of pyp2 mRNA in wild-type or csx1Δ cells after treatment with 1 mM H2O2. (C) Western blot analysis of activating phosphorylation of Spc1 in wild-type or csx1Δ cells after exposure to 1 mM H2O2. (D) Genetic interaction involving csx1Δ and atf1Δ (left panel) or csx1Δ and spc1Δ (right panel). Cells were treated in liquid medium with 1 mM H2O2 and plated in rich medium (no stress). Colonies were counted after 3–5 days incubation at 30°C.
We then tested whether the activating TGY motif phosphorylation of Spc1 carried out by Wis1 was dependent on Csx1 function. As shown in Figure 2C, H2O2 induced robust phosphorylation of Spc1 in csx1Δ cells. These findings indicated that Csx1 is not necessary for Spc1 activation.
Atf1 is required for induced expression of pyp2 mRNA in response to various forms of stress (Figure 2A). Data showing that Csx1 is also required for pyp2 mRNA induction prompted genetic epistasis studies of H2O2 survival. As shown in Figure 2D, the csx1Δ mutant was more sensitive to H2O2 than the atf1Δ mutant, whereas the csx1Δ atf1Δ double mutant was equivalent to the csx1Δ strain. These findings suggested that Atf1-dependent control of gene expression required Csx1, but that Csx1 has additional functions that did not involve Atf1. Similar epistasis studies were carried out with the spc1Δ mutation. The csx1Δ single mutant was less sensitive to H2O2 than the spc1Δ mutant, whereas the csx1Δ spc1Δ double mutant was more sensitive than the spc1Δ strain. These findings were consistent with the idea that Csx1 and Spc1 have independent functions in oxidative stress tolerance.
Stress-induced phosphorylation of Csx1
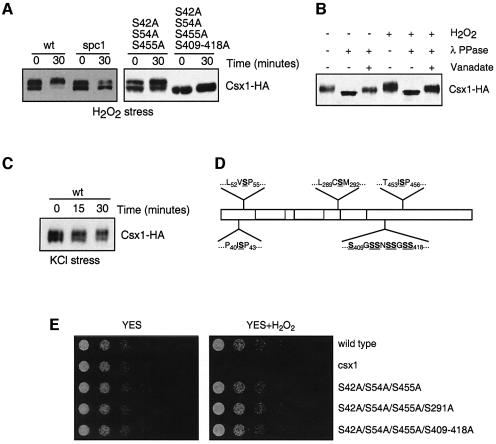
Immunoblot analysis detected multiple electrophoretic mobility species of Csx1 (Figure 3A and C). Osmotic stress had no effect on Csx1 electrophoretic mobility, but oxidative stress caused Csx1 protein to migrate with slower mobility, indicative of a possible phosphorylation (Figure 3A). Consistent with this possibility, the change in Csx1 mobility was abolished in an spc1Δ strain (Figure 3A). Treatment of Csx1 with λ phosphatase confirmed that the change in Csx1 electrophoretic mobility was caused by phosphorylation (Figure 3B). Mass spectrometry analysis of purified Csx1-TAP was used to identify phosphorylation sites. This analysis identified four specific serine residues (S42, S54, S291 and S455) that were phosphorylated and one region that contained multiple phosphorylation sites (S409–S418; see Materials and methods) that could not be mapped precisely. Serine residues at positions 42, 54 and 455 were followed by proline, and thus were potential Spc1 phosphorylation sites. Various single and multiple mutant combinations were made and used to replace the genomic copy of csx1+ (see Materials and methods and Figure 3D). One mutant that contained serine to alanine mutations at positions 42, 54, 455 and 409–418 almost completely abolished the electrophoretic mobility change induced by oxidative stress (Figure 3A). However, neither this mutant, nor any other mutant that had any other combination of single or multiple phosphorylation sites, had any obvious effect on survival of oxidative stress (Figure 3E). Thus, it appears that stress-induced phosphorylation of Csx1, controlled directly or indirectly by Spc1, is not vital for the function of Csx1 in the response to oxidative stress.
Fig. 3. Csx1 phosphorylation. (A) Wild-type (Csx1-HA) and spc1Δ (spc1Δ Csx1-HA) cultures were treated with 1 mM H2O2 and cells were collected after 30 min. Equal amounts of whole-cell extract were loaded per lane (25 µg). Detection of Csx1-HA was done using monoclonal α-HA antibodies. Mutants S42A/S54A/S455A and S42A/S54A/S455A/S409–418A were treated in the same way. (B) Wild-type cells (Csx1-HA) were treated with 1 mM H2O2 for 30 min, and whole-cell extracts were obtained. After immunoprecipitation with polyclonal α-HA antibodies, the pull-down was treated with λ phosphatase. The resulting reaction was resolved by SDS–PAGE, and Csx1-HA protein was detected by western blot using specific monoclonal α-HA antibodies. (C) Wild-type (Csx1-HA) and spc1Δ (spc1Δ Csx1-HA) cultures were treated with 0.6 M KCl and cells were collected after 15 and 30 min. Equal amounts of whole-cell extract were loaded per lane (25 µg). The detection was done using monoclonal α-HA antibodies. (D) Scheme of Csx1 proteins showing underlined the serine residues mutated to alanine in different mutants. (E) Serial dilutions of wild-type, csx1Δ, csx1-S42A/S54A/S455A, csx1-S42A/S54A/S455A/S291A and csx1-S42A/S54A/S455A/S409-S418A were plated in rich medium (YES) or rich medium with 0.4 mM H2O2 (oxidative stress). Pictures were taken after incubation of the plates for 3–5 days at 30°C.
Csx1 controls expression of atf1+ and pcr1+ mRNA during oxidative stress
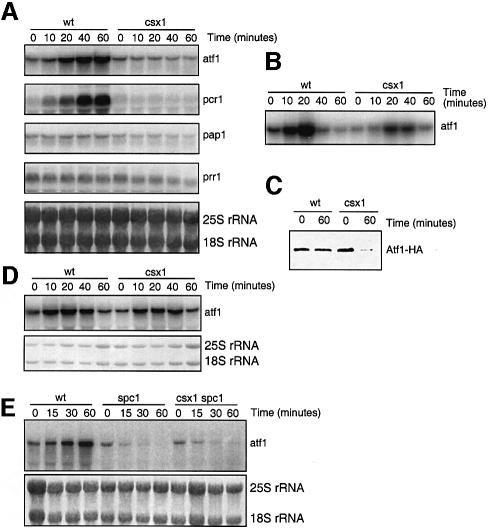
The defect in pyp2 mRNA expression in the csx1Δ mutant might be explained if Csx1 controls expression of atf1+ mRNA. To test this model, the atf1+ mRNA expression level was measured in csx1Δ cells exposed to H2O2 (Figure 4A and B). This analysis showed that the large increase in atf1+ mRNA that is induced by H2O2 in wild-type cells was abolished in csx1Δ cells. This decrease correlated with a large drop in the amount of Atf1 protein (Figure 4C). The H2O2-induced increase in expression of pcr1+ mRNA, which encodes a binding partner of Atf1, was similarly eliminated in csx1Δ cells (Figure 4A). The mRNA expression levels of two other transcription factor genes involved in oxidative stress, pap1+ and prr1+, were unaffected by the csx1Δ mutation (Figure 4A).
Fig. 4. Effect of csx1Δ mutation on the expression of stress transcription factors. (A) Northern blot of atf1+, pcr1+, pap1+ or prr1+ mRNAs after treatment of wild-type or csx1Δ cells with 1 mM H2O2. Aliquots were collected at the indicated times after addition of H2O2. (B) Northern blot of atf1+ mRNA after treatment of wild-type or csx1Δ cells with 0.3 mM H2O2. Aliquots were collected at the indicated times after addition of H2O2. (C) Western blot showing Atf1-HA in wild-type or csx1Δ cells after treatment with 1 mM H2O2. (D) Northern blot of atf1+ mRNA after treatment of wild-type or csx1Δ cells with 0.8 M KCl. Aliquots were collected at the indicated times after addition of KCl. (E) Northern blot of atf1+ mRNA after treatment of wild-type, spc1Δ or csx1Δ spc1Δ cells with 1 mM H2O2.
The csx1Δ mutation specifically affects survival of fission yeast cells in the presence of oxidative stress (Figure 1A–C). We analyzed the effect of csx1Δ mutation on atf1+ mRNA accumulation in response to osmotic stress. As shown in Figure 4D, osmotic stress triggers an accumulation of atf1+ mRNA. This accumulation was not perturbed in the csx1Δ mutant. Thus, the stress-sensitive phenotypes of csx1Δ mutants correlate with stress-specific defects in atf1+ mRNA accumulation.
The steady-state level of atf1+ mRNA in wild-type cells increase to 5–10 times above the non-stressed levels when treated with H2O2 for 1 h (Figure 4A). In the csx1Δ mutant, atf1+ mRNA levels decreased after 1 h of oxidation treatment (Figure 4A). In order to obtain more information about the functional relationship between Spc1 and Csx1, we analyzed the effect of the spc1Δ mutation on the atf1+ mRNA level. The level of atf1+ mRNA in a spc1Δ mutant dropped to half in only 15 min, being almost undetectable after 60 min of treatment (Figure 4E). The double mutant csx1Δ spc1Δ showed a profile of atf1+ mRNA expression almost identical to the single mutant spc1Δ (Figure 4E). These results show that both Csx1 and Spc1 contribute to the increased steady-state levels of atf1+ mRNA after oxidative stress, with Spc1 having a somewhat more profound effect.
Global patterns of gene expression controlled by Csx1 in response to oxidative stress
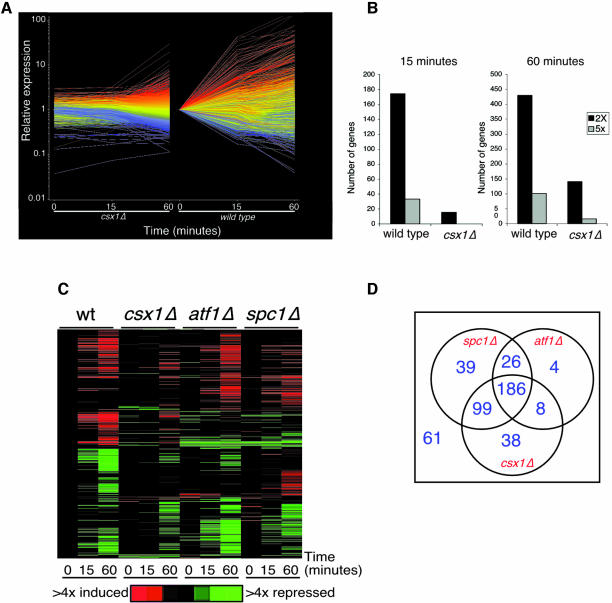
The evidence that Csx1 controls expression of atf1+ and pcr1+ mRNAs suggested that Csx1 was likely to exert global control of gene expression in response to oxidative stress. To explore this possibility, DNA microarrays were used to characterize changes in expression profiles of all known and predicted genes in the fission yeast genome. Microarrays were hybridized with probes derived from RNA harvested from wild-type and csx1Δ strains exposed to 1 mM H2O2 for 0, 15 or 60 min (see the Supplementary data available at The EMBO Journal Online). This analysis showed that the global pattern of gene expression induction in wild-type cells was severely dampened in csx1Δ cells (Figure 5A). We compared the number of genes that were induced 2- or 5-fold in wild-type or csx1Δ cells, 15 or 60 min after treatment with 1 mM H2O2. We observed a profound reduction in the number of genes that showed increased expression after oxidative stress in the csx1Δ mutant (Figure 5B).
Fig. 5. Global effect of Csx1 on gene expression after oxidative stress. (A) Relative expression levels of stress response genes are represented. Expression levels in wild-type and csx1Δ strains are compared at 0, 15 or 60 min after treatment with 1 mM H2O2. (B) The number of genes with two or five times increased expression after stress (when compared with wild-type without stress) is represented for each strain at 15 or 60 min after treatment with H2O2. (C) Hierarchical cluster analysis of genes after oxidative stress in wild-type, atf1Δ, csx1Δ and spc1Δ mutants, showing genes (1576 in total) with ≥2-fold change in expression in at least one of the experimental samples studied and that gave measurable data in at least 50% of all samples. The wild-type and csx1Δ data are from this study, while the atf1Δ and spc1Δ data are from Chen et al. (2003). (D) Venn diagram representing the genes with expression dependent on Spc1 (Chen et al., 2003), Atf1 (Chen et al., 2003) and/or Csx1 (this study). The numbers indicate the genes with expression of half or less in the mutants when compared with wild-type at the same time point (15 or 60 min). The number outside the circles (61) indicates the number of stress-induced genes that are not dependent on any of the three regulators.
This defect was also reflected in the hierarchical cluster analysis of gene expression after oxidative stress in csx1Δ, atf1Δ and spc1Δ mutants (Figure 5C). Venn diagram analysis was performed to evaluate dependency relationships among 461 genes that were induced at least 2-fold at one or both of the time points (15 or 60 min) after exposure to oxidative stress in wild-type cells. Of the 350 genes whose expression was increased at least 2-fold at 15 or 60 min in an Spc1-dependent manner, 212 were also regulated by Atf1 (Figure 5D). Of the 138 remaining genes, expression of 99 was regulated by Csx1. Almost all the genes that were regulated by Atf1 were also dependent on Csx1 (Figure 5D), but there were a significant number of genes that were regulated by Csx1 but not Atf1. These findings were consistent with the genetic epistasis analysis of atf1Δ and csx1Δ mutants (Figure 2D). There was a large overlap of the genes regulated by both Spc1 and Csx1, but there were also significant numbers of the genes that were regulated independently by Spc1 or Csx1. These findings were consistent with the synergistic interactions involving spc1Δ and csx1Δ mutations (Figure 2D).
Csx1 is a cytoplasmic protein that controls atf1+ mRNA turnover
As mentioned above, Csx1 contains three RRM motifs, suggesting that it could be involved in post-transcriptional regulation of mRNA. Newly synthesized mRNA is spliced and 3′ polyadenylated in the nucleus prior to its export to the cytoplasm where is it translated and eventually degraded. We determined the intracellular localization of Csx1 to evaluate whether Csx1 acts before or after nuclear export. The genomic copy of csx1 was modified to encode a protein with a C-terminal green fluorescent protein (GFP) tag. This strain survived oxidative stress with levels comparable with wild-type, indicating that the activity of Csx1–GFP was intact (data not shown). As shown in Figure 6A, Csx1–GFP was detected in the cytoplasm and appeared to be excluded from the nucleus. This pattern of Csx1–GFP localization was unaffected by oxidative stress.
Fig. 6. Effect of Csx1 and Spc1 on atf1+ mRNA turnover. (A) In vivo localization of Csx1–GFP expressed at endogenous levels, after treatment with 1 mM H2O2 (0–30 min). (B) Northern blot of atf1 and pcr1+ mRNAs from cells treated with RNA polymerase inhibitor 1,10-phenanthroline (300 µg/ml) with or without exposure to 1 mM H2O2 or 0.8 M KCl. (C) Quantification of atf1 and pcr1+ mRNA levels from (B) (only oxidative stress is represented).
These results suggested that Csx1 was likely to control atf1+ mRNA turnover. To evaluate this possibility, northern analysis of atf1+ and pcr1+ mRNAs was performed with cells that were treated with 1,10-phenanthroline, a potent transcriptional inhibitor (Parker et al., 1991; Gallagher et al., 1996). In the presence of 1,10-phenanthroline, the abundance of mRNAs is governed solely by the rate of turnover. In wild-type cells, atf1+ and pcr1+ mRNAs had a half-life of >60 min. In the presence of oxidative stress, the half-life of atf1+ and pcr1+ mRNAs was very similar (Figure 6B). In csx1Δ, spc1Δ or csx1Δ spc1Δ cells, the half-life of atf1+ or pcr1+ mRNAs in the absence of stress was >60 min, similar to wild-type. However, in the presence of oxidative stress, the half-life of atf1+ mRNA in csx1Δ, spc1Δ or csx1Δ spc1Δ was ∼12 min and the half-life of pcr1+ mRNA in the same mutants dropped to 9–12 min, much shorter than wild-type (Figure 6B and C). Osmotic stress-treated cells showed atf1+ or pcr1+ mRNAs half-lives very similar to non-treated cells (Figure 6B). These results showed that Csx1 and Spc1 are required to stabilize atf1+ and pcr1+ mRNAs in cells exposed to oxidative stress. Their effects are very similar and not additive, indicating either that their functions are dependent on each other or that each of them control an independent limiting step required to maintain the normal rate of atf1+ and pcr1+ mRNA turnover.
Csx1 binds to atf1+ mRNA under oxidative stress
To evaluate whether the stabilization of atf1+ mRNA by Csx1 involves a direct physical interaction between Csx1 and atf1+ mRNA, we performed northern blot analysis of mRNA associated with TAP-tagged Csx1 that had been affinity purified from fission yeast (see Materials and methods). In the absence of oxidative stress, we were unable to detect atf1+ mRNA associated with Csx1-TAP (Figure 7A). However, following exposure to H2O2, atf1+ mRNA was detected in the Csx1-TAP preparation and absent in the untagged control (Figure 7A). This binding was oxidative stress specific, since atf1+ mRNA was not detected with Csx1-TAP obtained from cells treated with osmotic stress (Figure 7A). Additionally, we tested the binding capacity of Csx1-TAP to leu1 mRNA that is not regulated by Csx1 activity. As shown in Figure 7B, Csx1-TAP did not bind leu1 mRNA under oxidative stress. These results suggested that regulation of atf1+ mRNA turnover was mediated by direct interaction with Csx1.
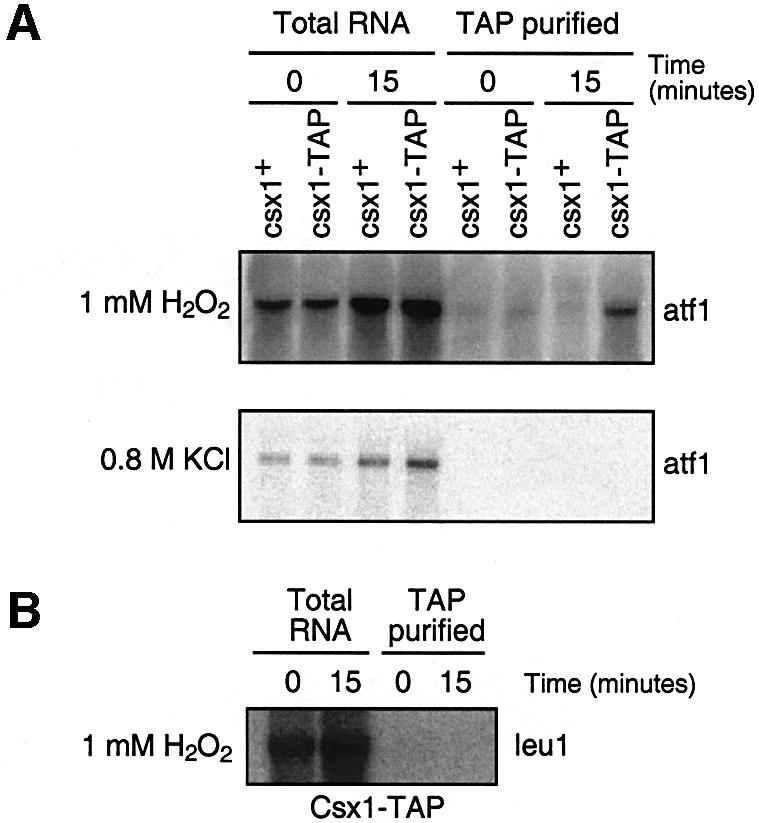
Fig. 7. Binding of Csx1 to atf1+ mRNA. (A) Northern blots of atf1+ mRNA co-immunoprecipitated with Csx1-TAP obtained from untreated or treated cells (oxidative stress 1 mM H2O2 or osmotic stress 0.8 M KCl for 15 min ). (B) Northern blot of leu1 mRNA co-immunoprecipitated with Csx1-TAP obtained from untreated or treated cells (oxidative stress 1 mM H2O2).
Discussion
Csx1 is an essential regulator of the fission yeast oxidative stress response
In this study, we have isolated fission yeast csx1+ as a gene essential for the cellular survival of oxidative stress. One of the key functions of csx1+ appears to be the regulation of Atf1, a bZIP transcription factor required for oxidative stress-induced expression of >200 genes. The Csx1 protein has RRMs and binds to atf1+ mRNA under oxidative stress conditions, stabilizing atf1+ mRNA and keeping normal levels of Atf1 protein after oxidative stress. Thus, in the csx1Δ mutant, a reduced level of Atf1 under oxidative stress results in significantly compromised expression of the Atf1-dependent genes, leading to the stress-sensitive phenotype.
On the other hand, csx1Δ mutants are more sensitive to oxidative stress than atf1Δ, indicating that Csx1 has other important functions in the cellular responses to oxidative stress other than the regulation of Atf1. Indeed, the microarray analysis of wild-type and csx1Δ cells under oxidative stress has identified a number of genes whose expression is dependent on csx1+ but not atf1+. We predict that these csx1+-dependent genes are regulated either by direct binding of Csx1 to their mRNAs or by Csx1-mediated stabilization of unknown transcription factor mRNAs.
Relationship between Csx1 and the stress-activated Spc1 MAPK pathway
Activation of Spc1 MAPK is crucial for cell viability under oxidative stress (Degols et al., 1996). Spc1 controls ∼75% of the genes with increased levels of expression after oxidative stress (Chen et al., 2003), many of which are regulated in an Atf1-dependent manner. Csx1 and Spc1 have an almost identical effect on the atf1+ mRNA half-life, and there is no synergistic effect in the csx1Δ spc1Δ double mutant, suggesting that both Spc1 and Csx1 are necessary to keep the levels of atf1+ mRNA turnover normal after oxidative stress. On the other hand, the effect of Spc1 on the cellular level of atf1+ mRNA after oxidative stress is more dramatic than the effect of Csx1 (Figure 4). It is possible that Spc1 controls atf1+ mRNA half-life as well as atf1+ transcription, and the spc1Δ mutation results in a significant decrease in the total amount of atf1+ mRNA.
Csx1 and Spc1 control the atf1+ mRNA turnover only under oxidative stress conditions. This may imply that only under oxidative stress is there an active mechanism of atf1+ mRNA degradation, or that the atf1+ mRNA degradation is compensated by other unknown stabilizing factor(s) under other stress conditions. Such a hypothetical stabilizing factor would not be regulated by Spc1, since spc1Δ cells have no defect in atf1+ mRNA stability under osmotic stress conditions.
The cis elements responsible for the effects of Csx1 on the stability of atf1+ and pcr1+ mRNAs are not known yet. The presence of AREs in their 3′-UTRs could indicate that those are the sites of regulation. However, the presence of similar AREs in the 3′-UTRs of pap1+ and prr1+, that are not regulated by Csx1, indicate that those AREs cannot be the only sites of regulation. Future experiments will address that question.
Consistent with the Csx1 function limited to oxidative stress conditions, Csx1 is phosphorylated in response to oxidative stress but not osmotic stress (Figure 3). The Csx1 phosphorylation is dependent on Spc1 MAPK, further implying the interaction between Csx1 and the Spc1 MAPK cascade. MAPKs have been shown to promote phosphorylation of several RNA-binding proteins such as TTP (Mahtani et al., 2001), nucleolin (Yang et al., 2002), hnRNP-K (Habelhah et al., 2001) and, more recently, Rnc1 in fission yeast (Sugiura et al., 2003). TTP is involved in ARE mRNA degradation (Lai et al., 1999) and it seems to be part of the p38 pathway (Carballo et al., 2001; Stoecklin et al., 2001). Nucleolin binds to several mRNAs involved in the response to genotoxic stress, but the function of this binding is unknown (Yang et al., 2002), and hnRNP-K accumulates in the cytoplasm after ERK phosphorylation and inhibits mRNA translation of some mRNAs (Habelhah et al., 2001). In all these cases, phosphorylation seems to have a role in the function of those RNA-binding proteins. However, we have not succeeded in determining the role of Csx1 phosphorylation. Different combinations of the phosphorylation site mutations in Csx1 show no apparent effect on the Csx1 function, when tested by its capacity to complement the oxidative stress sensitivity phenotype. It is possible that phosphorylation of other sites in the Csx1 protein is below the levels detectable by mass spectrometry analysis or that some important phosphorylation is lost during the protein purification process. We also cannot exclude the possibility that phosphorylation does not regulate Csx1 function; the stress specificity of the Csx1 activity might be achieved by other types of modification. Alternatively, Csx1 could have a constitutive activity that counteracts a mRNA degradation mechanism activated only under oxidative stress.
Our microarray data indicate that many genes are coordinately regulated by Csx1 and Spc1. On the other hand, there are many genes that are Csx1 dependent but Spc1 independent (46), and vice versa (65). These results are consistent with the genetic analysis of the csx1Δ spc1Δ double mutant, which showed higher sensitivity to H2O2 than either of the single mutants. It is likely that, in addition to the common role in the control of atf1+ mRNA stability, Csx1 and Spc1 have independent functions in oxidative stress-induced gene expression. The coordinated efforts of Csx1 and Spc1 increase the specificity of the response to oxidative stress, leading to increased survival in this condition.
RNA-binding proteins in stress response
To our knowledge, this is the first report describing a central and specific role for an RNA-binding protein in cellular resistance to oxidative stress. It was unexpected that global gene expression is regulated not only by transcription factors but also by a protein binding to mRNAs to modulate their stability.
One of the most important factors that will dictate the fate of a cell after being exposed to stressful conditions (e.g. oxidative stress) is how rapidly the protective responses are activated. MAPK cascade–transcription factor modules have been shown to react very rapidly and efficiently to stressful situations. Once the signal is transmitted to MAPK, activated MAPK enters the nucleus and activates transcription factors, bringing about synthesis de novo of many mRNAs necessary for survival. However, it is apparently advantageous for cells to have an additional system that, once the stress is sensed, can stabilize the pre-existing cytoplasmic mRNAs to increase rapidly the protein synthesis required for cellular protection. Such a mechanism would also be less energy demanding in comparison with transcription de novo, which might be particularly important under stressful conditions. Thus, binding and stabilization of pre-existing mRNAs by Csx1 in the cytoplasm may significantly increase the probability of cell survival upon acute oxidative stress.
The conservation of the responses to stress between fission yeast and mammalian cells, and the sequence conservation of Csx1 protein, suggest that Csx1-like proteins may have similar function in higher eukaryotes.
Materials and methods
Yeast strains, media and general methods
Basic cell growth and media conditions were described before (Moreno et al., 1991). Tagging or deletion of Csx1 was performed using the kanamycin resistance gene as described (Bähler et al., 1998).
The ura4+ insertion mutagenesis was performed as described (Chua et al., 2000; Tanaka and Russell, 2001). The Ura4 ORF was amplified by PCR, and wild-type S.pombe cells were transformed. The ura+ colonies were selected further for oxidative stress sensitivity and stability of the ura+ prototrophy. The positive clones (H2O2-sensitive and ura+ prototroph) were analyzed by reverse PCR to identify sites of insertion.
All strains used were h– ura4-D18 leu1-32: PR109 wild-type; MR3211 wis1-1F7; MR3212 csx1-9B8; MR3213 csx1::kanMX6; KS1497 atf1::ura4+ (Shiozaki and Russell, 1996); KS1366 spc1::ura4+ (Shiozaki and Russell, 1995); MR3219 csx1::kanMX6 atf1::ura4+; MR3218 csx1::kanMX6 spc1::ura4+; MR3217 csx1+::GFP(kanMX6); MR3216 csx1+::TAP(kanMX6); KS1376 spc1+::HA6His(ura4+) (Shiozaki and Russell, 1995); MR3215 csx1::kanMX6 spc1:HA6His(ura4+); MR3254 csx1+::HA(kanMX6); MR3255 csx1+::HA(kanMX6) spc1::ura4+; MR3256 csx1::kanMX6 leu1-32::csx1-HA(leu1+); MR3257 csx1::kanMX6 leu1-32::csx1-HA(leu1+) S42A/S54A/S455A; MR3258 csx1::kanMX6 leu1-32::csx1-HA(leu1+) S42A/S54A/S455A/S409–418A; KS1779 atf1::HA6His; and MR3390 csx1::kanMX6 atf1+::HA6His.
For plate survival assays, serial dilutions of yeast culture were plated in media containing either 1 M KCl (osmotic stress) or 0.4 mM H2O2 (oxidative stress). For liquid survival assays, cells were grown for different times in the presence of KCl or H2O2 and then plated in rich media and colonies counted after 3–4 days at 30°C. Mutants: S42A, S54A, S291A, S409–418A (S409GSSNSSGSS418 to A409GAANAAG AA418) and S455A.
RNA and microarray methods
Total RNA was obtained using Trizol reagent (Invitrogen) as recommended by the manufacturer. Total RNA (5–15 µg) was resolved by electrophoresis in agarose–formaldehyde gels. After transferring to hybond-N+ membranes (Amersham) and staining with methylene blue, the different mRNAs were detected using specific probes, commonly the ORF of the genes amplified by PCR. The probes were labeled with [α-32P]dCTP using Prime-It II (Stratagene).
RNA for microarray analysis was obtained as described in http://www.sanger.ac.uk/PostGenomics/S_pombe/protocols/. Microarray design, sample labeling and hybridization, as well as array data processing and analyses were performed as described before (Lyne et al, 2003). The complete normalized data set is available from http://www.sanger.ac.uk/PostGenomics/S_pombe/, and all raw data will be available from the ArrayExpress repository: www.ebi.ac.uk/arrayexpress.
For RNA extraction of TAP-purified material, we collected cells by filtration and washed them with ice-cold water. RNA extraction was performed as described by Gari et al. (2001) with several modifications. Cells were resuspended in lysis buffer II [10 mM HEPES pH 7.5, 3 mM MgCl2, 40 mM KCl, 5% glycerol, 1 mM dithiothreitol (DTT)] plus RNase/protease/phosphatase inhibitors. Cells were broken by vortexing in the presence of glass beads. Equal amounts of extract were incubated in the presence of IgG beads. After extensive washing with lysis buffer, IgG beads were treated with Trizol reagent and RNA extracted as described before.
mRNA half-life experiments were performed using 1,10-phenanthroline (Sigma) to inhibit transcription (Parker et al., 1991; Gallagher et al., 1996).
Protein methods
Detection of phosphorylated Spc1 using anti-phosphorylated p38 (Thr180/Tyr182) MAPK antibody (New England Biolabs) and immunoprecipitation were performed as described (Gaits et al., 1998). The hemagglutinin (HA) epitope was detected using mouse monoclonal antibodies (12CA5). Phosphatase treatment was carried out using λ phosphatase on immunoprecipitated Csx1-HA as substrate.
Microscopy
Cells were grown in minimal medium until the exponential phase of growth, and the presence of GFP-specific fluorescence was tested by microscopy (Nikon Eclipse E800 microscope).
Mass-spectrometry
Csx1-TAP protein was purified from fission yeast cells treated with 1 mM H2O2 using the method described before (Saitoh et al., 2002). Csx1 was proteolyzed and the peptide mixture analyzed by multidimensional protein identification technology (MudPIT) as described (MacCoss et al., 2002).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Toru Nakamura, Kazuhiro Shiozaki and Michel Toledano for critical reading of the manuscript, Chris J.Penkett for help with the preparation of data for ArrayExpress, and present and past members of the Russell laboratory for support and encouragement, especially Katsunori Tanaka and Antonia Lopez-Girona. The project described was supported by grant number ES10337 from the National Institute of Environmental Health Sciences, NIH (P.R.) and by grants from Cancer Research UK (J.B.), MERK-MGRI-241 (W.H.M.), NIH (EY1328801) and MERK-MGRI-241 (J.R.Y.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
References
- Bähler J., Wu,J.Q., Longtine,M.S., Shah,N.G., McKenzie,A.,3rd, Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Brennan C.M. and Steitz,J.A. (2001) HuR and mRNA stability. Cell. Mol. Life Sci., 58, 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck V., Quinn,J., Soto Pino,T., Martin,H., Saldanha,J., Makino,K., Morgan,B.A. and Millar,J.B. (2001) Peroxide sensors for the fission yeast stress-activated mitogen- activated protein kinase pathway. Mol. Biol. Cell, 12, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E., Lai,W.S. and Blackshear,P.J. (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science, 281, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Carballo E., Cao,H., Lai,W.S., Kennington,E.A., Campbell,D. and Blackshear,P.J. (2001) Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem., 276, 42580–42587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. and Karin,M. (2001) Mammalian MAP kinase signalling cascades. Nature, 410, 37–40. [DOI] [PubMed] [Google Scholar]
- Chen C.Y. and Shyu,A.B. (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci., 20, 465–470. [DOI] [PubMed] [Google Scholar]
- Chen C.Y. et al. (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell, 107, 451–464. [DOI] [PubMed] [Google Scholar]
- Chen D., Toone,W.M., Mata,J., Lyne,R., Burns,G., Kivinen,K., Brazma,A., Jones,N. and Bähler,J. (2003) Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell, 14, 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua G., Taricani,L., Stangle,W. and Young,P.G. (2000) Insertional mutagenesis based on illegitimate recombination in Schizosaccharomyces pombe. Nucleic Acids Res., 28, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols G. and Russell,P. (1997) Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol. Cell. Biol., 17, 3356–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols G., Shiozaki,K. and Russell,P. (1996) Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol., 16, 2870–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaria C.T. and Brewer,G. (1996) AUF1 binding affinity to A + U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem., 271, 12179–12184. [DOI] [PubMed] [Google Scholar]
- Fan J., Yang,X., Wang,W., Wood,W.H.,3rd, Becker,K.G. and Gorospe,M. (2002) Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl Acad. Sci. USA, 99, 10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. and Holbrook,N.J. (2000) Oxidants, oxidative stress and the biology of ageing. Nature, 408, 239–247. [DOI] [PubMed] [Google Scholar]
- Frevel M.A., Bakheet,T., Silva,A.M., Hissong,J.G., Khabar,K.S. and Williams,B.R. (2003) p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol., 23, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S.Y., Tappin,I. and Ronai,Z. (2000) Stability of the ATF2 transcription factor is regulated by phosphorylation and dephosphorylation. J. Biol. Chem., 275, 12560–12564. [DOI] [PubMed] [Google Scholar]
- Gaits F. and Russell,P. (1999) Active nucleocytoplasmic shuttling required for function and regulation of stress-activated kinase Spc1/StyI in fission yeast. Mol. Biol. Cell, 10, 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaits F., Degols,G., Shiozaki,K. and Russell,P. (1998) Phosphorylation and association with the transcription factor Atf1 regulate localization of Spc1/Sty1 stress-activated kinase in fission yeast. Genes Dev., 12, 1464–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher J., Perrin D.M., Chan L., Kwong E. and Sigman D. (1996). Recognition of tetrahedral 1,10-phenanthroline–cuprous chelates by transcriptionally active complexes does not depend on the sequence of the promoter. Chem. Biol. 3, 739–746. [DOI] [PubMed] [Google Scholar]
- Gallouzi I.E., Brennan,C.M., Stenberg,M.G., Swanson,M.S., Eversole,A., Maizels,N. and Steitz,J.A. (2000) HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl Acad. Sci. USA, 97, 3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari E. Volpe,T., Wang,H., Gallego,C., Futcher,B. and Aldea,M. (2001) Whi3 binds the mRNA of the G1 cyclin CLN3 to modulate cell fate in budding yeast. Genes Dev., 15, 2803–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueydan C., Droogmans,L., Chalon,P., Huez,G., Caput,D. and Kruys,V. (1999) Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem., 274, 2322–2326. [DOI] [PubMed] [Google Scholar]
- Guhaniyogi J. and Brewer,G. (2001) Regulation of mRNA stability in mammalian cells. Gene, 265, 11–23. [DOI] [PubMed] [Google Scholar]
- Habelhah H., Shah,K., Huang,L., Ostareck-Lederer,A., Burlingame,A.L., Shokat,K.M., Hentze,M.W. and Ronai,Z. (2001) ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nature Cell Biol., 3, 325–330. [DOI] [PubMed] [Google Scholar]
- Kato T. Jr, Okazaki,K., Murakami,H., Stettler,S., Fantes,P.A. and Okayama,H. (1996) Stress signal, mediated by a Hog1-like MAP kinase, controls sexual development in fission yeast. FEBS Lett., 378, 207–212. [DOI] [PubMed] [Google Scholar]
- Kawasaki H., Schiltz,L., Chiu,R., Itakura,K., Taira,K., Nakatani,Y. and Yokoyama,K.K. (2000) ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature, 405, 195–200. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Cho,M.R., Li,W., Yacono,P.W., Chen,S., Gilks,N., Golan,D.E. and Anderson,P. (2000) Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol., 151, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D., Kotlyarov,A., Carballo,E., Alexopoulou,L., Blackshear,P.J., Gaestel,M., Davis,R., Flavell,R. and Kollias,G. (2001) Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J., 20, 3760–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis J.M. and Avruch,J. (2001) Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev., 81, 807–869. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Carballo,E., Strum,J.R., Kennington,E.A., Phillips,R.S. and Blackshear,P.J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol., 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa M., Mahtani,K.R., Finch,A., Brewer,G., Saklatvala,J. and Clark,A.R. (2000) Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol. Cell. Biol., 20, 4265–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R., Burns G., Mata J., Penkett C.J., Rustici G., Chen D., Langford C., Vetrie D. and Bähler J. (2003) Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility and processing of array data. BMC Genomics, 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCoss M.J. et al. (2002) Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl Acad. Sci. USA, 99, 7900–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani K.R., Brook,M., Dean,J.L., Sully,G., Saklatvala,J. and Clark,A.R. (2001) Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor α mRNA stability. Mol. Cell. Biol., 21, 6461–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale J.L. and Holbrook,N.J. (2002) Cellular response to oxidative stress: signaling for suicide and survival. J. Cell. Physiol., 192, 1–15. [DOI] [PubMed] [Google Scholar]
- Millar J.B., Buck,V. and Wilkinson,M.G. (1995) Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev., 9, 2117–2130. [DOI] [PubMed] [Google Scholar]
- Ming X.F., Stoecklin,G., Lu,M., Looser,R. and Moroni,C. (2001) Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell. Biol., 21, 5778–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000) mRNA stability in eukaryotes. Curr. Opin. Genet. Dev., 10, 193–198. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Mukherjee D., Gao,M., O’Connor,J.P., Raijmakers,R., Pruijn,G., Lutz,C.S. and Wilusz,J. (2002) The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J., 21, 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.N., Lee,A., Place,W. and Shiozaki,K. (2000) Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell, 11, 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Herrick,D., Peltz,S.W. and Jacobson,A. (1991) Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol., 194, 415–423. [DOI] [PubMed] [Google Scholar]
- Piecyk M. et al. (2000) TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J., 19, 4154–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J., Findlay,V.J., Dawson,K., Millar,J.B., Jones,N., Morgan,B.A. and Toone,W.M. (2002) Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell, 13, 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.J. and Cobb,M.H. (1997) Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol., 9, 180–186. [DOI] [PubMed] [Google Scholar]
- Saitoh S., Takahashi,K., Nabeshima,K., Yamashita,Y., Nakaseko,Y., Hirata,A. and Yanagida,M. (1996) Aberrant mitosis in fission yeast mutants defective in fatty acid synthetase and acetyl CoA carboxylase. J. Cell Biol., 134, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S., Chabes,A., McDonald,W.H., Thelander,L., Yates,J.R. and Russell,P. (2002) Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell, 109, 563–573. [DOI] [PubMed] [Google Scholar]
- Samejima I., Mackie,S. and Fantes,P.A. (1997) Multiple modes of activation of the stress-responsive MAP kinase pathway in fission yeast. EMBO J., 16, 6162–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh J.C., Wilkinson,M.G., Buck,V., Morgan,B.A., Makino,K. and Millar,J.B. (1997) The Mcs4 response regulator coordinately controls the stress-activated Wak1–Wis1–Sty1 MAP kinase pathway and fission yeast cell cycle. Genes Dev., 11, 1008–1022. [DOI] [PubMed] [Google Scholar]
- Shieh J.C., Wilkinson,M.G. and Millar,J.B. (1998) The Win1 mitotic regulator is a component of the fission yeast stress-activated Sty1 MAPK pathway. Mol. Biol. Cell, 9, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K. and Russell,P. (1995) Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature, 378, 739–743. [DOI] [PubMed] [Google Scholar]
- Shiozaki K. and Russell,P. (1996) Conjugation, meiosis and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev., 10, 2276–2288. [DOI] [PubMed] [Google Scholar]
- Shiozaki K., Shiozaki,M. and Russell,P. (1997) Mcs4 mitotic catastrophe suppressor regulates the fission yeast cell cycle through the Wik1–Wis1–Spc1 kinase cascade. Mol. Biol. Cell, 8, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K., Shiozaki,M. and Russell,P. (1998) Heat stress activates fission yeast Spc1/StyI MAPK by a MEKK-independent mechanism. Mol. Biol. Cell, 9, 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Stoeckle,P., Lu,M., Muehlemann,O. and Moroni,C. (2001) Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA, 7, 1578–1588. [PMC free article] [PubMed] [Google Scholar]
- Sugiura R., Kita,A., Shimizu,Y., Shuntoh,H., Sio,S.O. and Kuno,T. (2003) Feedback regulation of MAPK signalling by an RNA-binding protein. Nature, 424, 961–965. [DOI] [PubMed] [Google Scholar]
- Takeda T., Toda,T., Kominami,K., Kohnosu,A., Yanagida,M. and Jones,N. (1995) Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J., 14, 6193–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. and Russell,P. (2001) Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nature Cell Biol., 3, 966–972. [DOI] [PubMed] [Google Scholar]
- Toda T., Shimanuki,M. and Yanagida,M. (1991) Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev., 5, 60–73. [DOI] [PubMed] [Google Scholar]
- Toone W.M., Kuge,S., Samuels,M., Morgan,B.A., Toda,T. and Jones,N. (1998) Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev., 12, 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam H., Wilhelm,D., Herr,I., Steffen,A., Herrlich,P. and Angel,P. (1995) ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J., 14, 1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S. and Peltz,S.W. (2001) Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell, 7, 1191–1200. [DOI] [PubMed] [Google Scholar]
- Wang W., Furneaux,H., Cheng,H., Caldwell,M.C., Hutter,D., Liu,Y., Holbrook,N. and Gorospe,M. (2000) HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol., 20, 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu,C.L., Storey,J.D., Tibshirani,R.J., Herschlag,D. and Brown,P.O. (2002) Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA, 99, 5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C., Gibson,S., Jarpe,M.B. and Johnson,G.L. (1999) Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev., 79, 143–180. [DOI] [PubMed] [Google Scholar]
- Wilkinson M.G., Samuels,M., Takeda,T., Toone,W.M., Shieh,J.C., Toda,T., Millar,J.B. and Jones,N. (1996) The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev., 10, 2289–2301. [DOI] [PubMed] [Google Scholar]
- Wilusz C.J., Wormington,M. and Peltz,S.W. (2001) The cap-to-tail guide to mRNA turnover. Nature Rev. Mol. Cell. Biol., 2, 237–246. [DOI] [PubMed] [Google Scholar]
- Winzen R. et al. (1999) The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J., 18, 4969–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Maiguel,D.A. and Carrier,F. (2002) Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res., 30, 2251–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wagner,B.J., Ehrenman,K., Schaefer,A.W., DeMaria,C.T., Crater,D., DeHaven,K., Long,L. and Brewer,G. (1993) Purification, characterization and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol., 13, 7652–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]