Abstract
Listeria monocytogenes is a food-borne pathogen with a clonal population structure and apparently limited gene flow between strains of different lineages. Strains of epidemic clone I (ECI) have been responsible for numerous outbreaks and invariably have DNA that is resistant to digestion by Sau3AI, suggesting methylation of cytosine at GATC sites. A putative restriction-modification (RM) gene cassette has been identified in the genome of the ECI strain F2365 and all other tested ECI strains but is absent from other strains of the same serotype (4b). Homologous RM cassettes have not been reported among L. monocytogenes isolates of other serotypes. Furthermore, conclusive evidence for the involvement of this RM cassette in the Sau3AI resistance phenotype of ECI strains has been lacking. In this study, we describe a highly conserved RM cassette in certain strains of serotypes 1/2a and 4a that have Sau3AI-resistant DNA. In these strains the RM cassette was in the same genomic location as in the ECI reference strain F2365. The cassette included a gene encoding a putative recombinase, suggesting insertion via site-specific recombination. Deletion of the RM cassette in the ECI strain F2365 and the serotype 1/2a strain A7 rendered the DNA of both strains susceptible to Sau3AI digestion, providing conclusive evidence that the cassette includes a gene required for methylation of cytosine at GATC sites in both strains. The findings suggest that, in addition to its presence in ECI strains, this RM cassette and the accompanying genomic DNA methylation is also encountered among selected strains of other lineages.
Listeria monocytogenes is a Gram-positive, facultative intracellular food-borne pathogen capable of causing severe disease (listeriosis) in animals and humans. Listeriosis most often affects pregnant women and their fetuses, neonates, the elderly, and immunocompromised individuals. The disease is predominantly transmitted via the consumption of contaminated foods and has a ca. 20% fatality rate (12, 27). Application of numerous genotyping methods has consistently shown that the organism has a clonal population structure with three major phylogenetic lineages: lineage I consists of strains of serotypes 1/2b, 3b, and 4b, while those of serotypes 1/2a, 1/2c, 3a, and 3c are clustered in lineage II; strains of serotypes 4a and 4c, along with certain serotype 4b strains, constitute lineage III (37, 38).
Most epidemics of human listeriosis have involved a small number of closely related strains (epidemic clones), predominantly of serotype 4b (7, 35). The earliest identified clone, epidemic clone I (ECI), has been responsible for several major outbreaks in Europe and North America. In addition, strains of this clonal group are frequently encountered in sporadic illness (10, 28, 29). ECI strains have also been found to comprise a significant portion of the serotype 4b strains from foods and from the environments of food processing plants (10, 11, 40).
Genomic DNA of ECI strains has been long known to resist digestion with Sau3AI, suggesting methylation of cytosine at GATC sites (41). Genome sequencing of the ECI strain F2365, implicated in the 1985 California outbreak of listeriosis, revealed a putative restriction-modification (RM) gene cassette with specificity for GATC sites (25). This RM cassette was harbored by all tested serotype 4b strains with Sau3AI-resistant DNA and was absent from those with DNA that could be digested with Sau3AI (40). These findings were in agreement with previous evidence that a fragment of the putative methyltransferase gene was specific to ECI and absent from other strains (14).
In spite of extensive documentation for the presence of this putative RM cassette in ECI strains, and its apparent absence among other serotype 4b strains, limited information is available about the possible presence of the cassette among other lineages of L. monocytogenes. Furthermore, conclusive evidence for involvement of the cassette in the resistance of the DNA of ECI strains to Sau3AI digestion has been lacking. In this study, we investigated a panel of food-derived serotype 1/2a strains with Sau3AI-resistant DNA and characterized the genetic content and genomic localization of the RM cassette harbored by these strains. Furthermore, we employed deletion mutagenesis to assess the involvement of the RM cassette in Sau3AI resistance of the DNA of the ECI strain F2365, as well as of a serotype 1/2a strain harboring the cassette.
MATERIALS AND METHODS
Bacterial strains and growth media.
L. monocytogenes isolates used in this work are listed in Table 1 and were from the Listeria strain collection at our laboratory at North Carolina State University. We additionally screened 163 serotype 1/2a isolates for resistance to Sau3AI; these included 143 isolates from turkey processing plants (23), three quality control strains (kindly made available by L. Wolf), and 17 from human listeriosis cases in North Carolina between 2001 and 2006, kindly made available by L. Wolf. L. monocytogenes F2365 was from the 1985 California outbreak, and its genome has been completely sequenced (25). All food isolates were originally obtained from the strain collections of the Food and Drug Administration. Bacteria were routinely grown overnight at 37°C in tryptic soy broth containing 0.7% yeast extract (TSB-YE; Becton Dickinson and Co., Sparks, MD) or on tryptic soy agar with 5% sheep blood (Remel, Lenexa, KS), and long-term storage was at −80°C in brain heart infusion medium (BHI; Becton Dickinson and Co.) with 20% glycerol (Fisher Scientific, Fairlawn, NJ). Antibiotics, used as needed, were nalidixic acid (30 μg/ml) and chloramphenicol (5 μg/ml) and were purchased from Sigma (St. Louis, MO). Escherichia coli strains S-17 and SM10 were grown at 37°C on Luria-Bertani medium (Difco) supplemented with ampicillin (100 μg/ml; Fisher Scientific) as needed.
TABLE 1.
L. monocytogenes strains used in this study
| Strain ID | Yr isolated | Origin (reference) | Serotype | Sau3AI digestion | MboI digestion | RM probe reactivity |
|---|---|---|---|---|---|---|
| F2365 | 1985 | California outbreak (25) | 4b | − | + | + |
| DS86 | This study; RM deletion mutant of strain F2365 | 4b | + | + | − | |
| A7 | 1999 | Guacamole | 1/2a | − | + | + |
| DS14 | This study; RM deletion mutant of strain A7 | 1/2a | + | + | − | |
| A8 | 1999 | Avocado pulp, Mexico | 1/2a | − | + | + |
| A11 | 2001 | Avocado pulp, Mexico | 1/2a | − | + | + |
| A29 | 2002 | Avocado pulp, Mexico | 1/2a | − | + | + |
| A33 | 2003 | Avocado pulp, Mexico | 1/2a | − | + | + |
| A36 | 2003 | Guacamole | 1/2a | − | + | + |
| SE076 | 1986 | Crab meat, frozen | 1/2a | − | + | + |
| A95 | 2004 | Avocado pulp | 1/2a | − | + | + |
| FSL N3-165 | 2002 | Soil, farm, New York (26a) | 1/2a | − | + | NTb |
| FSL J1-208 | 1998 | Goat, encephalitis (38a) | 4a | − | + | NTb |
| A35a | 2003 | Guacamole | 1/2a | − | + | − |
| 1a (255)a | Unknown | Lab QCc strain, North Carolina Dept. of Health | 1/2a | − | + | − |
| 10a | 2005 | Environment, turkey processing plant (23) | 1/2a | − | + | − |
| 2003-184Ra | 2003 | Human case, North Carolina | 1/2a | − | + | − |
| A22 | 2001 | Sliced turkey | 1/2a | + | + | − |
| A34 | 2003 | Cuttlefish | 1/2a | + | + | − |
| A38 | 2003 | Frozen snow crab clusters | 1/2a | + | + | − |
| A39 | 2003 | Cooked baby clam meat | 1/2a | + | + | − |
| A42 | 2001 | Mozzarella curd | 1/2a | + | + | − |
| A67 | 2001 | Frozen capelin roe | 1/2a | + | + | − |
| A74 | 2003 | Avocado puree for ice cream | 1/2a | + | + | − |
| A75 | 2003 | Smoked salmon | 1/2a | + | + | − |
| A94 | 2004 | Cheese chile relleno | 1/2a | + | + | − |
| A96 | 2004 | Breaded catfish | 1/2a | + | + | − |
| A97 | 2004 | Avocado puree | 1/2a | + | + | − |
| AT-05 | 1987 | Raw milk | 1/2a | + | + | − |
| AT-01 | 1986 | Brie cheese | 1/2a | + | + | − |
These isolates were resistant to Sau3AI, but their RM cassettes were not homologous to those of strain F2365; the isolates are designated “x-RM” in Fig. 4.
NT, not tested. Genome sequence analysis revealed the presence of the RM genes, as described in Results.
QC, quality control.
Bacteriological characterizations.
Cell morphology was determined by phase-contrast microscopy. Motility was assessed by phase microscopy and on soft (0.4%) agar plates following incubation over 36 h at 25 and 37°C. Growth rates at 4, 25, and 37°C were determined by monitoring the optical density at 600 nm (OD600), using a spectrophotometer (SmartSpec 3000; Bio-Rad, Hercules, CA) as well as by plate counts. Hemolytic activity was determined on sheep blood agar plates (Remel, Lenexa, KS) following incubation at 37°C over 36 h.
PCR and Southern blot assays.
Primers used in this study are listed in Table 2 and were designed using the Web-based program primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Primers were purchased from Qiagen (Valencia, CA). PCR employed ExTaq DNA polymerase (Fisher Scientific) and was performed as described previously (40). Probe 85M for the putative methyltransferase gene of the RM cassette of F2365 was constructed as described previously (40). Labeling of the PCR products with digoxigenin and Southern blot assays were performed as described previously (40).
TABLE 2.
Primers employed in this study
| Primer name | Sequence (5′-3′)a |
|---|---|
| LMO0321RMdelF | GCATTCTAGACATCCGAATGTCTTCCTC |
| LMO0324RMdelR | GCTCGGATCCCACTTCCAAAGTGGCAAATAA |
| LMO0328RMdelF | CACTGGATCCAACATGGTATAGAGATTG |
| LMO0329RMdelR | ATACGAATTCTTTTTCATCTGCTTTGACATC |
| LMO0325R | GCTGAAGTTACTTCTGAAG |
| 0325F | CTGAGATTTTGAAGCACCGG |
| 0327R | CGTCGTCCATATTCTGCCGCAT |
Underlined portions of sequences indicate restriction sites used for ligations: TCTAGA, XbaI; GGATCC, BamHI; and GAATTC, EcoRI.
Construction of RM cassette deletion mutants.
A DNA fragment harboring the desired deletion of the RM cassette was cloned in the temperature-sensitive plasmid pCON-1. This plasmid harbors genes conferring resistance to ampicillin and chloramphenicol, expressed in E. coli and L. monocytogenes, respectively (2). To construct the deletion, two ∼700-bp fragments flanking the gene cassette were amplified by PCR from the serotype 4b ECI strain L. monocytogenes F2365 and from the serotype 1/2a strain A7 with the primer pairs LMO0321RMdelF/LMO0324RMdelR and LMO0328RMdelF/LMO0329RMdelR (Table 2). The two PCR products, delRM(F2365) and delRM(A7), were double-digested with XbaI/BamHI and with BamHI/EcoRI, respectively, and the fragments were coligated in pCON-1 digested with XbaI and EcoRI. The recombinant plasmids were then electroporated into E. coli S17-1 or E. coli SM10, yielding ampicillin-resistant transformants designated E. coli DS4 [E. coli S17-1 with pCON-1::delRM(A7)] and E. coli DS58 [E. coli SM10 with pCON-1delRM(F2365)], harboring the deletion construct from strains A7 and F2365, respectively. These transformants were used as plasmid donors in conjugations with L. monocytogenes F2365 and A7, performed as described previously (19). Transconjugants were selected on BHI agar supplemented with nalidixic acid (30 μg/ml), to which L. monocytogenes is intrinsically resistant, and chloramphenicol (5 μg/ml) and grown at 42°C for 24 h in BHI with chloramphenicol. Putative integrants were serially transferred over three consecutive passages at 30°C followed by a last passage at 42°C (to ensure curing of any reconstituted free plasmid). Chloramphenicol-sensitive derivatives were identified and screened for the presence of the chromosomal deletions by PCR. Both deletions were confirmed by Southern blotting and sequencing.
DNA sequencing and analysis.
DNA sequencing was performed at the University of North Carolina Chapel Hill Genome Analysis Facility. Pair-wise alignments were done using BLAST2 (http://www.ncbi.nlm.nih.gov/), Align (http://www.ebi.ac.uk/), and EBI EMBOSS (http://www.ebi.ac.uk/Tools/emboss/align/index.html). Multiple sequence alignments were performed with CLUSTALW. Related sequences were detected in the NCBI databases with BLAST, BLASTX, BLASTP, and PSI-BLAST search tools. Codon frequency determinations used Web-based software (http://www.kazusa.or.jp/codon/) as described previously (24). The Artemis comparison tool, ACT (http://www.sanger.ac.uk/Software/ACT/), was used to compare equivalent genomic regions of different strains.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed as described previously (11) using ApaI (Roche Diagnostics, Indianapolis, IN) and AscI (New England Biolabs, Ipswich, MA). PFGE patterns were analyzed using BioNumerics software (Applied Maths, St. Martens-Latem, Belgium). Grouping was performed with the band position tolerance of the Dice coefficient at 1.5%, and the unweighted pair group method using arithmetic averages (UPGMA) was employed for clustering.
Nucleotide sequence accession number.
The nucleotide sequence of the RM cassette from L. monocytogenes A7 has been submitted to GenBank (accession no. DQ003209).
RESULTS
A restriction-modification cassette occupies genomically equivalent locations in ECI and serotype 1/2a strains with Sau3AI-resistant DNA.
Investigation of prevalence of Sau3AI resistance among 22 food-derived isolates of L. monocytogenes serotype 1/2a revealed that 9 (8 from processed avocado and 1 from seafood) had DNA that was resistant to digestion by Sau3AI but could be readily digested by MboI (Table 1). Since MboI also recognizes GATC sites but is not affected by cytosine methylation (it is instead inhibited by methylation of adenines) (21), the findings suggested methylation of cytosines at GATC sites in the genomes of these nine Sau3AI-resistant isolates. We identified three additional serotype 1/2a isolates with Sau3AI-resistant (but MboI-susceptible) DNA during routine screening of serotype 1/2a strains; these were isolates 10, 1a (255), and 2003-184R (Table 1). PCR of the 12 Sau3AI-resistant isolates with primer pairs flanking the RM cassette in strain F2365 and internal to it indicated that eight isolates (A7, A8, A11, A29, A33, A36, A95, and SE076), all food derived, harbored the cassette in the same location as F2365 (Fig. 1 and data not shown). No evidence for a cassette in this location could be obtained for isolates 10, A35, 1a (255), and 2003-184R (Fig. 1 and data not shown). Southern blot assays with a probe internal to the putative methyltransferase gene in the RM cassette of F2365 confirmed the presence of homologous sequences in all eight isolates which, on the basis of PCR, harbored the cassette in this location. In contrast, no hybridization signals could be obtained with 10, A35, 1a (255), and 2003-184R, even under low-stringency conditions (data not shown).
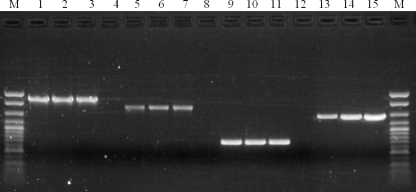
FIG. 1.
Serotype 1/2a isolates A7 and SE076 with Sau3AI-resistant DNA harbor an RM cassette homologous to that in the ECI strain F2365 and in a genomically equivalent location. PCR primers used were as follows: lanes 1 to 4, strains F2365, A7, SE076, and EGD-e, respectively, with primers 0325F and 0327R, amplifying the putative restriction endonuclease gene and putative methyltransferase gene of F2365; lanes 5 to 8, strains F2365, A7, SE076, and EGD-e, respectively, with primers LMO0321RMdelF and LMO0325R amplifying a fragment spanning a portion of ORFs LMOf2365_0321 and LMOf2365_0325 in F2365; lanes 9 to 12, strains F2365, A7, SE076, and EGD-e, respectively, with primers LMO0328RMdelF and LMO0329RMdelR, amplifying a fragment spanning a portion of ORFs LMOf2365_0329 and LMOf2365_0328 in F2365; lanes 13 to 15, strains EGD-e, A35, and 10, respectively, with primers LMO0321RMdelF and LMO0329RMdelR, flanking the RM cassette of F2365; lane M, DNA size markers (exACTGene cloning DNA ladder; Fisher Scientific). Strains A35 and 10 had Sau3AI-resistant DNA but lacked an RM cassette in this genomic location. PCR was done as described in Materials and Methods and using the primers listed in Table 2.
We determined the nucleotide sequence of a DNA fragment (ca. 5.1 kb) harboring the putative RM cassette of the serotype 1/2a strain L. monocytogenes A7. Sequence analysis confirmed that the RM cassette was in exactly the same location as in F2365 (between LMOf2365_0322 and the putative lipoprotein gene LMOf2365_0329), and the sequences within the cassette were found to be highly similar to those in F2365. With the exception of the pseudogene LMOf2365_0324 (93% identity between F2365 and A7), the other open reading frames (ORFs) had 97 to 98% identity between F2365 and A7 at the nucleotide sequence level. Furthermore, gene organization in this region was identical between strains A7 and F2365 (Fig. 2). The deduced amino acid sequence of the putative Sau3AI-like restriction endonuclease and its cognate methylase in L. monocytogenes A7 showed 97% and 98% identity, respectively, to their F2365 counterparts.
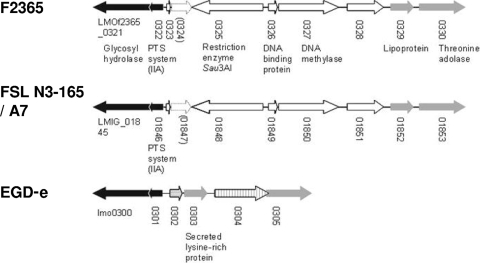
FIG. 2.
Genomic organization of the region harboring the RM cassette in L. monocytogenes strains F2365 (serotype 4b, ECI), FSL N3-165, A7 (serotype 1/2a), and EGD-e (serotype 1/2a). The arrows indicate the direction of transcription. The pseudogene (LMOf2365_0324 in F2365) is indicated by a stippled arrow. Conserved ORFs on the left and right of the RM cassette are shown as black and gray arrows, respectively. ORFs unique to EGD-e are indicated with horizontal and vertical striations.
Interestingly, analysis of recently sequenced Listeria genomes (http://www.broadinstitute.org/annotation/genome/listeria_group/GenomesIndex.html; accessed 04/28/2010) revealed that putative RM genes with similarly high (93 to 98%) homology to those of F2365 were also present in the genome of the serotype 1/2a strain FSL N3-165, derived from soil, and the serotype 4a strain FSL J1-208 (lineage III), from an animal listeriosis case. The genomic DNA of strain FSL N3-165 was indeed found to be resistant to digestion by Sau3AI but susceptible to MboI (Fig. 3), as was the genomic DNA of strain FSL J1-208 (data not shown). Accurate identification of the chromosomal location of the putative RM cassette in the serotype 4a strain FSL J1-208 was hindered by the fact that the genome sequence of this strain has not yet been annotated. However, analysis of the annotated genome sequence of strain FSL N3-165 (accession number, NZ_AARQ01000003) revealed that the RM cassette was in precisely the same chromosomal location in this strain as in F2365 and A7 (Fig. 2). Nucleotide sequences of the RM cassette were 99 to 100% identical between the two serotype 1/2a strains (A7 and FSL N3-165) and consistently higher than the 93 to 98% identity observed between them and their counterparts in F2365. The two genes flanking the RM cassette (corresponding to LMOf2365_0322 and LMOf2365_0329) were highly conserved (97 to 99%) between F2365, FSL N3-165, and all L. monocytogenes strains of serotype 1/2a for which genome sequences are available (13, 25; www.broadinstitute.org).
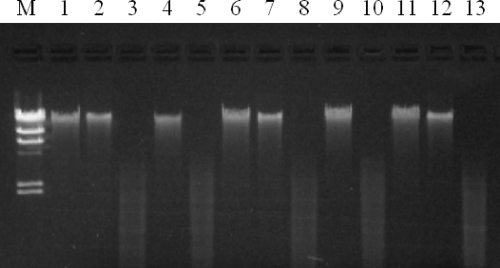
FIG. 3.
Evidence for cytosine methylation at GATC sites in wild-type strains and mutants with deletion of the RM cassette. Lanes: 1 to 3, DNA from strain F2365, uncut, digested with Sau3AI, and digested with MboI, respectively; 4 and 5, DNA from deletion mutant DS86 uncut and digested with Sau3AI, respectively; 6 to 8, DNA from strain A7, uncut, digested with Sau3AI, and digested with MboI, respectively; 9 and 10, DNA from deletion mutant DS14 uncut and digested with Sau3AI, respectively; 11 to 13, DNA from FSL N3-165, uncut, digested with Sau3AI, and digested with MboI, respectively; M, molecular weight marker II (Roche Diagnostics, Indianapolis, IN). DNA digestions were as described in Materials and Methods.
Comparative analysis of the gene content of the RM cassette in strains A7, FSL N3-165, and F2365 led to certain intriguing findings. (i) A small open reading frame (68 amino acids [aa], the putative product of LMOf2365_0326 in F2365) encoding a putative DNA binding protein was identified between the putative endonuclease and methylase genes in the genomes of all three strains, with the deduced polypeptides being identical at all 68 amino acid residues. The polypeptides harbored a helix-turn-helix motif and therefore were reminiscent of the so-called control (C) regulatory proteins of other RM gene complexes (22). The 68-aa polypeptide had 40 to 45% identity with transcriptional regulators of the Cro/CI family from several bacteria. (ii) All three strains harbored an ORF containing frameshifts near the left-most boundary of the cassette and convergent to the putative endonuclease (Fig. 2); in F2365, this ORF (LMOf2365_0324) was annotated as a pseudogene, i.e., containing a genuine frameshift (25). Interestingly, the sequences of this pseudogene were 100% identical between the two serotype 1/2a strains, A7 and FSL N3-165, but had only 93% identity (424/452 nucleotides) with the nucleotide sequence of the pseudogene in F2365. A 17-bp perfect inverted repeat (5′-AGCAAAATGCTTCATCT[N12]AGATGAAGCATTTTGCT-3′) surrounding the last stop codon of this pseudogene was detected in all three strains. (iii) A gene encoding a putative recombinase was identified downstream of the methyltransferase gene in all three strains. Taken together, these findings suggest that the RM cassette consists of a battery of four genes, including the predicted endonuclease and methyltransferase, and that certain serotype 1/2a strains with Sau3AI-resistant DNA, such as strains A7 and FSL N3-165, harbor all four genes.
The GC contents of the putative endonuclease, methyltransferase, DNA binding protein, and recombinase genes in the genomes of A7, FSL N3-165, and F2365 were noticeably lower (28 to 33%) than the average GC content for the genome of L. monocytogenes (38%), whereas the GC content of the pseudogene was 37% for strain F2365 and 39% for A7 and FSL N3-165, close to the genome average. A degree of codon bias was also noted in the putative RM cassette: most frequent codons in L. monocytogenes F2365 were, in decreasing order, GAA/AAA/AUU/GAU/CAA; in the RM gene cassette, the order was UUA/AUA/AUU/UUU/UCU. The GC content of the ORF upstream of the RM gene cassette was remarkably higher (43%) than the genome average; GC content of the two ORFs immediately downstream of the RM cassette (in F2365, LMOf2365_0329 and LMOf2365_0330) was 31% and 44%, respectively.
Deletion of the RM cassette renders genomic DNA of strains F2365 and A7 susceptible to digestion by Sau3AI.
In order to prove that methylation at the GATC sites was indeed associated with the RM cassette in F2365 and A7, RM cassette deletion mutants were constructed in both strains. The deletions were confirmed by PCR, Southern blotting, and nucleotide sequencing (data not shown). The deletion mutants were indistinguishable from their parental counterparts in cell and colony morphology, motility, hemolytic activity, and growth rate at 4, 25, and 37°C (data not shown). Unlike the parental strains, DNA of the deletion mutants of F2365 and A7 (designated DS86 and DS14, respectively) could be readily digested both by Sau3AI and by MboI (Fig. 3). Complementation experiments were pursued to confirm the involvement of the putative methyltransferase gene in cytosine methylation at GATC sites and resistance to digestion by Sau3AI. However, and in spite of numerous efforts, stable constructs with the methyltransferase gene could not be obtained in E. coli with two different shuttle vectors (pCON-1 and pLIV1) (9), suggesting toxicity of the cloned gene in E. coli (data not shown).
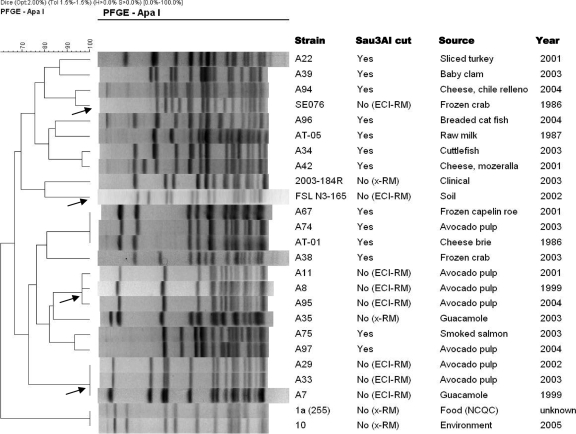
Serotype 1/2a strains harboring the RM gene cassette exhibit diverse PFGE profiles.
Four distinct ApaI PFGE profiles were identified among the serotype 1/2a isolates that had Sau3AI-resistant DNA and harbored the RM cassette: two of the profiles were exhibited by three isolates each (A7, A29, and A33, all from processed avocado but isolated in different years, and A8, A11, and A95, also from avocado and isolated in different years), whereas the other two profiles were exhibited by one strain each (SE076 and FSL N3-165, from seafood and soil, respectively) (Fig. 4). Similar clusters of PFGE profiles were identified following digestion with AscI (data not shown). We did not identify any isolates with indistinguishable PFGE profiles and differing in Sau3AI resistance and presence or absence of the RM cassette. The PFGE profiles of isolates 10, A35, 2003-184R, and 1a (255), which had Sau3AI-resistant DNA but lacked the RM cassette characterized here, were distinct from those of isolates found to harbor the RM cassette or from those of isolates with Sau3AI-susceptible DNA (Fig. 4).
FIG. 4.
PFGE fingerprints of serotype 1/2a strains with Sau3AI-resistant and Sau3AI-susceptible DNA. PFGE patterns generated following ApaI digestion were clustered using the BioNumerics program as described in Materials and Methods. RM indicates the presence of the RM cassette in the corresponding isolate with Sau3AI-resistant DNA; ECI-RM indicates isolates with Sau3AI-resistant DNA and harboring an RM cassette homologous to that in the ECI strain F2365 and in the same genomic location; x-RM indicates isolates with Sau3AI-resistant DNA but lacking an RM cassette with homology to that of F2365. Arrows indicate the four distinct PFGE profiles encountered among ECI-RM serotype 1/2a isolates.
DISCUSSION
In this study, we have shown that certain serotype 1/2a strains from foods (and one from soil) harbored a type II RM cassette highly similar to that previously thought to be unique to ECI strains. The RM cassette occupied a genomically equivalent location in these serotype 1/2a strains and in ECI. We further demonstrated that the Sau3AI resistance phenotype was abolished following the deletion of the cassette both in the serotype 1/2a strain A7 and the ECI strain F2365. This provided previously lacking evidence that the RM cassette mediates cytosine methylation at GATC sites, conferring resistance to digestion by Sau3AI.
In this study, the RM cassette was found among 8 of 22 serotype 1/2a isolates from food (7 from avocado and 1 from seafood) and in 1 soil isolate (FSL N3-165). A larger collection of food-derived serotype 1/2a strains of L. monocytogenes will need to be examined to accurately assess the prevalence of DNA methylation and of this specific RM cassette in strains of this serotype and from different foods. Nonetheless, the current findings suggest that ECI-like RM cassettes, and the accompanying methylation of cytosine at GATC sites of genomic DNA, are also encountered among selected strains of other lineages, such as the serotype 1/2a strains investigated here (lineage II) and the serotype 4a strain FSL J1-208 (lineage III).
The RM deletion mutants were indistinguishable from their parental counterparts in general bacteriological attributes. However, further studies (e.g., at the transcriptomic or proteomic level) will be needed to assess the potential impact of cytosine methylation at GATC sites on gene expression in ECI and in strains such as A7 and FSL N3-165. Investigations under an array of conditions are needed to assess the contribution of such methylation to competitive fitness of the organisms. At this time we also lack clear evidence for expression of the putative restriction endonuclease; preliminary experiments with phage P100 (5) propagated in strains F2365 and A7 versus the RM deletion mutants DS14 and DS86 failed to yield evidence for active endonucleases, possibly because this phage only harbors a single GATC site and may easily escape digestion through methylation by the methyltransferase of the RM cassette (phage A511 lacks GATC sites and could therefore not be used in these assays).
RM systems are well-known for their ability to be disseminated via horizontal gene transfer (15, 16, 18, 31, 39). Horizontal gene transfer has been postulated to mediate the observed lineage-specific distribution of RM systems in other bacterial pathogens (8, 32). In L. monocytogenes, the high conservation in nucleotide sequence and gene content of the putative RM cassettes in ECI and in strains of serotype 1/2a may reflect relatively recent dissemination of the RM cassette from ECI to selected strains of serotype 1/2a; the opposite direction of transfer seems less likely, since it would be then difficult to explain the fact that all tested ECI strains, without exception, harbored the RM cassette (14, 40, 41). It is also conceivable that the ancestor of ECI and selected strains of serotype 1/2a acquired the RM cassette through independent horizontal gene transfer events from a common donor. The conserved chromosomal location of the RM cassette in the different strains suggests site-specific recombination, possibly through the putative integrase of the RM cassette.
Although these possibilities cannot be excluded, we believe that the sequence data tend to favor an alternative hypothesis, according to which the RM cassette was harbored by an ancestral L. monocytogenes lineage but was subsequently only maintained by ECI (among serotype 4b strains) and by selected strains of other serotypes (e.g., 1/2a). Sequence identity of the cassette was higher between the two serotype 1/2a (lineage II) strains A7 and FSL N3-165 (99 to 100%) than between either of them and the ECI (lineage I) strain F2365 (93 to 98%). Such data agree with the well-documented genetic diversity between serotype 4b (lineage I) and serotype 1/2a (lineage II) strains (7, 38).
Regardless of the specific evolutionary scenario, the atypical GC content and codon usage of the genes in the cassette strongly suggest that this cassette was originally acquired by L. monocytogenes via horizontal gene transfer from another organism. The original source of this cassette in Listeria remains unknown. However, increasing evidence suggests the potential for horizontal gene transfer between L. monocytogenes and other Gram-positive bacteria, such as Staphylococcus aureus (3, 6). The highly conserved flanking sequences would provide an opportunity for recombination, facilitating incorporation of various “migratory genes,” as discussed by others (33).
The RM system that we characterized in this study has certain intriguing features. For instance, the putative DNA binding protein (encoded by LMOf2365_0326) may represent a novel member of the family of mobility-related genes in isoschizomeric RM systems, such as Sau3AI in S. aureus (30), LlacKR2I in Lactococcus lactis (36), Sth368I in Streptococcus thermophilus (4), and SsuDAT1I in Streptococcus suis (31, 32). Furthermore, the pseudogene in the RM cassette of F2365 (25) was also detected with the same frameshifts in the RM cassette of strains A7 and FSL N1-165. This suggests that this pseudogene is an authentic feature of the RM cassette and not just a peculiarity of F2365, which has been found to harbor significantly more frameshifts than other ECI strains (26). Pseudogenes usually arise from inactivation of paralogs or single-copy genes (20).
The deduced product of LMOf2365_0326 endonuclease and only 2 bp upstream from the start codon of the methyltransferase gene was reminiscent of a transcriptional regulator, “the control (C) protein,” due to the presence of a helix-turn-helix motif, which is a hallmark of activators and repressors (1). Several C proteins characterized before (e.g., PvuIIC and C AhdI) share a number of features, including sequence similarity, transcriptional orientation opposite to that of the endonuclease-encoding gene, and a conserved DNA sequence element immediately upstream of the gene, termed the C-box (16, 17, 22, 34). If indeed this gene in the L. monocytogenes RM cassette encodes a C protein, it would have novel features (i.e., lack of noticeable homology to other C proteins and lack of a clearly identifiable C-box) and would be worthy of further characterization.
In conclusion, we have provided evidence for highly conserved RM cassettes conferring cytosine methylation at GATC sites among strains derived from three L. monocytogenes lineages, i.e., ECI (serotype 4b, lineage I), serotype 1/2a (lineage II), and serotype 4a (lineage III). Thus, the RM cassettes are characteristic of specific clones and strains within diverse lineages of L. monocytogenes. Further studies are needed to elucidate the mechanisms underlying the observed distribution of these RM systems in L. monocytogenes and to assess their possible impact in the adaptation and evolution of L. monocytogenes.
Acknowledgments
Partial funding for this project was obtained from the USDA-National Alliance for Food Safety and Security through a project that involved collaboration between S. Kathariou and L.-A. Jaykus (North Carolina State University), K. Boor (Cornell University), and I. Wesley (USDA-ARS). The project was also partially funded by USDA grant 2006-35201-17377.
We thank L. Wolf and M. Wiedmann for some of the strains used in this study, Darren Higgins for the gift of pLIV1, and Steven Hagens for the gift of phage P100. We thank Sangmi Lee for assistance in preparation of Fig. 2 and all other members of our laboratory for discussions, encouragement, and support in the course of the project.
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Aravind, L., V. Anantharaman, S. Balaji, M. M. Babu, and L. M. Iyer. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231-262. [DOI] [PubMed] [Google Scholar]
- 2.Behari, J., and P. Youngman. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect. Immun. 66:3635-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brody, T., A. S. Yavatkar, Y. Lin, J. Ross, A. Kuzin, M. Kundu, Y. Fann, and W. F. Odenwald. 2008. Horizontal gene transfers link a human MRSA pathogen to contagious bovine mastitis bacteria. PLoS One 3:e3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrus, V., C. Bontemps, B. Decaris, and G. Guédon. 2001. Characterization of a novel type II restriction-modification system, Sth368I, encoded by the integrative element ICESt1 of Streptococcus thermophilus CNRZ368. Appl. Environ. Microbiol. 67:1522-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlton, R. M., W. H. Noordman, B. Biswas, E. D. de Meester, and M. J. Loessner. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 43:301-312. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., and R. P. Novick. 2009. Phage-mediated intergeneric transfer of toxin genes. Science 323:139-141. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, Y., R. M. Siletzky, and S. Kathariou. 2008. Genomic divisions/lineages, epidemic clones, and population structure, p. 337-358. In D. Liu (ed.), Handbook of Listeria monocytogenes. CRC Press, Inc., Boca Raton, FL.
- 8.Claus, H., A. Friedrich, M. Frosch, and U. Vogel. 2000. Differential distribution of novel restriction-modification systems in clonal lineages of Neisseria meningitidis. J. Bacteriol. 182:1296-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dancz, C. E., A. Haraga, D. A. Portnoy, and D. E. Higgins. 2002. Inducible control of virulence gene expression in Listeria monocytogenes: temporal requirement of listeriolysin O during intracellular infection. J. Bacteriol. 184:5935-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Bakker, H. C., E. D. Fortes, and M. Wiedmann. 2010. Multilocus sequence typing of outbreak-associated Listeria monocytogenes isolates to identify epidemic clones. Foodborne Pathog. Dis. 7:257-265. [DOI] [PubMed] [Google Scholar]
- 11.Eifert, J. D., P. A. Curtis, M. C. Bazaco, R. J. Meinersmann, M. E. Berrang, S. Kernodle, C. Stam, L. A. Jaykus, and S. Kathariou. 2005. Molecular characterization of Listeria monocytogenes of the serotype 4b complex (4b, 4d, 4e) from two turkey processing plants. Foodborne Pathog. Dis. 2:192-200. [DOI] [PubMed] [Google Scholar]
- 12.Farber, J. M., and P. I. Peterkin. 1990. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 14.Herd, M., and C. Kocks. 2001. Gene fragments distinguishing an epidemic-associated strain from a virulent prototype strain of Listeria monocytogenes belong to a distinct functional subset of genes and partially cross-hybridize with other Listeria species. Infect. Immun. 69:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeltsch, A., and A. Pingoud. 1996. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 42:91-96. [DOI] [PubMed] [Google Scholar]
- 16.Kita, K., J. Tsuda, T. Kato, K. Okamoto, H. Yanase, and M. Tanaka. 1999. Evidence of horizontal transfer of the EcoO1091 restriction-modification gene to Escherichia coli chromosomal DNA. J. Bacteriol. 181:6822-6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knowle, D., R. E. Lintner, Y. M. Touma, and R. M. Blumenthal. 2005. Nature of the promoter activated by C.PvuII, an unusual regulatory protein conserved among restriction-modification systems. J. Bacteriol. 187:488-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauer, P., M. Y. N. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, Y., P. M. Harrison, V. Kunin, and M. Gerstein. 2004. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 5:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClelland, M., M. Nelson, and E. Raschke. 1994. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 22:3640-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mruk, I., P. Rajesh, and R. M. Blumenthal. 2007. Regulatory circuit based on autogenous activation-repression: roles of C-boxes and spacer sequences in control of the PvuII restriction-modification system. Nucleic Acids Res. 35:6935-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mullapudi, S., R. M. Siletzky, and S. Kathariou. 2008. Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey processing plants. Appl. Environ. Microbiol. 74:1464-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nightingale, K. K., S. R. Milillo, R. A. Ivy, A. J. Ho, H. F. Oliver, and M. Wiedmann. 2007. Listeria monocytogenes F2365 carries several authentic mutations potentially leading to truncated gene products, including inlB, and demonstrates atypical phenotypic characteristics. J. Food Prot. 70:482-488. [DOI] [PubMed] [Google Scholar]
- 26a.Nightingale, K. K., Y. H. Schukken, C. R. Nightingale, E. D. Fortes, A. J. Ho, Z. Her, Y. T. Grohn, P. L. McDonough, and M. Wiedmann. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70:4458-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Painter, J., and L. Slutsker. 2007. Listeriosis in humans, p. 85-109. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis and food safety, 3rd ed. CRC Press, Boca Raton, FL.
- 28.Ragon, M., T. Wirth, F. Hollandt, R. Lavenir, M. Lecuit, A. Le Monnier, and S. Brisse. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauders, B. D., Y. Schukken, L. Kornstein, V. Reddy, T. Bannerman, E. Salehi, N. Dumas, B. J. Anderson, J. P. Massey, and M. Wiedmann. 2006. Molecular epidemiology and cluster analysis of human listeriosis cases in three U.S. states. J. Food Prot. 69:1680-1689. [DOI] [PubMed] [Google Scholar]
- 30.Seeber, S., C. Kessler, and F. Gotz. 1990. Cloning, expression and characterization of the Sau3AI restriction and modification genes in Staphylococcus carnosus TM300. Gene 94:37-43. [DOI] [PubMed] [Google Scholar]
- 31.Sekizaki, T., M. Osaki, D. Takamatsu, and Y. Shimoji. 2001. Distribution of the SsuDAT1I restriction-modification system among different serotypes of Streptococcus suis. J. Bacteriol. 183:5436-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekizaki, T., Y. Otani, M. Osaki, D. Takamatsu, and Y. Shimoji. 2001. Evidence for horizontal transfer of SsuDAT1I restriction-modification genes to the Streptococcus suis genome. J. Bacteriol. 183:500-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sibley, M. H., and E. A. Raleigh. 2004. Cassette-like variation of restriction enzyme genes in Escherichia coli C and relatives. Nucleic Acids Res. 32:522-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Streeter, S. D., I. Papapanagiotou, J. E. McGeehan, and G. G. Kneale. 2004. DNA footprinting and biophysical characterization of the controller protein C AhdI suggests the basis of a genetic switch. Nucleic Acids Res. 32:6445-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swaminathan, B., and P. Gerner-Schmidt. 2007. The epidemiology of human listeriosis. Microbes Infect. 9:1236-1243. [DOI] [PubMed] [Google Scholar]
- 36.Twomey, D. P., L. L. McKay, and D. J. O'Sullivan. 1998. Molecular characterization of the Lactococcus lactis LlaKR2I restriction-modification system and effect of an IS982 element positioned between the restriction and modification genes. J. Bacteriol. 180:5844-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Wiedmann, M., S. Mobini, J. R. Cole, Jr., C. K. Watson, G. Jeffers, and K. J. Boor. 1999. Molecular investigation of a listeriosis outbreak in goats caused by an unusual strain of Listeria monocytogenes. J. Am. Vet. Med. Assoc. 215:369-371. [PubMed] [Google Scholar]
- 39.Xu, Q., S. Stickel, R. J. Roberts, M. J. Blaser, and R. D. Morgan. 2000. Purification of the novel endonuclease, Hpy188I, and cloning of its restriction-modification genes reveal evidence of its horizontal transfer to the Helicobacter pylori genome. J. Biol. Chem. 275:17086-17093. [DOI] [PubMed] [Google Scholar]
- 40.Yildirim, S., W. Lin, A. D. Hitchins, L.-A. Jaykus, E. Altermann, T. R. Klaenhammer, and S. Kathariou. 2004. Epidemic clone I-specific genetic markers in strains of Listeria monocytogenes serotype 4b from foods. Appl. Environ. Microbiol. 70:4158-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng, W., and S. Kathariou. 1997. Host-mediated modification of Sau3AI restriction in Listeria monocytogenes: prevalence in epidemic-associated strains. Appl. Environ. Microbiol. 63:3085-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]