Abstract
Background
Most biological functions controlled by the brain and their related disorders are closely associated with activation in specific regions of the brain. Neuroproteomics has been applied to the analysis of whole brain, and the general pattern of protein expression in all regions has been elucidated. However, the comprehensive proteome of each brain region remains unclear.
Results
In this study, we carried out comparative proteomics of six regions of the adult rat brain: thalamus, hippocampus, frontal cortex, parietal cortex, occipital cortex, and amygdala using semi-quantitative analysis by Mascot Score of the identified proteins. In order to identify efficiently the proteins that are present in the brain, the proteins were separated by a combination of SDS-PAGE on a C18 column-equipped nano-liquid chromatograph, and analyzed by quadrupole-time of flight-tandem-mass spectrometry. The proteomic data show 2,909 peptides in the rat brain, with more than 200 identified as region-abundant proteins by semi-quantitative analysis. The regions containing the identified proteins are membrane (20.0%), cytoplasm (19.5%), mitochondrion (17.1%), cytoskeleton (8.2%), nucleus (4.7%), extracellular region (3.3%), and other (18.0%). Of the identified proteins, the expressions of glial fibrillary acidic protein, GABA transporter 3, Septin 5, heat shock protein 90, synaptotagmin, heat shock protein 70, and pyruvate kinase were confirmed by immunoblotting. We examined the distributions in rat brain of GABA transporter 3, glial fibrillary acidic protein, and heat shock protein 70 by immunohistochemistry, and found that the proteins are localized around the regions observed by proteomic analysis and immunoblotting. IPA analysis indicates that pathways closely related to the biological functions of each region may be activated in rat brain.
Conclusions
These observations indicate that proteomics in each region of adult rat brain may provide a novel way to elucidate biological actions associated with the activation of regions of the brain.
Background
The mammalian central nervous system regulates higher biological actions such as feelings and behaviors, which are known to associate with the activation of specific regions of brain as determined by positron-emission topography and MRI techniques [1-3]. A characterization of all components expressed in each region is essential to understand the mechanisms of higher actions and their functional properties. Recently, methods for the comprehensive analyses of gene expression such as DNA array and serial analysis gene expression have been developed in addition to the disclosure of genomic sequence information [4-7]. These applications have provided much information about gene expression in the brain [8-13]. A comprehensive account of the products of gene expression through protein profiling is needed even more than genome information to fully understand higher biological actions.
Neuroproteomics has been applied to the analysis of whole brain, and the general pattern of protein expression in all regions has been elucidated [14-16]. In specific regions, such as the hippocampus, thalamus, and striatum, the relationships between protein stimulations or diseases have been examined by proteomic analysis [17-20]. However, the comprehensive proteome of each brain region remains unclear despite the attention paid to the biological functions of each region, although comprehensive investigations of gene expression in each region of the brain have been undertaken using DNA array and in situ hybridization techniques [8].
We have undertaken proteomic analysis using a variety of protocols according to the research targets, such as the combination of two-dimensional electrophoresis with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and nano-liquid chromatography (LC)-quadrupole-time of flight (Q-TOF)-tandem mass spectrometry (MS/MS) after column concentration [21-27]. Here, we have divided the rat brain into six regions, thalamus, hippocampus, frontal cortex, parietal cortex, occipital cortex, and amygdala, and compared the proteome of each region by a combination of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and nano-LC-Q-TOF-MS/MS. We have identified a total of 2,909 peptides in all regions of the rat brain, and found proteins specifically expressed in each region of brain: 63 proteins in the thalamus, 38 in the hippocampus, 14 in the frontal cortex, 66 in the parietal cortex, 24 in the occipital cortex, and 36 in the amygdala by semi-quantitative analysis.
Results
Identification of proteins in each region of rat brain
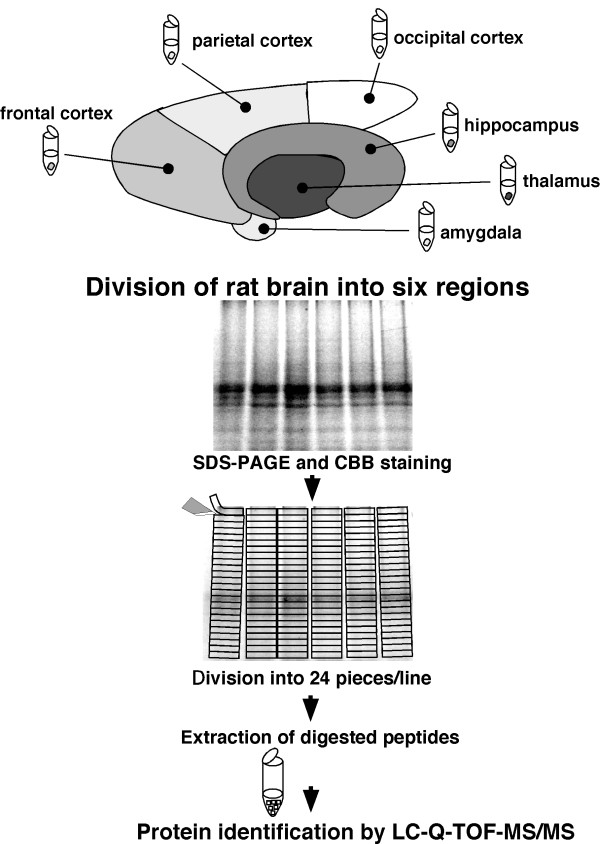
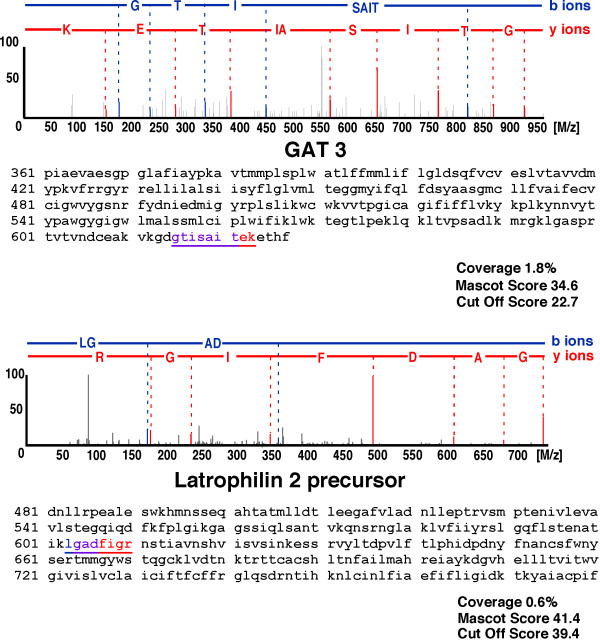
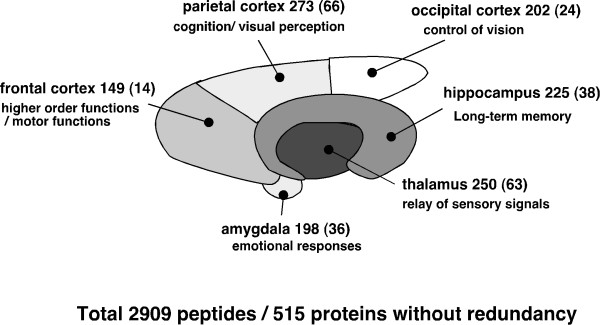
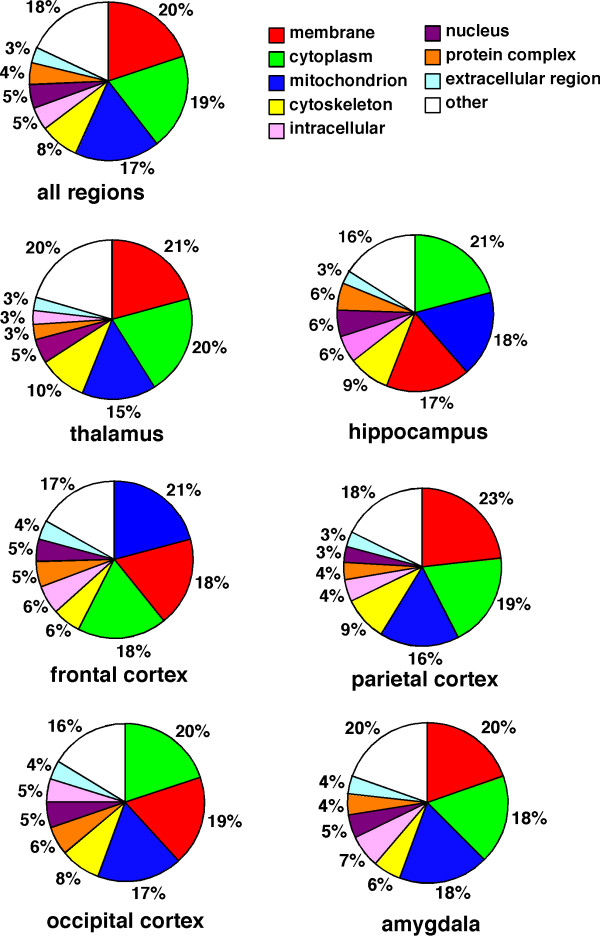
To identify proteins expressing in each region of rat brain, the removed brains were divided into six regions, thalamus, hippocampus, frontal cortex, parietal cortex, occipital cortex, and amygdala, according to the atlas of Paxinos and Watson. To test the efficiency of protein extraction by lysis buffer, we determined the protein concentrations extracted by lysis buffers included Triton X-100, CHAPS, NP-40, or SDS. The extracted proteins were determined by EZQ protein quantification kit (Molecular Probe), which is able to determine the protein concentrations in the presence of the detergents. The proteins concentration in the solution extracted by SDS sample buffer was significantly high compared with those in other lysis buffers (data not shown). The proteins in each region were extracted by lysis buffer including SDS, and separated by electrophoresis. Each gel lane was sliced into 24 pieces, and the proteins extracted from the gel pieces were applied to nano-LC-Q-TOF-MS/MS and analyzed by the Mascot search engine to identify amino acid sequences (Fig. 1). All together in all brain regions, 2,909 redundant peptides were assigned, and 515 proteins were identified with more than 95% confidence based on the amino acid sequences deduced from the MS/MS peaks (Additional file 1, Table S1). The proteins assigned by a single peptide were confirmed manually. Representative MS/MS spectra of the identified proteins GABA transporter 3 (GAT 3) and Latrophilin 2 are shown in Fig. 2. GAT 3 was identified based on the amino acid sequence GTISAITEK deduced by both b type ions and y type ions on the MS/MS spectrum, the sequence of which corresponds to 1.8% of the whole sequence. For the Latrophilin 2 precursor, the amino acid sequence deduced from the MS spectrum is LGADFIGR, which covers 0.6% of the full sequence. The peaks used for the identification of Latrophillin 2 were observed more prominently than other peaks on the spectrum, and the confidence level calculated from the peak data was greater than 95% by Mascot search. The numbers of proteins identified in each region without redundancy were 250, 225,149, 273, 202, and 198 for thalamus, hippocampus, frontal cortex, parietal cortex, occipital cortex, and amygdala, respectively (Fig. 3). Among the identified proteins, the number of region-abundant proteins was 63 in thalamus, 38 in hippocampus, 14 in frontal cortex, 66 in parietal cortex, 24 in occipital cortex, and 36 in amygdala (Fig. 3) by semi-quantitative analysis. Several functional molecules were observed in the specific regions, such as G protein coupled receptors (thyroid stimulating hormone receptor 1 and Latrophilin 2) in the parietal cortex, and olfactory receptors (olfactory receptor family 10 and olfactory Olr 436) in the occipital cortex (Table 1). Next we analyzed the intracellular localization of the identified proteins based on the component section in the NCBI Entrez Gene. In all regions, most proteins localized in the membrane fraction (20.0%), with other proteins classified into cytoplasm (19.5%), mitochondrion (17.1%), cytoskeleton (8.2%), nucleus (4.7%), extracellular region (3.3%), and other (18.0%) (Fig. 4).
Figure 1.
Flow diagram of the experimental design. Rat brains were divided into six regions: thalamus, hippocampus, frontal cortex, parietal cortex, occipital cortex, and amygdala. The divided samples were lysed in lysis buffer containing SDS, and subjected to SDS-PAGE with Coomassie Brilliant Blue staining. The gel lane was divided into 24 slices, and the slices were pre-treated by in-gel trypsin digestion. The amino acid sequences of all detected proteins were determined by nano-LC-Q-TOF-MS/MS.
Figure 2.
Representative MS/MS spectra of proteins identified in rat brain. The MS/MS spectrum for the peptide derived from GAT 3 (A), identified only in the thalamus of the rat brain, is shown; the amino acid sequence GTISAITEK deduced from the 5 b type ions (blue) and 8 y type ions (red) were assigned by Mascot search (upper panel). The identified sequence within the entire amino acid sequence of GAT 3 is indicated by the underline (lower panel). The MS/MS spectrum for the peptide derived from the Latrophilin 2 precursor (B), identified only in the parietal cortex of the rat brain, is shown; the amino acid sequence GADFIGR deduced from the 2 b type ions (blue) and 7 y type ions (red) were assigned by Mascot search (upper panel). The identified sequence within the entire amino acid sequence of Latrophilin 2 is indicated by the underline (lower panel).
Figure 3.
Schematic representation of proteins identified in six regions of rat brain. In total, 2,909 peptides including redundant peptides were identified by nano-LC-Q-TOF-MS/MS in all regions of the brain, leaving a total of 515 proteins. By region, 250 proteins were identified in the thalamus, 225 in the hippocampus, 149 in the frontal cortex, 273 in the parietal cortex, 202 in the occipital cortex, and 198 in the amygdala without redundancy. Sixty-three proteins in the thalamus, 38 in the hippocampus, 14 in the frontal cortex, 66 the in parietal cortex, 24 in the occipital cortex, and 36 in the amygdala were found in only that region of the brain.
Table 1.
Representative membrane proteins identified in each region of adult rat brain by Q-TOF-MS/MS
| NCBI ID | Protein Name | Region | Localization | Function |
|---|---|---|---|---|
| Receptor | ||||
| gi|94380294 | PREDICTED: similar to Carcinoembryonic antigen-related cell adhesion molecule 1 precursor (Biliary glycoprotein 1) (BGP-1) (Murine hepatitis virus receptor) (MHV-R) | T | ND | |
| gi|32401457 | opsin 5 | H, A | ND | G-protein coupled receptor |
| gi|38259186 | adiponectin receptor 1 | F | M | fatty acid metabolism |
| gi|62990176 | thyroid stimulating hormone receptor | P | M | G-protein coupled receptor |
| gi|6912464 | latrophilin 2 precursor | P | M | G-protein coupled receptor |
| gi|52317184 | olfactory receptor, family 10, subfamily X, member 1 | O | M | olfactory receptor |
| gi|47577367 | olfactory receptor Olr436 | O | M | olfactory receptor |
| Ion channel | ||||
| gi|6755963 | voltage-dependent anion channel 1 | T, H, F, P, O, A | M, Mt | voltage-gated ion-selective channel activity |
| Na Pump | ||||
| gi|21450321 | Na+/K+ -ATPase alpha 3 subunit | T, H, F, P, O, A | ND | |
| gi|30409956 | ATPase, Na+/K+ transporting, alpha 2 polypeptide | T, H, F, P, O | M | sodium:potassium-exchanging ATPase |
| gi|6978543 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | T, F, P, O, A | M | sodium:potassium-exchanging ATPase |
| gi|6978549 | ATPase, Na+/K+ transporting, beta 1 polypeptide | T, H, F, P, O, A | M | sodium:potassium-exchanging ATPase |
| Ca Pump | ||||
| gi|62234487 | plasma membrane calcium ATPase 1 | T, H, P, O | M | calcium-transporting ATPase |
| gi|48255951 | plasma membrane calcium ATPase 2 isoform a | P, O | M | calcium-transporting ATPase |
| Proton Pump | ||||
| gi|34856315 | PREDICTED: similar to ATPase, H+ transporting, V1 subunit B, isoform 1 | T, H, O, A | ND | |
| gi|62665162 | PREDICTED: ATPase, H+ transporting, V0 subunit D isoform 1 (predicted) | T, H, F, P, A | ND | |
| gi|34869154 | PREDICTED: similar to ATPase, H+ transporting, V1 subunit A, isoform 1 | T, H, F, P | ND | |
| gi|12025532 | ATPase, H+ transporting, lysosomal V0 subunit a isoform 1 | T, F, P | Cy, M, N | hydrogen ion transporter activity |
| gi|47717102 | ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H isoform 2 | T | Cy, M | hydrogen-transporting ATP synthase activity |
| gi|19913426 | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 | H, F, O, A | Cy, M, I | hydrogen-transporting ATPase |
| gi|124244102 | ATPase, H+ transporting, lysosomal V0 subunit a isoform 2 | P | Cy, Ex, M | |
| Transporter | ||||
| gi|78126167 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 isoform a | T, H, F, P, O, A | M | glutamate transport |
| gi|9507115 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | P, A | M | glutamate transport |
| gi|400626 | Sodium- and chloride-dependent GABA transporter 3 | T | M | neurotransmitter transport |
T, thalamus; H, hippocampus; F, frontal cortex; P, parietal cortex; O, occipital cortex; A, amygdala
M, membrane; Ex, extracellular region; Mt, mitochondrion; N, nucleus; Cy, cytoplasm; I, intracellular; ND, not described
Figure 4.
Intracellular localization of proteins identified by Q-TOF-MS/MS in rat brain. The intracellular localizations of all proteins identified by nano-LC-Q-TOF-MS/MS were classified based on the component section in NCBI Entrez Gene.
Confirmation of protein expression in each region of rat brain
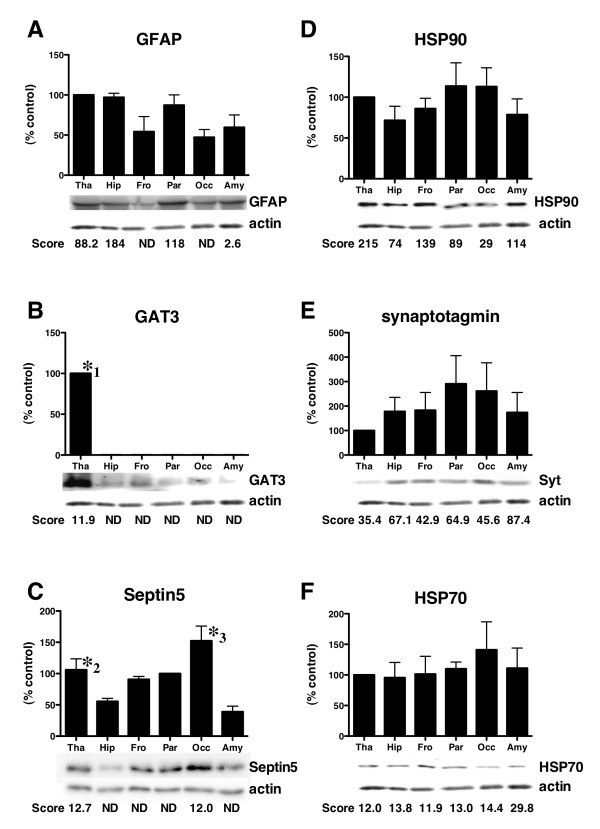
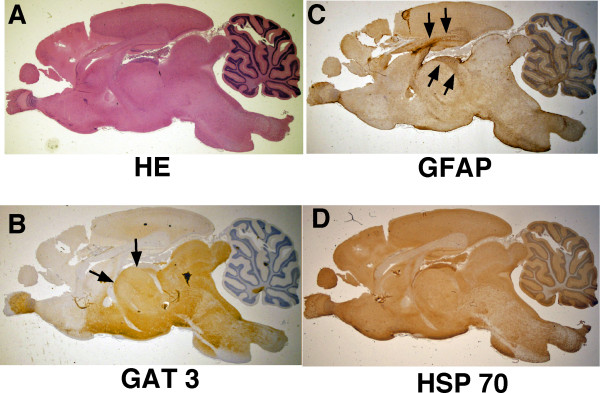
To confirm the brain distribution of proteins identified in the six regions by Q-TOF-MS/MS, we carried out immunoblotting using specific antibodies, and the results were compared with Mascot scores, since there is a correlation between the protein amounts on the gels and Mascot score [22]. Immunoblotting using the anti-glial fibrillary acidic protein (GFAP) antibody demonstrated that bands were observed in all regions, and the densities were increased in the thalamus and hippocampus, a finding that is almost the same as that obtained using Mascot scores (Fig. 5A). GAT 3 was only detected in the thalamus by immunoblotting, which is completely consistent with the results obtained by MS/MS analysis (Fig. 5B). Septin 5 detected in only Parietal cortex by mass analysis, was mainly observed as single band on the immunoblots in Parietal cortex fraction (Fig. 5C). The small amount of Septin 5 was observed in other regions. The difference of data might be due to the sensitivity between immunoblotting and mass analysis. The anti-heat shock protein (HSP) 90 antibody recognized a protein with an approximate molecular mass of 90 kDa in all regions of the brain on the immunoblots, but the band densities in the parietal cortex and occipital cortex were slightly reduced compared with other regions (Fig. 5D). The Mascot scores for HSP 90 were also low in both regions (Fig. 5D). The amount of synaptotagmin (Fig. 5E), HSP 70 (Fig. 5F), and pyruvate kinase (data not shown) were similar in all regions by immunoblotting using specific antibodies. Consistent with these blotting data, the proteins were detected ubiquitously in all regions with constant values in the Mascot scores (Fig. 5E and 5F). Furthermore, we carried out a distribution analysis with antibodies for GFAP, GAT 3, and HSP 70, useful for immunohistochemistry, to confirm the data obtained by the immunoblotting and MS/MS analysis. Immunohistochemical observations indicated that GAT 3 shows preferential staining in the thalamus, hypothalamus, medulla, and olfactory bulb, consistent with the MASCOT score data (Fig. 6A and 6B). GFAP staining was observed in all regions, and was enhanced in part of the hippocampus and thalamus (Fig. 6A and 6C). HSP 70 was ubiquitously detected in all regions of the brain by immunohistochemistry (Fig. 6A and 6D). These data are mostly consistent with the data obtained by immunoblotting and MS/MS analysis. Together with all of the observations, the data determined by nano-LC-Q-TOF-MS/MS are reliable for the identification of brain proteins.
Figure 5.
Immunoblotting analysis of proteins identified in six regions of rat brain. Rat brains were divided into six regions (thalamus, hippocampus, frontal cortex, parietal cortex, occipital cortex, and amygdala), and were extracted with lysis buffer. The samples (2.0 μg protein) were subjected to immunoblotting with anti-GFAP antibody (A), anti-GAT 3 antibody (B), anti-Septin 5 antibody (C), anti-HSP 90 antibody (D), anti-synaptotagmin antibody (E), and anti-HSP 70 antibody (F). Immunoblotting data using the indicated antibodies are shown in the upper panels; Mascot scores of the proteins determined by Q-TOF-MS/MS are shown in the lower panels. The immunoblotting data are shown as representative blots obtained in independent experiments from 3 rats. Data represent the means ± SE. Statistical significance was determined by ANOVA followed by Bonferroni's test. *1 p < 0.05 vs Hip, Fro, Par, Occ, and Amy, *2 p < 0.05 vs Amy, *3 p < 0.05 Amy and Hip.
Figure 6.
Immunohistochemical analysis of proteins identified by nano-LC-Q-TOF-MS/MS in rat brain. Rat brains were removed, fixed with 4% paraformaldehyde, and stained with hematoxylin and eosin (A), anti-GAT 3 antibody (B), anti-GFAP antibody (C), or anti-HSP 70 antibody (D) as described in "Experimental Procedures". The panels show representative photographs from independent experiments from 3 rats. Final magnification × 10.
IPA analysis in proteins identified in each region of rat brain
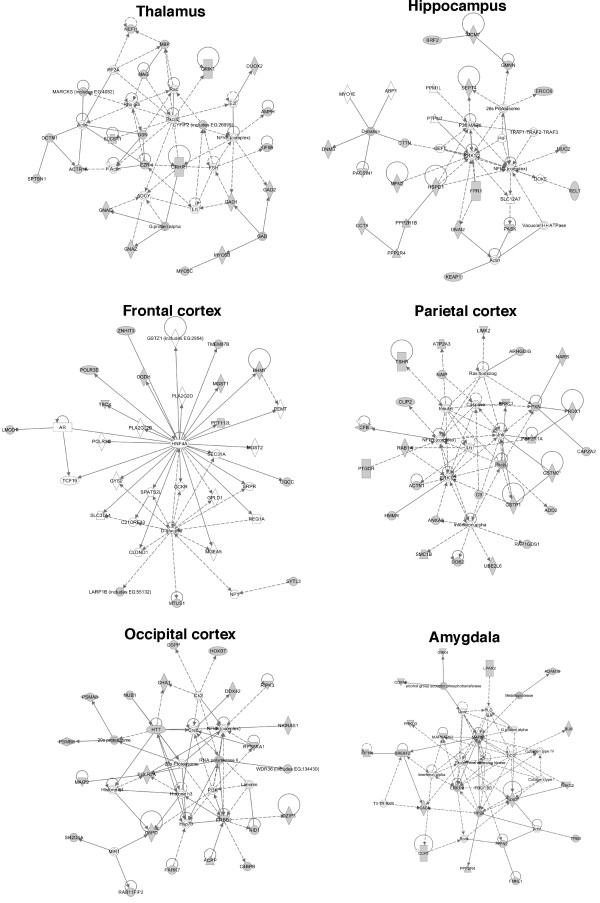
We carried out the pathway analysis by IPA analysis using the data of proteins identified in each region of rat brain. The representative networks in each region were shown in Fig.7 with Additional file 2, Table S2 and the canonical pathways were also revealed in Additional file 3, Fig. S1. The pathway through corticotropin releasing hormone receptor was detected in thalamus releasing corticotropin-releasing hormone under stress (Fig.7). In hippocampus, the pathways through ERK1 and S6 kinase were detected n Fig.7 and Additional file 3, Fig. S1, suggesting that the development and functions in nervous system may be activated through growth factors such as BDNF in the region. The pathway related to citrate cycle was observed in frontal cortex and the pathway involving in mitochondria dysfunction in amygdala was shown in Additional file 3, Fig. S1.
Figure 7.
Network analysis of proteins identified in each region of rat brain by Ingenuity pathway analysis. The networks were analyzed based on the data of proteins identified in the indicated regions of rat brain. The networks were revealed as circles (genes) and lines (biological relationship). Solid lines mean direct interaction, and dotted lines show indirect interactions between the genes.
Discussion
We have conducted a comprehensive analysis of the proteins expressing in six regions of rat brain following a proteomic approach using SDS-PAGE and nano-LC-Q-TOF-MS/MS. As a result, we identified 250 proteins in thalamus, 225 in hippocampus, 149 in frontal cortex, 273 in parietal cortex, 202 in occipital cortex, and 198 in amygdala. Furthermore, the localizations of several proteins identified by proteomics were confirmed by immunoblotting and immunohistochemistry using specific antibodies.
Membrane proteins including ion channels, receptors, and ion transporters play important roles in brain functions, with nerve cells in particular utilizing diverse functional proteins on the membrane for receiving, conducting and transmitting signals. In this report, the samples extracted directly by SDS from each region of the brain were separated using SDS-PAGE. The approach has advantages in identifying many proteins including membrane proteins in the brain because the SDS essential for electrophoresis is a strong detergent that lyses most proteins, and can be used directly for the preparation of brain samples, although the resolution cannot be improved. Using this protocol, proteins expressed in the brain were efficiently identified, and over 20% of the identified proteins were deduced to localize in membranes. In hippocampal samples prepared by two-dimensional electrophoresis, over 70% of the identified proteins were cytoplasm-localized, while only 7% were detected as membrane proteins [17].
By comparing the proteins found in each region, proteins closely related to the function of each region have been identified in this study. Of the identified proteins, GAT 3 was found only in the thalamus, and this specific expression was confirmed by immunoblotting. In the mammalian thalamus, GABA is a major inhibitory neurotransmitter, and the GABA transporter mediates GABA uptake into presynaptic terminals to terminate the effects of GABA [28,29]. Consistent with our data, ultrastructural investigations show that GABA transporter 3 is expressed most prominently in the thalamus [8,30]. The immunohistochemical investigation also detected GAT 3 in the olfactory bulb and hypothalamus, and GAT 3 may be expressed except for the regions used in this study [31]. In samples extracted from the occipital cortex, two types of olfactory receptors were specifically identified. Olfactory receptors are understood to function as molecular sensors for odorants [32]. Olfactory receptors are expressed mostly in pyramidal cells of the occipital cortex, which is consistent with our data [33]. Olfactory stimulation results in the activation of the bilateral occipital cortex as detected by positron-emission topography [34]. Improvements in extra-conversion and regional cerebral blood flow might be induced by smell stimulation through the activation of the occipital cortex [35]. The olfactory receptors expressed in the occipital cortex may be involved in biological functions, resulting in smell stimulation. Septin 5 participating in neuronal development was mainly detected in occipital cortex and thalamus. Septin 5 is colocalized in synaptic vesicles with SNARE proteins, and plays a role in neurotransmitter release [36]. However, Septin 5 mice showed no changes in synaptic transmission in hippocampus [37]. In our data, the protein amount of Septin 5 in hippocampus was much less than other regions as shown in Fig. 5 and mass analysis, which was consistent with data with Septin 5-/-mice. In the hippocampal region related to memory, we identified Glyoxalase 1, the localization of which was found to be mostly consistent with that determined by in situ hybridization [8]. The specific expression of Glyoxalase 1, as indicated in this study, suggests the involvement of Glyoxalase 1 in the biological functions of the hippocampus such as memory and long-term potentiation. In fact, Chen et al. indicate that Glyoxalase 1 plays a novel role in Alzheimer's disease and frontotemporal dementia [38].
Network analysis of the proteins expressed in each region of rat brain indicates that these proteins were linked in a pathway. Many of these proteins have associated with amino acid metabolism, molecular transport, and small molecular biochemistry in most regions, which is consistent with the previous observations that neuron activity in the brain was regulated by small molecules such as neurotransmitter and ion transport. In hippocampus playing an important role in long-term memory, the pathways involving in signal transduction such as ERK, NF-kB, S6 kinase, and mTOR were observed by IPA analysis. Long-term depression in hippocampus requires rapid protein synthesis, which is associated with mTOR, ERK, and S6-dependent signaling pathways [39]. Serum- and glucocorticoid-inducible kinase1 increased the acetylation and activation of NF-kB through phosphorylation of p300, and this also leads to the expression of N-methyl-d-aspartate receptor, NR2A and NR2B that is implicated in neuronal plasticity in hippocampus [40]. Dynamic chromatin remodeling in hippocampal neurons are associated with N-methyl-d-aspartate receptor-mediated activation of Bdnf gene promoter 1 [41]. BDNF activates ERK pathway in hippocampus neurons. The pathways shown by IPA analysis based on the data are closely related to the biological functions reported in each region of brain, suggesting that proteomic profiling of regions may be useful for the elucidation of biological functions.
Conclusions
A total of 2,909 peptides in all regions of the rat brain were identified, and we displayed proteins expressed in each region of brain: 250 proteins in the thalamus, 225 in the hippocampus, 149 in the frontal cortex, 273 in the parietal cortex, 202 in the occipital cortex, and 198 in the amygdala. Of the identified proteins, the expressions of GFAP, GAT3, Septin 5, HSP 90, synaptotagmin, HSP 70, and pyruvate kinase were confirmed by immunoblotting, and localizations of GAT3, GFAP, and HSP 70 were confirmed by immunohistochemistry. Proteomics analysis in each region of the brain reveals that protein compositions differ among the regions, and these differences in protein expression may be involved in distinct biological actions. Further investigations are needed to elucidate the molecules involved in the biological actions that take place in each region of the brain.
Experimental Procedures
Materials
Anti-GFAP, anti-GAT 3, and anti-synaptophysin antibodies were from Sigma-Aldrich (St. Louis, MO). Anti-HSP 70, anti-HSP 90, and anti-α-enolase antibodies were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). The anti-synaptotagmin antibody was from BD Transduction Laboratories (Mississauga, ON). The anti-Septin 5 antibody was from Abcam (Cambridge, MA). The anti-pyruvate kinase antibody was from Chemicon International, Inc (Temecula, CA). Sequencing grade-modified trypsin was from Promega Corp. (Madison, WI). All other chemicals were commercially available.
Animals
Sprague-Dawley rats were housed in individual plastic cages (40 × 25 × 25 cm) with wood chip bedding in a room with a 12 h light cycle (12:12 light-dark) maintained at 22°C. Animals had free access to food pellets and tap water. All experiments were accordance with the standards of the Committee for Ethics on Animal Experiments at Yamaguchi University School of Medicine. Rats were randomly assigned from a group, and were anesthetized with Nembutal (40 mg/kg, i.p.). After the anesthetization, rats were transcardially perfused with 0.9% chilled saline, and the brains were removed. In rat brains, cortex, hippocampus, thalamus, and amygdala were dissected, and the cortex were divided into 3 sections, frontal, parietal, and occipital cortexes. The division into six regions: thalamus, hippocampus, frontal cortex, parietal cortex, occipital cortex, and amygdala was carried out according to the atlas of Paxinos and Watson. Samples were homogenized individually in lysis buffer [150 mM Tris (pH 6.8), 12% (w/v) SDS, 36% (v/v) glycerol, and 6% (v/v) 2-mercaptoethanol]. The samples were centrifuged (15,000 × g, 30 min, 4°C), and the supernatants were stored at -20°C.
In-gel digestion with trypsin
Samples were subjected to SDS-PAGE, and stained lightly with Coomassie Brilliant Blue. In-gel digestion with trypsin was performed as described previously [22,42]. The protein bands in the lanes for samples from the six regions were excised from the gel and divided equally into 24 slices. Each gel slice was diced into small pieces and destained by rinsing in 30% acetonitrile containing 25 mM NH4HCO3. After the gel pieces were dehydrated in 100% acetonitrile, they were dried naturally at room temperature for 30 min. The proteins in the gel pieces were reduced by incubation with 10 mM dithiothreitol in 25 mM NH4HCO3 at 56°C for 1 hr, and alkylated with 55 mM iodoacetamide in 25 mM NH4HCO3 at room temperature for 45 min in the dark. The gel pieces were dehydrated in 50% acetonitrile containing 25 mM NH4HCO3 twice for 30 min, and then in 100% acetonitrile once for 5 min. After drying for 30 min at room temperature, the gel pieces were rehydrated in TPCK-treated trypsin solution (Trypsin Gold, Mass spectrometry grade, Promega; 10 ng/μl in 25 mM NH4HCO3) on ice for 30 min. After removing excess solution, digestion was performed overnight at 37°C. The resulting peptides were extracted twice with 50% acetonitrile containing 0.1% trifluoroacetic for 30 min. The extracts were dried in a vacuum concentrator and dissolved in a solution of 0.1% formic acid for mass spectrometric analysis.
Mass spectrometric analysis
The digested peptides were subjected to LC-MS/MS analysis as described previously [22,24]. Briefly, LC-MS/MS analysis was performed on a Q-Tof Micro (Micromass, Manchester, UK) interfaced with a capillary reverse-phase liquid chromatograph (Micromass CapLC™ system) as described previously [24]. A linear gradient of acetonitrile in 0.1% formic acid was produced and split at a 1:20 ratio. The gradient solution was then injected into a nano LC column (PepMap C18, 75 μm × 150 mm, LC Packings, Amsterdam, Holland) at 100 nl/min. The eluted peptides were sprayed directly into the mass spectrometer. The MS/MS data were acquired by MassLynx software (ver. 4.0; Micromass) using automatic switching between MS and MS/MS modes, and converted to a single text file (containing the observed m/z of the precursor peptide, the fragment ion m/z, and intensity values) by ProteinLynx software (ver. 2.0; Micromass). The files were analyzed by Mascot MS/MS Ions Search (ver. 3.5; Matrix Science Ltd.) to search and assign the obtained peptides to the NCBI Reference Sequence database (RefSeQ 20060317; 119764 sequences; selected species for the database were Homo sapiens, Mus musculus, and Rattus norvegicus). Peaks from porcine trypsin, human keratins and bovine serum proteins were removed. We set the parameters as follows: Parent mass error tolerance, ± 0.4 Da; Fragment mass error tolerance, ± 0.2 Da; Enzyme, trypsin; Maximum number of missed cleavages, 1; Fixed post-translational modifications, none; Variable post-translational modifications, oxidation (Met), carbamidomethyl (Cys), propionamide (Cys); Mass values, Monoisotopic. For peptide and protein identification, the search results were processed using a STEM software (STrategic Extractor for Mascot results) as follows [43]. (i) The candidate peptide sequences were screened with the probability-based MOWSE scores that exceeded their thresholds (p < 0.05) and with MS/MS signals for y- or b-ions > 3. (ii) Redundant peptide sequences were removed. (iii) Each peptide sequence was assigned to a protein that gave the maximal number of peptide assignments among the candidates. In this study, visual inspection was applied to individual MS/MS spectra for reliable identification of peptide sequences. Gene names followed the NCBI Gene ID.
Electrophoresis and immunoblotting
Electrophoresis and immunoblotting were carried out as described previously [21,44,45]. The brain extracts (400 ng) and molecular mass standards were subjected to electrophoresis in 10% (w/v) polyacrylamide gels in the presence of SDS, and transferred to nitrocellulose membranes. The blots were blocked with 5% non-fat dry milk in Tris-buffered saline containing 0.05% (w/v) Tween-20, and incubated with antibody. The blots were then washed, and the antigens were visualized by enhanced chemiluminescent detection reagents.
Histochemistry and immunohistochemistry
Rat brains were removed, washed with phosphate-buffered solution, and fixed with 4% paraformaldehyde. Then the paraffin-embedded samples were sliced into 4 μm pieces, and stained with hematoxylin and eosin [46,47]. For immunohistochemical observation, the specimens were incubated with 3% H2O2 in phosphate-buffered saline to quench the endogenous peroxidase activity, and then with blocking solution (Dako, Foster City, CA) to inhibit non-specific binding after deparaffinization. Antigen retrieval was performed in 100% formic acid for 1 min at room temperature, and the samples were incubated with antibodies in 1% bovine serum albumin in phosphate-buffered saline for 1 hr and immunostained by the avidin-biotin peroxidase complex method using a Vectastatin kit (Vector Laboratories, Burlingame, CA). The peroxidase label was visualized by exposing the sections to diaminobenzidine.
Ingenuity network analysis
The proteins identified in each region were carried out network analysis using Ingenuity Pathways analysis ver.8.6 (IPA, http://www.ingenuity.com). IPA analysis discerns molecular and cellular functions and canonical pathways on the basis of millions findings reported in the literatures, and the software is weekly updated. IPA uses a Fisher's exact test to determine whether the input genes were significantly related to pathways compared to the whole ingenuity knowledge base.
Abbreviations
LC: liquid chromatography; Q-TOF: quadrupole-time of flight; MS/MS: tandem mass spectrometry; SDS: sodium dodecyl sulfate; PAGE: polyacrylamide gel electrophoresis; GFAP: glial fibrillary acidic protein; GAT 3: GABA transporter 3; HSP: heat shock protein;
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TK, NH, TIzumi, TM, MS, and YM have made substantial contribution to the data acquisition and interpretation in proteomics. HK and TIshihara have participated in the acquisition of the immunohistochemical data. TK, MA, MO, and YM have carried out the experiments of western blotting. YL and SN have contributed to the sample preparations of rat brain. YM have preformed the design of the experiments and have been involved in writing the manuscript. All of authors have read and approved the final manuscript.
Supplementary Material
Table S1. Identification of proteins in each region of adult rat brain. Rat brains were divided into six regions, and extracted, separated by SDS-PAGE. The samples were subjected to nano-LC-Q-TOF-MS/MS, and analyzed by Mascot search. Mascot scores were subtracted cut off scores, and the localizations and biological functions were searched by NCBI Entrez Gene.
Table S2. Lists of ingenuity networks generated by proteins identified in each region of rat brain.
Figure S1. Canonical pathways analyzed by proteins identified in each region of rat brain.
Contributor Information
Tomoki Katagiri, Email: katagiri@mx81.tiki.ne.jp.
Naoya Hatano, Email: nhatano@kms.ac.jp.
Masamune Aihara, Email: n001uf@yamaguchi-u.ac.jp.
Hiroo Kawano, Email: khiroo@yamaguchi-u.ac.jp.
Mariko Okamoto, Email: okamotom@yamaguchi-u.ac.jp.
Ying Liu, Email: xi_liu@yahoo.com.
Tomonori Izumi, Email: izumi001@yamaguchi-u.ac.jp.
Tsuyoshi Maekawa, Email: tmaekawa@yamaguchi-u.ac.jp.
Shoji Nakamura, Email: snaka@yamaguchi-u.ac.jp.
Tokuhiro Ishihara, Email: tishiha@yamaguchi-u.ac.jp.
Mutsunori Shirai, Email: mshirai@yamaguchi-u.ac.jp.
Yoichi Mizukami, Email: mizukami@yamaguchi-u.ac.jp.
Acknowledgements
This work was supported in part by grants from the Ministry of Education, Science and Culture of Japan, Takeda Science Foundation, and NOVARTIS Foundation (Japan) for the Promotion of Science.
References
- Ethofer T, Pourtois G, Wildgruber D. Investigating audiovisual integration of emotional signals in the human brain. Prog Brain Res. 2006;156:345–361. doi: 10.1016/S0079-6123(06)56019-4. full_text. [DOI] [PubMed] [Google Scholar]
- Taylor SF, Welsh RC, Wager TD, Phan KL, Fitzgerald KD, Gehring WJ. A functional neuroimaging study of motivation and executive function. Neuroimage. 2004;21:1045–1054. doi: 10.1016/j.neuroimage.2003.10.032. [DOI] [PubMed] [Google Scholar]
- Allman JM, Hakeem A, Erwin JM, Nimchinsky E, Hof P. The anterior cingulate cortex. The evolution of an interface between emotion and cognition. Ann N Y Acad Sci. 2001;935:107–117. doi: 10.1111/j.1749-6632.2001.tb03476.x. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Coelho PS, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung KH, Sheehan A, Symoniatis D, Umansky L. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- Young RA. Biomedical discovery with DNA arrays. Cell. 2000;102:9–15. doi: 10.1016/S0092-8674(00)00005-2. [DOI] [PubMed] [Google Scholar]
- Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Sun T, Patoine C, Abu-Khalil A, Visvader J, Sum E, Cherry TJ, Orkin SH, Geschwind DH, Walsh CA. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science. 2005;308:1794–1798. doi: 10.1126/science.1110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- Ibrahim SM, Mix E, Bottcher T, Koczan D, Gold R, Rolfs A, Thiesen HJ. Gene expression profiling of the nervous system in murine experimental autoimmune encephalomyelitis. Brain. 2001;124:1927–1938. doi: 10.1093/brain/124.10.1927. [DOI] [PubMed] [Google Scholar]
- Mycko MP, Papoian R, Boschert U, Raine CS, Selmaj KW. cDNA microarray analysis in multiple sclerosis lesions: detection of genes associated with disease activity. Brain. 2003;126:1048–1057. doi: 10.1093/brain/awg107. [DOI] [PubMed] [Google Scholar]
- Sturzebecher S, Wandinger KP, Rosenwald A, Sathyamoorthy M, Tzou A, Mattar P, Frank JA, Staudt L, Martin R, McFarland HF. Expression profiling identifies responder and non-responder phenotypes to interferon-beta in multiple sclerosis. Brain. 2003;126:1419–1429. doi: 10.1093/brain/awg147. [DOI] [PubMed] [Google Scholar]
- Schuchhardt J, Glintschert A, Hartl D, Irmler M, Beckers J, Stephan C, Marcus K, Klose J, Meyer HE, Malik A. BrainProfileDB - a platform for integration of functional genomics data. Proteomics. 2008;8:1162–1164. doi: 10.1002/pmic.200700743. [DOI] [PubMed] [Google Scholar]
- Schindler J, Lewandrowski U, Sickmann A, Friauf E, Nothwang HG. Proteomic analysis of brain plasma membranes isolated by affinity two-phase partitioning. Mol Cell Proteomics. 2006;5:390–400. doi: 10.1074/mcp.T500017-MCP200. [DOI] [PubMed] [Google Scholar]
- Kim SY, Chudapongse N, Lee SM, Levin MC, Oh JT, Park HJ, Ho IK. Proteomic analysis of phosphotyrosyl proteins in morphine-dependent rat brains. Brain Res Mol Brain Res. 2005;133:58–70. doi: 10.1016/j.molbrainres.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Fountoulakis M, Tsangaris GT, Maris A, Lubec G. The rat brain hippocampus proteome. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;819:115–129. doi: 10.1016/j.jchromb.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Pan S, Shi M, Jin J, Albin RL, Lieberman A, Gearing M, Lin B, Pan C, Yan X, Kashima DT, Zhang J. Proteomics identification of proteins in human cortex using multidimensional separations and MALDI tandem mass spectrometer. Mol Cell Proteomics. 2007;6:1818–1823. doi: 10.1074/mcp.M700158-MCP200. [DOI] [PubMed] [Google Scholar]
- Paulson L, Martin P, Nilsson CL, Ljung E, Westman-Brinkmalm A, Blennow K, Davidsson P. Comparative proteome analysis of thalamus in MK-801-treated rats. Proteomics. 2004;4:819–825. doi: 10.1002/pmic.200300622. [DOI] [PubMed] [Google Scholar]
- Yeom M, Shim I, Lee HJ, Hahm DH. Proteomic analysis of nicotine-associated protein expression in the striatum of repeated nicotine-treated rats. Biochem Biophys Res Commun. 2005;326:321–328. doi: 10.1016/j.bbrc.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Iwamatsu A, Aki T, Kimura M, Nakamura K, Nao T, Okusa T, Matsuzaki M, Yoshida K, Kobayashi S. ERK1/2 regulates intracellular ATP levels through alpha-enolase expression in cardiomyocytes exposed to ischemic hypoxia and reoxygenation. J Biol Chem. 2004;279:50120–50131. doi: 10.1074/jbc.M402299200. [DOI] [PubMed] [Google Scholar]
- Kikuchi M, Hatano N, Yokota S, Shimozawa N, Imanaka T, Taniguchi H. Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J Biol Chem. 2004;279:421–428. doi: 10.1074/jbc.M305623200. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Hoshida Y, Kanai F, Tanaka Y, Tateishi K, Ikenoue T, Obi S, Sato S, Teratani T, Shiina S. Proteomic analysis of sera from hepatocellular carcinoma patients after radiofrequency ablation treatment. Proteomics. 2005;5:4287–4295. doi: 10.1002/pmic.200401287. [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Hatano N, Inuzuka H, Yokokura S, Nozaki N, Kobayashi R. Mechanism of the generation of autonomous activity of Ca2+/calmodulin-dependent protein kinase IV. J Biol Chem. 2004;279:40296–40302. doi: 10.1074/jbc.M406534200. [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Hatano N, Inuzuka H, Sueyoshi Y, Yokokura S, Ichimura T, Nozaki N, Kobayashi R. Phosphorylation of Numb family proteins. Possible involvement of Ca2+/calmodulin-dependent protein kinases. J Biol Chem. 2005;280:35108–35118. doi: 10.1074/jbc.M503912200. [DOI] [PubMed] [Google Scholar]
- Nunomura K, Nagano K, Itagaki C, Taoka M, Okamura N, Yamauchi Y, Sugano S, Takahashi N, Izumi T, Isobe T. Cell surface labeling and mass spectrometry reveal diversity of cell surface markers and signaling molecules expressed in undifferentiated mouse embryonic stem cells. Mol Cell Proteomics. 2005;4:1968–1976. doi: 10.1074/mcp.M500216-MCP200. [DOI] [PubMed] [Google Scholar]
- Nagano K, Taoka M, Yamauchi Y, Itagaki C, Shinkawa T, Nunomura K, Okamura N, Takahashi N, Izumi T, Isobe T. Large-scale identification of proteins expressed in mouse embryonic stem cells. Proteomics. 2005;5:1346–1361. doi: 10.1002/pmic.200400990. [DOI] [PubMed] [Google Scholar]
- Crunelli V, Leresche N. A role for GABAB receptors in excitation and inhibition of thalamocortical cells. Trends Neurosci. 1991;14:16–21. doi: 10.1016/0166-2236(91)90178-W. [DOI] [PubMed] [Google Scholar]
- Ulrich D, Huguenard JR. GABAB receptor-mediated responses in GABAergic projection neurones of rat nucleus reticularis thalami in vitro. J Physiol. 1996;493(Pt 3):845–854. doi: 10.1113/jphysiol.1996.sp021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasi S, Vitellaro-Zuccarello L, Brecha NC. Immunoreactivity for the GABA transporter-1 and GABA transporter-3 is restricted to astrocytes in the rat thalamus. A light and electron-microscopic immunolocalization. Neuroscience. 1998;83:815–828. doi: 10.1016/S0306-4522(97)00414-4. [DOI] [PubMed] [Google Scholar]
- Kawamoto M, Ohno K, Kuriyama K, Kubo T, Sato K. Developmental changes in GABA transporter (GAT1 and GAT3) mRNA expressions in the rat olfactory bulb. Brain Res Dev Brain Res. 2001;126:137–145. doi: 10.1016/S0165-3806(00)00137-1. [DOI] [PubMed] [Google Scholar]
- Wensley CH, Stone DM, Baker H, Kauer JS, Margolis FL, Chikaraishi DM. Olfactory marker protein mRNA is found in axons of olfactory receptor neurons. J Neurosci. 1995;15:4827–4837. doi: 10.1523/JNEUROSCI.15-07-04827.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaki JM, Yamamoto H, Firestein S. Odorant receptor expression in the mouse cerebral cortex. J Neurobiol. 2004;58:315–327. doi: 10.1002/neu.10272. [DOI] [PubMed] [Google Scholar]
- Qureshy A, Kawashima R, Imran MB, Sugiura M, Goto R, Okada K, Inoue K, Itoh M, Schormann T, Zilles K, Fukuda H. Functional mapping of human brain in olfactory processing: a PET study. J Neurophysiol. 2000;84:1656–1666. doi: 10.1152/jn.2000.84.3.1656. [DOI] [PubMed] [Google Scholar]
- Vaidya JG, Paradiso S, Andreasen NC, Johnson DL, Boles Ponto LL, Hichwa RD. Correlation between extraversion and regional cerebral blood flow in response to olfactory stimuli. Am J Psychiatry. 2007;164:339–341. doi: 10.1176/appi.ajp.164.2.339. [DOI] [PubMed] [Google Scholar]
- Amin ND, Zheng YL, Kesavapany S, Kanungo J, Guszczynski T, Sihag RK, Rudrabhatla P, Albers W, Grant P, Pant HC. Cyclin-dependent kinase 5 phosphorylation of human septin SEPT5 (hCDCrel-1) modulates exocytosis. J Neurosci. 2008;28:3631–3643. doi: 10.1523/JNEUROSCI.0453-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng XR, Jia Z, Zhang Y, Ware J, Trimble WS. The septin CDCrel-1 is dispensable for normal development and neurotransmitter release. Mol Cell Biol. 2002;22:378–387. doi: 10.1128/MCB.22.1.378-387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wollmer MA, Hoerndli F, Munch G, Kuhla B, Rogaev EI, Tsolaki M, Papassotiropoulos A, Gotz J. Role for glyoxalase I in Alzheimer's disease. Proc Natl Acad Sci USA. 2004;101:7687–7692. doi: 10.1073/pnas.0402338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antion MD, Hou L, Wong H, Hoeffer CA, Klann E. mGluR-dependent long-term depression is associated with increased phosphorylation of S6 and synthesis of elongation factor 1A but remains expressed in S6K-deficient mice. Mol Cell Biol. 2008;28:2996–3007. doi: 10.1128/MCB.00201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai DJ, Su CC, Ma YL, Lee EH. SGK1 phosphorylation of IkappaB Kinase alpha and p300 Up-regulates NF-kappaB activity and increases N-Methyl-D-aspartate receptor NR2A and NR2B expression. J Biol Chem. 2009;284:4073–4089. doi: 10.1074/jbc.M805055200. [DOI] [PubMed] [Google Scholar]
- Tian F, Hu XZ, Wu X, Jiang H, Pan H, Marini AM, Lipsky RH. Dynamic chromatin remodeling events in hippocampal neurons are associated with NMDA receptor-mediated activation of Bdnf gene promoter 1. J Neurochem. 2009;109:1375–1388. doi: 10.1111/j.1471-4159.2009.06058.x. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkawa T, Taoka M, Yamauchi Y, Ichimura T, Kaji H, Takahashi N, Isobe T. STEM: a software tool for large-scale proteomic data analyses. J Proteome Res. 2005;4:1826–1831. doi: 10.1021/pr050167x. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Kobayashi S, Uberall F, Hellbert K, Kobayashi N, Yoshida K. Nuclear mitogen-activated protein kinase activation by protein kinase czeta during reoxygenation after ischemic hypoxia. J Biol Chem. 2000;275:19921–19927. doi: 10.1074/jbc.M907901199. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Yoshioka K, Morimoto S, Yoshida K. A novel mechanism of JNK1 activation. Nuclear translocation and activation of JNK1 during ischemia and reperfusion. J Biol Chem. 1997;272:16657–16662. doi: 10.1074/jbc.272.26.16657. [DOI] [PubMed] [Google Scholar]
- Kiyama M, Hoshii Y, Cui D, Kawano H, Kanda T, Ishihara T. Immunohistochemical and immunochemical study of amyloid in liver affected by systemic Alambda amyloidosis with antibodies against three different regions of immunoglobulin lambda light chain. Pathol Int. 2007;57:343–350. doi: 10.1111/j.1440-1827.2007.02106.x. [DOI] [PubMed] [Google Scholar]
- Hoshii Y, Kiyama M, Cui D, Kawano H, Ishihara T. Immunohistochemical study of immunoglobulin light chain amyloidosis with antibodies to the immunoglobulin light chain variable region. Pathol Int. 2006;56:324–330. doi: 10.1111/j.1440-1827.2006.01953.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Identification of proteins in each region of adult rat brain. Rat brains were divided into six regions, and extracted, separated by SDS-PAGE. The samples were subjected to nano-LC-Q-TOF-MS/MS, and analyzed by Mascot search. Mascot scores were subtracted cut off scores, and the localizations and biological functions were searched by NCBI Entrez Gene.
Table S2. Lists of ingenuity networks generated by proteins identified in each region of rat brain.
Figure S1. Canonical pathways analyzed by proteins identified in each region of rat brain.