Abstract
Uracil DNA glycosylase (UDG), a ubiquitous and highly specific enzyme, commences the uracil excision repair pathway. Structural studies have shown that the tyrosine in a highly conserved GQDPY water-activating loop of UDGs blocks the entry of thymine or purines into the active site pocket. To further understand the role of this tyrosine (Y66 in Escherichia coli UDG), we have overproduced and characterized Y66F, Y66H, Y66L and Y66W mutants. The complexes of the wild-type, Y66F, Y66H and Y66L UDGs with uracil DNA glycosylase inhibitor (Ugi) (a proteinaceous substrate mimic) were stable to 8 M urea. However, some dissociation of the complex involving the Y66W UDG occurred at this concentration of urea. The catalytic efficiencies (Vmax / Km) of the Y66L and Y66F mutants were similar to those of the wild-type UDG. However, the Y66W and Y66H mutants were ∼7- and ∼173-fold compromised, respectively, in their activities. Interestingly, the Y66W mutation has resulted in an enzyme which is resistant to product inhibition. Preferential utilization of a substrate enabling a long range contact between the –5 phosphate (upstream to the scissile uracil) and the enzyme, and the results of modeling studies showing that the uracil-binding cavity of Y66W is wider than those of the wild type and other mutant UDGs, suggest a weaker interaction between uracil and the Y66W mutant. Furthermore, the fluorescence spectroscopy of UDGs and their complexes with Ugi, in the presence of uracil or its analog, 5-bromouracil, suggests compromised binding of uracil in the active site pocket of the Y66W mutant. Lack of inhibition of the Y66W UDG by apyrimidinic DNA (AP-DNA) is discussed to highlight a potential additional role of Y66 in shielding the toxic effects of AP-DNA, by lowering the rate of its release for subsequent recognition by an AP endonuclease.
INTRODUCTION
Damage to DNA is perilous to cells as it alters the genetic code or obstructs recognition by the proteins (1). In order to prevent such threats, cells possess a number of repair enzymes, which actively search for damage in DNA and correct it. The RNA base uracil arises in DNA either by erroneous incorporation by DNA polymerases or as a consequence of cytosine deamination by physiological or environmental factors (1–3). Uracil DNA glycosylase (UDG), a ubiquitous enzyme (4) recognizes uracils in DNA and pioneers the uracil the excision repair pathway by cleaving the N-glycosidic bond between the base and DNA backbone to generate uracil and apyrimidinic DNA (AP-DNA) as reaction products (5). UDGs are highly conserved not only in their primary structure but also in the overall architecture of the tertiary fold, and are extremely specific for uracil in DNA. Uracil analog 5-bromouracil in DNA is neither acted upon by UDGs nor does it, in its free form, inhibit the enzyme (5,6), suggesting that substitution of non-polar hydrogen at the uracil position 5 with electronegative and/or bulky atoms is unfavorable for binding into the active site pocket of UDGs. On the other hand, analogs such as 6-(p-n-octylanilino) uracil, 5-azauracil and 6-aminouracil are known to inhibit various UDGs (7–9).
UDGs are subject to product inhibition. The AP-DNA acts as a competitive inhibitor with a Ki of ∼1.2 µM (10). Similarly, 6-(p-n-octylanilino) uracil is a competitive inhibitor of HSV UDG (7). In contrast, uracil is known to be a non-competitive inhibitor of UDGs with a Ki of ∼0.2–5 mM (3,9–13). By definition, the non-competitive mode of inhibition means that the inhibitor binds to both the free enzyme and the enzyme–substrate complex. Since free uracil binds into the active site pocket of UDGs (14,15), the apparent non-competitive nature of inhibition may suggest additional site(s) of its binding on the enzyme. Yet another category of inhibitor is a phage encoded protein, uracil DNA glycosylase inhibitor (Ugi), a substrate mimic, which establishes an intricate network of interactions at the active site face of the UDGs to form a highly stable complex (16–18).
The co-crystal structures of UDG with uracil containing DNA as well as free uracil have shown that uracil binding in the active site pocket occurs by extensive shape and charge complementarity (14,15). Several hydrogen bonds are established from the conserved UDG residues such as histidine of the HPSPLS motif [H187 in Escherichia coli UDG (EcoUDG)] and asparagine of the GVLLLN motif (N123 in EcoUDG and N204 in human UDG) to positions 2, 3 and 4 of uracil. Specificity of these contacts avoids cytosine binding in the pocket. Furthermore, the side chain of tyrosine of the GQDPYH motif (Y66 in EcoUDG and Y147 in human UDG), which is in van der Waals’ contact with the C5 position of the uracil, excludes thymine with a methyl group at this position, or the purines with bulky rings (Fig. 1) (19–21). The significance of these interactions in the active site pocket as specificity determinants was highlighted in a protein- engineering quest wherein designed mutations (N204D, Y147A, Y147C and Y147S) in human UDG conferred detectable levels of altered substrate specificities of cytosine, and thymine DNA glycosylases, respectively, upon the mutant proteins (19). However, yet another pursuit in protein engineering is to design variants of the enzymes that are efficient in catalysis and that retain substrate specificity but become resistant to product inhibition (22).
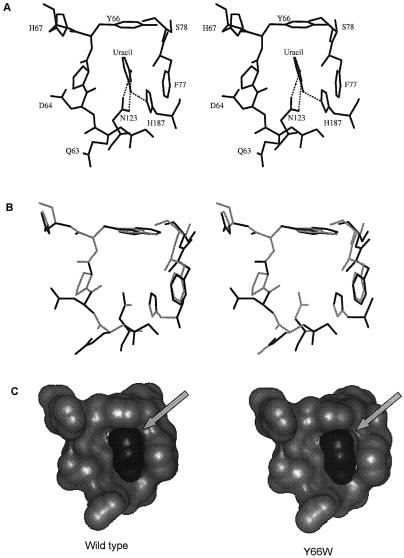
Figure 1.
(A) Uracil specificity pocket showing the interactions between the uracil residue, and the side chains of Y66, F77, N123 and H187 in the active site pocket of E.coli UDG (PDB code 1FLZ). (B) Stereo view of the superposition of uracil specificity pocket of the wild type (light gray) and the Y66W mutant (dark gray) subsequent to removal of short contacts in the Y66W mutant structure. (C) Accessible surface of the pocket (light gray) and the bound uracil (dark gray) illustrating the loosening of the fit in the Y66W mutant. H187 has been removed for clarity.
Recently, we proposed that the role of Y66 in EcoUDG is not restricted to merely preventing the entry of non-uracil residues into the active site pocket and that it also plays a role in catalysis of the glycosidic bond cleavage (23). To further our understanding of the significant role of Y66 in catalysis, product inhibition and interaction with Ugi, in this study, we have characterized additional mutants containing substitutions at the Y66 position.
MATERIALS AND METHODS
Oligodeoxyribonucleotides
DNA oligomers were obtained from Ransom Hill Bioscience, USA, and Microsynth, Switzerland. The oligomers SSU9, d(ctcaagtgUaggcatgcaagagct), and SSU4, d(agcUcatagtttacctgaagaatat), are single-stranded 24mer and 25mer DNA containing dU at the 9th and the 4th positions, respectively. The substrate AU9, d(ctcaagtgUaggcatgcttttgcatgcctacacttga), is a 37mer tetra T-loop hairpin containing dU in the stem region, in the same sequence context as the SSU9.
32P-labeling of oligodeoxyribonucleotides
DNA oligomers (10 pmol) were 5′ 32P-end-labeled using 10 µCi of [γ-32P]ATP (6000 Ci/mmol) and T4 polynucleotide kinase in 10 µl reaction volumes and purified by chromatography on Sephadex G-50 minicolumns (24).
Generation of the Y66 EcoUDG mutants and their over-expression constructs
The NcoI–HindIII DNA fragment containing UDG open-reading frame (ORF) from pTrcEcoUDG(Y66F) (23) was subcloned into the respective sites of pET11d to generate pETEcoUDG(Y66F). To create remaining mutations, quick-change mutagenesis was performed on pTrcEcoUDG and pETEcoUDG, using Pfu DNA polymerase with the following DNA oligomer sets, d(ccaggatcctcatcacggacc) and d(ggtccgtgatgaggatcctgg), d(ccaggatcctttacacggacc) and d(ggtccgtgtaaaggatcctgg), and d(ccaggatccttggcacggacc) and d(ggtccg tgccaaggatcctgg), to generate Y66H, Y66L and Y66W mutations, respectively (23). The plasmid mini-preparations were sequenced to ascertain the mutations (25).
Generation of UDG–Ugi bicistronic constructs
The UDG ORFs from the pTrc99C constructs were amplified by PCR using Pfu DNA polymerase by the gene-specific forward, d(cggaattccatggctaacgaattaacc), and reverse, d(ggaattcctattactcactctctgcc), primers, digested with NcoI and EcoRI and cloned into the same sites of the pTrcUDG (L191G)–Ugi construct by replacing the UDG(L191G) ORF (26). To generate bicistronic constructs in the T7 RNA polymerase-based expression vector, NcoI–HindIII fragments harboring the complete bicistron from the pTrc99c constructs were subcloned into pET11d (25).
Expression and purification of UDG and the UDG–Ugi complexes
The pET11d-derived expression constructs were introduced into E.coli BL21 (DE3) and the transformants inoculated into 1 l of 2YT medium (25). At mid-log phase, the cells were induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside for 3–4 h. Cells were harvested; UDG and UDG–Ugi complexes were purified and quantified (26–28).
Analysis of the in vivo formed UDG–Ugi complexes
The UDG–Ugi complexes from pET11d constructs were expressed as above in E.coli (BL-26, DE3) in 2 ml cultures, the cells were harvested by centrifugation, disrupted by sonication in 0.2 ml of TME buffer (25 mM Tris–HCl, pH 8.0, 2 mM β-mercaptoethanol, 1 mM Na2EDTA) and clarified by centrifugation at 20 000 g for 10 min. The cell-free extracts were analyzed on 15% polyacrylamide (19:1 cross-linking) gels with or without 2–8 M urea (26).
Determination of Km and Vmax
Reactions (15 µl) containing varying amounts of SSU9 along with 20 000 c.p.m. of the 5′ 32P-end-labeled counterpart and appropriate concentrations of UDG in the reaction buffer (50 mM Tris–HCl, pH 7.4, 1 mM Na2EDTA, 1 mM DTT and 25 µg/ml BSA) were incubated at 37°C for 10 min and stopped by adding 5 µl of 0.2 N NaOH. The reaction mixture was heated at 90°C for 30 min, dried in vacuo, taken up in 10 µl of loading dye, and half of the contents were electrophoresed on 15% polyacrylamide–8 M urea gels. The bands corresponding to the product and the leftover substrate were quantified using a BioImage Analyser (Fuji, FLA 2000). The percent product (P) and substrate (S) in each reaction were converted to [P] and [S] (29) and used to determine Km and Vmax from Hofstee plots of two independent experiments.
Time course of uracil excision from AU9
A reaction mixture (70 µl) was set up in UDG buffer containing 35 pmol of AU9 mixed with 20 000 c.p.m. of the same substrate (5′ 32P-end-labeled), as tracer in each reaction. The reaction was started by adding 5 µl of an appropriate dilution of the UDG (2.5 pg for wild type, Y66F, Y66L or Y66W; 250 pg for Y66H) at 37°C. Aliquots (10 µl) were removed at 2, 4, 6, 8 and 10 min and the reactions were terminated and processed as above and analyzed on 15% polyacrylamide–8 M urea gel. The bands corresponding to substrate and product were quantified by a BioImage Analyser to calculate the percent product formed, which was in turn used to calculate pmoles of uracil released per microgram of UDG and plotted against time.
Inhibition of UDG by uracil
The reaction mixture (20 µl) was set up in 2× UDG buffer with 5 pmol of SSU9 along with 20 000 c.p.m. of its 5′ 32P-end-labeled counterpart as tracer, in the absence or presence of different concentrations of uracil (2–12 mM). The reaction was started by addition of 5 µl of the same dilution of UDG to each tube of a set in the reaction buffer and incubated at 37°C for 10 min, terminated and processed as above and analyzed on 15% polyacrylamide–8 M urea gels. The bands corresponding to the product and the leftover substrate were quantified by using a BioImage Analyser.
Inhibition of UDG by uracil and AP-DNA
Reactions (5 µl) containing varying amounts of SSU9 (0, 1.25, 2.5 and 5 pmol) and appropriately diluted wild-type or mutant UDGs were incubated at 37°C for 30 min for complete excision of uracil for use as a source of AP-DNA. The reactions were added to another tube containing 5 pmol of SSU9 (along with 20 000 c.p.m. of the 5′ 32P-end-labeled counterpart) in 2× UDG buffer in 15 µl volumes in the absence or presence of 5 mM uracil, and incubated further for 10 min at 37°C; the reactions were then terminated and analyzed on 15% polyacrylamide urea gels. The AP-DNA so prepared contains submicromolar levels of free uracil. Since, the Ki of uracil as inhibitor of UDG is approximately two to three orders of magnitude higher than that of AP-DNA, the presence of such small amounts of uracil is insignificant.
Mixed substrate UDG assay
Reactions (10 µl) were set up in UDG buffer containing a total of 5 pmol (2.5 pmol each of SSU4 and SSU9) of quantitatively 5′ 32P-end-labeled (24) substrates. The UDG reaction was started by adding 5 µl of the appropriate dilution of UDG at 37°C for 3 min and stopped by adding 5 µl of 0.2 N NaOH. The reactions were analyzed on 15% polyacrylamide–8 M urea gels, and the counts in the product and the leftover substrate bands corresponding to SSU4 and SSU9 were estimated using a BioImage Analyser. Values of percent product formed (uracil excision) from each of the substrates were calculated as [P / (S + P) × 100], where P and S represent counts in the product and leftover substrate bands corresponding to each of the substrates. The ratios of the total counts (S + P) of SSU9 versus SSU4 provide the relative amount of the substrates taken in the reaction, and the ratios of the percent product formed provide a measure of their relative utilization by each of the mutants.
Changes in the intrinsic tryptophan fluorescence of UDG
UDGs or their complexes with Ugi (1 A280/ml) were incubated with urea (0, 2, 4, 6 or 8 M), uracil or 5-bromouracil (0–2 mM) for 2 h at room temperature and the fluorescence spectra were recorded using a spectrofluorimeter (Shimadzu RF-5301PC) (26). The excitation wavelength was 280 nm and the emission spectra were recorded between 300 and 400 nm.
Structural modeling of the UDG mutants
Five models each of free EcoUDG and EcoUDG–uracil complexes were generated from crystal structures with Protein Data Bank code 1EUG and 2EUG, respectively (30). The models were generated by replacing Y66 with phenylalanine, leucine, histidine and tryptophan. Two models containing alternate conformations for the nearly symmetrical side chain of Y66H with a dihedral angle difference of 180° about the Cβ–Cγ bond were constructed. All the histidines were kept neutral and were protonated at Nε. In the case of the Y66W mutant, of the two distinctly different conformers possible for the asymmetric side chain of tryptophan, only one was used. The other was ignored because of the larger exposed hydrophobic surface (by ∼34 Å2) and burial of the polar Nε1 atom. All the models, except that of Y66W, were sterically acceptable. However, in the Y66W mutant the larger side chain of tryptophan clashed with the main chain atoms of F77 and S78. This steric clash was removed by allowing residues 72–82 to move while keeping the rest of the structure fixed. This resulted in an acceptable model of Y66W with a slightly wider uracil pocket.
RESULTS
Purification of the UDG mutants
UDGs (wild type, Y66F, Y66H, Y66L and Y66W) were overproduced in E.coli BL21 (DE3) from T7 RNA polymerase-based high level expression constructs and purified to apparent homogeneity by using various column chromatography steps (26). Even though the E.coli BL21 (DE3) is wild type for ung, UDG preparations from such high level over-expression constructs are essentially free from the chromosomally encoded host UDG (31–33).
Analysis of UDG–Ugi complexes
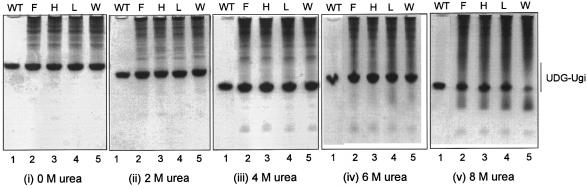
To check for proper folding of the mutant UDGs, we used a substrate mimic, Ugi, to form complexes with them in vivo by using the bicistronic constructs. As shown in Figure 2, the mutant UDGs formed complexes with Ugi, which co-migrated with the wild-type UDG–Ugi complex (i, compare lane 1 with lanes 2–5) suggesting their proper folding. Furthermore, when analyzed for their stability in urea, the complexes of Ugi with the mutant UDGs (Y66F, Y66H and Y66L), like its complex with the wild-type UDG, remained unperturbed in 0–8 M urea (ii–v, lanes 2–4). However, some dissociation of theY66W UDG–Ugi complex occurred in 8 M urea (v, lane 5).
Figure 2.
Stability of UDG–Ugi complexes. A Coomassie brilliant blue stained gel. Purified wild-type UDG–Ugi complex (lane 1) or the cell-free extracts (∼15 µg) of the transformants (lanes 2–5) expressing UDG–Ugi complexes from the bicistronic constructs, as shown, were subjected to electrophoresis on 15% polyacrylamide gels containing different concentrations of urea. (i) Native gel (no urea); (ii) 2 M urea; (iii) 4 M urea; (iv) 6 M urea; and (v) 8 M urea.
Kinetics of uracil excision
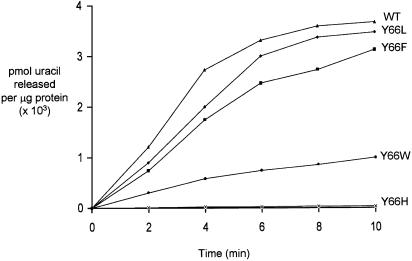
Kinetic parameters of uracil excision were determined to assess the effect of the mutations. Uracil excision from a single-stranded substrate, SSU9 (Table 1), showed that the wild-type and mutant UDGs have comparable Km but differ in their maximal velocity. The catalytic efficiencies (Vmax / Km) of uracil excision by the Y66L and Y66F mutants are similar to that of the wild-type UDG. However, the Y66W and Y66H mutants were compromised in their activities by ∼7- and ∼173-fold, respectively. Use of AU9 in a time course experiment (Fig. 3) suggested that, even for a double-stranded substrate, the Y66W and Y66H mutants were compromised to a similar extent as they were for SSU9. In addition, the Y66L and Y66F mutants excised uracil at a rate comparable to that of the wild-type UDG.
Table 1. Kinetic parameters of uracil excision from SSU9.
| UDG | Km (10–7 M) | Vmax (×103 pmol/min/µg) | Vmax / Km (×10–3) | Ratio (wild type/mutant) |
|---|---|---|---|---|
| Wild type | 3.2 | 58.3 | 182.19 | 1.0 |
| Y66L | 3.0 | 51.5 | 171.67 | 1.1 |
| Y66F | 1.3 | 29.1 | 223.85 | 0.8 |
| Y66W | 3.1 | 7.9 | 25.55 | 7.1 |
| Y66H | 3.3 | 0.35 | 1.05 | 173.5 |
Reactions (15 µl) containing 0.5–30 pmol of SSU9 along with 1.25 (wild type, Y66L, Y66F), 12.5 (Y66W) or 125 pg (Y66H) of UDGs were carried out as described in Materials and Methods. Average values of Km and Vmax were calculated from two independent sets of experiments whose values were within 20% of the average.
Figure 3.
Time course uracil excision by UDGs. Excision of uracil from a double-stranded DNA oligomer, AU9, by wild-type (triangles), Y66F (squares), Y66H (crosses), Y66L (diamonds) and Y66W (circles) UDGs. Reactions using appropriate dilutions (Materials and Methods) were carried out such that 30–50% of substrate was converted to product. The amount of product formed was calculated as pmoles of uracil released per microgram of UDG and plotted against time.
Inhibition of UDGs with uracil and AP-DNA
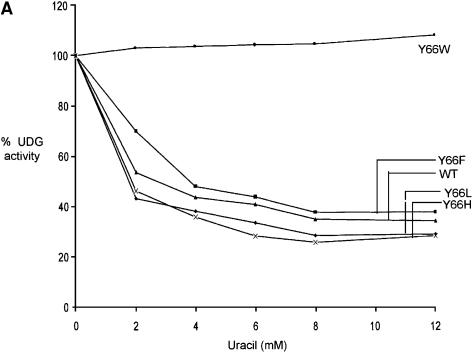
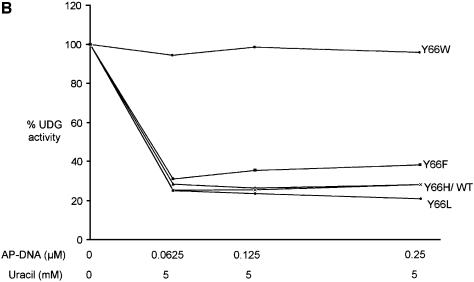
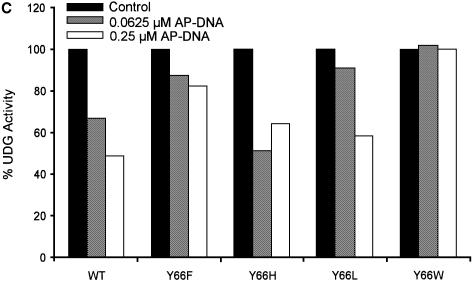
Uracil and AP-DNA are the products of the UDG reaction. Inclusion of uracil in the reactions (Fig. 4A) resulted in an inhibition of the wild-type, Y66L, Y66H and Y66F UDGs by ∼60%. These profiles of inhibition by uracil are identical to those obtained earlier with the UDGs from E.coli, human, HSV, mycoplasma and wheat germ (3,8–13). Interestingly, in these experiments, no inhibition of uracil excision by the Y66W UDG was seen (Fig. 4A). Also, upon inclusion of varying concentrations of AP-DNA (0.0625–0.25 µM) along with uracil (5 mM) in reactions, while the inhibition of the wild-type, Y66L, Y66H and Y66F UDGs increased to ∼80%, the Y66W UDG was still completely resistant to inhibition (Fig. 4B). Since the AP-DNA that we used in these reactions was prepared from the uracil containing DNA (Materials and Methods), to ensure that the preparation served as an inhibitor of UDGs, we carried out UDG assays in the absence of any externally added uracil. As shown in Figure 4C, at 0.25 µM concentration, the AP-DNA preparation inhibited the wild-type UDG by ∼50%. Interestingly, the Y66W mutant UDG was not inhibited at all. The other UDG mutants (Y66F, Y66H and Y66L) were inhibited to varying degrees. The Y66F protein was inhibited to a small extent by ∼20%. Even in the experiment shown in Figure 4B, upon addition of the AP-DNA in the reaction, this mutant showed the least increment in inhibition (over and above uracil inhibition). More importantly, these experiments identified the Y66W mutant as a UDG, which showed complete resistance to inhibition by both the products.
Figure 4.
Inhibition of UDGs by uracil and AP-DNA. (A) Inhibition by uracil: reactions were carried out using SSU9 with appropriately diluted UDGs, and the total product formed in the reactions with the wild-type, Y66F, Y66L, Y66H and Y66W UDGs was 2570, 2310, 3610, 2890 and 600 fmol, respectively, in the absence of externally added uracil, which was used as a reference for 100% activities. The products formed with the same dilutions of UDGs in the presence of uracil (2–12 mM) were calculated with respect to the reference values of the respective UDGs and plotted against uracil concentration (0–12 mM): wild-type (triangles), Y66F (squares), Y66H (crosses), Y66L (diamonds) and Y66W (circles) UDGs. (B) Inhibition by uracil and AP-DNA: UDG reactions were carried out in the presence of 5 mM uracil along with 0.062, 0.125 or 0.25 µM AP-DNA. The total amounts of product formed in the reactions with the wild-type, Y66F, Y66L, Y66H and Y66W UDGs were 1194, 999, 985, 1070 and 1870 fmol, respectively, in the absence of both the AP-DNA and uracil, which were used as reference for 100% activities. Activities in the presence of 5 mM uracil and the indicated amounts of AP-DNA were calculated with respect to these reference values for the respective UDGs and plotted against the AP-DNA concentration: wild-type (triangles), Y66F (squares), Y66H (crosses), Y66L (diamonds) and Y66W (circles) UDGs. (C) Same as (B) except that the experiment was carried in the absence of externally added uracil and in the presence of AP-DNA as inhibitor. The activities in the presence of AP-DNA were calculated with respect to the reference activities (100%) given in (B) and plotted as a histogram.
Y66W UDG shows differential utilization of SSU9 over SSU4
We have shown earlier that, with the addition of the –1, +1 and +2 phosphates (with respect to the scissile uracil), UDGs make an additional contact at the –5 phosphate (23,34). While this contact is not crucial for substrate utilization by the wild-type UDG in vitro, it is important for the activity of the mutant UDGs that are impaired in establishing a full complement of interactions with uracil in the active site pocket. The resistance of the Y66W UDG to uracil inhibition suggested its weak binding in the active site pocket. Hence, we checked for relative utilization of SSU9 and SSU4 by mutant UDGs in a mixed substrate (SSU9 and SSU4) assay. SSU9 contains the –5 phosphate, whereas the SSU4 lacks this phosphate even after 5′ end phosphorylation (Materials and Methods). The quantification of data (Fig. 5) shows that, while the wild-type, Y66F, Y66H and Y66L UDGs use the substrates with similar relative efficiencies (Fig. 5, ii–v), the Y66W UDG utilized SSU9 >4-fold better than the SSU4 (vi).
Figure 5.
Mixed substrate UDG assay: equimolar amounts of oligomers SSU4 and SSU9 (2.5 pmol each) were either incubated in buffer alone (i) or with UDGs [wild type (ii), Y66F (iii), Y66H (iv), Y66L (v) and Y66W (vi)] for 3 min at 37°C and analyzed on 15% polyacrylamide–8 M urea gels. Values of percent product formed (uracil excision) from each of the substrates were calculated as [P / (S + P) × 100], where P and S represent counts in the product and leftover substrate bands corresponding to each of the substrates. The ratios of the total counts (S + P) of SSU9 versus SSU4 provide the relative amount of the substrates in the reactions, whereas the ratios of the percent product formed from SSU9 versus SSU4 provide a measure of their relative utilization by each of the mutants. The mean values ± SD of three independent experiments are shown as histograms wherein the light gray bars and the dark gray bars represent the ratios of substrates taken in the reaction, and the ratios of the products formed, respectively. Representative autoradiograms have also been shown for each panel.
Widening of the uracil pocket in Y66W
As mentioned in Materials and Methods, a sterically acceptable model of the Y66W could be constructed only when the 72–82 polypeptide segment was allowed to move away from the center of the uracil binding cavity, to accommodate the larger size of the tryptophan side chain (Fig. 1B). The movement widens the uracil binding pocket, loosening the contacts between uracil and F77 (Fig. 1C). An analogous situation is observed in the case of mycobacterial RecA which has a lower affinity for nucleotides than its close homolog from E.coli. Modeling and crystal structures indicate that this difference occurs due to a wider nucleotide binding site in the case of mycobacterial RecA (35). In the case of the UDG mutants, the looseness of the fit in Y66W is quite conspicuous. While the shape complementarity coefficient between UDG and uracil varies between 0.78 and 0.81 for the WT, Y66F, Y66L and Y66H, it is 0.73 in the case of Y66W. These coefficients were computed using the Lawrence and Colman method (36), where a value of 1 on a scale of 0–1 indicated ideal compatibility between surfaces.
Taken together, the preferential utilization of SSU9 over SSU4 by the Y66W UDG, and the molecular modeling studies, support the view that the interaction of uracil in the active site pocket of the Y66W UDG is compromised. This, in turn, would explain the lack of inhibition of Y66W by uracil.
Changes in the intrinsic tryptophan fluorescence of UDGs and UDG–Ugi complexes in the presence of urea
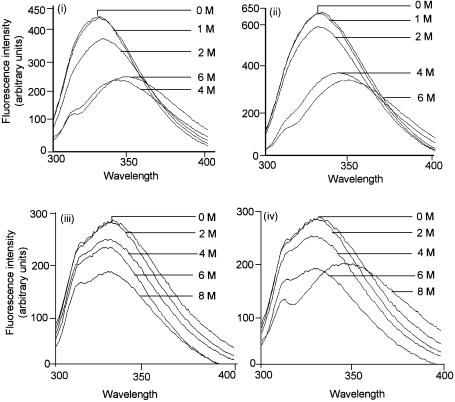
To further our understanding of the Y66W UDG, we subjected this and the wild-type UDGs to urea-mediated unfolding and monitored the changes in the intrinsic tryptophan fluorescence. In such experiments, unfolding of UDGs results in fluorescence quenching as well as in a red shift of the fluorescence spectra (26). As shown in Figure 6, the changes in the spectral profiles of both the wild-type and Y66W UDGs are very much alike and they both show a red shift upon treatment with 4 M urea (i and ii).
Figure 6.
Spectrofluorimetric analysis of UDGs and its complexes with Ugi in the presence of urea: UDGs (wild type and Y66W) or their complexes with Ugi (1 A280/ml) were incubated with different concentrations of urea at room temperature for 2 h and the fluorescence spectra (300–400 nm) were recorded upon excitation at 280 nm. (i) Wild-type UDG; (ii) Y66W UDG; (iii) complex of the wild-type UDG with Ugi; and (iv) complex of the Y66W UDG with Ugi.
Furthermore, as shown before (26), complexation of the wild-type UDG with Ugi makes it impervious to treatment even with 8 M urea, and in the intrinsic tryptophan fluorescence spectra, no red shifts [characteristic of decomposition of the complex (26)] are seen (Fig. 6, iii). However, when the complex of the Y66W UDG and Ugi was treated with 8 M urea (Fig. 6, iv), consistent with its dissociation on 8 M urea-containing gels (Fig. 2, v), it showed a red shift in the spectra. The observation that the free UDGs (wild type and Y66W) are alike in their resistance/susceptibility to urea, but among their complexes with Ugi, the UDG(Y66W)–Ugi, is slightly more susceptible to urea, suggests that the Y66W protein suffers a subtle structural change in the DNA binding/active site pocket which establishes contacts with Ugi, a substrate mimic (see Discussion).
Changes in the intrinsic tryptophan fluorescence of UDGs and/or the UDG–Ugi in the presence of uracil or 5-bromouracil
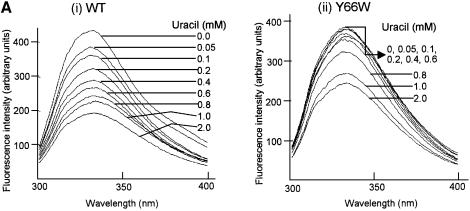
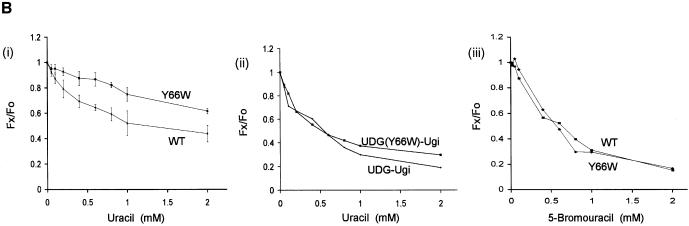
Treatment of UDGs or their complexes with uracil or 5-bromouracil led to changes in the intrinsic tryptophan fluorescence with no shifts in the spectra. As shown in the spectral recordings in Figure 7A, treatment of the wild-type UDG with increasing concentrations of uracil resulted in a gradual increase in fluorescence quenching (i). In contrast, under the identical assay conditions, treatment of the Y66W UDG did not result in significant fluorescence quenching at the lower concentrations of uracil (0–0.6 mM, ii). These observations suggest poor binding of uracil to the Y66W UDG.
Figure 7.
Spectrofluorimetric analyses. (A) Wild-type and Y66W UDGs (1 A280/ml) were incubated with different concentration of uracil (i and ii, respectively) at room temperature for 2 h and the fluorescence spectra (300–400 nm) were recorded upon excitation at 280 nm. (B) (i) The mean values of relative fluorescence (Fx / Fo) from three independent experiments [a representative set is shown in (A)] were calculated and plotted against the ligand concentration [Fo represents the fluorescence intensity at 332 nm (λmax) of the untreated control sample whereas Fx represents fluorescence intensity at 332 nm (λmax) at a given concentration of uracil or 5-bromouracil]. (ii and iii) Same as in (i), except that the data from single sets of experiments wherein the UDG–Ugi complexes or UDGs were treated with uracil (ii) or 5-bromouracil (iii), respectively, are shown.
To gain better insight into these effects, we compared the profiles of the relative ratios (Fx / Fo) of the fluorescence intensities (at λmax of 332 nm) of the uracil treated (Fx) and untreated (Fo) samples for both the UDGs. As shown in Figure 7B (i), treatment of the wild-type UDG with uracil resulted in a distinct biphasic profile of quenching. While a biphasic profile is discernible even for the Y66W UDG, the amplitude of the first phase in this profile is much decreased. Interestingly, a control experiment, where the complexes of the two UDGs with Ugi (wherein access of uracil to the active site pocket of UDGs is blocked by Ugi) were treated with uracil, revealed identical profiles for both the samples (ii). Thus, a more rapid dip in the profile of the wild-type UDG at lower concentrations of uracil (Fig. 7B, i) suggests its high affinity binding in the active site pocket of the wild-type UDG. In yet another approach, the wild-type and the Y66W UDGs were treated with 5-bromouracil, an analog of uracil possessing a substitution at C5 position unfavorable for interaction in the active site pocket. In this experiment also, the relative fluorescence quenching profiles of the two UDGs were alike (iii). Taken together, these experiments suggest that the binding of free uracil to the active site pocket of the Y66W UDG is weak.
DISCUSSION
An extreme specificity in detection and removal of uracil by UDGs is rendered by interaction of uracil with a number of residues in its active site pocket (Fig. 1A). Recent studies involving mutational analysis of the in vitro produced EcoUDG mutants at the Y66 position, suggested that in addition to its role in uracil base selectivity, the van der Waals’ interaction that it establishes with the C5 position of the uracil also facilitates the catalysis (23). In order to understand the role of Y66 further, we have overproduced several proteins mutated at this position (Y66F, Y66L, Y66H and Y66W). As summarized in Table 1, the catalytic efficiencies of the Y66L and Y66F proteins were very similar to those of the wild-type UDG. The observation that the mutant UDGs containing substitution of tyrosine with phenylalanine or leucine, but not with histidine (Table 1), cysteine or serine (23), were as active as the wild-type protein, suggests that a hydrophobic residue at this position is crucial for efficient catalysis.
All the conserved UDGs form a highly stable complex with the Bacillus subtilis phage, PBS-1, or -2, encoded proteinaceous inhibitor (Ugi). EcoUDG forms a complex with Ugi, which is physiologically irreversible and has been a good model system to understand the mechanism of protein–protein interaction. The Y66 side chain of EcoUDG forms a water-mediated hydrogen bond with E20 of Ugi. As shown in Figure 2, like the complex of the wild-type proteins, the complexes of mutant UDGs (Y66L, Y66H and Y66F) with Ugi were also stable in 8 M urea. Thus, the identity of tyrosine at this position is unimportant in forming a tight complex with Ugi. An earlier report showed that a mutant of Ugi (E20A) formed a weaker and reversible complex with UDG (37). However, it should be noted that E20 of Ugi, makes two more hydrogen bonds with S88 and S189 of EcoUDG (17,18), and a single mutation at this position in Ugi (E20A) will be expected to have a more severe phenotype, as observed (37). The presence of diminishing band of the UDG (Y66W)–Ugi complex in 8 M urea (Figs 2 and 6) is more likely a consequence of a subtle structural perturbation in the active site/DNA binding pocket, which is commonly utilized by both the Ugi and the substrate for their interaction with UDGs. Several other lines of evidence support this conclusion. (i) Co-crystal structures of uracil and UDG have shown that at least one site of uracil binding to UDGs is the active site pocket (14,15). Thus, unlike the uracil inhibition of the wild-type, Y66L, Y66F and Y66H UDGs, lack of uracil inhibition of the Y66W UDG indicates that uracil binding in its active site pocket is compromised. In fact, the modeled structures show that the active site pocket of the Y66W mutant is larger than those of the wild-type and the other mutant UDGs. (ii) In the co-crystals of human UDG with DNA, the equivalent of H67 (H148) in the neighbor of the equivalent of Y66 (Y147) forms hydrogen bonds with the uridine O4′ and the –2 phosphate (38). Since the three-dimensional structural folds of the UDGs are highly conserved, the lack of inhibition of the Y66W UDG by AP-DNA would again suggest a structural perturbation in the uridine binding pocket of this mutant. (iii) The observation that, unlike the wild-type UDG and other mutants studied here, the Y66W UDG utilizes SSU9 preferentially over SSU4 (Fig. 5), also suggests (23) that the Y66W UDG is deficient in establishing a full complement of interactions in the uracil binding pocket. (iv) The changes in the relative intrinsic fluorescences of UDG in the presence of uracil (Fig. 7) support the view that the Y66W mutation has resulted in poor binding of uracil in its active site. Earlier studies (39) suggested that the rate of catalysis by UDG is limited by the slower rate of product release. Since the affinity of the products to the Y66W UDG is highly compromised (no inhibition by the products), it is quite likely that the actual efficiency of the glycosidic bond cleavage by the Y66W UDG is compromised to an extent larger than the apparent decrease of ∼7-fold in its overall rate. Importantly, and consistent with our earlier report (23), this observation supports the role of the Y66 residue in UDGs in assisting transition state substrate binding in the active site pocket and in decreasing the rate of product release. As the unprotected AP sites in DNA are mutagenic and toxic, Y66 of this repair enzyme (UDG) may perform an important function in shielding the AP site in DNA (by decreasing product release) until its recognition by a relevant AP endonuclease (39).
The fluorescence quenching studies (Fig. 7) allow us to draw further insights into the complex mechanism of UDG inhibition kinetics by uracil (7). The crystal structures have clearly revealed that by the virtue of uracil binding in the active site pocket, it should be a competitive inhibitor of UDGs. However, a number of biochemical studies have shown that uracil is a non-competitive inhibitor (3,8–13). The observed biphasic nature of the uracil binding profile to the wild-type UDG (Fig. 7B, i) suggests that in addition to binding to the active site pocket, uracil binds to UDGs at some other site(s) as well. The lack of Y66W UDG inhibition suggests that uracil binding to this site alone does not lead to a direct inhibition of enzyme activity. However, the binding of uracil to the second site may still facilitate its channeling into the active site pocket to result in a complex mode of inhibition, which is apparently non-competitive. Interestingly, from a physiological perspective, the additional binding site(s) on the enzyme may help in rapid localization of the substrate into the active site pocket of UDG and in conferring processivity to this enzyme (40).
Finally, while an N-terminal deletion construct (UNG1 ΔN29) of human UDG1 (precursor of the mitochondrial UDG) is resistant to inhibition by AP-DNA, it is sensitive to inhibition by uracil (41). And, although the mutations in human UDG (Y147A and Y147C) have been described that are resistant to inhibition by uracil, these are >1000-fold compromised in their uracil excision activity (19). On the other hand, the Y66W UDG is only ∼7-fold compromised in its catalytic activity (compared with the wild-type counterpart). To our knowledge, the Y66W is the first characterized UDG mutant, which is an efficient enzyme and which shows no susceptibility to either of the reaction products. Furthermore, the observation that in the UDG assays, the only cleavage products that we detected corresponds to specific excision of uracils (Fig. 5), and the fact that the Y66W protein was hyperexpressed in vivo, clearly show that it retains high specificity towards uracils in DNA. The biochemical studies described here pave the way for determination of the structural basis of the fascinating properties of the Y66W UDG mutant.
Acknowledgments
ACKNOWLEDGEMENTS
We thank our laboratory colleagues for their suggestions. This work was supported in part by research grants from the Department of Biotechnology, Council of Scientific and Industrial Research (CSIR) and the Indian Council of Medical Research, New Delhi, India.
REFERENCES
- 1.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 2.Lindahl T. (1974) An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl Acad. Sci. USA, 71, 3649–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindahl T., Ljungquist,S., Siegert,W., Nybert,B. and Sperens,B. (1977) DNA N-glycosidase. J. Biol. Chem., 252, 3286–3294. [PubMed] [Google Scholar]
- 4.Aravind L. and Koonin,E.V. (2000) The α/β fold uracil DNA glyosylases: a common origin with diverse fates. Genome Biol., 1, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindahl T. (1982) DNA repair enzymes. Annu. Rev. Biochem., 51, 61–87. [DOI] [PubMed] [Google Scholar]
- 6.Duncan B.K. (1981) DNA Glycosylases. In Boyer,P. (ed.), The Enzymes. Academic Press, Orlando, pp. 565–586. [Google Scholar]
- 7.Focher F., Verri,A., Spadari,S., Manservigi,R., Gambino,J. and Wright,G.E. (1993) Herpes simplex virus type 1 uracil-DNA glycosylase: isolation and selective inhibition by novel uracil derivatives. Biochem. J., 292, 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krokan H.E. and Wittwer,C.U. (1981) Uracil DNA-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res., 9, 2599–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaisdell P. and Warner,H. (1983) Partial purification and characterization of a uracil-DNA glycosylase from wheat germ. J. Biol. Chem., 258, 1603–1609. [PubMed] [Google Scholar]
- 10.Domena J.D., Timmer,R.T., Dicharry,S.A. and Mosbaugh,D.W. (1988) Purification and properties of mitochondrial uracil-DNA glycosylase from rat liver. Biochemistry, 27, 6742–6751. [DOI] [PubMed] [Google Scholar]
- 11.Williams M.V. and Pollack,J.D. (1990) A mollicute (mycoplasma) DNA repair enzyme: purification and characterization of uracil-DNA glycosylase. J. Bacteriol., 172, 2979–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caradonna S.J. and Cheng,Y.C. (1980) Uracil DNA-glycosylase. Purification and properties of this enzyme isolated from blast cells of acute myelocytic leukemia patients. J. Biol. Chem., 255, 2293–2300. [PubMed] [Google Scholar]
- 13.Borle M.T., Campagnari,F. and Creissen,D.M. (1982) Properties of purified uracil DNA glycosylase from calf thymus. J. Biol. Chem., 257, 1208–1214. [PubMed] [Google Scholar]
- 14.Krokan H.E., Standal,R. and Slupphaug,G. (1997) DNA glycosylases in the base excision repair of DNA. Biochem. J., 325, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearl L.H. (2000) Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res., 460, 165–181. [DOI] [PubMed] [Google Scholar]
- 16.Cone R., Bonura,T. and Friedberg,E.C. (1980) Inhibitor of uracil-DNA glycosylase induced by bacteriophage PBS2. Purification and preliminary characterization. J. Biol. Chem., 255, 10354–10358. [PubMed] [Google Scholar]
- 17.Ravishankar R., Bidya Sagar,M., Roy,S., Purnapatre,K., Handa,P., Varshney,U. and Vijayan,M. (1998) X-ray analysis of a complex of E. coli uracil-DNA glycosylase (EcUDG) with its proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG. Nucleic Acids Res., 26, 4880–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Putnam C.D., Shroyer,M.J.N., Lundquist,A.J., Mol,C.D., Arvai,A.S. and Mosbaugh,D.W. (1999) Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol., 287, 331–346. [DOI] [PubMed] [Google Scholar]
- 19.Kavli B., Slupphaugh,G., Mol,C.D., Arvai,S.A., Petersen,S.B., Tainer,J.A. and Krokan,H.E. (1996) Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J., 15, 3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 20.Mol C.D., Arvai,A.S., Slupphaug,G., Kavli,B., Alseth,I., Krokan,H.E. and Tainer,J.A. (1995) Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell, 80, 869–878. [DOI] [PubMed] [Google Scholar]
- 21.Savva R., McAuley-Hecht,K., Brown,T. and Pearl,L. (1995) The structural basis of specific base excision repair by uracil-DNA glycosylase. Nature, 373, 487–493. [DOI] [PubMed] [Google Scholar]
- 22.Wada M., Awano,N., Haisa,K., Takagi,H. and Nakamori,S. (2002) Purification, characterization and identification of cysteine desulfhydrase of Corynebacterium glutamicum and its relationship to cysteine production. FEMS Microbiol. Lett., 217, 103–107. [DOI] [PubMed] [Google Scholar]
- 23.Handa P., Acharya,N. and Varshney,U. (2002) Effects of mutations at tyrosine 66 and asparagine 123 in the active site pocket of Escherichia coli uracil DNA glycosylase on uracil excision from synthetic DNA oligomers: evidence for the occurrence of long-range interactions between the enzyme and substrate. Nucleic Acids Res., 30, 3086–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar N.V. and Varshney,U. (1994) Inefficient excision of uracil from loop regions of DNA oligomers by E. coli uracil-DNA glycosylase. Nucleic Acids Res., 18, 3737–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 26.Handa P., Roy,S. and Varshney,U. (2001) The role of leucine 191 of Escherichia coli uracil DNA glycosylase in forming a stable complex with a substrate mimic, Ugi and in uracil excision from synthetic substrates. J. Biol. Chem., 276, 17324–17331. [DOI] [PubMed] [Google Scholar]
- 27.Roy S., Purnapatre,K., Handa,P., Boyanapalli,M. and Varshney,U. (1998) Use of coupled transcriptional system for consistent overexpression and purification of UDG-Ugi complex and Ugi from E. coli. Protein Expr. Purif., 13, 155–162. [DOI] [PubMed] [Google Scholar]
- 28.Sedmak J.J. and Grossberg,S.E. (1977) A rapid, sensitive and versatile assay for protein using Coomassie brilliant blue G250. Anal. Biochem., 79, 544–552. [DOI] [PubMed] [Google Scholar]
- 29.Kumar N.V. and Varshney,U. (1997) Contrasting effects of single stranded DNA binding protein on the activity of uracil-DNA glycosylase from Escherichia coli towards different DNA substrates. Nucleic Acids Res., 25, 2336–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao G., Tordova,M., Jagadeesh,J., Drohat,A.C., Stivers,J.T. and Gilliland,G.L. (1999) Crystal structure of Escherichia coli uracil-DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited. Proteins: Struct. Funct. Genet., 35, 13–24. [PubMed] [Google Scholar]
- 31.Shroyer M., Putnam,C.D., Tainer,J.A. and Mosbaugh,D.W. (1999) Mutation of an active site residue in Escherichia coli uracil-DNA glycosylase: effect of DNA-binding, uracil inhibition and catalysis. Biochemistry, 38, 4834–4845. [DOI] [PubMed] [Google Scholar]
- 32.Stivers J.T., Pankiewicz,K.W. and Watanabe,K.A. (1999) Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil-DNA glycosylase. Biochemistry, 38, 952–963. [DOI] [PubMed] [Google Scholar]
- 33.Sartori A.A., Fitz-Gibbon,S., Yang,H., Miller,J.H. and Jiricny,J.A. (2002) Novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J., 21, 3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varshney U. and van de Sande,J.H. (1991) Specificities and kinetics of uracil excision from uracil containing DNA oligomers by Escherichia coli uracil-DNA glycosylase. Biochemistry, 30, 4055–4061. [DOI] [PubMed] [Google Scholar]
- 35.Datta S., Prabu,M.M., Vaze,M.B., Ganesh,N., Chandra,N.R., Muniyappa,K. and Vijayan,M. (2000) Crystal structures of Mycobacterium tuberculosis RecA and its complexes with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation. Nucleic Acids Res., 28, 4964–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence R.A. and Colman,P.M. (1993) Shape complementarity at protein/protein interfaces. J. Mol. Biol., 234, 946–950. [DOI] [PubMed] [Google Scholar]
- 37.Lundquist A.J., Beger,R.D., Bennett,S.E., Bolton,P.H. and Mosbaugh,D.W. (1997) Site-directed mutagenesis and characterization of uracil-DNA glycosylase inhibitor protein. Role of specific carboxylic amino acids in complex formation with Escherichia coli uracil-DNA glycosylase. J. Biol. Chem., 272, 21408–21419. [DOI] [PubMed] [Google Scholar]
- 38.Slupphaug G., Mol,C.D., Kavli,B., Arvai,A.S., Krokan,H.E. and Tainer,J.A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- 39.Parikh S.S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.A. (1998). Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higley M. and Lloyd,R.S. (1993) Processivity of uracil-DNA glycosylase. Mutat. Res., 294, 109–116. [DOI] [PubMed] [Google Scholar]
- 41.Bharati S., Krokan,H.E., Kristiansen,L., Otterlei,M. and Slupphaug,G. (1998) Human mitochondrial uracil-DNA glycosylase preform (UNG1) is processed to two forms one of which is resistant to inhibition by AP sites. Nucleic Acids Res., 26, 4953–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]