Abstract
Prokaryotic transcription elongation factors GreA and GreB stimulate intrinsic nucleolytic activity of RNA polymerase (RNAP). The proposed biological role of Gre-induced RNA hydrolysis includes transcription proofreading, suppression of transcriptional pausing and arrest, and facilitation of RNAP transition from transcription initiation to transcription elongation. Using an array of biochemical and molecular genetic methods, we mapped the interaction interface between Gre and RNAP and identified the key residues in Gre responsible for induction of nucleolytic activity in RNAP. We propose a structural model in which the C-terminal globular domain of Gre binds near the opening of the RNAP secondary channel, the N-terminal coiled-coil domain (NTD) protrudes inside the RNAP channel, and the tip of the NTD is brought to the immediate vicinity of RNAP catalytic center. Two conserved acidic residues D41 and E44 located at the tip of the NTD assist RNAP by coordinating the Mg2+ ion and water molecule required for catalysis of RNA hydrolysis. If so, Gre would be the first transcription factor known to directly participate in the catalytic act of RNAP.
Keywords: endonucleolytic activity/RNA polymerase/transcript cleavage factors Gre
Introduction
Regulation of transcription elongation is an important mechanism for controlling gene expression in the cell (Uptain et al., 1997; Gnatt, 2002). In both eukaryotes and prokaryotes, the regulation may be exerted by factors that interact with RNA, DNA or RNA polymerases (RNAPs) of ternary elongation complexes (TCs). Prokaryotic transcript cleavage factors GreA and GreB (Sparkowski and Das, 1990; Borukhov et al., 1993) and their eukaryotic analog, elongation factor TFIIS (Izban and Luse, 1992; Reines, 1992), affect the efficiency of transcription elongation in vitro and in vivo by stimulating the intrinsic nucleolytic activity RNAPs (reviewed by Fish and Kane, 2002). The endonucleolytic reaction occurs in backtracked TCs, 2–18 bases upstream from RNA 3′-terminus. After the 3′-proximal fragment dissociates, transcription resumes from newly generated 3′-terminus of the transcript (Surratt et al., 1991). GreA induces hydrolysis of mostly di- and trinucleotides, whereas GreB and TFIIS induce cleavage of fragments of various lengths (Izban and Luse, 1992; Reines, 1992; Borukhov et al., 1993; Feng et al., 1994) depending on the extent of RNAP backtracking (Komissarova and Kashlev, 1997; Nudler et al., 1997). The cleavage of nascent RNA is an evolutionarily conserved function among all multisubunit RNAPs, including prokaryotic, eukaryotic, archael and viral enzymes (Fish and Kane, 2002). Presumably, the same catalytic center in RNAP is responsible for both RNA synthesis and hydrolysis reactions (Rudd et al., 1994; Orlova et al., 1995; Sosunov et al., 2003).
The biological role for factor-induced endonucleolytic reactions may include the following: (i) to enhance transcription fidelity by promoting excision of misincorporated nucleotides by RNAP (Erie et al., 1992; Thomas et al., 1998); (ii) to suppress transcriptional pausing and arrest by the ‘cleavage and restart’ mechanism during reversible and irreversible backtracking of TC (Borukhov et al., 1993; Nudler, 1999; Marr and Roberts, 2000; Toulme et al., 2000); and (iii) to stimulate RNAP promoter escape, and transition from initiation to elongation stage (Hsu et al., 1995), by suppressing early RNA release in favor of extension by the ‘cleavage and restart’ mechanism during abortive syntheses. The biological significance of Gre and TFIIS factors is underscored by their ubiquity in nature; Gre-homologs are found in >60 organisms including Mycoplasma genitalium, an organism with the smallest known genome (Hutchison et al., 1999), and in extremophiles such as Thermus and Deinococcus species (White et al., 1999; Laptenko et al., 2000; Hogan et al., 2002), while TFIIS-like factors are found in all known eukaryotic and archael genomes (Fish and Kaine, 2002).
All members of the Gre family are homologous polypeptides of ∼160 amino acids. The structures of GreA and GreB revealed by X-ray crystallography (Stebbins et al., 1995) and computer modeling (Koulich et al., 1997) show that the two factors have similar structural organization and surface charge distribution, and are made up of two domains: an N-terminal extended coiled-coil domain (NTD) and a C-terminal globular domain (CTD). The NTD is responsible for the induction of type-specific nucleolytic activity by Gre factors, while the CTD is responsible for the high affinity binding of Gre to RNAP (Koulich et al., 1997, 1998). The surface of the NTD contains a cluster of positively charged residues (basic patch) that contact the nascent RNA in backtracked TCs (Stebbins et al., 1995; Koulich et al., 1997). Gre–RNA interactions are essential for efficient readthrough and anti-arrest activities in vitro and in vivo, but not for induction of transcript cleavage reactions (Kulish et al., 2000).
The electron microscopy (EM) studies of the Gre–RNAP complex revealed that the CTD is likely to bind near the opening of RNAP secondary channel (Polyakov et al., 1998), which is made mostly of β′ subunit (Zhang et al., 1999; Vassylyev et al., 2002). The channel is presumed to serve as a port of entry for NTPs during synthesis, and as an exit channel for the RNA 3′-terminus during backtracking (Korzheva et al., 2000; Severinov, 2000; Epstein et al., 2003). We have proposed earlier that the NTD allosterically induces the catalytic center of RNAP through its interactions with β′E and β′G in the secondary channel (Kulish et al., 2000). And in recent analysis of the intrinsic exonucleolytic activity of RNAP it was hypothesized that Gre may indirectly help RNAP recruit a Mg2+ ion through the D814 residue of the β subunit (Sosunov et al., 2003).
Here we report detailed mapping of the interaction interface between Gre and RNAP, and the identification of the key residues in Gre factors responsible for the induction of nucleolytic activity in RNAP. Based on our results, we propose a structural model of the Gre–RNAP complex in which: (i) the CTD of Gre binds to the β′ coiled-coil region that forms the rim of the secondary channel opening; (ii) the NTD of Gre is protruded inside the channel, making contacts with several conserved regions of β and β′; and (iii) the tip of the NTD is brought into the immediate vicinity of the RNAP catalytic center. We propose that two conserved acidic residues of the NTD tip, D41 and E44, assist RNAP in coordinating the Mg2+ ion and water molecule required for catalysis of RNA hydrolysis.
Results
Site-specific GreB–RNAP photocrosslinking
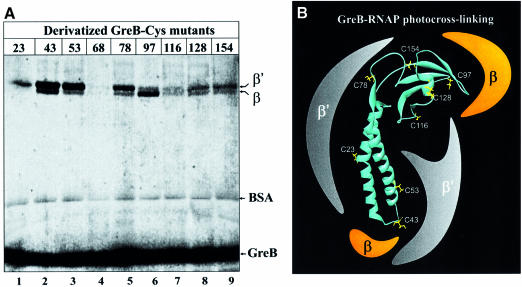
To define the areas in RNAP that may contact specific regions of Gre in native conformation, we applied site-specific protein–protein photocrosslinking (Chen et al., 1995). We chose GreB as a probe because of its higher binding affinity and crosslinking efficiency to RNAP compared with GreA (Koulich et al., 1997). Eight surface-exposed residues not involved in intramolecular interactions were selected and individually substituted with Cys in the GreB-C68S mutant background. None of the Cys-mutations affected the functional activity of GreB (data not shown). The GreB-Cys mutants were modified with a thiol-specific, radiolabeled photoactive reagent 4-azidosalicyl-2,2-dithiopyridyl-[35S]cysteine (ASDPC), which has ∼15 Å of radius of action. The derivatized Gre were incubated with RNAP core, UV-irradiated and analyzed by SDS–PAGE under reducing conditions.
The results (Figure 1) show that the most efficient crosslinks occur from GreBNTD, from C43 at the tip and C53 in the middle of the basic patch, predominantly in the β′-subunit. Some minor crosslinking occurs in β from C43. The crosslink from C23 on the acidic side of GreBNTD was relatively weak but specific to β′. From GreBCTD, C97 and C78 specifically and selectively crosslink to β and β′, respectively (Figure 1A). The crosslink from C116 from the interdomain region is weak and non-specific. The crosslink from C128 and C154 on the basic side of GreBCTD was more efficient and occurred in both β and β′. No crosslinking was observed with σ, α or ω subunits. The specificity of the reaction was verified by the use of an internal control, BSA, and the wild-type (wt) GreB with C68, which was used as a negative control. C68 is not a surface-exposed residue and it did not produce any crosslinks to RNAP.
Fig. 1. Site-specific GreB–RNAP photocrosslinking. (A) Autoradiogram of 8% Tris–glycine SDS–PAGE after UV-irradiation of nine [35S]ASDPC-derivatized GreB-Cys mutants in the presence of RNAP core and BSA as a carrier protein. Numbers on top of the gel indicate the position of the Cys substitution in GreB. GreB-68 is a wt GreB used as a negative control. Positions of free β, β′, BSA and GreB are indicated by arrows. (B) Location of eight derivatized Cys on the model structure of GreB (Koulich et al., 1997), shown as ribbons and a schematic representation of their crosslinking targets.
We conclude that most of the GreBNTD is surrounded by β′, except the tip, which may contact β. GreBCTD is in proximity to both β and β′, and therefore is likely to bind RNAP where β and β′ are joined.
Hydroxyl radical footprinting and mapping of Gre–RNAP complexes
For more detailed analysis of the Gre–RNAP complex, we used hydroxyl radical footprinting by Fe–EDTA (Price and Tullius, 1992; Heyduk et al., 1996; Wang et al., 1997), and localized hydroxyl radical cleavages by an Fe2+ ion placed in the catalytic center of RNAP (Zaychikov et al., 1996; Mustaev et al., 1997). We used the first method to identify areas on the solvent-accessible surface of RNAP that interact with GreA and GreB, and the second method to detect local Gre–RNAP interactions within 5–20 Å from Mg2+-binding site. With both methods, Gre–RNAP interactions are inferred by changes in the protection/enhancement patterns of hydroxyl radical cleavages, which may occur as either a direct or indirect consequence of Gre binding.
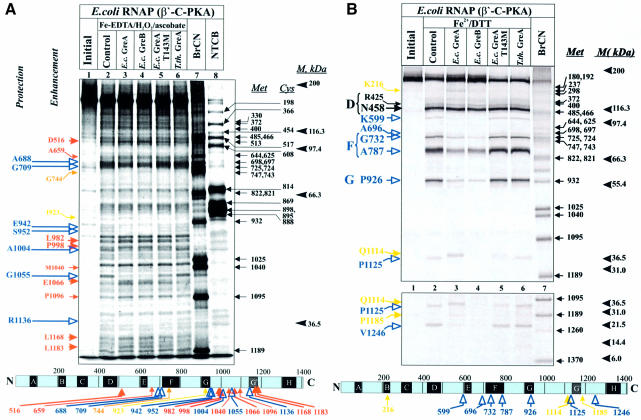
Two types of target molecules were used: (i) Escherichia coli RNAP core carrying either N-terminally radiolabeled β or C-terminally radiolabeled β′ and (ii) N-terminally radiolabeled E.coli GreA and GreB. Three proteins were used as negative controls: a mutant E.coli GreA-T143M defective in RNAP binding (Borukhov et al., 2001); non-cognate GreA homolog from Thermus thermophilus (Tth GreA1), which does not bind E.coli RNAP; and non-cognate Tth RNAP, which does not bind E.coli GreA (Laptenko et al., 2000). The radiolabeled targets were subjected to hydroxyl radical cleavage alone or in the presence of ∼15-fold molar excess of ‘cognate’ or ‘non-cognate’ partners. The cleavage products were analyzed by SDS–PAGE followed by autoradiography (Figure 2) and quantitative densitometric analysis using a phosphorimager and LabWorks program (see Supplementary figure 2 available at The EMBO Journal Online).
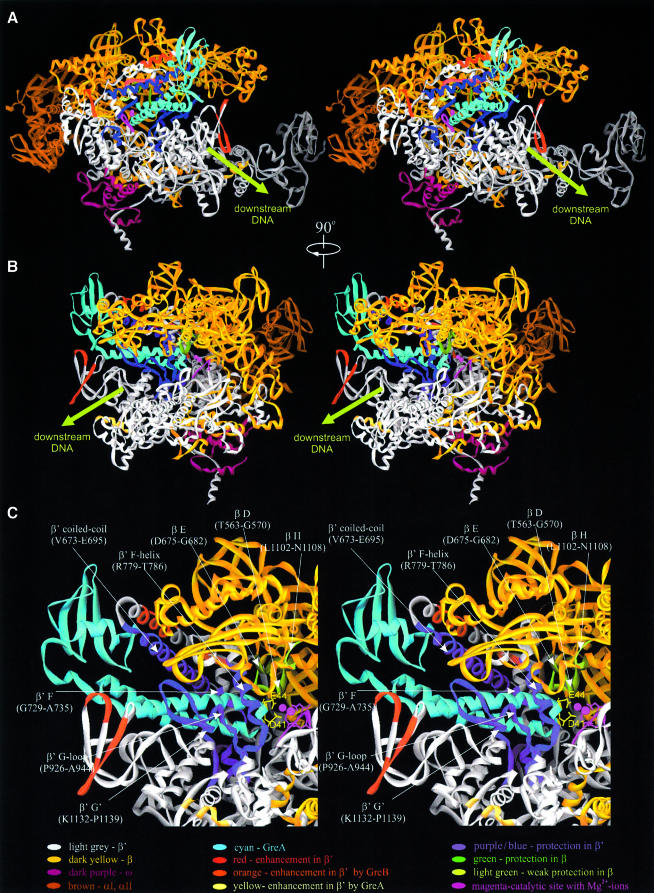
Fig. 2. Hydroxyl radical footprinting and mapping of RNAP–Gre complexes. (A–C) Top panels are autoradiograms of Tris–glycine SDS–7%-PAGE showing the patterns of protection/enhancement of hydroxyl radical cleavages in Gre–RNAP complexes 33P-labeled at the C-terminus of β′ (A and B) or the N-terminus of β (C). (A) General Fe–EDTA protein footprinting. (B and C) Localized hydroxyl radical mapping. Lower panel in (B) is an autoradiogram of a gradient 10–20% Bis–Tris/MES SDS–PAGE. (A, B and C) On the left of each panel, colored arrows indicate cleavage sites in β and β′ affected by Gre strongly (large arrow) or weakly (small arrow). Black arrows (B and C) indicate cleavage sites unaffected by Gre. Strong protection sites in β′ and β are shown in blue and green, respectively. Weak protection in β is shown in light green. Cleavage enhancements by GreA, GreB and both Gre are shown in yellow, orange and red, respectively. Large capital letters at left refer to conserved regions where cleavages occur. Size markers, products of chemical cleavages of β and β′ at Met and Cys, are indicated on the right by small black arrows with a number corresponding to the residue position in non-tagged β and β′. Molecular weight standards are shown on the right under the letter M. At the bottom, sites of cleavage protection and enhancement are indicated on horizontal bars representing β′ (A and B) and β (C) with conserved regions symbolized by lettered boxes. (D) Visualization of protection and enhancement sites on the 3-D structure of Tth RNAP (Vassylyev et al., 2002) with σ not shown. The backbone of RNAP subunits are shown as white ribbons with color coding indicated. Top panel is a view roughly parallel with the main axis of the RNAP secondary channel, with β on top and β′ at the bottom. The catalytic site with two Mg2+ ions (magenta) is seen through the secondary channel. Bottom panel showing the main channel view is obtained by rotation of the top view by 90° clockwise about the vertical axis.
Fe–EDTA hydroxyl radical footprinting
Both GreA and GreB protected three major areas in β′ (Figure 2A, compare lanes 3 and 4 with the controls in lanes 2, 5 and 6; also see Supplementary figure 2A): around residues 688 and 710, near conserved region F; around positions 942, 952 and 1136, within region G or the ‘G-loop’ element (Vassylyev et al., 2002) and β′G′ region (Epstein et al., 2003); around positions 1004 and 1055 within the large non-conserved ‘dispensable’ region of E.coli β′ between regions G and H (Severinov et al., 1996). The binding of Gre factors also caused cleavage enhancement in several areas of β′, mostly in non-conserved regions. The most affected areas are around residue D516, between regions D and E; around L982, P998 and E1066, within or near the dispensable region; and around L1168 and L1183, within region G′. Minor cleavage enhancement was also observed near residues A659, M1040 and P1096. Two areas of cleavage enhancement by Gre were type-specific: only GreA increased cleavage at I923 in region G, while only GreB increased cleavage at G744 in region F (compare lanes 3 and 4). Unlike the non-cognate Tth GreA control, E.coli GreA-T143M displayed a similar, but much weaker, pattern of protection and enhancement in the cleavage of β′ as wt GreA (compare lanes 3 and 5). Analysis of footprinting patterns near N- and C-termini of β′ by high- and low-percentage SDS–PAGE did not reveal any discernible alterations by GreA or GreB (data not shown). Under the same conditions, neither GreA nor GreB affected the cleavage pattern of β (data not shown).
Localized Fe2+-induced hydroxyl radical cleavages of RNAP
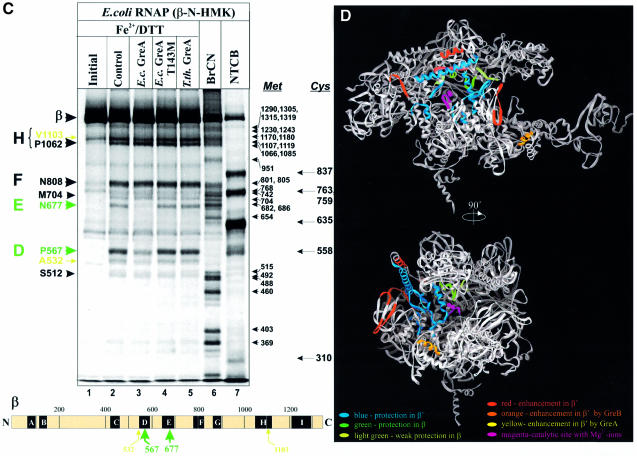
Hydroxyl radicals generated locally by a Fe2+ ion placed in the catalytic center of RNAP induce four major cleavages in β′ (Figure 2B, lane 2) in conserved regions D (N458), F (G732 and A787) and G (P926) and five major cleavages in β (Figure 2C, lane 2) in regions D (P567), E (N677), F (N808) and H (P1062 and V1103), consistent with the previously reported data (Mustaev et al., 1997). Some minor cleavages occurred in non-conserved regions of β (A532, S525 and M717) and β′ (K599, A696, P1125 and V1246). Neither E.coli GreA-T143M nor Tth GreA affected the patterns of localized cleavages in β and β′ (Figure 2B, lanes 5 and 6; Figure 2C, lanes 4 and 5; also Supplementary figure 2B and C). GreA and GreB, on the other hand, strongly protected all cleavage sites in β′, except two in region D (Figure 2B, lanes 3 and 4), the major site at N456 and the minor site at R425, representing respectively, the metal-binding site of the catalytic center and a conserved loop immediately adjacent to it. In the presence of GreA three new cleavage sites emerged: β′K216 in the conserved region B, β′Q1114 in the dispensable region and β′P1185 in region G′.
The effect of GreA and GreB on localized cleavages in β was less pronounced than in β′. Both Gre proteins specifically protected two major sites, βP567 in region D and βN677 in region F (only data for GreA are shown; Figure 2C, lane 3). We also observed a weak but reproducible protection at a major cleavage site in region H (βV1103) and at a minor site in the non-conserved region (βA532).
Taken together, these results indicate that Gre factors interact mostly with sites in β′ located within conserved regions F, G and G′, and within non-conserved segments between regions E and F, and between G and H. Consistent with our crosslinking data (Figure 1A), both Gre factors contact sites in regions D and E of β. Most of the sites protected in β and β′ represent either the active center of RNAP or its proximate surrounding.
3-D representation of the regions in β and β′ that are affected by Gre binding
In Figure 2D, the regions of protection and enhancement deduced from Fe–EDTA footprinting and localized radical cleavage experiments are superimposed on a high- resolution structure of Tth RNAP. Although protected regions are scattered throughout the primary sequences of β and β′, they converge in and around the secondary channel of RNAP in 3-D representation. Two major areas of protection are visible on the RNAP structure. The first is a 30-residue long α-helix (V673–E704) in the β′ coiled-coil structure that forms the rim of the secondary channel (Figure 2D). The second area of protection includes: G-loop and F-helix of β′, two highly conserved structural elements that form the inner wall of the secondary channel and part of the active center; two loops from regions D and H of β; and an α-helix from region E of β. The last three elements are located where the secondary channel intersects the main DNA-binding channel of RNAP, and are part of the catalytic center (Korzheva et al., 2000). These results suggest that Gre interacts with parts of β and β′ that form the wall of the secondary channel and the active center of RNAP.
The enhancement of cleavages at sites located 35–60 Å from the catalytic center may reflect allosteric effect(s). Binding of GreA, and to lesser extent GreB, enhanced cleavages in two areas of β′: one in the non-conserved region (K1167–P1191) immediately after G′ and another in region B (E211–K222). In 3-D structure, the first area is visible as a three β-strand bundle, and the second area as α-helix in region B. Both of these elements participate in the formation of a downstream DNA-binding site. These data suggest that Gre binding induces conformational changes in the downstream DNA-binding region of RNAP.
One major area that included both the protected and enhanced sites of β′ (see Figure 2A) could not be visualized on the structure. This is a 190 residue-long non-conserved segment of the G-loop structure of E.coli β′ (residues 944–1132), which is absent in the Tth RNAP β′. This region has been previously implicated in interactions with E.coli GreB (Zakharova et al., 1998).
Localized hydroxyl radical cleavages of GreA and GreB in complex with RNAP
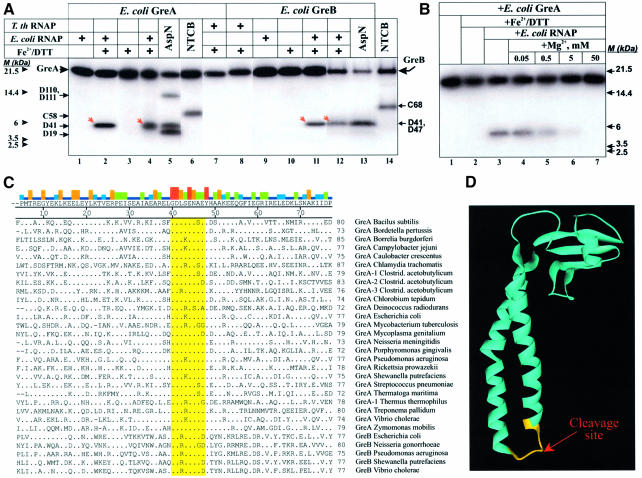
To determine the orientation of Gre in the Gre–RNAP complex, and to identify Gre region(s) that interacts with the RNAP active center, we subjected radiolabeled Gre to localized Fe2+-mediated cleavages (Figure 3). In the presence of E.coli RNAP, both GreA (Figure 3A, lanes 2 and 4) and GreB (lanes 11 and 12) were efficiently cleaved by hydroxyl radicals. Neither Gre protein was cleaved in the absence of RNAP (lanes 3 and 10), or in the presence of non-cognate Tth RNAP (lanes 7 and 8). Mg2+ ions competitively inhibited the cleavage of GreA (Figure 3B), GreB and RNAP β and β′ (Mustaev et al., 1997). The cleavages in both Gre and RNAP were also completely inhibited by EDTA (data not shown). Based on these data we conclude that Gre factor cleavage originates from Fe2+ in the catalytic center and not by Fe2+ in solution.
Fig. 3. Localized hydroxyl radical cleavages in Gre–RNAP complexes. (A and B) Autoradiograms of gradient 10–20% Bis–Tris/MES SDS–PAGE. (A) Fe2+-mediated cleavages of GreA and GreB (red arrows) upon binding RNAP. (B) Inhibition of localized Fe2+-mediated cleavages in GreA by Mg2+. Size markers, products of chemical cleavages of Gre at Asp and Cys, are indicated by small black arrows. The proteolytic sites for AspN were established by N-terminal amino acid sequencing of all cleavage products. Molecular weight markers are shown under M (kDa). (C) Amino acid sequence alignment of GreNTD from 30 bacterial organisms (Koulich et al., 2000 and references within; Hogan et al., 2002). Consensus sequence is shown above the alignment. Numbering is according to E.coli GreA. The colored bars on top indicate sequence conservation: red, 100% identity; dark blue, <20%; orange, green, and light blue represent intermediate levels. The conserved tip is highlighted. (D) Location of the cleavage site (in yellow) on the 3-D structure of GreA (Stebbins et al., 1995), shown as ribbons.
The cleavage sites in GreA and GreB were mapped using molecular size markers obtained by limited proteolysis of radiolabeled Gre at D and C (Figure 3A, lanes 5, 13 and 6, 14), using protease AspN and NTCB, respectively. The site for hydroxyl radical cleavage in both Gre is located within the conserved peptide G40–E47 at the tip of the NTD. This segment comprises a loop connecting two α-helices of the NTD coiled-coil (Figure 3D), and contains highly conserved residues, including invariant G40, D41 and E44 (Figure 3C).
We conclude that in the Gre–RNAP complex, the NTD tip is at or very near the catalytic center of RNAP.
Structural model of the Gre–RNAP complex
The results described in this and previous studies were used to build a 3-D model (Figure 4) of Gre–RNAP complex structure. To achieve the best possible fit we performed manual docking by WebLab ViewerPro program using high-resolution structures of E.coli GreA (Stebbins et al., 1995) and Tth RNAP holoenzyme (Vassylyev et al., 2002). The following restrictions were applied: (i) there should be minimal steric clashes between polypeptide backbones of GreA and RNAP; (ii) the Gre–RNAP interface should include areas revealed by hydroxyl radical cleavages; (iii) the CTD should be near the secondary channel (Polyakov et al., 1998), with residues around 97 and 78 facing β and β′, respectively; (iv) the CTD should interact mostly with β′ through residues around 121–123 (Loizos and Darst, 1999) and 143 (Borukhov et al., 2001); (v) the basic patch residues 37 and 52 of Gre should be within 18 Å of the catalytic site (Stebbins et al., 1995; Koulich et al., 1997, 2000); and (vi) the tip of the GreNTD should contact β′ and, partially, β, and be near the catalytic center.
Fig. 4. 3-D structural model of Gre–RNAP complex. (A–C) Three views of the complex shown in ribbon diagram with color coding indicated. Views A and B are the same as two views in Figure 2D. (C) The blown up view of B with proposed regions of Gre–RNAP interactions indicated. The side chains of two essential residues of GreANTD tip, D41 and E44, are shown in yellow.
In the resulting model (Figure 4), the GreCTD binds to the rim of the secondary channel formed by a 13-residue-long α-helical segment (V673–E685) of the β′ coiled-coil. In E.coli RNAP, the CTD would also interact with the non-conserved, dispensable region of the β′ G-loop. The NTD protrudes into the secondary channel towards the catalytic center. The first α-helix of the NTD contacts the middle section of an α-helix of the β′ coiled-coil (A688–K695) and a 7-residue-long loop in region F (G729–A735). The second α-helix of NTD contacts the β′ G-loop (P926–A944), G′-region (K1132–P1139), and the middle section of the F-helix (R779–T786). The tip of the NTD interacts with three conserved structural elements of β: region D loop (T563–G570), region E α-helical segment (D675–G682) and region H loop (S1105–M1107). This spatial arrangement brings two highly conserved acidic residues of the NTD tip, D41 and E44, within 3–4 Å of the catalytic Mg2+.
Mutational analysis of the GreNTD tip
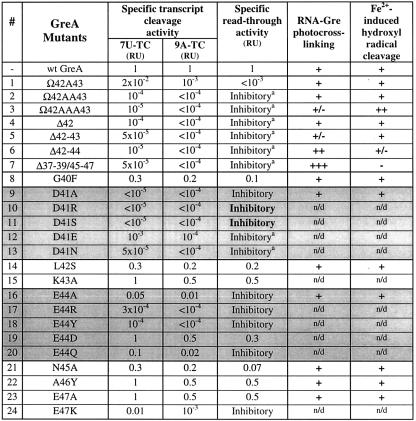
The results of our modeling suggest that the NTD tip may be involved in modulation of RNAP catalytic function. Single or multiple deletions and Ala insertions in the tip, or shortening of the NTD coiled-coil by one helical turn caused two to five orders of magnitude decrease in the functional activities of Gre (Table I, mutants 1–7). CD spectroscopy revealed that Gre mutants were structurally intact (data not shown). They were also similar to wt GreA in their amenability to RNA crosslinking and/or to localized Fe2+-induced cleavages. Thus, all deletion/insertion mutants were competent in binding RNAP and positioning their NTD in the secondary channel. These results indicate that the NTD tip is crucial for the functional activity of Gre. Moreover, the NTD should be of specific length and orientation in order for its tip to act on the catalytic center of RNAP.
Table I. Functional activity of GreANTD mutants.
To determine the function of each of the eight residues of the tip they were individually substituted as follows: D, N, K and E to A (to disrupt possible hydrogen bonds and/or electrostatic interactions); L to S (to disrupt possible hydrophobic or van der Waals interactions), G and A to F and Y, respectively (to destroy the possible steric fit between Gre and RNAP). Of the eight substitutions, K43A, A46Y and E47A (15, 22, 23) had no effect, while G40F, L42S and N45A (8, 14, 21) decreased the activity of mutant proteins 2- to 5-fold (Table I). The cleavage activity of E44A (16) was 20–100× lower than that of wt GreA, while the activity of D41A (9) was undetectable. All mutants behaved as wt GreA in RNA crosslinking and localized Fe2+-induced radical cleavage assays, excluding the possibility that protein denaturation or misfolding caused their loss of activity. We conclude that the functional importance of the NTD tip resides principally with two residues, D41 and E44. The contribution of E44 appears to be less significant than that of D41.
We examined the effects of altering the charge, geometry and polar property of the side chains of D41 and E44 (Table I). Substitutions of D41 to R and S (10 and 11) led to a complete loss of transcript cleavage activity, while change to N caused ∼104-fold decrease in activity. The conservative substitution D41E (12) decreased the activity 103–104-fold. Substitutions of E44 to R and Y (17 and 18) resulted in ∼104-fold loss of activity. The E44Q substitution (20), analogous to D42N, led to only 10- to 50-fold decrease in activity. Finally, conservative substitution E44D (19) had little or no effect. In conclusion, the presence of aspartate at position 41 of GreA is almost absolutely required for its function. The requirement for glutamate at position 44 is less stringent. Neither position 41 nor 44 tolerates either positively charged or bulky hydrophobic residues.
Functionally inactive D41 and E44 mutants inhibit RNAP catalytic activity
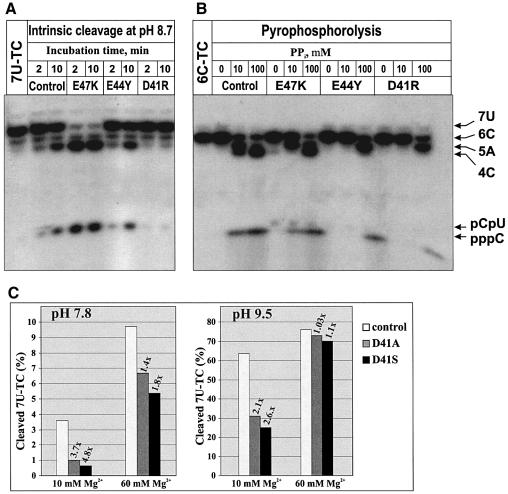
At high concentrations, all D41 and E44 mutants (except E44D) inhibited transcription elongation (Table I). The strength of inhibitory effect was in the following order: D41R>D41S>D41A, D41N, E44R, E44Y>E41D>E44A, E44Q. The effect was more pronounced at low concentrations of Mg2+ (2 mM versus standard 10 mM, data not shown). In contrast to partially active E47K, which was used as a control, three D41 mutants, D41R, D41S and D41A, but none of the E44 mutants, inhibited the intrinsic nucleolytic activity of RNAP induced by mild alkaline (Figure 5A, only data for representative mutants are shown). Interestingly, increasing the concentration of Mg2+ from 10 to 60 mM mitigated the inhibitory effect of D41 mutants (Figure 5C, only data for D41S and D41A are shown). In addition, E44R, E44Y and all D41 mutants but not E447 inhibited pyrophosphorolysis (Figure 5B, only data for three representative mutants are shown). The inhibition of RNAP catalytic activities by functionally defective D41 and E44 mutants, and the mitigating effect of Mg2+ on this inhibition, suggest that in the wt Gre, D41 and E44 may participate in the binding of the catalytic Mg2+ ion(s) required for RNA hydrolysis.
Fig 5. Inhibitory effects of GreA-D41 and -E44 mutations. Autoradiograms show (A) a time course of 7U-TC cleavage into pentamer (5A) and dimer (pCpU) at pH 8.7 with and without Gre mutants, and (B) pyrophorolysis of 6C-TC into 5A, tetramer (4C) and CTP at pH 7.5 with and without Gre mutants at different concentrations of pyrophosphate. 6C- and 7U-TC carry transcripts internally radiolabeled at C. (C) Effect of Mg2+ on the inhibition of intrinsic transcript cleavage activity by GreA-D41A and -D41S mutants. Bar graphs show the decrease in the amount of the 7U-TC after 10 min incubation at 30°C under indicated conditions. The % of 7U-TC cleaved was calculated based on the amount of 5A formed in the course of the reaction. Numbers on top of the bars indicate fold inhibition by D41A (gray) and D41S (black) compared to control (white).
D41 and E44 residues are part of the RNAP catalytic center
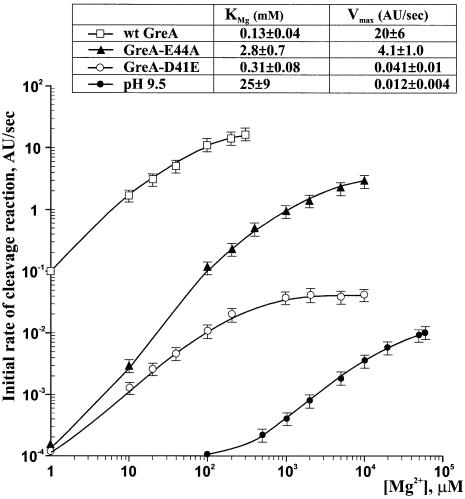
To investigate the role of D41 and E44 in Mg2+ binding, we analyzed the dependence of intrinsic and factor-induced transcript cleavage reactions on Mg2+ concentration (Figure 6; also see Supplementary figure 6). The results show that, compared to pH 9.1-induced intrinsic cleavage, wt GreA lowered the Mg2+ concentration required for maximum rate of the reaction by two orders of magnitude and accelerated the rate of RNA hydrolysis by more than three orders of magnitude. Thus, the mechanism by which Gre induces nuclease activity of RNAP comprises two distinct components: the increase of the affinity of RNAP catalytic center to Mg2+, and the lowering of the activation energy (Vmax) of phosphodiester bond hydrolysis. These two components represent distinct aspects of Gre action since D41E substitution affects the requirement for Mg2+ in the cleavage reaction by only 2-fold, while it reduces the Vmax value by 1000-fold. On the other hand, E44A affects both parameters of Gre action as evidenced by its 20-fold increase in the apparent dissociation constant for Mg2+ (KMg) and 8-fold decrease in Vmax for cleavage reaction.
Fig. 6. Dependence of the rate of endonucleolytic reaction on Mg2+ concentration. The curves in the graph show the initial rates of 5A formation from 7U-TC at 2°C as a function of Mg2+ concentration at pH 9.5 or at pH 7.5 in the presence of different GreA mutants. Horizontal bars above and below each data point indicate experimental errors. The apparent dissociation constant for Mg2+ ion (KMg) and maximum rate of nucleolytic reaction (Vmax) were derived from double reciprocal plots (see Supplementary figure 6) of the data shown in the graph.
Discussion
Structural overview of the Gre–RNAP complex
The most notable feature of the presented model of the Gre–RNAP complex is the positioning of the GreNTD inside the secondary channel of RNAP. Although the model was built with minimum steric clashes between the polypeptide backbones of Gre and RNAP, it is evident from the structures of core and holoenzyme (Zhang et al., 1999; Vassylyev et al., 2002) that the side chains of the residues in the secondary channel will hinder the entry of GreNTD. Therefore, upon binding RNAP, GreCTD must trigger a conformational change in RNAP to widen the channel. Our previous finding that the CTD polypeptide stimulates the cleavage activity of NTD >100-fold (Koulich et al., 1998) corroborates this hypothesis. Because the secondary channel is elliptic in shape, the backtracked RNA or NTP substrates can still occupy its internal space even with GreNTD bound.
There is also an inherent topological constraint posed by the unusual shape of the Gre molecule. The binding of GreCTD to the β′ coiled-coil does not afford Gre enough room to maneuver its extended (∼45 Å-long) rigid NTD. For Gre to anchor the CTD to the β′ coiled-coil and simultaneously insert the NTD into the channel, either the β′ coiled-coil, Gre, or both must be made flexible. In the first case, the β′ coiled-coil must swing 12–15 Å from the main body of RNAP; in the second case, the NTD must initially occupy an alternative site on β′, parallel to the β′ coiled-coil, then pivot towards the secondary channel. The internal flexibility of Gre has previously been implied from GreB–RNAP footprinting data (Loizos and Darst, 1999). That the NTD may occupy two binding sites on RNAP was earlier proposed based on the stimulatory effect of the NTD on Gre–RNA crosslinking (Koulich et al., 1998). It is also consistent with our observation that the areas of weak protection against hydroxyl radical cleavages in β′ include regions that are located 10–20 Å from the proposed Gre-binding interface (Figure 4): namely, the loop Q595–K605 and the lower segment of the β′ coiled-coil A696–E705 (Figure 2).
Our model suggests that Gre allosterically affects two functionally important areas in RNAP; part of the downstream DNA-binding clamp (β′ regions K1167–P1191 and E211–K222), and the wall in the DNA/RNA hybrid binding site (β region I530–P535) (Figures 2 and 4). This follows from the observation that Gre binding affects the protection/enhancement pattern of these areas in hydroxyl radical cleavage although they are located 20–60 Å from the Gre-binding site (Figures 2 and 4). Thus, Gre binding may be implicated in stabilization or destabilization of interactions between downstream DNA and the DNA-binding clamp, and between RNA–DNA hybrid and its binding site, both of which could affect abortive initiation. This could explain the stimulatory effect of Gre on RNAP transition from initiation to elongation and on RNAP promoter escape (Hsu et al., 1995).
While this paper was in review, two articles appeared in Cell describing the structures of E.coli GreB–RNAP core (Opalka et al., 2003) and Saccharomyces cerevisiae TFIIS–Pol II complexes (Kettenberger et al., 2003), respectively. The former is a 15 Å resolution structure reconstructed from cryo-EM and image processing, and the latter is a 3.8 Å resolution structure determined from X-ray crystallographic data. Remarkably, in both structures, the extended parts of the transcript cleavage factors (NTD of GreB and domain III of TFIIS) occupy the functionally analogous ‘secondary channel’ and ‘pore’ of bacterial RNAP and Pol II, respectively, consistent with our data. Moreover, TFIIS-induced changes in Pol II including the repositioning of the jaws, the clamp and the RPB1 cleft and foot domains (Kettenberger et al., 2003), are equivalent to GreA-induced changes in the downstream DNA-binding clamp of E.coli RNAP observed in our footprinting experiments, even though the structural conservation in this region of RNAPs is very low. The placement of Gre Cα backbone is almost identical between our structural model of GreA–RNAP and the cryo-EM model of GreB–RNAP, which are at comparable levels of resolution and reliability. The two models differ slightly in the placement of GreNTD tip relative to conserved regions of β at the active center. In the GreB–RNAP complex the tip is located near βF (812–814) and βH (1105–1106) (Opalka et al., 2003) whereas in our model, besides βH, the tip interacts with βD (563–570) and βE (675–682) (see Figure 2C and D and Figure 4D).
Mechanism of Gre action
It is postulated that all reactions catalyzed by RNAP (RNA synthesis, pyrophosphorolysis, and RNA hydrolysis, both intrinsic and factor-dependent) require two Mg2+ ions (Sosunov et al., 2003). The first Mg2+ ion (Mg-I) is chelated by three invariant aspartates of RNAP β′, and is tightly bound (Zaychikov et al., 1996). The second Mg2+ ion (Mg-II) is coordinated by two of the same aspartates, and is presumably less stably bound (Cramer et al., 2001; Vassylyev et al., 2002; Sosunov et al., 2003). It was recently proposed that Mg-II is recruited to the catalytic center by NTP substrates during synthesis or by other ligands such as non-complementary NTPs or pyrophosphate during exonucleolytic hydrolysis and pyrophosphorolysis, respectively (Sosunov et al., 2003). Our structural and functional data suggest that Mg-II required for endonucleolytic hydrolysis in TC is chelated by D41 and E44 of Gre. The striking proximity of the NTD tip to the catalytic center of RNAP, the near absolute requirement of aspartate at position 41, and the strong dependence of the activities of D41/E44 mutants on Mg2+ concentration all support this hypothesis.
In pre-translocated TC, in the absence of NTP, Mg-II is presumably chelated by two catalytic aspartates and the non-bridging oxygen of the 3′-proximal phosphate of RNA. We speculate that during Gre-induced hydrolysis, the carboxyls of E44 and D41 and the oxygen of the attacking water molecule complete the octahedral coordination of Mg-II (Steitz and Steitz, 1993). Furthermore, D41 may simultaneously coordinate the attacking water molecule to stabilize the pentavalent transition state of the scissile phosphate. This would explain the far more deleterious effects of D41 mutations compared with those of E44. Similar roles are proposed for the two invariant acidic residues of TFIIS, D290 and E291 (S.cerevisiae nomenclature) based on the 3-D crystal structure of the TFIIS–Pol II complex (Kettenberger et al., 2003). These residues are located at the tip of its RNA-binding Zn-ribbon domain, and are absolutely essential for transcript cleavage activity (Fish and Kane, 2002). Thus, Gre and TFIIS may represent the first known examples of transcription factors that modify RNAP activity by directly participating in the catalytic processes.
Our experimental data do not rule out the possibility that D41/E44 are involved in protein–protein interactions critical for the nucleolytic functions of RNAP. Several conserved basic residues of β and β′ are located in the catalytic cleft, and may represent the interacting partners for D41 and E44. It should also be noted that double D41A/E44A mutant (data not shown), as well as mutants with deletions, insertions and multiple Ala-substitutions in the tip, all induce very low but detectable cleavage activity in TC (Table I), indicating that Gre may not act entirely through coordination of Mg2+ and water. The interaction of NTD with the β′ G-loop, F-helix (see Figure 4), and RNA (through Gre basic patch) may stabilize the backtracked conformation of RNA in TC, and align the scissile phosphodiester bond in an orientation favorable to hydrolysis, contributing to an increased rate of reaction.
Materials and methods
Cloning, expression and purification of Gre factors and RNAPs carrying phosphorylation sites
Escherichia coli RNAP core carrying heart muscle kinase (HMK) recognition site either at the N-terminus of β or at the C-terminus of β′ was purified as described (Orlova et al., 1995) from E.coli strain CAG3016 transformed with corresponding plasmids (Borukhov et al., 2001). pCYB2β′C-PKA (derivative of pMKa201, a gift of K.Severinov) carries rpoC, encoding β′-subunit with a C-terminal HMK recognition site (LRRASV) and a hexahistidine (6×His) tag, under the control of an IPTG-inducible tac promoter. pTRC99Aβ-S531Y-NPH (derivative of pMKSe250) carries mutant rpoB, which encodes β-S531Y with the N-terminal HMK site and 6×His tag. The HMK and 6×His tags were introduced into rpoB-S531Y, by PCR amplification using appropriate primers and pMKSer carrying rpoB-S531Y (gift of K.Severinov) as the template, followed by cloning into pTRC99A vector. The S531Y mutation eliminates the intrinsic phosphorylation site for HMK in wt β.
The N-terminally HMK- and 6×His-tagged E.coli greA gene was constructed by PCR-mutagenesis using appropriate primers and pMO1.1 (Koulich et al., 1997) as the template, and cloned into pET19b (Novagen). The resulting pET19b[NPH]greA was transformed into E.coli BL21(DE-3)pLysS cells for overexpression. All other E.coli and Tth Gre genes were constructed similarly. All greA and greB mutations were introduced either into pET19b[NPH]greA or pDKB(GreB) (Kulish et al., 2000) background, respectively, using three-step PCR or QuickChange site-directed mutagenesis kit (Stratagene). Primers used in all PCR-based cloning were obtained from IDT and their sequences are available upon request. The sequences of all resulting plasmids were verified by DNA sequencing (GeneWiz). The resulting Gre proteins were purified and stored as described (Borukhov and Goldfarb, 1996; Laptenko and Borukhov, 2003). CD spectroscopy was performed at Rutgers University CD Spectroscopy Facility.
The E.coli and Tth RNAPs were purified from greA–/greB– E.coli strain AD8571 (Orlova et al., 1995) and Tth HB8 strain, respectively (Borukhov and Goldfarb, 1993; Vassylyev et al., 2002).
Fe–EDTA protein footprinting and localized Fe2+-induced hydroxyl radical mapping
HMK-tagged RNAP core (10 µg) or Gre proteins (1.5 µg) were radiolabeled in 30 µl reaction as described (Heyduk et al., 1996; Loizos and Darst, 1999), mixed with 1 µl of 20 mg/ml BSA, and purified by G-50 QuickSpin column (Roche) pre-equilibrated with chelated buffer A: 20 mM Na-HEPES pH 7.5, 150 mM NaCl, yielding ∼8.0 µg (0.15∼0.2 µCi/pmol) of core and ∼1 µg (0.04∼0.05 µCi/pmol) of Gre.
For Fe–EDTA footprinting, the radiolabeled RNAP core (0.6 µg), unlabeled Gre (0.6 µg) and BSA (3 µg) were mixed in 7 µl of buffer A containing 1 mM MgCl2. Then 1 µl each of freshly prepared 10× solutions of Fe–EDTA [10 mM (NH4)2Fe(SO4)2 + 20 mM EDTA], ascorbate (200 mM) and H2O2 (10 mM) were added, the reaction was incubated for 15 min at 25°C, and terminated by addition of 3.4 µl of 4× NuPAGE sample loading buffer (SLB, Novex) with 50 mM β-ME. The samples were separated by 8 × 10 cm long gradient 4–12% Bis–Tris/MES SDS–PAGE (Mark 12, Novex), or by a standard 20 × 20 cm long 8% Tris–Glycine SDS–PAGE. The reaction products were visualized by autoradiography and quantified by phosphorimager (Molecular Dynamics).
For localized hydroxyl radical mapping of RNAP–Gre complex 8 µl-reactions containing 160 nM of radiolabeled RNAP and 1.6 µM of unlabeled Gre, 7 µg BSA in buffer A were pre-incubated for 5 min at 25°C, then mixed with 1 µl of freshly made 500 µM (NH4)2Fe(SO4)2 (Sigma) and 1 µl of 10 mM DTT (Borukhov et al., 2001), and incubated for 10 min at 25°C. The localized radical cleavages of Gre in complex with RNAP were performed similarly, except 0.1 µM of radiolabeled Gre and 0.6 µM of unlabeled RNAP were used (Laptenko and Borukhov, 2003). All reactions were terminated and analyzed as above.
The position of the cleavage sites indicated in the figures and in the text correspond to the center 7- to 10-residue-long segment, which represents the limit of precision of mapping. All results presented were reproduced at least three times.
Visualization of protein 3-D structures and molecular modeling was done using WebLab ViewerPro program (Accelris).
Enzymatic and residue-specific chemical degradation of proteins
N-terminally radiolabeled Gre proteins were subjected to limited degradation by AspN protease (Koulich et al., 1997). Briefly, 40 ng of radiolabeled Gre was mixed with 2 µg of unlabeled protein and incubated with 40–60 ng of AspN in 20 µl of 40 mM Tris–HCl pH 8.2, 40 mM NaCl, 1 mM DTT, 5 mM CaCl2 for 30–60 min at 30°C. The reaction was terminated by the addition of 5 µl SLB containing 20 mM EDTA and analyzed by 4–12% Bis–Tris/MES SDS–PAGE (Novex) as above. Alternatively, the gels were stained by Coomassie Blue R-450 and the protein bands were subjected to N-terminal amino acid sequencing performed by SUNY Downstate Protein Facility.
β, β′ and Gre molecular weight markers were prepared by subjecting radiolabeled proteins to residue-specific chemical cleavages either at Met by BrCN, or at Cys by NTCB as described (Koulich et al., 1997; Mustaev et al., 1997).
GreB–RNAP photocrosslinking
[S35]ASDPC was synthesized as described (Chen et al., 1994) using [S35]cysteine (1000 Ci/mmol, Amersham-Pharmacia Biotech) as starting material. For derivatization, 3 µg of each purified GreB-Cys mutant were immobilized on 5 µl of Ni–NTA agarose (Qiagen), incubated in buffer B (40 mM Tris–HCl pH 8.0, 0.2 M NaCl) containing 25 mM β-mercaptoethanol for 1 h at 25°C, washed 4 times with 0.5 ml of degassed buffer B, and incubated with 0.4 nmol of [S35]ASDPC (10 Ci/mmol) in 10 µl buffer B containing 10% dimethylformamide for 4 h at 25°C under argon. The extent of Cys modification in GreB was determined spectrophotometrically at 314 nm (ε1M = 6500) and by transfer of radioactivity to GreB. The derivatized proteins (15–20% yield of modification) were washed 4 × 0.5 ml with buffer B, then mixed with 20 µg BSA and 60 µg of E.coli RNAP in 50 µl of buffer B, and UV-irradiated at 302 nm for 10 min on ice. Reactions were washed as above and analyzed by SDS–PAGE. Under reducing conditions, the radioactive moiety of ASDPC is transferred from GreB to the target RNAP and the crosslink is visualized by autoradiography.
Transcriptional assays
The transcript cleavage and readthrough assays were performed using 202 bp E.coli rrnB P1 DNA fragment (Koulich et al., 1997). The TCs carrying radiolabeled hexamer CpApCpCpApC (6C-TC), heptamer CpApCpCpApCpU (7U-TC) and nonamer CpApCpCpApCpUpGpA (9A-TC) were prepared as described (Borukhov and Goldfarb, 1996). In all assays, each reaction contained ∼1 nM TC and various concentrations of Gre factors in standard transcription buffer (STB): 40 mM Tris–HCl pH 7.5, 0.1 mM DTT, 60 mM NaCl, 10 mM MgCl2. For the readthrough assays, all reactions contained 25 µM NTPs. For all assays, samples were incubated for 10 min at 30°C, unless otherwise indicated. The intrinsic transcript cleavage reactions (Orlova et al., 1995) were carried out in STB or in 40 mM Tris–HCl pH 9.1 or 40 mM CAPS-Na pH 9.5 containing 0.1 M NaCl, 0.1 mM DTT with or without MgCl2. Pyrophosphorolysis was performed using 6C-TC in STB with or without Gre factors and/or MgCl2 at various concentrations. All RNA products were analyzed by urea–23%-PAGE, autoradiographed and quantified by phosphorimager.
Gre–RNA photocrosslinking was performed using radiolabeled 9A-TC carrying 8-azidoadenosine monophosphate (N3AMP) placed at the RNA 3′-terminus as described (Koulich et al., 1997, 2000).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to O.Khodova for help in synthesis of crosslinking reagent, K.Severinov for providing β- and β′-plasmids and for helpful discussions, and N.J.Greenfield for help in CD spectroscopy data interpretation. This work was supported, in part, by NIH Grant GM54098 to S.B.
References
- Borukhov S. and Goldfarb,A. (1993) Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr. Purif., 4, 503–511. [DOI] [PubMed] [Google Scholar]
- Borukhov S. and Goldfarb,A. (1996) Purification and assay of Escherichia coli transcript cleavage factors GreA and GreB. Methods Enzymol., 274, 315–326. [DOI] [PubMed] [Google Scholar]
- Borukhov S., Sagitov,V. and Goldfarb,A. (1993) Transcript cleavage factors from E.coli. Cell, 72, 459–466. [DOI] [PubMed] [Google Scholar]
- Borukhov S., Laptenko,O. and Lee,J. (2001) Escherichia coli transcript cleavage factors GreA and GreB: functions and mechanisms of action. Methods Enzymol., 342, 64–76. [DOI] [PubMed] [Google Scholar]
- Chen Y., Ebright,Y.W. and Ebright,R.H. (1994) Identification of the target of a transcription activator protein by protein–protein photocrosslinking. Science, 265, 90–92. [DOI] [PubMed] [Google Scholar]
- Cramer P., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science, 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- Epstein V., Mustaev,A., Markovtsov,V., Bereshchenko,O., Nikiforov,V. and Goldfarb,A. (2002) Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol. Cell, 10, 623–634. [DOI] [PubMed] [Google Scholar]
- Erie D.A., Hajiseyedjavadi,O., Young,M.C. and von Hippel,P.H. (1993) Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science, 262, 867–873. [DOI] [PubMed] [Google Scholar]
- Feng G.H., Lee,D.N., Wang,D., Chan,C.L. and Landick,R. (1994) GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J. Biol. Chem., 269, 22282–22294. [PubMed] [Google Scholar]
- Fish R.N. and Kane,C.M. (2002) Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta, 1577, 287–307. [DOI] [PubMed] [Google Scholar]
- Gnatt A. (2002) Elongation by RNA polymerase II: structure–function relationship. Biochim. Biophys. Acta, 1577, 175–190. [DOI] [PubMed] [Google Scholar]
- Heyduk T., Heyduk,E., Severinov,K., Tang,H. and Ebright,R.H. (1996) Determinants of RNA polymerase alpha subunit for interaction with beta, beta’, and sigma subunits: hydroxyl-radical protein footprinting. Proc. Natl Acad. Sci. USA, 93, 10162–10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B.P., Hartsch,T. and Erie,D.A. (2002) Transcript cleavage by Thermus thermophilus RNA polymerase. Effects of GreA and anti-GreA factors. J. Biol. Chem., 277, 967–975. [DOI] [PubMed] [Google Scholar]
- Hsu L.M., Vo,N.V. and Chamberlin,M.J. (1995) Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc. Natl Acad. Sci. USA, 92, 11588–11592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C.A., Peterson,S.N., Gill,S.R., Cline,R.T., White,O., Fraser,C.M., Smith,H.O. and Venter,J.C. (1999) Global transposon mutagenesis and a minimal Mycoplasma genome. Science, 286, 2165–2169. [DOI] [PubMed] [Google Scholar]
- Izban M.G. and Luse,D.S. (1992). The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′-5′ direction in the presence of elongation factor SII. Genes Dev., 6, 1342–1356. [DOI] [PubMed] [Google Scholar]
- Kettenberger H., Armache,K.J. and Cramer,P. (2003) Architecture of the RNA polymerase II–TFIIS complex and implications for mRNA cleavage. Cell, 114, 347–357. [DOI] [PubMed] [Google Scholar]
- Komissarova N. and Kashlev,M. (1997) Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl Acad. Sci. USA, 94, 1755–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzheva N., Mustaev,A., Kozlov,M., Malhotra,A., Nikiforov,V., Goldfarb,A. and Darst,S.A. (2000). A structural model of transcription elongation. Science, 289, 619–625. [DOI] [PubMed] [Google Scholar]
- Koulich D., Orlova,M., Malhotra,A., Sali,A., Darst,S.A. and Borukhov,S. (1997) Domain organization of Escherichia coli transcript cleavage factors GreA and GreB. J. Biol. Chem., 272, 7201–7210. [DOI] [PubMed] [Google Scholar]
- Koulich D., Nikiforov,V. and Borukhov,S. (1998) Distinct functions of N and C-terminal domains of GreA, an Escherichia coli transcript cleavage factor. J. Mol. Biol., 276, 379–389. [DOI] [PubMed] [Google Scholar]
- Kulish D., Lee,J., Lomakin,I., Nowicka,B., Das,A., Darst,S., Normet,K. and Borukhov,S. (2000) The functional role of basic patch, a structural element of Escherichia coli transcript cleavage factors GreA and GreB. J. Biol. Chem., 275, 12789–12798. [DOI] [PubMed] [Google Scholar]
- Laptenko O. and Borukhov,S. (2003) Methods Enzymol., in press. [Google Scholar]
- Laptenko O., Hartsch,T., Bozhkov,A.I. and Borukhov,S. (2000) Gre-homologic transcription factors GreA-1 and GreA-2 from Thermus thermophilus: cloning, purification and analysis of functional activity. Biologicheskiy Vestnik, 4, 3–14 (in Russian). [Google Scholar]
- Loizos N. and Darst,S.A. (1999) Mapping interactions of Escherichia coli GreB with RNA polymerase and ternary elongation complexes. J. Biol. Chem., 274, 23378–23386. [DOI] [PubMed] [Google Scholar]
- Marr M.T. and Roberts,J.W. (2000) Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell, 6, 1275–1285. [DOI] [PubMed] [Google Scholar]
- Mustaev A., Kozlov,M., Markovtsov,V., Zaychikov,E., Denissova,L. and Goldfarb,A. (1997) Modular organization of the catalytic center of RNA polymerase. Proc. Natl Acad. Sci. USA, 94, 6641–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E. (1999) Transcription elongation: structural basis and mechanisms. J. Mol. Biol., 288, 1–12. [DOI] [PubMed] [Google Scholar]
- Nudler E., Mustaev,A., Lukhtanov,E. and Goldfarb,A. (1997) The RNA–DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell, 89, 33–41. [DOI] [PubMed] [Google Scholar]
- Opalka N., Chlenov,M., Chacon,P., Rice,W.J., Wriggers,W. and Darst,S.A. (2003) Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell, 114, 335–345. [DOI] [PubMed] [Google Scholar]
- Orlova M., Newlands,J., Das,A., Goldfarb,A. and Borukhov,S. (1995) Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl Acad. Sci. USA, 92, 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyakov A. Richter,C. Malhotra,A. Koulich,D. Borukhov,S. and Darst,S.A. (1998) Visualization of the binding site for the transcript cleavage factor GreB on Escherichia coli RNA polymerase. J. Mol. Biol., 281, 465–473. [DOI] [PubMed] [Google Scholar]
- Price M.A. and Tullius,T.D. (1992) Using hydroxyl radical to probe DNA structure. Methods Enzymol., 212, 194–219. [DOI] [PubMed] [Google Scholar]
- Reines D. (1992). Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J. Biol. Chem., 267, 3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Severinov K. (2000) RNA polymerase structure–function: insights into points of transcriptional regulation. Curr. Opin. Microbiol., 3, 118–125. [DOI] [PubMed] [Google Scholar]
- Severinov K., Mustaev,A., Kukarin,A., Muzzin,O., Bass,I., Darst,S.A. and Goldfarb,A. (1996) Structural modules of the large subunits of RNA polymerase. Introducing archaebacterial and chloroplast split sites in the beta and beta’ subunits of Escherichia coli RNA polymerase. J. Biol. Chem., 271, 27969–27974. [DOI] [PubMed] [Google Scholar]
- Sparkowski J. and Das,A. (1990) The nucleotide sequence of greA, a suppressor gene that restores growth of an Escherichia coli RNA polymerase mutant at high temperature. Nucleic Acids Res., 18, 6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins C.E., Borukhov,S., Orlova,M., Polyakov,M., Goldfarb,A. and Darst,S.A. (1995) Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature, 373, 636–640. [DOI] [PubMed] [Google Scholar]
- Steitz T.A. and Steitz,J.A. (1993) A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA, 90, 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulme F., Mosrin-Huaman,C., Sparkowski,J., Das,A., Leng,M. and Rahmouni,A.R. (2000) GreA and GreB proteins revive backtracked RNA polymerase in vivo by promoting transcript trimming. EMBO J., 19, 6853–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.J., Platas,A.A. and Hawley,D.K. (1998) Transcriptional fidelity and proofreading by RNA polymerase II. Cell, 93, 627–637. [DOI] [PubMed] [Google Scholar]
- Uptain S.M., Kane,C.M. and Chamberlin,M.J. (1997) Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem., 66, 117–172. [DOI] [PubMed] [Google Scholar]
- Vassylyev D.G., Sekine,S., Laptenko,O., Lee,J., Vassylyeva,M.N., Borukhov,S. and Yokoyama,S. (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature, 417, 712–719. [DOI] [PubMed] [Google Scholar]
- Wang Y., Severinov,K., Loizos,N., Fenyo,D., Heyduk,E., Heyduk,T., Chait,B.T. and Darst,S.A. (1997) Determinants for Escherichia coli RNA polymerase assembly within the β subunit. J. Mol. Biol., 270, 648–662. [DOI] [PubMed] [Google Scholar]
- White O. et al. (1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science, 286, 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharova N., Bass,I., Arsenieva,E., Nikiforov,V. and Severinov,K. (1998) Mutations in and monoclonal antibody binding to evolutionary hypervariable region of Escherichia coli RNA polymerase beta′ subunit inhibit transcript cleavage and transcript elongation. J. Biol. Chem., 273, 24912–24920. [DOI] [PubMed] [Google Scholar]
- Zaychikov E. et al. (1996) Mapping of catalytic residues in the RNA polymerase active center. Science, 273, 107–109. [DOI] [PubMed] [Google Scholar]
- Zhang G., Campbell,E.A., Minakhin,L., Richter,C., Severinov,K. and Darst,S.A. (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell, 98, 811–824. [DOI] [PubMed] [Google Scholar]