Abstract
WASH is the Arp2/3 activating protein that is localized at the surface of endosomes, where it induces the formation of branched actin networks. This activity of WASH favors, in collaboration with dynamin, the fission of transport intermediates from endosomes, and hence regulates endosomal trafficking of several cargos. We have purified a novel stable multiprotein complex containing WASH, the WASH complex, and we examine here the evolutionary conservation of its seven subunits across diverse eukaryotic phyla. This analysis supports the idea that the invention of the WASH complex has involved the incorporation of an independent complex, the CapZ α/β heterodimer, forming the so-called Capping Protein (CP), as illustrated by the yeasts S. cerevisiae and S. pombe, which possess the CP heterodimer but no other subunits of the WASH complex. The alignements of the orthologous genes that we have generated give a view on the conservation of the different subunits and on their organization into domains. Moreover, we propose here a unique nomenclature for the different subunits to prevent future confusions in the field.
Key words: Arp2/3 complex, endosome, VPEF, KIAA0592, KIAA1033, strumpellin, KIAA0196, CP, CapZ, Ccdc53
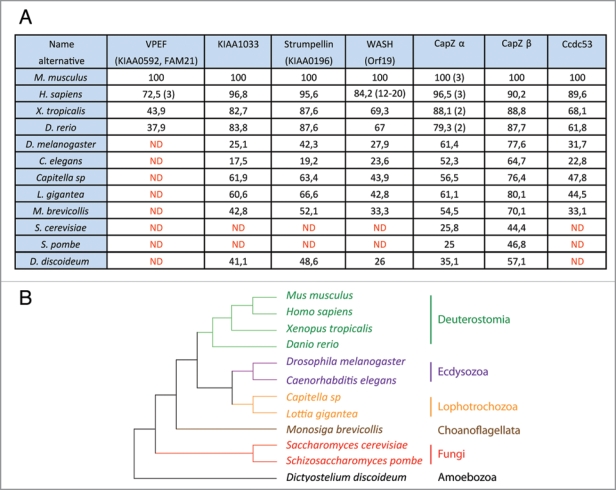
The WASH complex has been purified from human cells by tandem affinity purification.1 WASH was found associated with VPEF, KIAA1033, Strumpellin, Ccdc53, CapZ α and β. The native WASH protein is found in a large complex characterized by a Stokes’ radius of 88 Å and a sedimentation coefficient of 12.5 S.1 Moreover, depletion of any of the abovementioned proteins associated with WASH destabilizes WASH. Together these properties are compatible with a stable complex containing one molecule of each of these proteins. One expects to find all the genes encoding the 7 subunits of this molecular machine in species where this organization into a multiprotein complex is conserved. So using the BLAST algorithm, we looked into public databases for proteins homologous to the ones we had purified. Genes encoding subunits of the WASH complex were detected in genomes of animal, fungi and amoeba species. But none of the subunits were detected in bacterial and plant genomes. The orthologous subunits were retrieved and compared to the mouse subunits chosen as a reference instead of the human ones, to avoid the complexity of the human WASH family.2,3
The two subunits forming the CP cap actin filaments and hence blocks filament elongation. The 2 CP subunits, but no other subunits of the WASH complex, were detected in the S. cerevisiae and S. pombe yeasts. The same was true for other sequenced fungi. Since the amoeba D. discoideum possesses most WASH complex subunits, the ancestor of fungi and amoeba likely possessed these genes, suggesting that the WASH complex has been subsequently lost in fungi. In fungi genomes, the occurrence of CP without the WASH complex is in line with a free heterodimer representing the major pool of CP independently of the WASH complex.1,4 The CP heterodimer is also an integral component of the Dynactin complex.1,4 The many interactions of CP with actin, the WASH complex and the dynactin complex must exert a strong selective pressure on the CP genes, probably explaining why these genes encoding subunits of the WASH complex are among the most conserved ones.
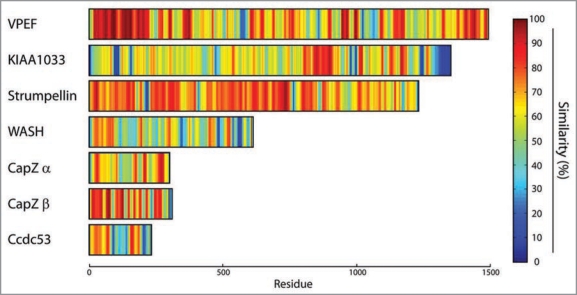
The WASH subunit activates the Arp2/3 complex through its VCA domain at its C-terminus. This domain is about 100 amino-acid long. WASH stands for Wiskott-Aldrich Syndrome Protein and SCAR Homolog.3 This descriptive name has imposed itself and replaced the previously used Orf19.2 The organization of WASH is indeed very similar to the ones of WASP and SCAR/WAVE activators of the Arp2/3 complex. Based on this analogy, the N-terminal WAHD domain covering the first 300 amino-acids, is likely to be integral to the multiprotein complex similar to the SCAR/WAVE N-terminal domain in the pentameric SCAR/WAVE complex and to the N-terminal domain of N-WASP, which complexes N-WASP with WIP family proteins.5 The rest of WASH, including the VCA domain, is predicted to be unstructured (Suppl. Fig. 2).
The VPEF subunit is of major interest, but confusing. VPEF was first characterized as a factor required for Vaccinia virus entry into mammalian cells, hence its name Vaccinia PEnetration Factor.6 Unfortunately, this publication has been overlooked in the two recent publications reporting VPEF association with WASH. We used the name of the cDNA, KIAA0592, to refer to this protein.1 Gomez and Billadeau used the name FAM21, standing for FAMily number 21 coined in automatic annotations of protein families.7 We recommend to use the name VPEF, which is anterior and descriptive. The major pool of the WASH complex and hence of VPEF is associated with endosomes.1,7 Nonetheless, the WASH complex is recruited to the plasma membrane upon infection with pathogens. WASH is detected at attachment sites of Salmonella, and is involved in the entry of this pathogenic bacterium into the cell.8 Similarly, VPEF downregulation impairs the entry of Vaccinia virus.6 A surprising result, however, was that entry of Vaccinia is also blocked by extracellular antibodies directed against VPEF or by soluble VPEF in the culture medium, as if VPEF played the role of an extracellular receptor for the virus.6 These observations remain to be explained, because the WASH complex is cytosolic and no transmembrane domain is predicted in the VPEF protein. VPEF is overall poorly conserved, and the percentage of identity drops so rapidly in vertebrates that invertebrate orthologs are not detected (Fig. 1). The N-terminal 350 amino-acids of VPEF are necessary and sufficient for building the WASH complex. 7 This result is in line with the conservation concentrated in the N-terminal domain (Fig. 2). Right after this domain, VPEF is predicted to be disordered with high probability (Suppl. Fig. 2). This lack of intrinsic structure for the most part of the protein might explain why VPEF is highly sensitive to proteases and does not migrate at its expected size after purification of the WASH complex.1
Figure 1.
(A) Percentage of identity of orthologous subunits of the WASH complex in different species. More than a dozen of WASH genes are present in Homo sapiens and the exact number varies between individuals.2,3 Reference has thus been set to the Mus musculus ortholog. The number of CapZ α is indicated in parentheses when more than one. The reference was set to murine CapZ α1, and the % of identity was calculated from the closest homolog in species possessing more than one paralogous CapZ α genes. ND : Not Detected. Accession numbers of the genes can be found in Supplementary Figure 1. (B) Known established relationships between the selected species from different eukaryotic phyla.11
Figure 2.
Percentage of similarity among orthologous subunits of the WASH complex displayed in a color-coded manner to highlight domain organization. Hot colors represent the highest level of conservation. The similarity score at each position was calculated from each multiple alignment including all subunit orthologs described in Figure 1 using Jalview. These scores were then averaged using a gliding window of 10 residues and colorcoded using the MAT LAB software. This representation gives an overview of the organization of the subunits into domains, but cannot be used to compare the conservation between subunits since the number of detected orthologous genes is variable. The VPEF plot, for example, is derived from only four orthologous genes.
The Strumpellin subunit is mutated in patients affected by hereditary spastic paraplegia. It is named after the physician Strümpell, who first described the disease more than 100 years ago.9 Three missense mutations have been identified in unrelated families.10 Strumpellin is one of the most conserved protein of the WASH complex, and these mutations are likely to affect the structure of Strumpellin. The mutant alleles of Strumpellin are dominant, suggesting that half a dose of WT Strumpellin is not sufficient to maintain a WT phenotype or that mutant Strumpellin blocks the assembly of functional WASH complexes despite the presence of WT Strumpellin in a dominant negative manner.
The KIAA1033 subunit is also well conserved, similarly to Strumpellin. The last 100 amino-acids, however, correspond to a poorly conserved region, which is predicted to be intrinsically unstructured (Suppl. Figs. 2 and 4).
The Ccdc53 subunit is the second less conserved subunit after VPEF. It cannot be detected in D. discoideum, even though a functional ortholog is likely to exist in this species, where 5 out of 7 subunits are detected (Fig. 1). Its name comes from Coiled-coil domain containing protein 53, because of a short and conserved coiledcoil predicted in the N-terminal domain. In this protein, two blocks of conserved residues alternate with two variable regions, predicted to be highly disordered (Suppl. Fig. 2).
Understanding how these 7 subunits are assembled into a functional actin polymerization machine is of major fundamental importance and might also provide an explanation for why hereditary spastic paraplegia arises in patients affected by Strumpellin mutations.
Acknowledgements
We thank Ferdinand Marlétaz for critically reading the manuscript and Agence Nationale pour la Recherche for funding (ANR-08-BLAN-0012-03 and ANR-08-PCVI-0010-03).
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/11185
Supplementary Material
References
- 1.Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Gianfrancesco F, Falco G, Esposito T, Rocchi M, D’Urso M. Characterization of the murine orthologue of a novel human subtelomeric multigene family. Cytogenet Cell Genet. 2001;94:98–100. doi: 10.1159/000048796. [DOI] [PubMed] [Google Scholar]
- 3.Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ. Human Subtelomeric WASH Genes Encode a New Subclass of the WASP Family. PLoS Genet. 2007;3:237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. Int Rev Cell Mol Biol. 2008;267:183–206. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derivery E, Gautreau A. Generation of branched actin networks: assembly and regulation of the N-WASP and WAVE molecular machines. BioEssays. 2010. In press. [DOI] [PubMed]
- 6.Huang CY, Lu TY, Bair CH, Chang YS, Jwo JK, Chang W. A novel cellular protein, VPEF, facilitates vaccinia virus penetration into HeLa cells through fluid phase endocytosis. J Virol. 2008;82:7988–7999. doi: 10.1128/JVI.00894-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanisch J, Ehinger J, Ladwein M, Rohde M, Derivery E, Bosse T, et al. Molecular dissection of Salmonella-induced membrane ruffling versus invasion. Cell Microbiol. 2009. Epub ahead of print. [DOI] [PubMed]
- 9.Strümpell A. Die primaere Seitenstrangsklerose (spastische Spinalparalyse) Deutsche Zeitschrift für Nervenheilkunde. 1904;27:291–339. [Google Scholar]
- 10.Valdmanis PN, Meijer IA, Reynolds A, Lei A, MacLeod P, Schlesinger D, et al. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am J Hum Genet. 2007;80:152–161. doi: 10.1086/510782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.