Abstract
Replication and assembly of adenovirus occurs in the nucleus of infected cells, requiring the nuclear import of all viral structural proteins. In this report we show that nuclear import of the major capsid protein, hexon, is mediated by protein VI, a structural protein located underneath the 12 vertices of the adenoviral capsid. Our data indicate that protein VI shuttles between the nucleus and the cytoplasm and that it links hexon to the nuclear import machinery via an importin α/β-dependent mechanism. Key nuclear import and export signals of protein VI are located in a short C-terminal segment, which is proteolytically removed during virus maturation. The removal of these C-terminal transport signals appears to trigger a functional transition in protein VI, from a role in supporting hexon nuclear import to a structural role in virus assembly.
Keywords: adenovirus/assembly/hexon/nucleo-cytoplasmic transport/protein VI
Introduction
Proteolytic processing or degradation is a key event in numerous unidirectional cellular pathways including mitosis, differentiation and apoptosis. Similarly, the replication cycle of most viruses involves proteolysis, which can occur during disassembly of the infecting virus as well as during assembly of the newly replicated virion (Krausslich and Wimmer, 1988). Here we report that proteolysis act in an unprecedented way as a functional switch in adenovirus assembly.
Adenovirus is a non-enveloped icosahedral virus with an ∼36 kb double-stranded DNA genome (Philipson, 1995). It consists of an outer capsid surrounding an inner nucleoprotein core. The capsid, the major structural protein of which is hexon, is stabilized by a number of relatively minor components. These include protein VI, which helps to link the nucleoprotein core to the capsid (Stewart and Burnett, 1995). During infection, adenovirus enters the cell by receptor-mediated endocytosis (Nemerow and Stewart, 1999), which is followed by virus escape into the cytosol. During the early stages of infection, the virus undergoes stepwise dismantling, in part through the action of the virally encoded cysteine protease (Greber et al., 1993; Russell and Kemp, 1995). Ultimately the viral nucleocapsid, still containing the hexon protein, docks at the nuclear pore complex (NPC) (Lonberg-Holm and Philipson, 1969; Chardonnet and Dales, 1970; Greber et al., 1993, 1997; Whittaker et al., 2000). The particle undergoes further disassembly at the NPC, culminating with the translocation of the DNA into the nucleoplasm, where viral transcription and DNA replication take place (Greber et al., 1993, 1996; Whittaker et al., 2000).
Assembly of new adenovirus particles occurs in the nucleus, and is tightly connected to the expression and nuclear import of viral structural proteins. Virus assembly is believed to involve insertion of the genome into an empty capsid (Sunquist et al., 1973; D’Halluin, 1995) and/or assembly of the capsid around the viral nucleoprotein core (Zhang and Imperiale, 2003). Following formation of the genome-containing capsid, a maturation process renders the progeny virion infectious. During this maturation, several viral proteins, including protein VI, are processed by the viral cysteine protease (Anderson et al., 1973; Bhatti and Weber, 1978; Weber, 1995).
The nuclear import of viral proteins (Whittaker et al., 2000) is believed to involve cellular pathways of signal-mediated transport through the NPC, which are mediated by nucleocytoplasmic shuttling receptors of the karyopherin/importin β superfamily. Karyopherins interact with nuclear localization signals (NLSs) or nuclear export signals (NESs) on the cargo molecules and carry them through the NPC (Kuersten et al., 2001; Macara, 2001; Bednenko et al., 2003). NLSs are commonly short stretches enriched in basic amino acid residues. This class of NLS has been found in a number of adenoviral proteins. However, a NLS has not to date been identified on the hexon (Russell and Kemp, 1995).
In this report we show that nuclear import of hexon in cultured cells is mediated by protein VI, which shuttles between the nucleus and the cytoplasm and appears to provide an adaptor for hexon import. Interestingly, both the major NLS and NES of protein VI are contained within a short C-terminal peptide that is proteolytically cleaved during virus maturation. This peptide forms a complex with the import receptors importin α/β in vitro and is sufficient to direct the nuclear import of a carrier protein in permeabilized cells in an importin α/β-dependent pathway. We propose that proteolysis of protein VI changes its function from a shuttling transport adapter to a viral structural protein.
Results
Expression of hexon in cultured cells from transfected cDNA
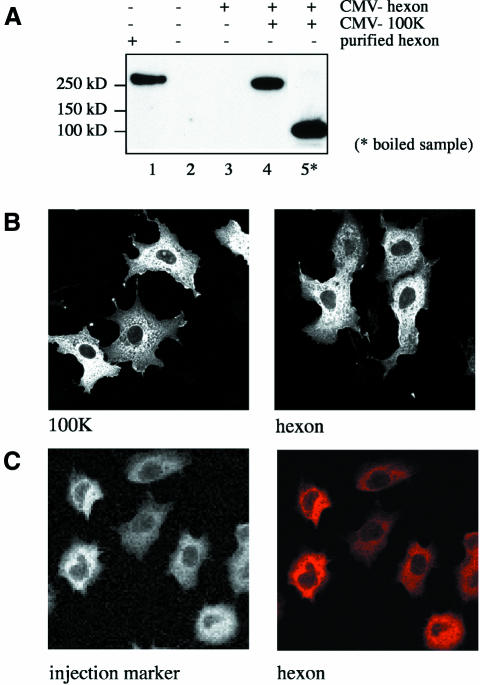
Hexon, the major adenovirus capsid protein, is a 320 kDa trimer (Roberts et al., 1986) that is present at 240 copies/virion (van Oostrum and Burnett, 1985). Hexon accumulates in the nucleus late in viral infection for assembly of new virions. To analyze the nuclear import of hexon, we first established a virus-independent system for expression of hexon in cultured cells. It was previously shown that efficient expression and trimerization of hexon during viral infection requires the ‘100K’ protein, a chaperone that is encoded by all adenoviruses (Frost and Williams, 1978; Cepko and Sharp, 1982). Cos-7 cells were transfected with a hexon expression vector in presence or absence of an expression vectors for the 100K protein, and transfected cells were analyzed for hexon expression by western blot analysis. The ∼320 kDa hexon trimer was obtained when the hexon and 100K expression vectors were co-transfected (Figure 1A, compare lanes 1 and 4). In contrast, no hexon was detectable in cells transfected with the hexon plasmid without the 100K plasmid (Figure 1A, compare lanes 2 and 3). Boiling the sample caused dissociation of the trimer to the monomer (Figure 1A, lane 5). In most cells, both proteins were concentrated in the cytoplasm and showed little or no nuclear staining (Figure 1B) after immunofluorescence staining to detect the epitope tag.
Fig. 1. Expression of hexon in transfected cells and its localization to the cytoplasm. (A) Cos-7 cells were transfected with CMV promoter-driven plasmids encoding the Ad5 hexon (lanes 3–5) or 100K proteins (lanes 4 and 5), and hexon expression was detected by immunoblot analysis with a rabbit polyclonal antiserum against hexon. Hexon migrates as a trimer when gel samples are not boiled (lane 4), having a mobility similar to purified hexon that was mixed with a cell lysate before sample preparation without boiling (lane 1). In contrast, the hexon migrates as the monomeric form after the gel sample is boiled (lane 5). (B) Cos-7 cells transfected with a plasmid expressing the HA-tagged 100K protein (left panel) or HA-tagged 100K protein and myc-tagged hexon (right panel) were labeled for immunofluorescence with antibodies to the epitope tags to detect the 100K (left) and hexon (right). (C) Adherent NRK cells were injected in the cytoplasm with purified hexon, and hexon localization was determined 30 min after injection by immunofluorescent staining with a monoclonal anti-hexon antibody (right panel). The integrity of the nuclei in the injected cells was verified by coinjection of fluorescently labeled BSA (left panel). The microinjection was repeated twice, each time including >15 cells with consistent cytoplasmic localization of hexon.
The predominantly cytoplasmic localization of hexon seen in the transfection experiments was unexpected, since previous work in other experimental systems indicated that hexon efficiently localizes to the nucleus (Saphire et al., 2000; Carlisle et al., 2001). To investigate the nuclear import capacity of hexon with a second experimental approach, hexon trimer purified from infected cell lysates was microinjected into the cytoplasm of NRK cells. Hexon localized exclusively to the cytoplasm in all of the injected cells (Figure 1C, right panel, and injection marker left panel). Therefore, whether it is introduced into cells by microinjection or transfection, hexon by itself is not localized to the nucleus in most cells.
Protein VI strongly promotes nuclear import of hexon
Circumstantial evidence has suggested that adenovirus protein VI might be involved in hexon nuclear import. The ts147 strain of adenovirus, which shows degradation of protein VI at the non-permissive temperature, is defective for the nuclear import of hexon (Kauffman and Ginsberg, 1976; Praszkier and Ginsberg, 1987). In addition, direct interactions between protein VI and hexon have been reported (Boulanger et al., 1979; Russell et al., 1981; Matthews and Russell, 1994, 1995).
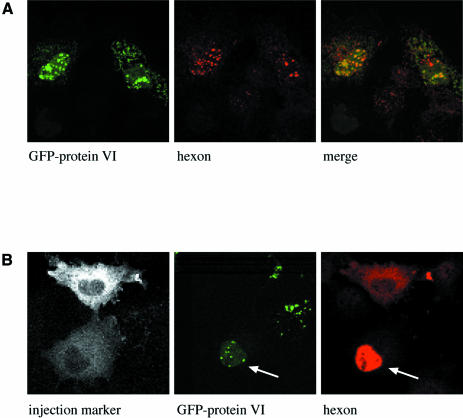
To test whether protein VI can promote the nuclear import of hexon, we constructed an expression vector in which protein VI was fused to the C-terminus of green fluorescent protein (GFP). This vector was co-transfected with the hexon and 100K protein expression vectors into Cos-7 cells (Figure 2A). Interestingly, both proteins were significantly more concentrated in the nucleus than in the cytoplasm of the transfected cells as seen by fluorescence microscopy (Figure 2A). The strong nuclear localization of hexon in cells transfected with both hexon and protein VI contrasts sharply with the mostly cytoplasmic localization of hexon in the absence of protein VI (Figure 1B).
Fig. 2. Nuclear localization of hexon promoted by adenovirus protein VI. (A) Cos-7 cells were co-transfected with plasmids encoding the 100K protein, hexon and GFP–protein VI fusion. The localization of protein VI was determined by GFP fluorescence (left panel), and hexon localization was determined by staining with a monoclonal anti-hexon antibody (middle panel). The merged images are indicated in the right panel. (B) Cos-7 cells were transfected with an expression vector for protein VI fused to GFP. Twenty-four hours after transfection, clusters of cells were microinjected in the cytoplasm with purified hexon together with a fluorescent injection marker. Thirty minutes after injection, cells were fixed and hexon localization was detected with a monoclonal anti-hexon antibody. Localization of the injection marker (left panel), protein VI (middle panel) and hexon (right panel) are depicted. Whereas hexon localizes to the cytoplasm in the upper cell that does not express GFP–protein VI, it becomes concentrated in the nucleus (indicated by an arrow in the lower cell expressing protein VI). The microinjection was repeated twice, each time including at least 10 cells. Nuclear accumulation of hexon was consistently observed in all cells transfected with the GFP–protein VI fusion.
To examine the role of protein VI in nuclear import of hexon by a separate approach we microinjected purified hexon trimers into the cytoplasm of cells transfected with GFP–protein VI without the 100K protein. All of the microinjected cells expressing GFP–protein VI showed strong localization of hexon in the nucleus (lower injected cell in Figure 2B), whereas none of the non-transfected cells showed nuclear accumulation of hexon (e.g. upper injected cell in Figure 2B). These data not only confirmed that protein VI strongly promotes the nuclear import of hexon, they also indicated that the100K protein is not essential for hexon import.
Nuclear import signals in protein VI
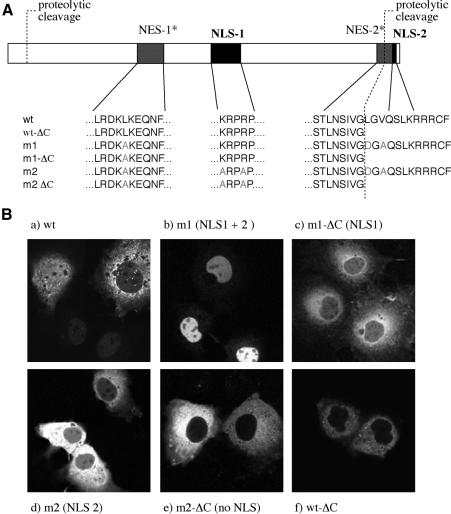
The finding that protein VI can promote the nuclear import of hexon suggests that protein VI might be an NLS-containing adapter that links hexon to the nuclear import machinery. An examination of the primary sequence of protein VI revealed two basic amino acid-rich, classical NLS-like sequences, ‘NLS-1’ between amino acids K131–P135 and ‘NLS-2’ between amino acids K245–R248 (Figure 3A). In addition, two hydrophobic regions that are potential NESs were seen (NES-1 and NES-2; Figure 3A). Both potential NESs contain sequence homologies to known leucine-rich NES such as found in the HIV-1 Rev protein and PKI (Supplementary figures 1 and 2, available at The EMBO Journal Online) (Fischer et al., 1995; Wen et al., 1995; Bogerd et al., 1996; Fukuda et al., 1996; Taagepera et al., 1998). Interestingly, NLS-2 is located at the very C-terminus of protein VI, and is removed upon maturation by proteolysis together with an essential part of the potential NES-2 (Figure 3A) (Mangel et al., 1993; Webster et al., 1993; Weber, 1995).
Fig. 3. Nuclear localization signals and nuclear export signals in protein VI. (A) Potential NLSs and NESs in protein VI, and the mutants constructed to analyze the functions of these regions. Potential NLSs and NESs are indicated as solid and hatched boxes, respectively. Cleavage sites for the viral protease at the N- and C-termini of protein VI are indicated. Note that the C-terminal cleavage disrupts NES-2 and removes NLS-2. (B) Cos-7 cells were transfected with plasmids expressing wild-type or mutant forms of protein VI [according to nomenclature of (A)], fused to the C-terminus of GFP–PK. Localization of fusion proteins was monitored by the GFP signal. Potential transport signals present in each construct are given in parenthesis (see text for details).
To investigate whether the NLS-like sequences influence the nuclear localization of protein VI, we transfected Cos-7 cells with an expression vector in which protein VI was fused to the C-terminus of a GFP–chicken muscle pyruvate kinase (GFP–PK) fusion protein. GFP–PK is a cytoplasmic protein of ∼90 kDa, which exceeds the limit for passive diffusion into the nucleus (Sherman et al., 2001). While GFP–PK localized entirely to the cytoplasm (unpublished observation; Sherman et al., 2001), the GFP–PK–protein VI fusion protein was localized to both the nucleus and the cytoplasm (Figure 3B, a). In cells expressing a low level of protein, the GFP fusion was more concentrated in the nucleus, whereas in cells expressing a high level of protein, the fusion protein was more concentrated in the cytoplasm.
The partially cytoplasmic localization obtained with the GFP–PK–protein VI fusion in many cells suggests that the putative NES(s) in protein VI might functionally antagonize the NLSs. To analyze the nuclear import of protein VI in the absence of competing nuclear export, we generated the mutant m1 in which the two putative NESs were altered to impair their potential export activity. For this, amino acids 70–72 were changed from Lys-Leu-Lys to Lys-Ala-Lys, and amino acids 239–241 were changed from Leu-Gly-Val to Asp-Gly-Ala (Figure 3A, m1). In contrast to the wild-type fusion protein, the fusion protein containing the protein VI-m1 mutant accumulated exclusively in the nucleus in all transfected cells (Figure 3B, compare a and b).
Starting with protein VI-m1, we removed the putative NLS-2 by deleting residues downstream of amino acid 239, generating m1-ΔC, which contained only an intact putative NLS-1. The truncated C-terminus of protein VI in the ΔC mutant is identical to that generated in wild-type protein VI by the viral protease. In a second construct, we changed two basic amino acid residues in putative NLS-1 to alanines to impair the predicted NLS-1 function, generating mutant m2, which contained an intact NLS-2 (Figure 3A). Inactivation of either NLS-1 (in mutant m2; Figure 3B, d) or NLS-2 (in m1-ΔC; Figure 3B, c) resulted in a shift of the fusion protein from the exclusively nuclear localization seen with the m1 mutant to a mostly cytoplasmic localization in all transfected cells (Figure 3B, compare b with c and d). Low, but detectable, levels of nuclear localization of the fusion protein was seen with both the m2 and the m1-ΔC mutants, as evidenced by nucleolus-excluded fluorescence in the nucleoplasm (Figure 3B, c and d). Lastly, we inactivated both putative NLSs within a single construct by generating a C-terminal deletion of the m2 mutant to create the m2-ΔC construct. The fusion protein containing m2-ΔC localized exclusively to the cytoplasm. Together, these data indicate that both NLS-1 and NLS-2 contribute to the nuclear localization of protein VI.
The above analysis, carried out with the mutant m1 form of protein VI, suggested that the NLS-2 cleaved from protein VI during virus maturation might contribute significantly to the nuclear localization of the unprocessed wild-type protein VI. To examine this further, we constructed a fusion protein containing wild-type protein VI that is C-terminally deleted at residue 239 (Figure 3A, wt ΔC). As predicted, the fusion protein had a strong cytoplasmic localization in all cells independent of the expression levels (Figure 3B, e), confirming that NLS-2 has substantial nuclear localization activity.
Protein VI is a nucleo-cytoplasmic shuttling protein
The point mutagenesis experiments discussed above suggested that putative NES-1 and/or NES-2 serve as NESs, because a combination of mutations in these two regions (mutant m1) shifted the GFP–protein VI fusion from a mixed nuclear + cytoplasmic localization to an exclusively nuclear localization (Figure 3B). The presence of an NES(s) in protein VI would enable it to shuttle between the nucleus and the cytoplasm, allowing a single molecule of protein VI to promote the nuclear import of multiple hexons.
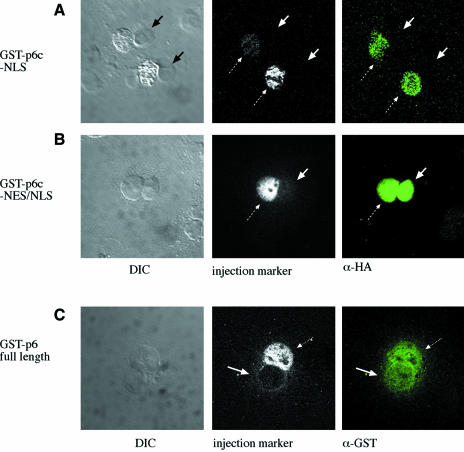
We directly examined the nuclear export activity of the C-terminal putative NES-2. This region was of particular interest because its activity is predicted to be modulated by the cleavage of the C-terminal 12 amino acids of protein VI by the 23 kDa adenovirus cysteine protease, which removes hydrophobic residues that are critical for the activity of leucine-rich NESs (Supplementary figure 1; Fischer et al., 1995). To test this possibility, we expressed and purified a recombinant GST construct fused at its C-terminus with the last 25 amino acids of protein VI, which includes both NLS-2 and putative NES-2, followed by an HA tag (termed GST-p6c-NES/NLS; Figure 4B). As a control, a GST fusion protein was expressed that contains only the last 12 amino acids of protein VI, which include NLS-2, followed by an HA tag (termed GST-p6c-NLS; Figure 4A). Each purified fusion protein, together with a fluorescent microinjection marker, was microinjected into one nucleus of bi- or multinucleated cells, and the ability of the GST fusion protein to shuttle from one nucleus to another via the common cytoplasm was monitored. GST without any fusion partner (unpublished observation), or GST fused to the C-terminal 12 amino acids of protein VI (dotted arrows in Figure 4A, middle panel), remained in the injected nucleus and thus was incapable of nucleo-cytoplasmic shuttling. In contrast, GST fused to the 25 C-terminal amino acids of protein VI rapidly shuttled from the injected nucleus (dotted arrow in Figure 4B) to the non-injected nucleus (solid arrows in Figure 4B), reaching the same concentration in the two nuclei at the earliest time measured (30 min). These experiments clearly established that the C-terminal segment of protein VI contains an NES, in addition to NLS-2 described above. When the constructs were injected into the cytoplasm instead of the nucleus, both GST fusion proteins accumulated in the nucleus, suggesting that the NLS at the C-terminus of protein VI is dominant over the NES (data not shown)
Fig. 4. Nucleo-cytoplasmic shuttling by protein VI. Bi- or multinucleated cells (left panel) were microinjected into one nucleus with GST-p6c-NLS (A) or GST-p6c-NES/NLS (B), together with fluorescently labeled BSA as an injection marker (middle panel). After 1 h cells were fixed and injected protein was detected with a monoclonal antibody against the HA tag (right panel). Dotted arrows point to the injected nucleus, solid arrows point to non-injected nuclei. (C) Bi- or multinucleated Cos-7 cells (left panel) were microinjected into one nucleus with GST–protein VI and fluorescently labeled BSA as an injection marker (middle panel). One hour after injection, cells were fixed and stained for immunofluorescence with antibodies to GST (right panel). Dotted arrows point to the injected nucleus, solid arrows point to non-injected nuclei.
We found that full-length protein VI fused to GST also was able to shuttle from nucleus to nucleus in a similar microinjection assay (Figure 4C, injected nucleus, dotted arrow, and noninjected nucleus, solid arrow). We believe that the lower signal intensity in the non-injected nucleus is due to the fact that we detected the fusion protein with antibodies to GST, and that our GST–protein VI preparation was partially degraded (see Materials and methods for details). Taken together, the above results indicate that protein VI is a nucleo-cytoplasmic shuttling protein, and suggest that the C-terminus containing NLS-2 and NES-2 plays an active role in this process.
The C-terminal segment of protein VI promotes nuclear import via importin α/β
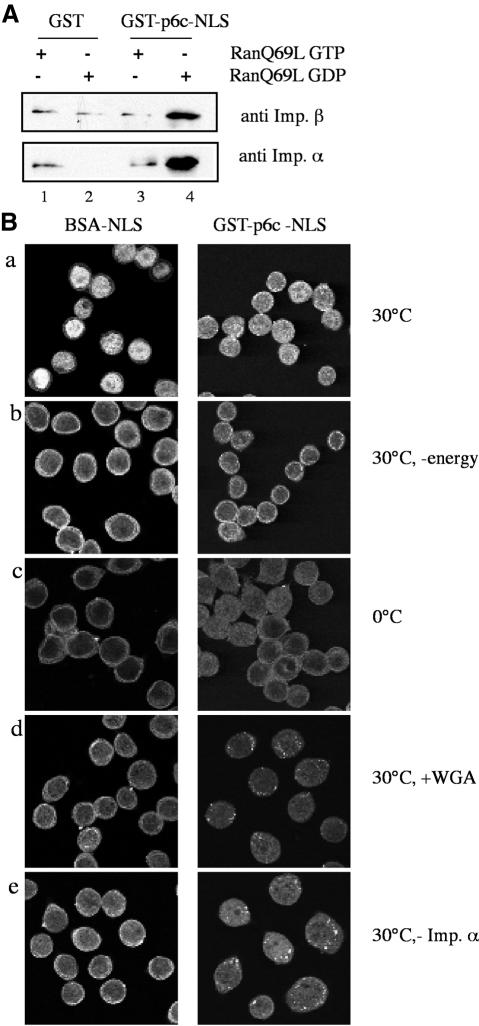
Since the C-terminal peptide of protein VI appears to strongly contribute to protein VI import, we investigated the nuclear import pathway used by this region. Therefore we incubated the GST-p6c-NLS (described above) with cytosol in a pull-down assay to define nuclear transport receptors that bind to it. As a specificity control, some binding reactions were carried out in the presence of RanQ69L preloaded with either GDP or GTP. The RanQ69L mutant is impaired in GTP hydrolysis and promotes import complex dissociation when present in its GTP-bound state (Macara, 2001). Using immunoblotting, we found that significant amounts of importin α/β in cytosol bound to the fusion protein containing the C-terminal peptide of protein VI, as compared with the background levels of binding obtained with GST alone (Figure 5A, compare lane 4 with 2). Whereas binding was normal with RanQ69L-GDP, essentially all binding was abolished with RanQ69L-GTP (Figure 5A, compare lane 3 with 4). Specific binding of recombinant importins α/β to the NLS fusion protein was also obtained (unpublished observation). Thus, NLS-2 in the C-terminal peptide of protein VI binds specifically to classical import receptors.
Fig. 5. C-terminal domain of protein VI mediates import via importin α/β. (A) Western blot analysis of material bound to GST-p6c-NLS after GST pull-down from cytosol. Glutathione beads were saturated with either free GST (lane1 and 2) or GST-p6c-NLS (lanes 3 and 4) and incubated with cytosol in the presence of RanQ69L preloaded with GTP (lanes 1 and 3) or GDP (lanes 2 and 4). Bound material was examined by western blotting with antibodies against importin α (lower row) or importin β (upper row). (B) In vitro import of GST-p6c-NLS in permeabilized cells. Confocal images are shown. Detection of BSA-NLS-FITC was performed directly after fixation of cells (first column). To detect GST-p6c-NLS, indirect immunofluorescence with a specific antibody against the HA tag was carried out after fixation (second column). All reactions contained an energy regenerating system (except the reaction shown in b), and the recombinant transport factors Ran, NTF2, importin β and importin α (except the reaction shown in e, which lacks recombinant importin α). In b the reaction was depleted of energy by addition of hexokinase/glucose.
We next examined whether we could reconstitute nuclear import of the C-terminal domain of protein VI fused to GST in permeabilized HeLa cells using recombinant importin α/β. Nuclear accumulation of the fusion protein was detected by immunofluorescence to detect the HA tag. As a control, we examined the import of BSA coupled to the classical SV40 T antigen NLS, known to use the importin α/β pathway. Like the BSA-NLS control, GST-p6c-NLS accumulated in the nucleus during a 20 min import reaction (Figure 5B, a). Nuclear accumulation of the fusion protein was temperature sensitive (Figure 5B, c). It also was energy dependent, as determined by the addition of hexokinase/glucose to deplete the endogenous pool of NTPs from the cells (Figure 5B, b). Furthermore, the import was inhibited by addition of the lectin wheat germ agglutinin (WGA), a selective inhibitor of nuclear import (Figure 5B, d). Consistent with the results from our pull-down experiments, nuclear accumulation of GST-p6c-NLS was significantly impaired in the absence of recombinant importin α, indicating that NLS-2 of protein VI does not directly bind to importin β, as seen for certain importin β-dependent import substrates (Macara, 2001). The residual import activity in this case most likely is a consequence of endogenous importin α that was not depleted from the permeabilized cells. In summary, these experiments directly show that the C-terminal domain of protein VI contains a classical NLS that promotes nuclear import by binding to importin α/β.
C-terminal processing of protein VI abolishes nuclear import of hexon
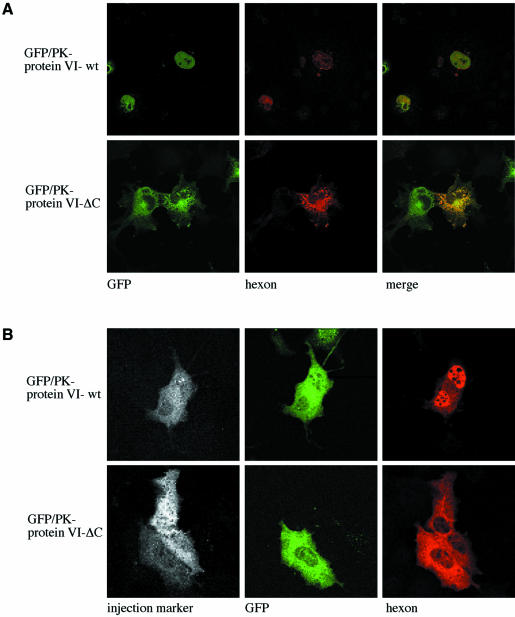
Having established the role of the C-terminal domain of protein VI in nuclear import, we investigated whether its removal by proteolytic processing would abrogate the ability to promote nuclear import of hexon by protein VI. Cos-7 cells were transfected with expression plasmids for hexon, the 100K protein and GFP–PK fused to either wild-type protein VI or to the ΔC mutant, which mimics the naturally processed form. Consistent with the findings discussed above (Figure 2), we found that both protein VI and hexon were strongly localized to the nucleus when the unprocessed (wild-type) protein VI sequence was co-transfected (Figure 6A, upper row). In contrast, when the processed form of protein VI (ΔC) was co-transfected, the protein VI mutant was localized to the cytoplasm as noted above (Figure 3), and hexon failed to accumulate in the nucleus (Figure 6A, lower row).
Fig. 6. Requirement of the C-terminus of protein VI for nuclear import of hexon. (A) Cos-7 cells were transfected with plasmids expressing the 100K protein, hexon and GFP–PK fused to either full-length protein VI (upper row) or to protein VI-ΔC lacking the C-terminus (lower row). Localization of protein VI was determined by GFP fluorescence (left panels) and hexon localization was detected with a monoclonal anti-hexon antibody (middle panels). The merged images are shown in the right-hand panels. (B) Cos-7 cells were transfected with GFP–PK fusions containing wild-type protein VI (upper row) or protein VI-ΔC lacking the C-terminal peptide (lower row). Twenty-four hours after transfection, cells were microinjected in the cytoplasm with a fluorescent injection marker and purified hexon. Thirty minutes after injection, the cells were fixed and hexon was detected using a monoclonal anti-hexon antibody. The localization of the injection marker (left panel), protein VI (middle panel) and hexon (right panel) are indicated. All microinjection experiments were carried out at least twice, each time including an average of 10–15 cells. All cells transfected with wild-type protein VI fused to GFP–PK showed nuclear accumulation of hexon as shown in the upper row, while none of the cells transfected with protein VI-ΔC showed nuclear accumulation of hexon after microinjection (lower row).
In a complementary approach, Cos-7 cells were transfected with GFP–PK fused to wild-type protein VI or the ΔC mutant. Subsequently, purified hexon was microinjected into the cytoplasm of the transfected cells (see Figure 1), and the localization of hexon was determined. As shown in Figure 6B, hexon was strongly localized to the nucleus in all of the microinjected cells transfected with the wild-type protein VI (Figure 6B, upper row), consistent with the above results (Figures 3 and 6A). In contrast, hexon remained entirely cytoplasmic in all of the microinjected cells transfected with the ΔC mutant of protein VI (Figure 6B, lower row). This result provides strong support for the hypothesis that C-terminal processing of protein VI abrogates its ability to promote hexon import.
Discussion
Our data indicate that the nuclear import of adenoviral hexon involves the structural protein VI, which appears to serve as a nucleo-cytoplasmic shuttling adapter that links hexon to the nuclear transport machinery. To achieve this role, protein VI encodes NLS and NES that are evolutionary conserved among mastadenoviral serotypes (Supplementary figure 1). Moreover, our results indicate that the C-terminal proteolytic processing of protein VI triggers a functional transition in this protein, from a role in supporting hexon transport to an exclusively structural role in virus assembly.
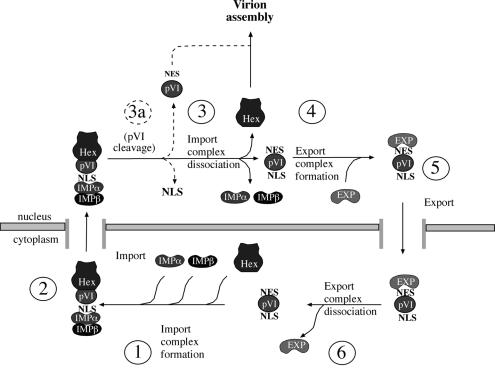
Based on our results and on previous studies, we propose that protein VI links the processes of hexon import and virus assembly, as outlined in the model in Figure 7. We suggest that protein VI and hexon form a complex in the cytoplasm, which binds to the importin α/β complex via NLSs on protein VI (step 1). Subsequently the import complex is translocated into the nucleus (step 2), and the import receptors are dissociated by the action of RanGTP (Macara, 2001; Bednenko et al., 2003). At this point, hexon dissociates from protein VI and remains nuclear (step 3), whereas protein VI binds to an exportin (step 4) and is transported back to the cytoplasm (step 5), where it dissociates from the export receptor (step 6) and engages in a new round of hexon import. Since the association between hexon and protein VI involves hydrophobic regions of protein VI, including the region containing NES-1 (Supplementary figure 2; Matthews and Russell, 1995), we suggest that the NESs on protein VI are masked by binding to hexon. This would ensure that only protein VI, and not the hexon–protein VI complex, is exported back to the cytoplasm after these two proteins are transported into the nucleus.
Fig. 7. Model for involvement of protein VI in the nuclear import of hexon. Protein VI and hexon from a complex in the cytoplasm via hydrophobic interactions involving the NESs present in protein VI (step 1). Exposed NLSs within protein VI recruit the import receptors α and β and the complex is imported into the nucleus (step 2), where the complex dissociates (step 3) leaving hexon inside the nucleus. Complex dissociation exposes the NES on protein VI, which in turn binds to export receptor(s) (step 4). An export complex is formed and protein VI is exported into the cytoplasm (step 5). Dissociation of the export complex releases protein VI (step 6), which can engage in a new round of hexon nuclear transport. Late in infection protein VI is processed at its C-terminus by the viral protease, removing the transport signals and enhancing the affinity for hexon. As a consequence protein VI is converted into a structural component of the maturing new assembled viral particle (step 3b) (see text for details).
We propose that protein VI undergoes a transition from a nuclear transport function to a virus assembly role by a combination of mass-action effects and proteolytic processing of protein VI. Whereas the affinity of unprocessed protein VI for hexon is relatively low (Matthews and Russell, 1994), the strength of the interaction is increased by the N- and C-terminal proteolytic processing of protein VI (Matthews and Russell, 1994). Since hexon is the major viral binding partner for protein VI, we suggest that the concentration of intranuclear unprocessed protein VI increases co-ordinately with a rise in the concentration of intranuclear hexon. This is because binding of protein VI to intranuclear hexon, either free or present in partially assembled virions, would compete with export of protein VI from the nucleus (see above). The increased nuclear concentration of protein VI in turn would trigger the activation of the viral cysteine protease over its basal level, because the C-terminal peptide released from protein VI by the protease considerably enhances protease activity (Mangel et al., 1993; Webster et al., 1993; Weber, 1995). Owing to this positive feedback loop, the level of proteolytically processed protein VI would increase. This would promote assembly of protein VI into the virus due to the enhanced affinity of processed protein VI for hexon and trigger virus maturation. Thus, the C-terminal processing of protein VI would help to convert it from a recycling import adapter into a structural component of the virus (step 3a). To our knowledge, the presence of a proteolytically cleavable NLS and an NES in immediate proximity on a protein, as we have described at the C-terminus of protein VI (with an ∼16 amino acid stretch), is unprecedented. This cleavable ‘shuttling’ region provides an economic mechanism to trigger a shift in the functions of protein VI.
Genetic evidence for a pivotal role of protein VI in hexon nuclear import comes from the Ad5 ts147 mutant, which is characterized by a strong impairment of hexon nuclear import at the non-permissive temperature, accompanied by instability of protein VI (Kauffman and Ginsberg, 1976). The ts147 mutation was mapped to the region of the adenovirus genome containing the adjacent hexon and protein VI open reading frames (ORFs) (Kauffman and Ginsberg, 1976). We sequenced the hexon ORF from the ts147 virus strain, and detected five point mutations (Supplementary figure 3), of which only the G776D mutation was predicted to have a profound effect on protein VI binding. G776 is located at the central cavity of hexon, the proposed binding site for protein VI (Stewart et al., 1993). Our molecular modeling analysis suggests that the G776D mutation has the potential to extend helix V2-α1, which in turn could alter the local protein structure and potentially inhibit protein VI binding, which would explain the loss of hexon import in cells infected with Ad5 ts147 (Supplementary figure 3; Kauffman and Ginsberg, 1976). This tertiary rearrangement of the hexon cavity is predicted to be more pronounced at the non-permissive temperature, consistent with the enthalphic nature of helix folding (Creigthon, 1993)
Proteolysis of protein VI is not absolutely required for assembly of protein VI-containing particles. The temperature-sensitive mutant Ad2 ts1, which contains a mutation that inactivates the viral protease, assembles virus particles with morphology resembling the wild-type virus at the non-permissive temperature (Weber, 1976). Nuclear import of hexon and the level of protein VI are not altered in ts1-infected cells and protein VI is not cleaved, although the assembled particles are non-infectious due to the lack of virus maturation (Begin and Weber, 1975; Weber, 1976; Bhatti and Weber, 1978). These results are consistent with our model, since accumulation of hexon to high levels in the nucleus would promote protein VI assembly into the virus even in the absence of processing.
In contrast to our results, previous work suggested that hexon could be transported into the nucleus in a permeabilized cell import assay (Saphire et al., 2000) or by microinjection into cells (Carlisle et al., 2001). However, we have determined that the hexon prepared by the methods described in these studies contains a significant amount of co-purified protein VI (our unpublished observations). For the present study, we obtained hexon devoid of protein VI by using an additional purification step involving gel filtration.
A recent report showed that the nucleocapsid of the infecting adenovirus docks at the NPC via hexon (Trotman et al., 2001). The authors of that study suggest that nuclear import of some hexon occurs at this point by direct interaction with histone H 1.2, which is proposed to serve as an import adapter providing the requisite import signal (Trotman et al., 2001). This raises the interesting possibility that different nuclear import pathways for hexon have evolved, one of which is linked to capsid disassembly early in infection in the context of a docked nucleocapsid, and a second of which occurs during assembly when high-level import of hexon trimers involving protein VI takes place.
In summary, our data support the notion that a key event late in adenovirus replication is the conversion of protein VI from a nuclear transport adapter to a structural component of the mature viral particle. This transition is promoted by the proteolytic processing of protein VI by the adenoviral cysteine protease, which co-ordinately removes key transport signals from the protein and increases its affinity for hexon. Analogous to these findings with adenovirus, the removal of nuclear transport signals from cellular proteins by proteolysis might be used in the regulation of other directional processes such as the cell cycle, apoptosis and differentiation.
Materials and methods
DNA cloning and sequencing
Expression vectors for hexon, 100K and protein VI were amplified from the pAdEasy vector (He et al., 1998). The hexon ORF was amplified using the 5′ primer (GGGAATTCAAGatggctaccccttcgatgatgcc), which introduced an EcoRI site (underlined), and the 3′ primer (ggtctaga ctattaCAAGTCTTCTTCAGAAATAAGCTTTTGCTCtgttgtggcgttgccgg), which introduced a myc epitope tag and an XbaI site (underlined). The EcoRI–XbaI fragment was cloned into pcDNA3.1 (Invitrogen) to generate pcDNA3.1-hex-myc. The 100K protein was amplified using the 5′ primer (GGGAATTCAAGatggagtcagtcgagaagaagg) and the 3′ primer (GCTCT AGAttagtcgagtgcgtagtctggtacgtcgtacggatacggttgggtcggcgaacg), which introduced the same restriction sites and an HA tag. The EcoRI–XbaI fragment was cloned into pcDNA3.1 to generate expression vector pcDNA3.1-100K-HA. The expression vector for the GFP–protein VI fusion protein was cloned by amplifying the protein VI ORF with the 5′ primer (GAATTCGCGGCCGCAAGatggaagacatcaactttgcg) to introduce an EcoRI site and the 3′ primer (GTGGATCCCGgaagcatcgtcggcgcttcagg) to introduce a BamHI site. The EcoRI–BamI fragment was cloned into pEGFP-N1 (Invitrogen). For the analysis of nuclear transport signals, the protein VI coding sequence was amplified with the 5′ primer (ccGGTACC gcatggaagacatcaactttgcg) and the 3′ primer as above and cloned as a KpnI–BamHI fragment downstream of the GFP–PK ORF in the vector pEGFP-CMPK (Sherman et al., 2001).
Introduction of mutations into protein VI was performed using an in vitro mutagenesis kit (Stratagene). GST expression vectors are based on pGEX-KG (Novagen). The protein VI ORF was amplified and cloned in-frame to the C-terminus of GST. GST-p6c-NES/NLS and GST-p6c-NLS were constructed by amplifying amino acids 226–251 (NES/NLS) and 239–251 (NLS) of protein VI followed by HA tagging and cloning to the C-terminus of GST. Expression of proteins was confirmed by transfection and western blotting. The hexon gene was amplified from Ad5 ts147 DNA and subcloned for sequencing.
Cell culture and transfection
NRK cells and Cos-7 cells were maintained in complete Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal calf serum (FCS). Cos-7 cells (2 × 105) were seeded into 6-well dishes the day before transfection. The medium was replaced by serum-reduced Opti-MEM (Gibco) 1 h prior to transfection. Four micrograms of DNA were suspended in 1 ml of Opti-MEM and 5 µl of Targetfect F2 reagent (Targetsys, San Diego, CA) was added. The solution was mixed and incubated for 10 min at room temperature before addition to a well of the 6-well plates. In co-transfection assays, the total amount of DNA was maintained at a constant value by including appropriate amounts of empty vector DNA. Three hours after transfection, the medium was replaced with fresh Dulbecco’s medium. Protein expression was analyzed 24–48 h after transfection.
Immunofluorescence microscopy
Cells grown on coverslips were rinsed with PBS, fixed for 10 min in PBS 4% paraformaldehyde, and pre-blocked and permeabilized with 0.2% Saponin in PBS containing 10% FCS. Staining for hexon was performed with a monoclonal antibody specific for the hexon trimer (clone141; Biodesign) at a dilution 1:100 in PBS containing 10% FCS and 0.2% saponin. A Texas Red-coupled secondary donkey anti rabbit antibody (Jackson Immunoresearch, West Grove, PA) was used at a dilution 1:50 in the same solution. 100K protein was detected using an anti-HA mAb (HA.11; Babco, Richmond,) at a dilution of 1:500 and a Texas Red-coupled anti-mouse secondary at a dilution of 1:50. Stained cells were mounted in Slow Fade (Molecular Probes, Eugene, OR). GFP fluorescence was monitored by fixing the cells in 4% paraformaldehyde and mounting them in Slow Fade. Cells were examined using Bio-Rad 1024 laser scanning confocal microscopy.
Western blot analysis
Cells were harvested 24–48 h after transfection. Cell lysates were analyzed by SDS–PAGE on 12.5% gels and proteins were transferred to nitrocellulose membranes as described previously (Wodrich et al., 2000). Hexon was detected using polyclonal rabbit anti-hexon antiserum (Saphire et al., 2000) at a dilution 1:250. Secondary horseradish peroxidase-conjugated anti-rabbit antibodies (Jackson Immunoresearch) were used at a dilution 1:5000. The signal was detected by chemiluminescence.
Microinjection
For microinjection of NRK cells, hexon purified from infected cells was injected into the cytoplasm of subconfluent cells at a concentration of 1 mg/ml in transport buffer (Adam et al., 1991), together with Alexa-460 labeled BSA as an injection marker. After injection cells were incubated for 30 min at 37°C, and were fixed and labeled for immunofluorescence (see above). The same protocol was used for microinjection of Cos-7 cells, except that the Cos-7 cells were transfected 24 h prior to injection, as described above. For the shuttling experiments a preparation containing GST–protein VI at a concentration of 0.2 mg/ml (see below) was injected into one nucleus of bi- or multinucleated Cos-7 cells, and after 1 h incubation at 37°C, the cells were fixed and stained for immunofluorescence with antibodies to GST. For microinjection, GST-p6c-NES/NLS-HA and GST-p6c-NLS-HA were concentrated to 0.5 mg/ml. Injected cells were immunolabeled with a monoclonal antibody against the HA tag to detect only intact protein. Each microinjection experiment included an average of 10–15 cells and was repeated at least twice.
In vitro import assay in permeabilized cells
Nuclear import assays in digitonin-permeabilized HeLa cells were carried out essentially as in Kehlenbach et al. (2001), except that cells were preincubated in TB for 15 min at 30°C before import to deplete endogenous transport factors. FITC labeling and NLS peptide conjugation to BSA was performed as in Melchior et al. (1993). Unlabeled GST-p6c-NLS was added to the reaction at ∼500 nM and detected with a monoclonal antibody against the HA tag. Import reactions contained an energy regenerating system and recombinant factors: importin β at 250 nM, importin α at 330 nM, Ran at 300 nM and NTF2 at 500 nM. In control reactions, WGA was added to the reaction at a final concentration of 200 ng/µl. In the absence of an energy regenerating system, hexokinase/glucose was added to deplete endogenous cellular NTPs.
Protein purification
Hexon was purified according to a protocol from Waris and Halonen (1987). Briefly, 293-T cells were infected with Ad5 at an m.o.i. of 100 and incubated for additional 48–60 h until the cells started to detach. Cells were harvested and washed once with PBS. Cells were disrupted by three alternating freeze–thaw cycles in 2 vol. of 20 mM Bis-Tris propane pH 6.5 and 1 vol. of 1,1,2-trichlorotrifluoroethane (Freon 113; Aldrich). Lysates were cleared by low-speed centrifugation (1000 g for 20 min) and high-speed centrifugation (100 000 g for 30 min). The cleared supernatant was applied to a MonoQ column (Pharmacia). Elution of hexon from the MonoQ column was performed with a 0–0.5 M NaCl salt gradient in 20 mM Bis-Tris propane pH 6.5. The pool of peak hexon fractions was further purified by molecular sieving on a S-200 column. Fractions containing trimeric hexon were pooled, concentrated to 1 mg/ml and frozen in liquid nitrogen.
GST fusion proteins were expressed in the Escherichia coli strain BL 21–. Cells were grown to an OD of 0.6 at 37°C and induced with 50 µM IPTG. When expressing full-length GST–protein VI together with the IPTG, NaCl to a concentration of 300 mM was added to the growth media to prevent hypotonic cell lysis, due to high toxicity of the expressed protein VI. Otherwise, cell lysis occurred within minutes as described previously (Matthews and Russell, 1995). Induction was continued for 2 h and cells were disrupted by sonication in the presence of 10 mg/ml RNAse and protease inhibitors (2 mM PMSF).
Purification was performed using glutathione–Sepharose according to the manufacturer’s protocol (Amersham). In the GST–protein VI preparation the eluate from the glutathione column contained some full-length GST–protein VI, but also had many degradation products that migrated at the size of GST and larger. The protein was concentrated to ∼0.2 mg/ml and frozen in liquid nitrogen. GST-p6c-NLS and GST-p6c-NES/NLS were concentrated to 0.5 mg/ml and stored in liquid nitrogen.
GST pull-down assays
Glutathione beads (Amersham) were loaded to saturation with GST or GST-p6c-NLS in transport buffer (see above) containing 1% BSA. Loaded beads were incubated with cytosol diluted to 2 mg/ml in transport buffer in the presence of protease inhibitors. Cytosol was obtained from HeLa cells by digitonin lysis (Melchior et al., 1993). The sample was rotated for 3 h at 4°C in the presence of RanQ69L, which was preloaded either with GTP or GDP (Kehlenbach et al., 2001). The beads were collected by centrifugation and washed three times with transport buffer. Bound material was analyzed by western blotting with antibodies to importin α and importin β.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are thankful to Warner Greene for providing plasmids GFP–PK and to Philomena Opstachuck for providing Ad5 ts147. We are also grateful to our laboratory members for many helpful comments and suggestions.This work was supported by NIH grant GM41955 to L.G. and grants HL54352 and EY11431 to G.N., and by a fellowship from the Deutsche Forschungsgemeinschaft (DFG) (WO 805/1-1) to H.W.
References
- Adam S.A., Sterne-Marr,R. and Gerace,L. (1991) In vitro nuclear protein import using permeabilized mammalian cells. Methods Cell Biol., 35, 469–482. [DOI] [PubMed] [Google Scholar]
- Anderson C.W., Baum,P.R. and Gesteland,R.F. (1973) Processing of adenovirus 2-induced proteins. J. Virol., 12, 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednenko J., Cingolani,G. and Gerace,L. (2003) Nucleocytoplasmic transport: navigating the channel. Traffic, 4, 127–135. [DOI] [PubMed] [Google Scholar]
- Begin M. and Weber,J. (1975) Genetic analysis of adenovirus type 2. I. Isolation and genetic characterization of temperature-sensitive mutants. J. Virol., 15, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatti A.R. and Weber,J. (1978) Protease of adenovirus type 2. In vitro processing of core protein. Biochem. Biophys. Res. Commun., 81, 973–979. [DOI] [PubMed] [Google Scholar]
- Bogerd H.P., Fridell,R.A., Benson,R.E., Hua,J. and Cullen,B.R. (1996) Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol., 16, 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger P., Lemay,P., Blair,G.E. and Russell,W.C. (1979) Characterization of adenovirus protein IX. J. Gen. Virol., 44, 783–800. [DOI] [PubMed] [Google Scholar]
- Carlisle R.C., Bettinger,T., Ogris,M., Hale,S., Mautner,V. and Seymour,L.W. (2001) Adenovirus hexon protein enhances nuclear delivery and increases transgene expression of polyethylenimine/plasmid DNA vectors. Mol. Ther., 4, 473–483. [DOI] [PubMed] [Google Scholar]
- Cepko C.L. and Sharp,P.A. (1982) Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell, 31, 407–415. [DOI] [PubMed] [Google Scholar]
- Chardonnet Y. and Dales,S. (1970) Early events in the interaction of adenoviruses with HeLa cells. I. Penetration of type 5 and intracellular release of the DNA genome. Virology, 40, 462–477. [DOI] [PubMed] [Google Scholar]
- Creigthon T.E. (1993) Proteins, Structures and Molecular Properties. W.H.Freeman and Company, New York, NY. [Google Scholar]
- D’Halluin J.C. (1995) Virus assembly. Curr. Top. Microbiol. Immunol., 199, 47–66. [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Luhrmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Frost E. and Williams,J. (1978) Mapping temperature-sensitive and host-range mutations of adenovirus type 5 by marker rescue. Virology, 91, 39–50. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Gotoh,I., Gotoh,Y. and Nishida,E. (1996) Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem., 271, 20024–20028. [DOI] [PubMed] [Google Scholar]
- Greber U.F., Willetts,M., Webster,P. and Helenius,A. (1993) Stepwise dismantling of adenovirus 2 during entry into cells. Cell, 75, 477–486. [DOI] [PubMed] [Google Scholar]
- Greber U.F., Webster,P., Weber,J. and Helenius,A. (1996) The role of the adenovirus protease on Virus entry into cells. EMBO J., 15, 1766–1777. [PMC free article] [PubMed] [Google Scholar]
- Greber U.F., Suomalainen,M., Stidwill,R.P., Boucke,K., Ebersold,M.W. and Helenius,A. (1997) The role of the nuclear pore complex in adenovirus DNA entry. EMBO J., 16, 5998–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T.C., Zhou,S., da Costa,L.T., Yu,J., Kinzler,K.W. and Vogelstein,B. (1998) A simplified system for generating recombinant adenoviruses. Proc. Natl Acad. Sci. USA, 95, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman R.S. and Ginsberg,H.S. (1976) Characterization of a temperature-sensitive, hexon transport mutant of type 5 adenovirus. J. Virol., 19, 643–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach R.H., Assheuer,R., Kehlenbach,A., Becker,J. and Gerace,L. (2001) Stimulation of nuclear export and inhibition of nuclear import by a Ran mutant deficient in binding to Ran-binding protein 1. J. Biol. Chem., 276, 14524–14531. [DOI] [PubMed] [Google Scholar]
- Krausslich H.G. and Wimmer,E. (1988) Viral proteinases. Annu. Rev. Biochem., 57, 701–754. [DOI] [PubMed] [Google Scholar]
- Kuersten S., Ohno,M. and Mattaj,I.W. (2001) Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol., 11, 497–503. [DOI] [PubMed] [Google Scholar]
- Lonberg-Holm K. and Philipson,L. (1969) Early events of virus–cell interaction in an adenovirus system. J. Virol., 4, 323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I.G. (2001) Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev., 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangel W.F., McGrath,W.J., Toledo,D.L. and Anderson,C.W. (1993) Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature, 361, 274–275. [DOI] [PubMed] [Google Scholar]
- Matthews D.A. and Russell,W.C. (1994) Adenovirus protein–protein interactions: hexon and protein VI. J. Gen. Virol., 75, 3365–3374. [DOI] [PubMed] [Google Scholar]
- Matthews D.A. and Russell,W.C. (1995) Adenovirus protein–protein interactions: molecular parameters governing the binding of protein VI to hexon and the activation of the adenovirus 23K protease. J. Gen. Virol., 76, 1959–1969. [DOI] [PubMed] [Google Scholar]
- Melchior F., Paschal,B., Evans,J. and Gerace,L. (1993) Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J. Cell Biol., 123, 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemerow G.R. and Stewart,P.L. (1999) Role of alpha(v) integrins in adenovirus cell entry and gene delivery. Microbiol. Mol. Biol. Rev., 63, 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipson L. (1995) Adenovirus—an eternal archetype. Curr. Top. Microbiol. Immunol., 199, 1–24. [DOI] [PubMed] [Google Scholar]
- Praszkier J. and Ginsberg,H.S. (1987) Isolation and characterization of temperature-sensitive mutants of adenovirus type 7. J. Virol., 61, 3089–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M.M., White,J.L., Grutter,M.G. and Burnett,R.M. (1986) Three-dimensional structure of the adenovirus major coat protein hexon. Science, 232, 1148–1151. [DOI] [PubMed] [Google Scholar]
- Russell W.C. and Kemp,G.D. (1995) Role of adenovirus structural components in the regulation of adenovirus infection. Curr. Top. Microbiol. Immunol., 199, 81–98. [DOI] [PubMed] [Google Scholar]
- Russell W.C., Patel,G., Precious,B., Sharp,I. and Gardner,P.S. (1981) Monoclonal antibodies against adenovirus type 5: preparation and preliminary characterization. J. Gen. Virol., 56, 393–408. [DOI] [PubMed] [Google Scholar]
- Saphire A.C., Guan,T., Schirmer,E.C., Nemerow,G.R. and Gerace,L. (2000) Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. J. Biol. Chem., 275, 4298–4304. [DOI] [PubMed] [Google Scholar]
- Sherman M.P., de Noronha,C.M., Heusch,M.I., Greene,S. and Greene,W.C. (2001) Nucleocytoplasmic shuttling by human immunodeficiency virus type 1 Vpr. J. Virol., 75, 1522–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P.L. and Burnett,R.M. (1995) Adenovirus structure by X-ray crystallography and electron microscopy. Curr. Top. Microbiol. Immunol., 199, 25–38. [DOI] [PubMed] [Google Scholar]
- Stewart P.L., Fuller,S.D. and Burnett,R.M. (1993) Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J., 12, 2589–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunquist B., Pettersson,U., Thelander,L. and Philipson,L. (1973) Structural proteins of adenoviruses. IX. Molecular weight and subunit composition of adenovirus type 2 fiber. Virology, 51, 252–256. [DOI] [PubMed] [Google Scholar]
- Taagepera S., McDonald,D., Loeb,J.E., Whitaker,L.L., McElroy,A.K., Wang,J.Y. and Hope,T.J. (1998) Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc. Natl Acad. Sci. USA, 95, 7457–7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman L.C., Mosberger,N., Fornerod,M., Stidwill,R.P. and Greber,U.F. (2001) Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol., 3, 1092–1100. [DOI] [PubMed] [Google Scholar]
- van Oostrum J. and Burnett,R.M. (1985) Molecular composition of the adenovirus type 2 virion. J. Virol., 56, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waris M. and Halonen,P. (1987) Purification of adenovirus hexon protein by high-performance liquid chromatography. J. Chromatogr., 397, 321–325. [DOI] [PubMed] [Google Scholar]
- Weber J. (1976) Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J. Virol., 17, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.M. (1995) Adenovirus endopeptidase and its role in virus infection. Curr. Top. Microbiol. Immunol., 199, 227–235. [DOI] [PubMed] [Google Scholar]
- Webster A., Hay,R.T. and Kemp,G. (1993) The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell, 72, 97–104. [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth,J.L., Tsien,R.Y. and Taylor,S.S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Whittaker G.R., Kann,M. and Helenius,A. (2000) Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol., 16, 627–651. [DOI] [PubMed] [Google Scholar]
- Wodrich H., Schambach,A. and Krausslich,H.G. (2000) Multiple copies of the Mason–Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res., 28, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. and Imperiale,M.J. (2003) Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol., 77, 3586–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]