Abstract
In eukaryotes, changes in chromatin structure regulate the access of gene regulatory sequences to the transcriptional machinery and play important roles in the repression of transposable elements, thereby protecting genome integrity. Chromatin dynamics and gene expression states are highly correlated, with DNA methylation and histone post-translational modifications playing important roles in the establishment or maintenance of chromatin states in plants. Histones can be covalently modified in a variety of ways, thereby affecting nucleosome spacing and/or higher-order nucleosome interactions directly or via the recruitment of histone-binding proteins. An extremely important group of chromatin modifying enzymes are the histone lysine methyltransferases (HKMTs). These enzymes are involved in the establishment and/or maintenance of euchromatic or heterochromatic states of active or transcriptionally repressed sequences, respectively. The vast majority of HKMTs possess a SET domain named for the three Drosophila proteins that are the founding members of the family: Suppressor of variegation, Enhancer of zeste and Trithorax. It is the SET domain that is responsible for HKMT enzymatic activity. Mutation of Arabidopsis HKMT genes can result in phenotypic abnormalities due to the improper regulation of important developmental genes. Here, we review the different classes of HKMTs present in the model plant Arabidopsis thaliana and discuss what is known about their biochemical and biological functions.
I. INTRODUCTION
In eukaryotes, nuclear DNA is organized by histone proteins to form the fundamental unit of chromatin, the nucleosome. Each nucleosome is composed of 147 base pairs of DNA that is wrapped not quite twice around a histone octamer composed of two copies each of histone H2A, H2B, H3 and H4 (Luger et al., 1997). It is now clear that chromatin assembly exerts a major influence on gene expression by affecting the accessibility of the transcriptional machinery, including RNA polymerase complexes and transcription factors, to the DNA. As a consequence, changes in chromatin structure accompany a broad spectrum of important processes during development, including differentiation, embryonic stem cell maintenance and senescence (Baroux et al., 2007; He and Amasino, 2005; Hochedlinger and Plath, 2009; Kouzarides, 2007).
Nucleosome positioning is highly dependent on the genome sequence itself (Kaplan et al., 2009; Segal et al., 2006). The accessibility of DNA sequences within each nucleosome is further modulated by covalent modifications of the histones, methylation of cytosines in the DNA that is wrapped around the histones and differential use of histone variants. In vertebrates and plants, post-translational modification of histones and DNA methylation regulate or reflect the chromatin condensation and transcriptional status of the associated DNA. Genes located in a condensed chromatin context (heterochromatin) are generally inactive or silenced, whereas those found in a decondensed chromatin context (euchromatin) are more likely to be transcribed (Jenuwein and Allis, 2001; Kouzarides, 2007).
Heterochromatin is typically enriched in repetitive DNA, including transposable elements, centromeric repeats and excess, inactive ribosomal RNA (rRNA) gene repeats. Unlike constitutive heterochromatin, which remains condensed throughout the cell cycle, euchromatic regions undergo dynamic changes in chromatin condensation state and include intervals, such as intergenic sequences, that are often characterized by the presence heterochromatic marks (Bender, 2004a).
Changes in DNA methylation or histone modification states are mediated by specific enzymes. With regard to histone post-translational modifications, the enzymes modifying histones H3 and H4, particularly within their N-termini that protrude from the nucleosome core, are best understood (Kouzarides, 2007). The modifications carried out by these enzymes include methylation, acetylation, phosphorylation, ubiquitination, sumoylation, citrullination and ADP-ribosylation. The large variety of histone modifications, potentially conferring regulatory information, have been hypothesized to constitute a so-called ‘histone code’ (Jenuwein and Allis, 2001; Kouzarides, 2007).
Histone methylation occurs at both lysine and arginine amino acids and is used to mark both active and inactive chromatin, depending on context (Lachner and Jenuwein, 2002; Wang et al., 2007; Yu et al., 2006). For instance, Histone 3 Lysine 4 (H3K4) that is mono-, di- or trimethylated is present in nucleosomes associated with the promoter regions of active genes, whereas Histone 3 Lysine 9 (H3K9) mono-, di- and trimethylation occurs in nucleosomes associated with inactive genes located in euchromatic region and within highly condensed constitutive heterochromatin (Bernatavichute et al., 2008; Gendrel et al., 2002; Zhang et al., 2009).
Repressive histone modifications and DNA methylation are mechanistically linked (Richards, 2002). For example, mutations in the cytosine methyltransferase, MET1, lead to decreased H3K9 dimethylation, whereas mutations disrupting the functions of the H3K9 methyltransferase, Kryponite/SUVH4 (SU(VAR)3–9 homologues 4) results in decreased cytosine methylation (Jackson et al., 2002, 2004; Tariq et al., 2003). Genome-wide analyses have also revealed correlations between patterns of histone modification and cytosine methylation (Bernatavichute et al., 2008; Cokus et al., 2008; Gendrel et al., 2002; Lister et al., 2008; Zhang et al., 2009).
In Arabidopsis, a family of genes encode putative histone methyltransferases. Some of these enzymes function as arginine methyltransferases, but the majority are believed to be histone lysine methyltransferases (HKMTs) (Baumbusch et al., 2001; Ng et al., 2007; Niu et al., 2007; Wang et al., 2007). Five lysine methylation sites have been identified so far in plants, namely lysines 4, 9, 27 and 36 of Histone 3 and lysine 20 of Histone 4 (Pfluger and Wagner, 2007; Zhang et al., 2007b). In other eukaryotes, methylation of H3K79, H4K59 and H1BK26 has also been reported (Trojer et al., 2007; Zhang et al., 2003). All known HKMTs in plants have a so-called SET (Suppressor of variegation, Enhancer of zeste and Trithorax) domain that is responsible for the catalytic activity of the enzymes. Thus, these proteins are members of the SET Domain Group (SDG) protein super-family (Gendler et al., 2008). Our review focuses on those classes of SDG proteins that are known, or thought, to possess HKMT activity.
II. HKMT CLASSIFICATION IN ARABIDOPSIS
A. GENE ORGANIZATION AND EVOLUTION
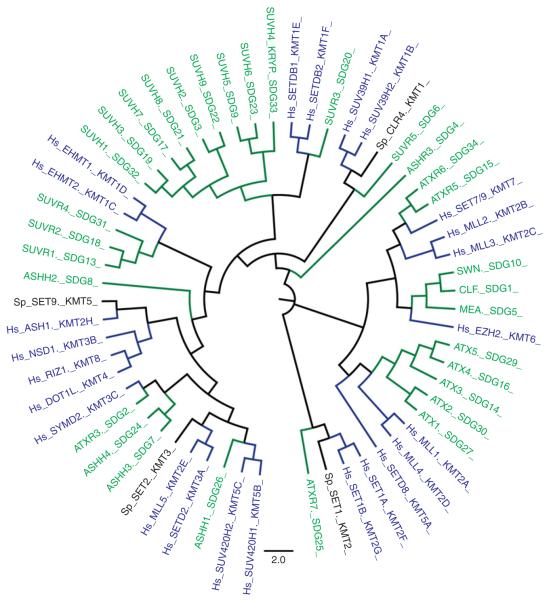
In Arabidopsis thaliana, 49 genes encoding putative SET domain-containing proteins have been identified (www.chromDB.org; Baumbusch et al., 2001; Ng et al., 2007). Similarly, the human genome encodes 50 SDG proteins, including 24 HKMTs. By contrast, the budding yeast (Saccharomyces cerevisiae) genome encodes only four SDG proteins (Fig. 1) (Allis et al., 2007). Of the 49 A. thaliana SDG proteins, 31 are known, or thought, to have HKMT activity and can be divided into five classes (I to V), based on their domain architectures (Fig. 2) and/or differences in enzymatic activity (Fig. 3) (Baumbusch et al., 2001; Ng et al., 2007; Springer et al., 2003). A class VI, which contain SDG proteins with a disrupted SET domain, and a class VII, which include SDG proteins that methylate non-histone proteins, have also been described but are not discussed in this review (Ng et al., 2007).
Fig. 1.
HKMTs organization and evolution. Relationships among HKMT Arabidopsis HKMT (grey), Human (Hs) HKMT and fission yeast (Sp) HKMT (dark) protein sequences determined by clustalW multiple alignment, followed by Neighbour Joining and Boostrap analysis.
Fig. 2.
Domain architecture of histone HKMTs in A. thaliana. Abbreviations: EZD, E(Z) domain; SANT, SWI3, ADA2, N-CoR and TFIIIB” DNA-binding domain; CXC, cysteine-rich region; PHD, plant homeodomain; zf-CW, a zinc finger with conserved Cys and Trp residues; PWWP, domain named after a conserved Pro–Trp–Trp–Pro motif; FYRN, F/Y-rich N-terminus; FYRC, F/Y-rich C-terminus.
Fig. 3.
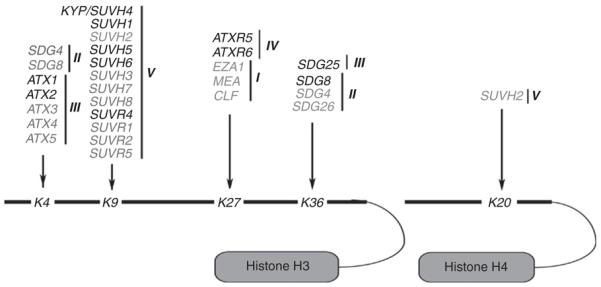
Diagram representing the H3 and H4 Lysines targeted by Arabidopsis HKMT proteins. The HKMTs whose activity has been biochemically demonstrated are highlighted using in black, whereas the HKMTs whose specificities have not been confirmed are shown in gray.
Phylogenetic analyses of A. thaliana and Zea mays genes have indicated that most of the gene duplication and functional diversification events that gave rise to the SDG protein family occurred prior to the divergence of monocotyledonous and dicotyledonous plants (Ng et al., 2007; Springer et al., 2003). However, there are exceptions, as exemplified by class III SDG genes that encode Trithorax-like (ATX) proteins. The Arabidopsis genome encodes five ATX genes, whereas rice and maize have only two or three, respectively (Ng et al., 2007; Springer et al., 2003). This observation suggests that duplication and diversification of some ATX genes occurred after the divergence of monocots and dicots, resulting in two sub-groups: sub-group 1 including ATX1 and ATX2 in Arabidopsis and two ATX2-like in maize and subgroup 2 including ATX3, ATX4 and ATX5 in Arabidopsis and a single copy of ATX4-like in maize (Fig. 1) (Avramova, 2009; Ng et al., 2007).
B. THE SET DOMAIN OF HKMT ENZYMES
Lysines can be mono-, di- or trimethylated, with differences in methylation state impacting or reflecting chromatin structure and gene transcriptional activity (Lachner and Jenuwein, 2002; Pfluger and Wagner, 2007). All known lysine methylation modifications, with the exception of Histone 3 Lysine 79 methylation (which has not been reported in plants), are carried out by methyltransferases that contain a SET domain. The SET domain encompasses approximately 130–150 amino acids that is thought to have evolved from an ancient motif found in bacterial proteins (Alvarez-Venegas et al., 2007). Structural and functional analyses of the SET domain of the human protein SUV39H1, which methylates H3K9, identified a series of key amino acids that are conserved in all SET-domain group proteins (Rea et al., 2000). Crystal structures of SET domains of several HKMTs have been solved, providing insights into their catalytic mechanisms and protein substrate specificities (Couture et al., 2005, 2006a, b; Xiao et al., 2003a, 2005). The SET domain possesses a unique fold dominated by 12 β-strands (Couture and Trievel, 2006). Two others domains, the pre-SET and the post-SET domains, sometimes flank the SET domain and may facilitate interactions with specific histone substrates. The hydroxyl group of a highly conserved tyrosine in the SET domain interacts with the substrate and transfers a methyl group to the lysine using S-adenosylmethionine (AdoMet) as the methyl group donor (Couture and Trievel, 2006; Rea et al., 2000; Xiao et al., 2003b).
III. CLASS I—IV HKMTS AND THEIR ROLES IN PLANT DEVELOPMENT
A. CLASS I HKMT ENZYMES
Class I HKMTs are homologues of Enhancer of Zeste E(Z) from Drosophila that have H3K27 methyltransferase activity (Jones and Gelbart, 1993; Muller et al., 2002). The Arabidopsis genome encodes three E(Z)-like proteins: CURLY LEAF (CLF), MEDEA (MEA) and SWINGER (SWN), each of which contain a SET domain, two E(Z) domains, a SANT (SWI3, ADA2, N-CoR and TFIIIB DNA-binding) domain and a CXC (cysteine-rich) region. E(Z)-like proteins are components of Arabidopsis Polycomb Repressive Complex 2 (PRC2)-like complexes that function astranscriptional repressors in diverse eukaryotes (Baroux et al., 2007). Analysis of clf, mea and swn mutants suggests that PRC2 complexes involving these proteins are required for H3K27 trimethylation, but direct biochemical evidence is currently lacking (Fig. 3) (Gehring et al., 2006; Makarevich et al., 2006).
CLF and MEA were the first HKMT genes described in plants, and helped underscore the importance of chromatin modification for proper plant development. CLF is required to repress FLOWERING LOCUS C (FLC) (Jiang et al., 2008; Wood et al., 2006). FLC, in turn, is a repressor of flowering, which is a process requiring a number of chromatin modifications, including histone methylation (He and Amasino, 2005). CLF is required for the methylation of H3K27 among histones associated with the FLC gene, as well as other developmentally important genes. Thus, clf mutations induce pleiotropic phenotypic defects in addition to altered flowering time, including altered leaf shape – hence the gene name (Katz et al., 2004; Makarevich et al., 2006; Schubert et al., 2006).
MEA is necessary for proper seed development. Maternally inherited loss of function mea alleles cause embryo abortion and endosperm over-proliferation (Grossniklaus et al., 1998; Kiyosue et al., 1999). Phylogenetic and molecular analyses have shown that MEA arose through duplication of an ancestral E(Z) homologue within the Brassicaceae family, indicating that MEA function came about relatively recently in angiosperm evolutionary history (Fig. 1) (Spillane et al., 2007).
SWN, the third E(Z)-like protein in Arabidopsis, also appears to participate in trimethylation of H3K27 at loci important for flower development, including AGAMOUS and SHOOTMERISTEMLESS (STM)(Katz et al., 2004; Schubert et al., 2006). MEA, CLF and SWN are probably responsible for the regulation of many genes and it was shown that more than 4000 genes carry H3K27 trimethylation marks (Makarevich et al., 2006; Zhang et al., 2007a). The specific role of each of them might potentially be identified through genome-wide comparisons of histone modifications in wild-type plants and mea, clf and swn mutants, and it should be considered that CLF and SWN regulate genes involved in many different processes of the plant life cycle.
B. CLASS II HKMT ENZYMES
Class II HKMTs are essentially implicated in the methylation of H3K36 (Fig. 3), a chromatin modification that is enriched within the region of actively transcribed genes (Lee and Shilatifard, 2007). Except for SDG4, class II HKMTs have a SET domain preceded by an AWS (Associated with SET) motif (Ng et al., 2007). The function of the AWS motif is unknown, but it is also found in mammalian class II HKMTs.
Functional insights have been obtained for two class II HKMTs: SDG4 and SDG8. Mutation of SDG8 affects FLC expression and induces an early flowering phenotype (Zhao et al., 2005). SDG8 is implicated in H3K36 di- and trimethylation, but sdg8 mutations do not affect H3K4, H3K9 or H3K27 methylation at the FLC locus (Zhao et al., 2005). Thus, changes in H3K36 methylation are sufficient to affect FLC expression. Several metabolic pathways are also perturbed in sdg8 mutants (Cazzonelli et al., 2009; Dong et al., 2008; Xu et al., 2008; Zhao et al., 2005). sdg8 mutants show altered expression of SPS/BUS (Supershoot/Bushy) and UGT74E2 genes, both of which affect shoot branching, a key process for plant biomass and seed production (Dong et al., 2008). Expression of CAROTENOID ISOMERASE, a gene required for carotenoid synthesis, is also perturbed in sdg8 mutants. Consequently, a lower accumulation of lutein is observed in sdg8 mutants, a carotenoid implicated in photosynthesis and photoprotection (Cazzonelli et al., 2009).
SDG4 was recently shown to be involved in pollen and stamen development (Cartagena et al., 2008; Thorstensen et al., 2008). Deficiency of SDG4 leads to reduced expression of multiple genes, probably due to defects in H3K4 dimethylation and H3K36 trimethylation. The sdg4 mutation also affects fertility (Cartagena et al., 2008).
C. CLASS III HKMT ENZYMES
Like other HKMTs, class III HKMTs have also been shown to be involved in flowering time regulation. Class III HKMTs consist of five Arabidopsis genes that encode homologues of Trithorax; they have therefore been named Arabidopsis Trithorax-like proteins 1-5 (ATX1-5) (Fig. 1) (Avramova, 2009). Class III proteins contain both SET and a post-SET domains, as well as PHD (plant homeodomain), PWWP (proline–tryptophane– tryptophane–proline), FYRN (F/Y-rich N-terminus) and FYRC (F/Y-rich C-terminus) domains (Fig. 2) (Alvarez-Venegas and Avramova, 2001). The PHD domain is thought to interact with trimethylated H3K4 (Peña et al., 2006). The PWWP domain is present in diverse proteins involved in chromatin function, including histone-modifying enzymes, DNA-modifying enzymes and transcription factors and have been found to interact with both histone and DNA (Laue et al., 2008; Qiu et al., 2002; Stec et al., 2000; Wang et al., 2009).
ATX1 and ATX2 form a protein sub-group: ATX1 mediates H3K4 trimethylation, whereas ATX2 mediates H3K4 dimethylation (Fig. 3) (Pien et al., 2008; Saleh et al., 2008). atx1 mutants display an early flowering phenotype and altered leaf morphogenesis (Alvarez-Venegas et al., 2003; Saleh et al., 2008). Intriguingly, the double atx1 atx2 mutant has an even more severe early flowering phenotype than atx1, suggesting that ATX1 and ATX2 activities overlap for proper expression of genes implicated in flowering time regulation (Pien et al., 2008; Saleh et al., 2008).
Despite the evidence for partial redundancy in controlling flowering time, ATX1 and ATX2 do not appear to regulate the same pool of genes (Saleh et al., 2008). Transcriptome analysis revealed that 7% of overall gene expression is affected in atx1. By contrast, only 0.7% of all genes display a different pattern of expression in atx2 mutants compared with controls (Alvarez-Venegas et al., 2006).
To date, no functions have been reported for ATX3, ATX4 or ATX5. These proteins have very conserved amino acid sequences, suggesting that they may have redundant functions. Double and triple mutant combinations of these three genes might potentially reveal their functional significance in A. thaliana.
ATXR3 and ATXR7 also belong to the class III AtKMTs; however, no data are available on the putative role and/or activity of ATXR3. However, ATXR7 (also known as SDG25) is able to specifically methylate Histone 3 in vitro and loss of function of SDG25 promotes flowering through reduction of FLC expression (Berr et al., 2009). Further analyses suggest that ATXR7 might be implicated in H3K36 dimethylation and its role might overlap with the class II HKMT, SDG26 (Berr et al., 2009).
D. CLASS IV HKMT ENZYMES
There are two class IV HKMTs in Arabidopsis, ATXR5 and ATXR6. Both proteins possess a PHD domain associated with their SET domain. Class IV proteins have an additional motif that allows them to interact with proliferating cell nuclear antigen (PCNA) (Raynaud et al., 2006). PCNA is a processivity factor for DNA polymerase delta during DNA replication, which suggests a role for class IV HKMTs in cell cycle regulation (Raynaud et al., 2006).
ATXR5 and ATXR6 were recently shown to carry out monomethylation of H3K27 (Fig. 3) (Jacob et al., 2009). ATXR5 and ATXR6 appear to act redundantly, because depletion of H3K27 monomethylation is only detectable in the atxr5 atxr6 double mutant (Jacob et al., 2009). Genome-wide analyses have revealed the presence of H3K27 monomethylation in heterochromatic chromocentres, whereas H3K27 di- and trimethylation are mainly present in euchromatic regions (Jacob et al., 2009; Zhang et al., 2007a). This suggests that distinct H3K27 methylation states correlate with different chromatin states. Interestingly, derepression of repetitive elements occurs in atxr5 atxr6 double mutants (Jacob et al., 2009) and is correlated with reduced H3K27 monomethylation but not reduced DNA methylation or H3K9 methylation, confirming a key role for H3K27 monomethylation in gene silencing and genome stability (Jacob et al., 2009; Mathieu et al., 2005).
IV. CLASS V HKMTS MARK INACTIVE CHROMATIN VIA HISTONE 3 LYSINE 9 METHYLATION
A. SUVH PROTEINS
1. Discovery of SUVH proteins
In 2002, two independent mutant screens revealed roles for SUVH4/KRYPTONITE in H3K9 methylation and gene silencing in Arabidopsis (Jackson et al., 2002; Malagnac et al., 2002). In these screens, suvh4 mutations were identified by their effects on the expression of the SUPERMAN locus, thereby affecting the number of floral organs (Jackson et al., 2002), or by the derepression of silenced PHOSPHOANTHRINILATE ISOMERASE (PAI) genes (Bender, 2004b; Luff et al., 1999; Malagnac et al., 2002). Importantly, cytosine methylation patterns were also perturbed at the loci where H3K9 dimethylation was lost, revealing a link between histone methylation and DNA methylation (Jackson et al., 2002). In other studies, depletion of cytosine methylation in the methyltransferase mutant met1 correlates with altered H3K9 methylation (Tariq et al., 2003), further indicating a functional relationship between DNA methylation and H3K9 modification.
2. Characteristics of SUVH proteins
SUVH proteins have a SET domain, a pre-SET domain and a post-SET domain. An additional motif, named the SET and RING finger-associated (SRA) domain, is also a characteristic of SUVH proteins (Fig. 2). There are 10 members of the SUVH protein family, which appears to be plant specific (Fig. 1).
The SRA domain serves as a methylcytosine-binding motif in both animals and plants (Citterio et al., 2004; Kraft et al., 2008; Unoki et al., 2004; Woo et al., 2007, 2008). In Arabidopsis, point mutations in the SRA domain of SUVH2 or SUVH4 leads to reduced H3K9 dimethylation, and deletion of the motif in SUHV4 or SUVH6 results in a failure of the protein to bind methylated DNA in vitro (Johnson et al., 2007). Interestingly, different SRA domains preferentially bind methylated cytosines, in particular, DNA sequence contexts. In vitro, the SUVH2 SRA domain has highest affinity for symmetric, CG methylation, whereas the SUVH9 SRA domain has highest affinity for asymmetric, CHH methylation (Johnson et al., 2008). HKMT binding to DNA sequences displaying different cytosine methylation contexts would provide an elegant mechanism for transducing epigenetic information encoded by DNA methylation patterns into altered histone methylation states.
3. Activity of SUVH proteins
The H3K9 methyltransferase activity of SUVH4 was first inferred from molecular genetic studies, and then confirmed by mass spectrometric analysis of in vitro methylated histones (Jackson et al., 2002, 2004; Johnson et al., 2004; Malagnac et al., 2002). In vitro activities for SUVH1, SUVH5 and SUVH6 indicate that these proteins are H3K9 methyltransferases, which raises the possibility that all SUVH proteins methylate H3K9 (Fig. 3) (Ebbs and Bender, 2006; Ebbs et al., 2005; Naumann et al., 2005).
SUVH2 and SUVH9 arose via a recent duplication in the Arabidopsis genome (Blanc et al., 2000, 2003). Initial evidence suggested that SUVH2 possesses H4K20 and H3K9 methyltransferase activity (Naumann et al., 2005). However, a more recent study found that suvh2 and suvh9 mutants, and suvh2 suvh9 double mutants do not display detectably altered histone methylation patterns (Johnson et al., 2008). Moreover, unlike SUVH4, SUVH5 or SUVH6, no HKMT activity was detected for SUVH2 or SUVH9 in vitro (Johnson et al., 2008). Furthermore, the methyl group donor AdoMet does not bind to recombinant SUVH2 or SUVH9, whereas AdoMet binding to recombinant SUVH4, SUVH5 and SUVH6 is observed (Johnson et al., 2008). One possibility is that the biological functions of SUVH2 and SUVH9 depend primarily on SRA domain interactions with methylated DNA, rather than on putative HKMT activity.
4. Functions of SUVH proteins
SUVH4 is involved mainly in maintenance of CHG methylation controlled by the cytosine methyltransferase CMT3 (chromomethylase 3), such that a loss of DNA methylation is observed in suvh4 mutants (Jackson et al., 2002; Malagnac et al., 2002). Jackson et al. (2002) showed that CMT3 does not interact with H3K9 dimethylation directly, and suggested that LHP1 was necessary to link SUVH4 and CMT3 activities (Jackson et al., 2002). However, subsequent work cast doubt on this hypothesis (Malagnac et al., 2002). It is now clear that SUVH4 is responsible for the majority of H3K9 dimethylation in heterochromatin (Jackson et al., 2004; Jasencakova et al., 2003; Johnson et al., 2002). Mutations in SUVH4 do not lead to a significant reactivation of repetitive elements, as is observed for cytosine methyltransferase mutants. Several HKMT proteins may act redundantly to silence these loci. Indeed, SUVH5 and SUVH6 have been shown to work together with SUVH4 to silence inverted repeats (Ebbs and Bender, 2006; Ebbs et al., 2005). Moreover, the triple mutant suvh2 suvh4 suvh9 shows altered expression of an F-box gene SUPPRESSOR of DRM1 DMR2 CMT3 (SDC), which induces a curly leaf phenotype, whereas no changes are observed in single mutants (Johnson et al., 2008). Over-expressing SUVH2 induces a general increase in heterochromatization mediated by an increase in repressive histone marks, including H3K9 dimethylation, H3K27 di- and trimethylation and H4K20 dimethylation (Naumann et al., 2005). This chromatin re-organization results in development changes, such as delayed leaf senescence (Ay et al., 2008).
The function of SUVH proteins in mediating H3K9 methylation is linked to cytosine methylation. A close correlation between these two heterochromatic marks is observed genome-wide (Bernatavichute et al., 2008; Gendrel et al., 2002). Furthermore, similar molecular defects are observed in mutants deficient in DNA methyltranferases or histone methyltransferases (Cao et al., 2003; Chan et al., 2006; Ebbs and Bender, 2006; Ebbs et al., 2005; Jackson et al., 2002; Johnson et al., 2007, 2008; Malagnac et al., 2002). For example, the triple mutant drm1 drm2 cmt3 shows depletion of DNA methylation at CHG and CHH sites, as well as some reduction in CG methylation. A decrease in H3K9 dimethylation is also observed in drm1 drm2 cmt3, similar to the effect of suvh4 mutations (Johnson et al., 2007). Similarly, suhv4 suvh2 suvh9 and drm1 drm2 cmt3 triple mutants display similar derepression of the SDC locus that correlates with a loss of DNA methylation and H3K9 methylation (Johnson et al., 2008).
Collectively, the available data reveal an important role for SUVH proteins in regulating the activity of loci present both in euchromatic and heterochromatic regions of the Arabidopsis genome. A recent study also analysed telomere length in the suvh4 mutant. Compared to wild-type plants, suvh4 plants have shorter telomeres, which suggest that heterochromatin maintenance affects telomere stability (Grafi et al., 2007).
B. SUVR PROTEINS
1. Characteristics of SUVRs
Few studies have investigated the properties and functions of SUVR (SU (VAR)3–9 related) proteins. Like SUVH proteins, SUVR proteins have a SET domain which is associated with a pre-SET domain and a post-SET domain (Baumbusch et al., 2001; Thorstensen et al., 2006). However, in contrast to SUVH proteins, SUVRs lack an SRA domain. A novel N-terminal plant-specific domain, named WIYLD based on conserved residues, has been identified in SUVR1, SUVR2 and SUVR4 proteins (Fig. 2) (Thorstensen et al., 2006). The WIYLD domain is a conserved region (residues 21–77 in SUVR4) that possesses structural similarity to the C-terminal domain of RuvA, a DNA-binding protein implicated in homologous recombination in bacteria (Rice et al., 1997; Thorstensen et al., 2006). Five genes encoding SUVR proteins (SUVR1 to SUVR5) are present in the A. thaliana genome (Fig. 2). SUVR4 possesses H3K9 mono- and dimethylation activities, which suggests that this class of HKMTs is responsible for repressive chromatin marks (Fig. 3) (Thorstensen et al., 2006).
2. Functions of SUVR proteins
The functions of SUVR proteins remain unclear. However, SUVR1, SUVR2 and SUVR4 proteins are most similar to HKMTs in humans that are implicated in heterochromatin formation. A recent report found that Arabidopsis SUVR5 (also known as AtCZS) interacts with the remodelling factor AtSWP1 and that both SUVR5 and AtSWP1 are required to down-regulate FLC expression. This repression correlates with H3K9 dimethylation and H3K27 dimethylation at the FLC promoter (Krichevsky et al., 2007). As yet, no functions have been described for the other four SUVR proteins.
SUVR proteins all display at least partial nucleolar localization, which has not been observed for other classes of HKMTs (Thorstensen et al., 2006). The nucleolus is best known as the site of ribosome biogenesis, but it is now clear that myriad aspects of RNA metabolism occur in the nucleolus (Boisvert et al., 2007), including processing of siRNAs involved in RNA-directed DNA methylation (RdDM) (Li et al., 2006; Pontes et al., 2006). This nucleolar processing centre generates heterochromatic, 24-nt small RNAs that target both RdDM and H3K9 methylation to specific genomic regions. The presence in the nucleolus of SUVR proteins that catalyse H3K9 methylation could reflect a role in rRNA gene modification or a potential link to the heterochromatic siRNA production machinery.
V. CONCLUSIONS AND PERSPECTIVES
The analysis of Arabidopsis HKMTs remains challenging because of the large number of genes in this family. Of the 31 predicted histone HKMTs, half have assigned functions or lysine specificities (Fig. 3). Because different HKMTs can help activate or silence gene expression, they can have antagonistic roles in modulating gene activity. For instance, CLF is implicated in H3K27 methylation, a repressive chromatin mark, whereas ATX1 methylates H3K4, a mark of active chromatin. CLF and ATX1 can both modify the same chromatin region, with opposing effects on gene activity. Single clf and atx1 mutants show abnormal leaf development, which is rescued in the double mutant clf atx1, indicating that this antagonism is biologically significant (Saleh et al., 2007).
Much effort is now focused on discovering specific functions for HKMTs. There are already several examples of HKMTs that seem to act preferentially at a specific tissue and time in development. An example is MEA, which is only expressed in the endosperm and the embryo (Grossniklaus et al., 1998; Kinoshita et al., 1999). Creating double and triple mutants for members of each Arabidopsis HKMT sub-group could reveal functions, and functional redundancies, among these proteins. However, this is a time-consuming approach that requires loss-of-function mutations for all HKMT genes, which are not yet available for all of the genes. Artificial microRNAs that target one or more genes simultaneously might be a useful strategy for analysing the functions of potentially redundant HKMTs (Schwab et al., 2006). An important consideration for the interpretation of in vivo data concerning HKMT functions is that the enzymes may modify non-histone targets that are responsible for the observed phenotypes. For example, the plant protein Rubisco is targeted by a methyltransferase that possesses a SET domain (Trievel et al., 2002; Ying et al., 1999). Moreover, non-histone targets of HKMTs have been described in mammals, including transcription factors (Chuikov et al., 2004; Kouskouti et al., 2004).
Genome-wide DNA methylation and histone modification data have revealed heterogeneity in H3K9 dimethylation distribution across the genome. H3K9 dimethylation tends to occupy larger regions in pericentromeric regions than it does in the chromosome arms, suggesting that distinct H3K9 methyltransferases, possibly the SUVR and SUVH proteins, could regulate the level of histone methylation in these different regions (Bernatavichute et al., 2008). Moreover, a genome-wide analysis of H3K27 trimethylation patterns also revealed that perhaps 4400 A. thaliana genes are impacted by this specific type of histone modification (Zhang et al., 2007b). This observation is consistent with the hypothesis that H3K27 methylation is a major silencing mechanism in plants. It is known that H3K27 and H3K9 methylation function independently as repressive chromatin marks (Mathieu et al., 2005). Recent data obtained for class IV HKMTs ATXR5 and ATXR6 confirm this hypothesis (Jacob et al., 2009).
H3K9 methylation and DNA methylation are both critical for epigenetic regulation of gene expression in plants, but the mechanisms linking these interdependent processes remain to be fully determined. However, the SRA domain of SUVH proteins helps explain how H3K9 methyltransferase activity can be recruited to regions characterized by methylated DNA, as does the presence of a chromodomain in the DNA methyltransferase (cf. SUVHs proteins). CMT3 can potentially explain the recruitment of DNA methyltransferase activity to nucleosomal regions enriched for histones methylated on H3K9 or H3K27.
Intriguingly, neither SUVH2 nor SUVH9 displays HKMT activity but both can interact with methylated DNA. Alignment of SUVH proteins reveals amino acids substitutions in the SUVH2 and SUVH9 SET domains that might explain their lack of activity. However, they retain conserved pre-SET, SET and post-SET domains. It would be interesting to test whether their SET domains can interact with methylated H3K9. If this were the case, it might be possible that SUVH2 and SUVH9 recognize methylated DNA, and, at the same time, protect methylated H3K9 from histone lysine demethylase activities.
Acknowledgements
The authors would like to thank Yannick Jacob for critical reading of the manuscript. Research in the Pikaard lab is supported by United States NIH grants GM60380 and GM077590. TB is supported by a Ruth L. Kirschstein National Research Service Award from the NIH. The content of this paper is solely the responsibility of the authors and does not necessarily reflect the views of the NIH.
REFERENCES
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Avramova Z. Two Arabidopsis homologs of the animal trithorax genes: a new structural domain is a signature feature of the trithorax gene family. Gene. 2001;271(2):215–221. doi: 10.1016/s0378-1119(01)00524-8. [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 2003;13(8):627–637. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Sadder M, Hlavacka A, Baluska F, Xia Y, Lu G, et al. The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc. Natl. Acad. Sci. U.S.A. 2006;103(15):6049–6054. doi: 10.1073/pnas.0600944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Sadder M, Tikhonov A, Avramova Z. Origin of the bacterial SET domain genes: vertical or horizontal? Mol. Biol. Evol. 2007;24(2):482–497. doi: 10.1093/molbev/msl184. [DOI] [PubMed] [Google Scholar]
- Avramova Z. Evolution and pleiotropy of TRITHORAX function in Arabidopsis. Int. J. Dev. Biol. 2009;53(2-3):371–381. doi: 10.1387/ijdb.082664za. [DOI] [PubMed] [Google Scholar]
- Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G, Humbeck K. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 2009;58(20):333–346. doi: 10.1111/j.1365-313X.2008.03782.x. [DOI] [PubMed] [Google Scholar]
- Baroux C, Pien S, Grossniklaus U. Chromatin modification and remodeling during early seed development. Curr. Opin. Genet. Dev. 2007;17(6):473–479. doi: 10.1016/j.gde.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, et al. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29(21):4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J. Chromatin-based silencing mechanisms. Curr. Opin. Plant Biol. 2004a;7(5):521–526. doi: 10.1016/j.pbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Bender J. DNA methylation of the endogenous PAI genes in Arabidopsis. Cold Spring Harb. Symp. Quant. Biol. 2004b;69:145–153. doi: 10.1101/sqb.2004.69.145. [DOI] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE. 2008;3(9):e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Xu L, Gao J, Cognat V, Steinmetz A, Dong A, et al. SET DOMAIN GROUP 25 encodes a histone methyltransferase and is involved in FLC activation and repression of flowering. Plant Physiol. 2009;151(3):1476–1485. doi: 10.1104/pp.109.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell. 2000;12(7):1093–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13(2):137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8(7):574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003;13(24):2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- Cartagena JA, Matsunaga S, Seki M, Kurihara D, Yokoyama M, Shinozaki K, et al. The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen. Dev. Biol. 2008;315(2):355–368. doi: 10.1016/j.ydbio.2007.12.016. [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, et al. Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell. 2009;21(1):39–53. doi: 10.1105/tpc.108.063131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet. 2006;2(6):e83. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432(7015):353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, et al. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 2004;24(6):2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 lys-20 methyltransferase. Genes Dev. 2005;19(12):1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Hauk G, Trievel RC. Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 2006a;13(2):140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- Couture JF, Hauk G, Thompson MJ, Blackburn GM, Trievel RC. Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases. J. Biol. Chem. 2006b;281(28):19280–19287. doi: 10.1074/jbc.M602257200. [DOI] [PubMed] [Google Scholar]
- Couture JF, Trievel RC. Histone-modifying enzymes: encrypting an enigmatic epigenetic code. Curr. Opin. Struct. Biol. 2006;16(6):753–760. doi: 10.1016/j.sbi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Dong G, Ma DP, Li J. The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem. Biophys. Res. Commun. 2008;373(4):659–664. doi: 10.1016/j.bbrc.2008.06.096. [DOI] [PubMed] [Google Scholar]
- Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell. Biol. 2005;25(23):10507–10515. doi: 10.1128/MCB.25.23.10507-10515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18(5):1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124(3):468–470. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler K, Paulsen T, Napoli C. ChromDB: the chromatin database. Nucleic Acids Res. 2008;36:D298–D302. doi: 10.1093/nar/gkm768. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297(5588):1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- Grafi G, Ben-Meir H, Avivi Y, Moshe M, Dahan Y, Zemach A. Histone methylation controls telomerase-independent telomere lengthening in cells undergoing dedifferentiation. Dev. Biol. 2007;306(2):838–846. doi: 10.1016/j.ydbio.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280(5362):446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- He Y, Amasino RM. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10(1):30–35. doi: 10.1016/j.tplants.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136(4):509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, et al. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112(6):308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416(6880):556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jacob Y, Feng S, Leblanc CA, Bernatavichute YV, Stroud H, Cokus S, et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 2009;16(7):763–768. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z, Soppe WJ, Meister A, Gernand D, Turner BM, Schubert I. Histone modifications in Arabidopsis – high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 2003;33(3):471–480. doi: 10.1046/j.1365-313x.2003.01638.x. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE. 2008;3(10):e3404. doi: 10.1371/journal.pone.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 2007;17(4):379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 2002;12(16):1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRAdomain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4(11):e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Mollah S, Garcia BA, Muratore TL, Shabanowitz J, Hunt DF, et al. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32(22):6511–6518. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol. Cell. Biol. 1993;13(10):6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458(7236):362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A, Oliva M, Mosquna A, Hakim O, Ohad N. FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 2004;37(5):707–719. doi: 10.1111/j.1365-313x.2003.01996.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL. Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell. 1999;11(10):1945–1952. doi: 10.1105/tpc.11.10.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, et al. Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 1999;96(7):4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell. 2004;14(2):175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kraft E, Bostick M, Jacobsen S, Callis J. ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases. Plant J. 2008;56(5):704–715. doi: 10.1111/j.1365-313X.2008.03631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A, Gutgarts H, Kozlovsky SV, Tzfira T, Sutton A, Sternglanz R, et al. C2H2 zinc finger-SET histone methyltransferase is a plantspecific chromatin modifier. Dev. Biol. 2007;303(1):259–269. doi: 10.1016/j.ydbio.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14(3):286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Laue K, Daujat S, Crump JG, Plaster N, Roehl HH, Consortium, T.S. et al. The multidomain protein brpf1 binds histones and is required for hox gene expression and segmental identity. Development. 2008;135(11):1935–1946. doi: 10.1242/dev.017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat. Res. 2007;618(1-2):130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson I, Bernatavichute Y, Chan SW, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126(1):93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luff B, Pawlowski L, Bender J. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell. 1999;3(4):505–511. doi: 10.1016/s1097-2765(00)80478-5. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, et al. Different polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006;7(9):947–952. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21(24):6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24(15):2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Naumann K, Fischer A, Hofmann I, Krauss V, Phalke S, Irmler K, et al. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 2005;24(7):1418–1429. doi: 10.1038/sj.emboj.7600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC. Plant SET domain-containing proteins: structure, function and regulation. Biochim. Biophys. Acta. 2007;1769(5-6):316–329. doi: 10.1016/j.bbaexp.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L, Lu F, Pei Y, Liu C, Cao X. Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep. 2007;8(12):1190–1195. doi: 10.1038/sj.embor.7401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442(7098):100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J, Wagner D. Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 2007;10(6):645–652. doi: 10.1016/j.pbi.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inze D, Avramova Z, et al. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell. 2008;20(3):580–588. doi: 10.1105/tpc.108.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, et al. The arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126(1):79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Qiu C, Sawada K, Zhang X, Cheng X. The PWWP domain of mammalian DNA methyltransferase dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 2002;9(3):217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C, Sozzani R, Glab N, Domenichini S, Perennes C, Cella R, et al. Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J. 2006;47(3):395–407. doi: 10.1111/j.1365-313X.2006.02799.x. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rice DW, Rafferty JB, Artymiuk PJ, Lloyd RG. Insights into the mechanisms of homologous recombination from the structure of RuvA. Curr. Opin. Struct. Biol. 1997;7(6):798–803. doi: 10.1016/s0959-440x(97)80149-2. [DOI] [PubMed] [Google Scholar]
- Richards EJ. Chromatin methylation: who’s on first? Curr. Biol. 2002;12(20) doi: 10.1016/s0960-9822(02)01208-3. R. [DOI] [PubMed] [Google Scholar]
- Saleh A, Al-Abdallat A, Ndamukong I, Alvarez-Venegas R, Avramova Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish “bivalent chromatin marks” at the silent AGAMOUS locus. Nucleic Acids Res. 2007;35(18):6290–6296. doi: 10.1093/nar/gkm464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Yilmaz M, Le O, Hou G, Sadder M, et al. The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell. 2008;20(3):568–579. doi: 10.1105/tpc.107.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, et al. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25(19):4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18(5):1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, et al. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane C, Schmid KJ, Laoueillé-Duprat S, Pien S, Escobar-Restrepo JM, Baroux C, et al. Positive darwinian selection at the imprinted MEDEA locus in plants. Nature. 2007;19(448 (7151)):349–352. doi: 10.1038/nature05984. [DOI] [PubMed] [Google Scholar]
- Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, et al. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132(2):907–925. doi: 10.1104/pp.102.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000;473(1):1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. U.S.A. 2003;100(15):8823–8827. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstensen T, Fischer A, Sandvik SV, Johnsen SS, Grini PE, Reuter G, et al. The Arabidopsis SUVR4 protein is a nucleolar histone methyltransferase with preference for monomethylated H3K9. Nucleic Acids Res. 2006;34(19):5461–5470. doi: 10.1093/nar/gkl687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstensen T, Grini PE, Mercy IS, Alm V, Erdal S, Aasland R, et al. The Arabidopsis SET-domain protein ASHR3 is involved in stamen development and interacts with the bHLH transcription factor ABORTED MICROSPORES (AMS) Plant Mol. Biol. 2008;66(1-2):47–59. doi: 10.1007/s11103-007-9251-y. [DOI] [PubMed] [Google Scholar]
- Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111(1):91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- Trojer PLG, Sims RJ, III, Vaquero A, Kalakonda N, Boccuni P, Lee D, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129(5):915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23(46):7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JRR, et al. Regulation of set9-mediated H4K20 methylation by a PWWP domain protein. Mol. Cell. 2009;33(4):428–437. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang Y, Ma Q, Zhang Z, Xue Y, Bao S, et al. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 2007;26(7):1934–1941. doi: 10.1038/sj.emboj.7601647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Dittmer TA, Richards EJ. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in arabidopsis. PLoS Genet. 2008;4(8):e1000156. doi: 10.1371/journal.pgen.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21(3):267–277. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood C, Robertson M, Tanner G, Peacock W, Dennis E, Helliwell C. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. U.S.A. 2006;130(39):14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, et al. Specificity and mechanism of the histone methyltransferase pr-set7. Genes Dev. 2005;19(12):1444–1454. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003a;421(6923):652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- Xiao B, Wilson JR, Gamblin SJ. SET domains and histone methylation. Curr. Opin. Struct. Biol. 2003b;13(6):699–705. doi: 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, et al. Diand tribut not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 2008;28(4):1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z, Mulligan RM, Janney N, Houtz RL. Rubisco small and large subunit N-methyltransferases. Bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J. Biol. Chem. 1999;274(51):36750–36756. doi: 10.1074/jbc.274.51.36750. [DOI] [PubMed] [Google Scholar]
- Yu MC, Lamming DW, Eskin JA, Sinclair DA, Silver PA. The role of protein arginine methylation in the formation of silent chromatin. Genes Dev. 2006;20(23):3249–3254. doi: 10.1101/gad.1495206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10(6):R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007a;5(5):e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112(2):77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- Zhang K, Sridhar VV, Zhu J, Kapoor A, Zhu JK. Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE. 2007b;2(11):e1210. doi: 10.1371/journal.pone.0001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 2005;7(12):1256–1260. doi: 10.1038/ncb1329. [DOI] [PubMed] [Google Scholar]