Abstract
“Candidatus Accumulibacter” and total bacterial community dynamics were studied in two lab-scale enhanced biological phosphorus removal (EBPR) reactors by using a community fingerprint technique, automated ribosomal intergenic spacer analysis (ARISA). We first evaluated the quantitative capability of ARISA compared to quantitative real-time PCR (qPCR). ARISA and qPCR provided comparable relative quantification of the two dominant “Ca. Accumulibacter” clades (IA and IIA) detected in our reactors. The quantification of total “Ca. Accumulibacter” 16S rRNA genes relative to that from the total bacterial community was highly correlated, with ARISA systematically underestimating “Ca. Accumulibacter” abundance, probably due to the different normalization techniques applied. During 6 months of normal (undisturbed) operation, the distribution of the two clades within the total “Ca. Accumulibacter” population was quite stable in one reactor while comparatively dynamic in the other reactor. However, the variance in the clade distribution did not appear to affect reactor performance. Instead, good EBPR activity was positively associated with the abundance of total “Ca. Accumulibacter.” Therefore, we concluded that the different clades in the system provided functional redundancy. We disturbed the reactor operation by adding nitrate together with acetate feeding in the anaerobic phase to reach initial reactor concentrations of 10 mg/liter NO3-N for 35 days. The reactor performance deteriorated with a concomitant decrease in the total “Ca. Accumulibacter” population, suggesting that a population shift was the cause of performance upset after a long exposure to nitrate in the anaerobic phase.
Enhanced biological phosphorus removal (EBPR) has been widely applied to reduce phosphorus (P) levels in wastewater treatment effluents, through the transformation of soluble inorganic phosphate (Pi) to intracellular polyphosphate [poly(P)] by poly(P)-accumulating organisms (PAOs) under alternating anaerobic/aerobic conditions. Anaerobically, PAOs take up organic substrates such as acetate, coupled to P release, as a result of intracellular poly(P) degradation. Lacking an external electron acceptor, acetate is converted to polyhydroxybutyrate (PHB), which is depolymerized and oxidized under subsequent aerobic conditions, leading to ATP generation and poly(P) regeneration (21).
A currently uncultured bacterial group in Betaproteobacteria, named “Candidatus Accumulibacter phosphatis” (13), was found to be the primary PAO in lab-scale and some full-scale EBPR systems (6, 37). Based on the phylogeny of polyphosphate kinase genes (ppk1), the “Ca. Accumulibacter” lineage is comprised of two major types, and each type contains a number of coherent clades (11, 23). Several studies suggested that these clades differ in their ability to reduce nitrate (5, 9) and the involvement of the tricarboxylic acid cycle in EBPR metabolism (33).
Although “Ca. Accumulibacter” clades other than IA and IIA have been found in several lab-scale sequencing batch reactors (SBRs) (33), in our previous study we only detected IA and IIA in two acetate-fed SBRs operated under similar conditions but at different geographical locations with different inoculation sludge sources (11). In addition, we found that the identity of the dominant clade switched between two sampling events (i.e., changed from IIA to IA). This raises intriguing questions, such as, how frequently the population shift occurs, how the clade dynamics influences the reactor performance, and how reactor operating conditions affect the clade composition.
To answer these questions, we studied “Ca. Accumulibacter” population composition and dynamics on a fine time scale. Since “Ca. Accumulibacter” clades may interact positively or negatively with each other, or with other bacterial groups, we searched for bacterial community composition patterns associated with shifts in the relative abundances of the two clades. For this purpose, we applied a community fingerprint method, automated ribosomal intergenic spacer analysis (ARISA) (8), which had been used to study bacterial community composition and dynamics in freshwater lakes (22, 27), activated sludge (34), and even environments with comparatively more complex microbial communities, such as soils (24). ARISA relies on the length heterogeneity of the internal transcribed spacer (ITS) region between 16S and 23S rRNA to distinguish different operational taxonomic units (OTUs). “Ca. Accumulibacter” clades IA and IIA detected in our reactors have distinct ITS lengths (12), thus allowing their unique detection by ARISA.
In this study, we first evaluated the quantitative capability of ARISA, compared to quantitative real-time PCR (qPCR) assays previously developed (11), and then used ARISA to monitor bacterial community composition dynamics. We analyzed samples collected weekly from two lab-scale SBRs during a 6-month period, when both reactors were operated under undisturbed and nearly identical conditions. We also evaluated samples obtained under disturbed conditions, when nitrate was introduced in the anaerobic phase for a period of 35 days, as well as those collected from time points when the reactors experienced “Ca. Accumulibacter” clade shift or poor performance, in order to explore potential relationships between “Ca. Accumulibacter” clade dynamics, total bacterial community composition patterns, operating conditions, and reactor performance. An understanding of such relationships should bring us closer to a mechanistic understanding of EBPR ecology and therefore more rational process design and operation.
MATERIALS AND METHODS
Reactor operation and chemical analysis.
Two acetate-fed sequencing batch reactors (R1 and R2) were operated in parallel under nearly identical conditions as described by He et al. (12), except for some periods with intentional manipulation of operational parameters in one reactor to study the effects of a specific parameter on the microbial community. The reactor sludge was originally inoculated from the Nine Springs wastewater treatment plant (Madison, WI), where “Ca. Accumulibacter” usually accounts for ∼20% of total cells (11, 37). The two reactors had a working volume of 2 liters and were operated under a 12-hour hydraulic retention time and a 4-day solid retention time (SRT). The influent contained (mg/liter of total feed indicated in parentheses): CH3COONa·3H2O (425), Casamino Acids (31), yeast extract (8.6), NaH2PO4·H2O (62.3), and other mineral salts to achieve the chemical oxygen demand:P (COD:P) mass ratio of 14 g/g. Allyl-thiourea was added to a 4-mg/liter concentration to inhibit nitrification, and pH was controlled at 7.0 to 7.5. These conditions were referred to as “normal operating conditions,” in comparison to conditions such as adding nitrate to the anaerobic phase. Soluble Pi was measured by using an ascorbic acid method (standard method 4500-P E) (3). Acetate, NO3-N, and NO2-N were measured by a Shimadzu high-performance liquid chromatograph (Kyoto, Japan) with an Alltech Previal organic acid column (Deerfield, IL) at the detector setting of 210 nm for acetate and 214 nm for NO3-N and NO2-N.
Nitrate addition in the anaerobic phase.
To investigate the effects of nitrate in the anaerobic phase on reactor performance and community composition, nitrate was added to R2, during steady-state R2 operation (indicated by stable reactor performance and relatively constant biomass concentrations for 12 days, i.e., three SRTs). Sodium nitrate was added together with the acetate feeding in the anaerobic phase to reach an initial reactor concentration of 10 mg/liter NO3-N for 35 consecutive days.
Sample collection and DNA extraction.
Samples for DNA extraction were collected by centrifuging 2 ml of mixed liquor at 8,000 × g for 3 min, and the resulting pellets were transferred to a −80°C freezer immediately for long-term storage. DNA was extracted using the PowerSoil DNA isolation kit (Mo Bio, Carlsbad, CA) following the manufacturer's instructions, except that cell disruption was performed by a 3-min bead beating at a speed setting of 3.5 on a minibeadbeater (Biospec Products, Bartlesville, OK). This extraction method provided qPCR results comparable to the previously used extraction method (11), with negligible PCR inhibition (data not shown). DNA concentration was estimated by using microspectrophotometry (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE). A total of 95 samples were analyzed, including 56 samples during a 6-month monitoring of population dynamics, 13 samples from the nitrate addition experiment, and 26 samples from other time points when the reactors experienced dominant “Ca. Accumulibacter” clade shifts or poor performance.
Quantitative real-time PCR.
To determine the relative abundance of “Ca. Accumulibacter” clades IA and IIA, qPCR with ppk1-targeted primers specific for “Ca. Accumulibacter” clades IA and IIA was performed, as described by He et al. (11). PCR with primers targeting ppk1 from clades IIB, IIC, and IID was also performed to test if other “Ca. Accumulibacter” clades ever became detectable in the SBRs. The 16S rRNA gene abundance levels from total “Ca. Accumulibacter” and total bacteria were measured by qPCR with the primer set 518f/PAO 841r (11) and bacteria-specific forward primer 341f and universal reverse primer 534r (36), respectively, with the reaction conditions described previously (11). Two 16S rRNA gene primer sets specific to clades IA and IIA were also designed to verify the quantification obtained by ppk1-targeted qPCR (Table 1). qPCR conditions for these two primer pairs were the same as described previously (11), except that primers and betaine were added to 400 nM and 0.5 M, respectively, and the annealing temperature (Ta) was 63°C.
TABLE 1.
16S rRNA primers for “Ca. Accumulibacter” clades IA and IIA
| Target | Primer | Primer sequence (5′-3′)a | Amplicon length (bp) | Ta (°C) |
|---|---|---|---|---|
| Clade IA | 16S-Acc-IA-f | TTGCTTGGGTTAATACCCTGAG | 211 | 63 |
| 16S-Acc-IA-r | CTGCCAAACTCCAGTCTTGC | |||
| Clade IIA | 16S-Acc-IIA-f | TTGCACGGGTTAATACCCTGTG | 213 | 63 |
| 16S-Acc-IIA-r | CTCTGCCAAACTCCAGCCTG |
Bases shown in bold indicate mismatches to the other clade.
ARISA.
PCR was conducted on genomic DNA to amplify the intergenic spacer region between the 16S and 23S rRNA genes. Each 25-μl reaction mixture consisted of 1× Idaho Tech buffer (Salt Lake City, UT), 200 nm deoxynucleoside triphosphates, 400 nM 6-carboxy fluorescein-labeled forward universal primer 1406f (5′-TGYACACACCGCCCGT-3′), 400 nM reverse bacteria-specific primer 23Sr (5′-GGGTTBCCCCATTCRG-3′), 1.25 U of GoTaq Flexi DNA polymerase (Promega, Madison, WI), and 5 to 10 ng of DNA template (27). The reaction was conducted in a Mastercycler gradient thermal cycler (Eppendorf, New York, NY), with an initial 2-min denaturation at 94°C, 25 cycles of 94°C for 35 s, 55°C for 45 s, and 72°C for 2 min, and a final extension of 2 min at 72°C.
Subsequently, 1 μl of distilled deionized H2O-diluted PCR product was added to a mixture containing 0.4 μl of X-rhodamine-labeled custom ROX (6-carboxyl-X-rhodamine) internal size standard 100-2000 (BioVentures, Murfreesboro, TN) and 9.6 μl of 10% formamide to run the denaturing capillary electrophoresis. The ARISA profiles were visualized, quality checked, and analyzed using GeneMarker software, version 1.51 (SoftGenetics, LLC, State College, PA). Each OTU was defined as an ARISA PCR amplicon by its size (ARISA size, in bp). The raw data were run through an R script (14, 25) to statistically set up an appropriate baseline for each profile by reiterated removal of the “true” peaks with heights greater than 4 standard deviations of all peaks until no peak was higher than 4 standard deviations of the remaining peaks (1). An individual peak height was normalized to the sum of heights from all detected peaks in that profile. The normalized peak height with the correction of rrn operon copy number per genome was used as a proxy of the relative abundance of each OTU in a sample (the correction method is described further in Results).
Statistical analysis.
Correspondence analysis was conducted as an exploratory tool to search for patterns in bacterial community composition, using Canoco for Windows, version 4.5.3. A species matrix containing the normalized and rrn operon copy number-corrected peak heights from each OTU in each sample and an environmental variable matrix containing appropriate variables from these samples were used in a correspondence analysis. The species scores and sample scores, which specified the locations of species and samples in an ordination plot, were calculated. A biplot was created, representing sample locations in the ordination, where the first and second axes explained the largest variances, indicated by the eigenvalues from the axes. The biplot arrows represent variables pointing in the direction of maximum correlation between that variable and species composition, and the length of the arrow is related to the strength of the correlation.
Analysis of similarities (ANOSIM) was conducted to test the statistical significance of the community composition difference between the classified sample groups, using the software PRIMER 6 (version 6.1.9). The R statistic and the significance level were calculated, with an R value of 0 indicating completely random grouping and an R value of 1 indicating complete separation among sample groups.
RESULTS
Linkage of ppk1 and 16S rRNA phylogenies.
Previously, “Ca. Accumulibacter” clade structure in the 16S rRNA phylogeny was inferred from the coherent clades defined by ppk1 (11). To verify the linkage between 16S rRNA and ppk1 phylogenies for clades IA and IIA, 16S rRNA primers specific to clades IA and IIA were designed, with a total 5-bp mismatch with each other (Table 1). When the clade IA positive control (purified clade IA PCR product) was used as the negative control for clade IIA (and vice versa), less than 0.001% of the negative control was amplified, confirming that the 16S rRNA primers were specific to their targets.
The relative abundance of clades IA and IIA within the “Ca. Accumulibacter” lineage (percent IA and IIA) was quantified by both 16S rRNA- and ppk1-targeted qPCR in three samples. The slope from the linear regression between the two quantifications of percent IA was 0.874 (R2 = 0.986) or 0.999 (R2 = 0.995), assuming the ratio of rrn operon copy number per genome from “Ca. Accumulibacter” clade IA and IIA is 1:1 or 1:2, respectively. Metagenomic analysis of clade IIA-dominated EBPR sludge determined that clade IIA had two copies of the rrn operon per genome and the ITS region was 100% identical between these two operons (10). Using dot blot hybridization on genomic DNA, it was estimated that the ratio of type I (later defined as clade IA, more specifically) ppk1 to the “Ca. Accumulibacter” 16S rRNA gene in type I-dominated sludge was approximately 1 (19). Therefore, it is likely that clade IA has one copy of the rrn operon per genome while IIA has two copies. By using this assumption, we achieved a linear regression slope closer to 1, which indicated comparable quantifications using ppk1- and 16S rRNA-targeted qPCRs. Therefore, clades IA and IIA defined by ppk1 are the same populations as defined by 16S rRNA, confirming the linkage between the ppk1 and 16S rRNA phylogenies. This linkage was also later confirmed from the completed genome of clade IIA (GenBank accession no. CP001715).
Comparison of ARISA and qPCR.
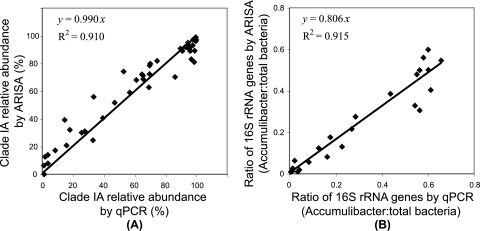
Quantification of the percent IA was also compared for the ppk1 qPCR and ARISA. The lengths of ARISA fragments for IA and IIA are 818 to 819 bp and 800 bp, respectively, as determined by conducting ARISA with the corresponding 16S plus ITS rRNA clones generated previously (12). When linear regression was performed between the ppk1 qPCR- and ARISA-derived quantifications, slopes of 0.990 (R2 = 0.910) (Fig. 1 A) and 0.867 (R2 = 0.937) were obtained, with the assumption of one or two copies of the rrn operon per clade IA genome, respectively. The close-to-1 slope under the former assumption agrees with the comparison between the ppk1- and 16S rRNA-targeted qPCRs mentioned above. Therefore, when interpreting ARISA-derived population abundances, “Ca. Accumulibacter” abundances were corrected for rrn operon copy number by using one copy per IA genome and two copies per IIA genome. In addition, an average of 4.2 copies of the rrn operon per genome was assumed for the flanking bacterial community during population abundance estimation of each OTU (15, 29). This assumption was based on the average of all bacterial rrn operons currently available in the rrnDB (http://ribosome.mmg.msu.edu/rrndb/index.php) and provides a best estimate, because the flanking community in our sludge is often comprised of Bacteroidales, Flavobacteriales, Burkholderia, Acidovorax, Cytophaga, and Dechloromonas (10) (unpublished data), which have averages of three to six copies of the rrn operon per genome, based on information in rrnDB.
FIG. 1.
Comparison of quantifications by qPCR and ARISA. (A) Relative abundance of clade IA within the “Ca. Accumulibacter” lineage as measured by ppk1-targeted qPCR and by ARISA, respectively, assuming that clade IA has one copy of the rrn operon per genome. (B) The ratio of “Ca. Accumulibacter” 16S rRNA genes to those from the total bacterial community as determined by qPCR and ARISA, respectively.
It is worth mentioning that three data points were removed from Fig. 1A. For these three samples, both clades IA and IIA ARISA peaks were very low (<0.1% of the sum of peak heights in a profile), and thus quantifications did not agree well between the two methods due to typical random baseline noise in ARISA profiles. Generally, qPCR has a very low detection limit, while random noise in ARISA makes the estimation of clade distribution less trustworthy when total “Ca. Accumulibacter” abundance is very low. Therefore, qPCR is superior to ARISA in such cases.
The ratio of “Ca. Accumulibacter” 16S rRNA genes to those from the total bacterial community was also quantified by qPCR and ARISA, respectively. Based on qPCR, this ratio was calculated by dividing the 16S rRNA gene copies determined with the primer set targeting total “Ca. Accumulibacter” (518f and PAO 841r) by that determined with a bacterial general primer set (341f and 534r). Based on ARISA, this ratio was calculated as the sum of normalized peak heights from IA and IIA. The quantifications by qPCR and ARISA were highly correlated (R2 = 0.915) (Fig. 1B). However, ARISA generated a systematically lower quantification than qPCR, as indicated by the slope (0.806), likely due to the difference in target spectra in sludge by the primer sets used in these two methods.
Population dynamics under normal operating conditions.
ARISA was conducted on samples from two lab-scale EBPR reactors (R1 and R2, respectively) collected at least once per week during a 6-month period, when both reactors were operated under normal and essentially identical conditions (28 samples from each reactor). On day 1, R2 was inoculated with biomass from R1. Therefore, during this study period, the two reactors started with similar bacterial community composition, as confirmed by their ARISA profiles.
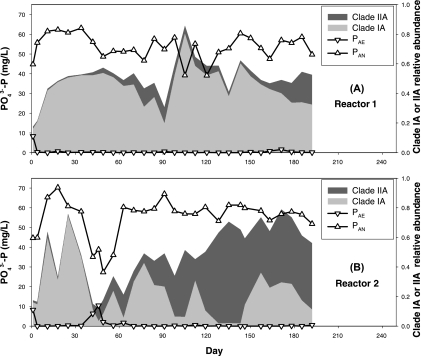
Figure 2 shows reactor performance as indicated by the soluble PO43−-P concentration at the end of the anaerobic (PAN) and aerobic (PAE) phases. At the beginning of the time series, both reactors were performing poorly. After a week of operation, good EBPR activity was recovered in R1 and established in R2. After that, R1 exhibited reliable and good P removal while R2 experienced an unexplained performance upset during days 42 to 63. The “Ca. Accumulibacter” population abundance and composition estimated by ARISA are also shown in Fig. 2. Both reactors were dominated by clade IA (IA, ≥60% of total “Ca. Accumulibacter”) during the first 34 days, and the “Ca. Accumulibacter” clade composition remained relatively stable in R1, with clade IA dominating during the entire 6 months. In R2, “Ca. Accumulibacter” clade composition was more dynamic, and sometimes the dominant “Ca. Accumulibacter” population shifted from one clade to the other within a week, without apparent explanation. There was no consistent association between the reactor performance and the clade distribution. During the performance upset on days 42 to 63 in R2, the total “Ca. Accumulibacter” abundance decreased sharply (Fig. 2), while the relative abundance of OTUs 942, 954, and 986 increased (see Fig. S1 in the supplemental material).
FIG. 2.
Reactor performance, total “Ca. Accumulibacter” abundance, and clade composition from R1 and R2 during the 6-month normal operating period. (PAN and PAE indicate the soluble PO43−-P concentration at the end of an anaerobic or aerobic phase, respectively.) Relative abundances were measured by semiquantitative ARISA as described in the text.
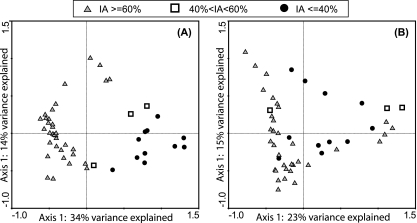
To further search for community composition patterns associated with variations in clade distribution, correspondence analysis was conducted without samples exhibiting poor performance (days 1 to 4 from both reactors and days 42 to 63 from R2) to exclude communities with very low total “Ca. Accumulibacter” abundance. Samples were plotted on an ordination with the first two axes explaining the largest variance and the distance between samples indicating the difference in bacterial community composition. In Fig. 3 A, all OTUs were included in the analysis. Bacterial communities were grouped into three categories based on “Ca. Accumulibacter” clade composition: dominated by IA (IA ≥ 60%), dominated by IIA (IA ≤ 40%), or coexisting with two relatively comparable clades (40% < IA <60%). The clustering pattern indicated that the three community types were significantly different from each other, as supported by ANOSIM (R = 0.802, P < 0.001). However, after bioinformatically removing clades IA and IIA from the community and renormalizing peak heights from individual OTUs to the sum of remaining OTUs, the clustering pattern was less strong (Fig. 3B) (ANOSIM, R = 0.151, P = 0.041), suggesting no signature of flanking community composition associated with either clade.
FIG. 3.
Correspondence analysis of ARISA profiles collected from R1 and R2 during the 6-month normal operating period, without including the samples collected during poor reactor performance. Gray triangles are samples with clade IA as ≥60% of total “Ca. Accumulibacter,” open squares are samples with clade IA as 40 to 60% of the total “Ca. Accumulibacter,” and black circles are samples with clade IA as ≤40% of total “Ca. Accumulibacter.” All 61 OTUs were included in analysis in panel A. Clades IA and IIA were not included in analysis in panel B; they were removed, and peak heights were renormalized from individual OTUs to the sum of remaining OTUs.
Nitrate addition in the anaerobic phase.
To investigate how the microbial community responded to abnormal operating conditions, nitrate was added to R2, together with acetate feeding to reach an initial reactor concentration of 10 mg/liter of NO3-N in the anaerobic phase for 35 days. During the first 2 days of nitrate addition, only 20 to 30% of the nitrate was consumed within the anaerobic phase and remained unchanged during the subsequent aerobic phase. However, by the eighth day, nitrate was completely consumed in the anaerobic phase (data not shown), suggesting an establishment of denitrification populations.
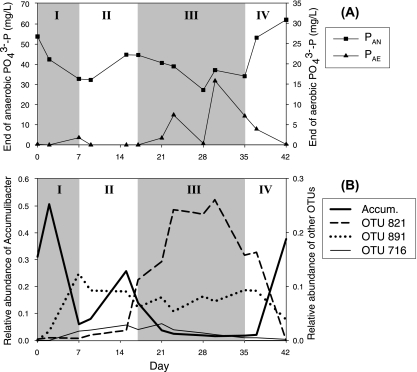
Figure 4 A shows the reactor performance levels, indicated by PAN and PAE during the nitrate experiment period, including 1 day prior to nitrate addition (day 0), the 35 days during nitrate addition (days 1 to 35), and 7 days post-nitrate addition (days 36 to 42). ARISA was used to monitor the community composition dynamics during the 43-day study period (Fig. 4B). Based on reactor performance, days 1 to 42 can be divided into four phases. During phase I (days 1 to 7), which we call the initial deterioration phase, PAN decreased significantly and PAE increased slightly, concomitant with a decrease in total “Ca. Accumulibacter.” During phase II (days 8 to 17), which we call the transient recovery phase, the reactor exhibited relatively good EBPR activity; the “Ca. Accumulibacter” abundance started to recover, but neither “Ca. Accumulibacter” abundance nor PAN had yet reached levels observed under normal conditions. During phase III (days 18 to 35), which we call the severe deterioration phase, the performance was comparatively unstable and poor, with decreased PAN and very high PAE; these changes were coincident with extremely low “Ca. Accumulibacter” abundance. During phase IV, which we called the final recovery phase, nitrate was no longer added to the reactor and performance recovered, along with “Ca. Accumulibacter” abundance reaching the same levels as prior to nitrate addition. We searched ARISA profiles during the nitrate addition and found that the increases in relative abundance of OTUs 716, 821, and 891 were concurrent with the decrease of EBPR activity (Fig. 4B).
FIG. 4.
Reactor performance and population dynamics during the nitrate addition experiment. (A) Performance measured as the soluble PO43−-P concentration at the end of the anaerobic phase (PAN; left axis) and at the end of the aerobic phase (PAE; right axis). (B) Relative proportion of total “Ca. Accumulibacter” measured using semiquantitative ARISA (left axis) and relative proportion of other ARISA OTUs (right axis) over time. Day 35 was the last day when nitrate was added to the reactor.
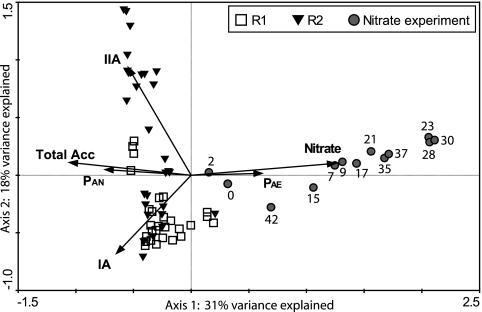
Correspondence analysis was conducted to investigate the total bacterial community response to nitrate addition (Fig. 5). Upon longer exposure to nitrate, the community shifted, generally along with the direction of the nitrate (Fig. 5, arrow), and exhibited distinct differences from the communities without nitrate exposure. This was supported by ANOSIM (R = 0.775, P < 0.001), in which a 2-day lag was assumed for the community response to adding or excluding nitrate.
FIG. 5.
Correspondence analysis of ARISA profiles collected during the 6-month normal operating period and the nitrate addition experiment. Gray circles are samples collected for the nitrate addition experiment, labeled by the experimental day indicated in Fig. 4. The arrow labels IA, IIA, and Total Acc stand for abundance levels of IA, II and total “Ca. Accumulibacter” relative to the total bacterial community; PAN and PAE are the soluble PO43−-P concentrations at the end of the anaerobic and aerobic phases; and “nitrate” represents results in samples with nitrate addition during the anaerobic phase.
The relative abundance levels of clades IA and IIA within the “Ca. Accumulibacter” lineage were also quantified during this study period by using ppk1-targeted qPCR, since the ARISA-based quantification method may not be accurate when total “Ca. Accumulibacter” abundance is low. Clade IA started as 66% of total “Ca. Accumulibacter” on day 0 and changed to 20%, 95%, 92%, and 100% at the end of phases I, II, III, and IV, respectively.
Searching for community patterns.
In addition, 26 samples collected from other time points when the reactors experienced dominant “Ca. Accumulibacter” clade shifts or poor performance were also investigated to search further for bacterial community patterns associated with these events. A total of 61 OTUs were detected within the total 95 samples analyzed, with an average of ∼41 OTUs per sample. A total of 18 OTUs, including clades IA and IIA, persisted (present in more than 90% of all samples). The correlation coefficient (r) between each individual OTU and “Ca. Accumulibacter” clades was low, except for OTU 770, a persisting clade, which exhibited a relatively high positive correlation to clade IIA (r = 0.61, P < 0.001) but not to clade IA (r = −0.047, P = 0.65). It is unclear if this population is beneficial to clade IIA or vice versa.
Correlation coefficients were also calculated between each OTU and the EBPR activity, as represented by the amount of P aerobically taken up (PAN − PAE). Table 2 is a summary of OTUs that were numerically important (average relative peak height of >2% among all 95 samples) or OTUs with significant correlations to EBPR activity (P < 0.01). As might be expected, total “Ca. Accumulibacter” had the strongest positive correlation to EBPR activity. Indeed, in Fig. 5, the gradient of total “Ca. Accumulibacter” abundance was oriented with a similar direction as the PAN gradient and opposite to the PAE gradient, confirming that “Ca. Accumulibacter” abundance, but neither clade IA nor IIA alone, was the most important factor in determining reactor performance. Several OTUs exhibited negative correlations to EBPR activity. Among them, OTUs 716, 821, and 891 were associated with deteriorated performance during the nitrate addition experiment; OTUs 954 and 986 were associated with R2 performance upsets during the full 6-month monitoring period. Some populations (e.g., OTU 730) did not seem to have obvious relationships to reactor performance. Some populations (e.g., OTU 542), although present at low levels in the ARISA profiles, exhibited a significant correlation with EBPR activity.
TABLE 2.
Abundant OTUs or OTUs with a significant correlation to EBPR activity
| OTU | Abundance (%)a |
Correlation coefficientb | −Log10Pc | |
|---|---|---|---|---|
| Max | Mean | |||
| Total Accd | 88.1 | 39.8 | 0.55 | 8.0 |
| IA | 80.0 | 27.0 | 0.35 | 3.2 |
| IIA | 67.3 | 12.8 | 0.29 | 2.3 |
| 407 | 15.1 | 3.4 | 0.20 | 1.3 |
| 692 | 30.4 | 7.1 | −0.41 | 4.4 |
| 730 | 7.1 | 2.0 | 0.06 | 0.2 |
| 745 | 33.1 | 10.0 | −0.23 | 1.6 |
| 780 | 14.1 | 3.4 | −0.23 | 1.6 |
| 786 | 16.2 | 2.9 | 0.17 | 1.0 |
| 828 | 14.4 | 3.2 | −0.25 | 1.8 |
| 954 | 23.7 | 3.6 | −0.19 | 1.2 |
| 986 | 19.7 | 2.4 | −0.21 | 1.4 |
| 542 | 1.8 | 0.2 | 0.30 | 2.5 |
| 716 | 3.0 | 0.3 | −0.41 | 4.5 |
| 821 | 25.3 | 1.5 | −0.46 | 5.5 |
| 852 | 1.6 | 0.3 | −0.37 | 3.7 |
| 856 | 1.6 | 0.2 | −0.36 | 3.4 |
| 865 | 2.1 | 0.3 | −0.44 | 5.2 |
| 891 | 13.5 | 1.7 | −0.53 | 7.6 |
| 942 | 7.9 | 0.6 | −0.26 | 2.0 |
| 973 | 1.3 | 0.3 | −0.43 | 4.9 |
The normalized ARISA peak height with the correction of rrn operon copy number per genome was used as a proxy to calculate percent abundance, as described in the text.
Pearson product-moment correlation coefficient of OTU abundance to EBPR activity.
Values in bold indicate significant correlations (P < 0.01 or −log10[P] > 2).
Total “Ca. Accumulibacter.”
DISCUSSION
Quantitative ARISA?
ARISA provides a community composition profile that is assumed to consist of nearly all amplifiable bacterial groups present in a sample. However, it was believed that the amplicon peak height or area in an ARISA profile could not provide even semiquantitative information (35). Here, we found very high correlations between quantifications obtained by qPCR and ARISA, indicating quantitative information in the ARISA profile.
Like any PCR-based methods, both qPCR and ARISA are subject to potential sources of bias, such as DNA extraction and differential amplification (32). Furthermore, when the rRNA gene is used as the genetic marker, quantification is difficult to interpret due to the different copy numbers of the rrn operon per genome among different bacterial species (or strains) (2, 16). However, even with these inherent biases, the quantification of “Ca. Accumulibacter” clades provided by ARISA profiles was strikingly accurate and revealed important patterns that may have been missed if fewer samples were analyzed due to cost limitations associated with qPCR (e.g., the decrease of “Ca. Accumulibacter” and the emergence of new populations during reactor upset). Therefore, ARISA can serve as an exploratory tool to identify the important populations or samples of particular interest, which can then be further analyzed by qPCR and/or clone library construction to identify the phylogenetic affiliations of target populations (22).
It should be noted that the ARISA amplicon sizes of different strains within a “Ca. Accumulibacter” clade may vary (12). The detectable “Ca. Accumulibacter” clades in our reactors exclusively contained strains with coherent ARISA sizes within each clade, making clade detection and quantification possible by ARISA. The range of ARISA amplicon sizes associated with “Ca. Accumulibacter” strains should be thoroughly assessed before using ARISA to detect “Ca. Accumulibacter” in other systems.
Population dynamics under normal operating conditions.
In this study, EBPR reactor performance was positively associated with total “Ca. Accumulibacter” abundance. Similar correlations were also previously observed by comparing biomass poly(P) content to the abundance of total “Ca. Accumulibacter” as determined by using fluorescent in situ hybridization (FISH) (6). However, surprisingly, we found that the “Ca. Accumulibacter” clade distribution can be quite dynamic even under relatively constant operating conditions. The cause(s) of the shifts in dominance between one or the other clade is still unknown. Seemingly random shifts occurred occasionally without relevance to detectable environmental changes. Therefore, we suspected that in addition to chemical and physical environmental variables, microbial interactions may have a more important influence on the clade distribution. Using ARISA, we attempted to search for community composition patterns and bacterial groups that had strong positive or negative correlations to the two clades. However, we did not discover clear patterns in the flanking community dynamics or particularly strong correlations between “Ca. Accumulibacter” clades and individual OTUs. Previously, bacterial viral gene expression was detected in our reactors, together with the detection of “Ca. Accumulibacter” defense mechanisms against viral invasion (18), even though the biomass was performing well. If one “Ca. Accumulibacter” clade is more resistant to viral invasion but may have a lower growth rate or other competitive disadvantage compared to the other clade, viruses can mediate “Ca. Accumulibacter” clade dynamics, as has been observed in experiments with Escherichia coli (4) and pelagic marine bacteria (20). Future studies using metagenomics and metatranscriptomics are needed to test this hypothesis.
Although the two “Ca. Accumulibacter” clades shifted their relative abundance several times over the time series, there was no obvious EBPR activity variation associated with the clade dynamics. Therefore, we hypothesize that the presence of different clades in the system provides functional redundancy, which could increase the system robustness. Besides the change within “Ca. Accumulibacter,” the flanking community was also quite dynamic (Fig. 3B). Fernandez et al. found that a stable function can be maintained by an extremely dynamic community in a methanogenic reactor (7). Therefore, there might be an extent within which the populations can fluctuate without affecting the overall function, thus creating a system buffering capacity.
Sludge response to nitrate in the anaerobic phase.
In wastewater treatment plants, ammonia is oxidized to nitrate in the aerobic zone. With internal recycling, nitrate resulting from the aerobic zone can be introduced into the anaerobic zone, especially in plants that are operated in anaerobic/aerobic (A/O) or anaerobic/anoxic/aerobic (A2O) configurations. Nitrate in the anaerobic zone has been shown to decrease the P release rate and negatively influence EBPR performance (28). This may be explained by (i) competition for organic carbon from other microorganisms that can use nitrate as electron acceptor (30); (ii) an inhibitory effect of nitrate or denitrification intermediates, such as NO, influencing PAO metabolism (31); (iii) PAOs shifting their metabolism to use nitrate for acetate oxidation, leading to simultaneous P release and uptake by PAO, ultimately resulting in a decrease in net P release (17).
In our study, complete denitrification occurred after the biomass had become “acclimated” to nitrate, concurrent with a decrease in total “Ca. Accumulibacter” to very low levels. Thus, “Ca. Accumulibacter” was likely not responsible for the denitrification activity observed, and we can conclude that a “Ca. Accumulibacter” metabolic shift was not the cause of EBPR deterioration. To exclude the possibility that “Ca. Accumulibacter” clades other than IA or IIA might be selected by nitrate, qPCR was performed using ppk1 primers targeting other clades, as well as 16S rRNA primers targeting total “Ca. Accumulibacter,” all of which confirmed the decrease of the total “Ca. Accumulibacter” population with no detection of other clades (data not shown).
A search for populations that increased during the nitrate addition identified OTUs 716, 821, and 891. These three OTUs seemed unique to nitrate-amended samples among all 95 samples studied, as indicated by a 42-, 96-, and 10-fold increase, respectively, of their average representation in the ARISA profiles during the nitrate exposure, compared to other samples. Their uniqueness and their population boost upon nitrate exposure suggested they may have contributed to the denitrification activity observed. Besides the competition with “Ca. Accumulibacter” for acetate in the anaerobic phase, other mechanisms may be involved, such as the inhibitory effects of denitrification intermediates. For example, nitrite has been shown to inhibit both aerobic and anoxic P uptake (26) and nitric oxide (NO) has been reported to inhibit anaerobic P release (31), and these potential causes remain to be tested thoroughly.
Supplementary Material
Acknowledgments
The project was funded by the National Science Foundation (BES 0332136).
We thank Andrew A. Tokelson, Nick Bartolerio, and Tess Treutelaar for bioreactor operation, Stuart Jones and Ashley Shade for help in ARISA, and Jason Flowers and Philip Hugenholtz for thoughtful discussions.
Footnotes
Published ahead of print on 2 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdo, Z., U. M. E. Schüette, S. J. Bent, C. J. Williams, L. J. Forney, and P. Joyce. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8:929-938. [DOI] [PubMed] [Google Scholar]
- 2.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Public Health Association. 1995. Standard methods for the examination of water and waste water, 19th ed. American Public Health Association, American Water Works Association, and Water Environment Federation. APHA, Washington, DC.
- 4.Bohannan, B. J. M., B. Kerr, C. M. Jessup, J. B. Hughes, and G. Sandvik. 2002. Trade-offs and coexistence in microbial microcosms. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 81:107-115. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho, G., P. C. Lemos, A. Oehmen, and M. A. M. Reis. 2007. Denitrifying phosphorus removal: linking the process performance with the microbial community structure. Water Res. 41:4383-4396. [DOI] [PubMed] [Google Scholar]
- 6.Crocetti, G. R., P. Hugenholtz, P. L. Bond, A. Schuler, J. Keller, D. Jenkins, et al. 2000. Identification of polyphosphate accumulating organisms and the design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 66:1175-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez, A., S. Huang, S. Seston, J. Xing, R. Hickey, C. Criddle, et al. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flowers, J. J., S. He, S. Yilmaz, D. R. Noguera, and K. D. McMahon. 2009. Denitrification capabilities of two biological phosphorus removal sludges dominated by different ‘Candidatus Accumulibacter’ clades. Environ. Microbiol. Rep. 1:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia Martin, H., N. Ivanova, V. Kunin, F. Warnecke, K. W. Barry, A. C. McHardy, et al. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24:1263-1269. [DOI] [PubMed] [Google Scholar]
- 11.He, S., D. L. Gall, and K. D. McMahon. 2007. “Candidatus Accumulibacter” population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl. Environ. Microbiol. 73:5865-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, S., A. Z. Gu, and K. D. McMahon. 2006. Fine-scale differences between Accumulibacter-like bacteria in enhanced biological phosphorus removal activated sludge. Water Sci. Technol. 54:111-117. [DOI] [PubMed] [Google Scholar]
- 13.Hesselmann, R. P. X., C. Werlen, D. Hahn, J. R. van der Meer, and A. J. B. Zehnder. 1999. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 22:454-465. [DOI] [PubMed] [Google Scholar]
- 14.Jones, S. E., and K. D. McMahon. 2009. Species-sorting may explain an apparent minimal effect of immigration on freshwater bacterial community dynamics. Environ. Microbiol. 11:905-913. [DOI] [PubMed] [Google Scholar]
- 15.Kaetzke, A., D. Jentzsch, and K. Eschrich. 2005. Quantification of Microthrix parvicella in activated sludge bacterial communities by real-time PCR. Lett. Appl. Microbiol. 40:207-211. [DOI] [PubMed] [Google Scholar]
- 16.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuba, T., A. Wachtmeister, M. C. M. van Loosdrecht, and J. J. Heijnen. 1994. Effect of nitrate on phosphorus release in biological phosphorus removal systems. Water Sci. Technol. 30:263-269. [Google Scholar]
- 18.Kunin, V., S. He, F. Warnecke, S. B. Peterson, H. Garcia Martin, M. Haynes, et al. 2008. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 18:293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMahon, K. D., D. Jenkins, and J. D. Keasling. 2002. Polyphosphate kinase genes from activated sludge carrying out enhanced biological phosphorus removal. Water Sci. Technol. 46:155-162. [PubMed] [Google Scholar]
- 20.Middelboe, M., A. Hagstrom, N. Blackburn, B. Sinn, U. Fischer, N. H. Borch, et al. 2001. Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb. Ecol. 42:395-406. [DOI] [PubMed] [Google Scholar]
- 21.Mino, T., M. C. M. Van Loosdrecht, and J. J. Heijnen. 1998. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32:3193-3207. [Google Scholar]
- 22.Newton, R. J., A. D. Kent, E. W. Triplett, and K. D. McMahon. 2006. Microbial community dynamics in a humic lake: differential persistence of common freshwater phylotypes. Environ. Microbiol. 8:956-970. [DOI] [PubMed] [Google Scholar]
- 23.Peterson, S. B., F. Warnecke, J. Madejska, K. D. McMahon, and P. Hugenholtz. 2008. Environmental distribution and population biology of Candidatus Accumulibacter, a primary agent of biological phosphorus removal. Environ. Microbiol. 10:2692-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranjard, L., F. Poly, J. C. Lata, C. Mougel, J. Thioulouse, and S. Nazaret. 2001. Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl. Environ. Microbiol. 67:4479-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 26.Saito, T., D. Brdjanovic, and M. C. M. van Loosdrecht. 2004. Effect of nitrite on phosphate uptake by phosphate accumulating organisms. Water Res. 38:3760-3768. [DOI] [PubMed] [Google Scholar]
- 27.Shade, A., S. E. Jones, and K. D. McMahon. 2008. The influence of habitat heterogeneity on freshwater bacterial community composition and dynamics. Environ. Microbiol. 10:1057-1067. [DOI] [PubMed] [Google Scholar]
- 28.Shehab, O., R. Deininger, F. Porta, and T. Wojewski. 1996. Optimizing phosphorus removal at the Ann Arbor wastewater treatment plant. Water Sci. Technol. 34:493-499. [Google Scholar]
- 29.Simmons, S. L., D. A. Bazylinski, and K. J. Edwards. 2007. Population dynamics of marine magnetotactic bacteria in a meromictic salt pond described with qPCR. Environ. Microbiol. 9:2162-2174. [DOI] [PubMed] [Google Scholar]
- 30.Stensel, H. D. 1991. Principles of biological phosphorus removal, p. 141-166. In R. I. Sedlak (ed.), Phosphorus and nitrogen removal from municipal wastewater: principles and practices, 2nd ed. Lewis Publishers, New York, NY.
- 31.van Niel, E. W. J., K. J. Appeldoorn, A. J. B. Zehnder, and G. J. J. Kortstee. 1998. Inhibition of anaerobic phosphate release by nitric oxide in activated sludge. Appl. Environ. Microbiol. 64:2925-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 33.Wexler, M., D. J. Richardson, and P. L. Bond. 2009. Radiolabelled proteomics to determine differential functioning of Accumulibacter during the anaerobic and aerobic phases of a bioreactor operating for enhanced biological phosphorus removal. Environ. Microbiol. 11:3029-3044. [DOI] [PubMed] [Google Scholar]
- 34.Xin, G., H. L. Gough, and H. D. Stensel. 2008. Effect of anoxic selector configuration on sludge volume index control and bacterial population fingerprinting. Water Environ. Res. 80:2228-2240. [DOI] [PubMed] [Google Scholar]
- 35.Yannarell, A. C., A. D. Kent, G. H. Lauster, T. K. Kratz, and E. W. Triplett. 2003. Temporal patterns in bacterial communities in three temperate lakes of different trophic status. Microb. Ecol. 46:391-405. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida, N., N. Takahashi, and A. Hiraishi. 2005. Phylogenetic characterization of a polychlorinated-dioxin-dechlorinating microbial community by use of microcosm studies. Appl. Environ. Microbiol. 71:4325-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zilles, J. L., J. Peccia, M.-W. Kim, C.-H. Hung, and D. R. Noguera. 2002. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl. Environ. Microbiol. 68:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.