Abstract
The lactic acid bacterial (LAB) community dynamics of two wheat and two spelt sourdough fermentations that were daily back-slopped were monitored during a period of 10 days by hybridizing time-related RNA samples, representing the metatranscriptome, to an LAB functional gene microarray. To indicate the species present in each hybridized sample, annotation information for the 2,269 oligonucleotides on the microarray was used. The overall hybridization data revealed that after a transition phase of 5 days, in which atypical sourdough LAB species, including Enterococcus species, were found, a stabilized ecosystem was established with Lactobacillus plantarum and Lactobacillus fermentum as the dominating LAB species. Compared with the combined outcome of culture-dependent and culture-independent identification techniques, the microarray data revealed a functional role for Lactococcus lactis in the early stage ecosystem and the dominance of Pediococcus pentosaceus in most of the fermentations, besides L. plantarum and L. fermentum. Consequently, metatranscriptome hybridization data obtained using an LAB functional gene microarray was shown to be an interesting alternative to microbiological analysis of the community dynamics of complex food ecosystems.
Fermented food ecosystems consist of lactic acid bacteria (LAB), acetic acid bacteria, and other Gram-positive and Gram-negative bacteria and/or fungi that contribute to several beneficial characteristics of fermented food products, such as an extended shelf life, improved texture, and enhanced organoleptic properties (36). In most of the cases, fermented food ecosystems are rapidly dominated by LAB, whether or not in conjunction with yeasts, that cause acidification of the food matrix by the production of lactic acid and/or acetic acid (20, 32). In some fermentations, the ecosystem is even characterized by a targeted, temporal succession of microorganisms during the course of the fermentation (21, 30). Stable sourdough ecosystems typically consist of LAB and yeasts, wherein the LAB species diversity depends to a large extent on the type of flour and process parameters and comprises mainly Lactobacillus species, among which are Lactobacillus plantarum, Lactobacillus fermentum, Lactobacillus sanfranciscensis, Lactobacillus paralimentarius, and Lactobacillus brevis (13).
In the past, techniques based on physiological tests were used to identify microbial members of fermented food ecosystems. During the last decades, DNA-based identification techniques have taken over, thereby mainly relying on comparative analysis of partial or full 16S rRNA gene sequences (19, 23). In a culture-dependent approach, genomic DNA from single isolates is used to sequence the partial or complete 16S rRNA gene for identification purposes (15). In a culture-independent approach, metagenomic DNA that is directly extracted from the food ecosystem is subjected to a community PCR, usually targeting the 16S rRNA gene. The resulting pool of PCR amplicons is subsequently separated by denaturing gradient gel electrophoresis (DGGE) (17) or temperature gradient gel electrophoresis (TGGE) (9) and can be followed by extraction, purification, and sequencing of specific DNA bands in the resulting ecosystem fingerprint. Other culture-independent fingerprinting techniques encompass, for instance, terminal restriction fragment length polymorphism (T-RFLP) (27). When linked to sequence data, these methods allow inventory of the predominant microbiota present in a given fermented food ecosystem and provide a solid basis for the development of more targeted molecular tools such as microarrays.
To be able to identify members of bacterial ecosystems in a broad and high-throughput way, several microarrays have been developed recently (1, 22, 26, 34, 35, 37). Actually, two types of microarrays can be distinguished: (i) microarrays that contain partial 16S rRNA gene sequences as targets, also referred to as phylogenetic oligonucleotide microarrays; and (ii) microarrays that contain integral genomic DNA as targets, also referred to as community genome microarrays (38). Of the latter type, one example is known that can be applied to food fermentation ecosystems, namely, a so-called “genome-probing microarray” to monitor community dynamics of LAB (1). This microarray has been used to monitor community dynamics and biological activity during a 12-day kimchi fermentation, thereby using the metagenome as well as the metatranscriptome of 10 time-related samples (25).
The aim of the present study was to reveal the LAB community dynamics of two wheat and two spelt sourdough fermentations that were back-slopped during a period of 10 days, by hybridizing time-related RNA samples, representing the metatranscriptome, to a recently developed LAB functional gene microarray (35). These results were compared with culture-dependent and culture-independent identification data to assess future implementation of this microarray for analysis of bacterial community dynamics of complex fermented food ecosystems.
MATERIALS AND METHODS
Sourdough fermentations.
Two wheat (D12W and D13W) and two spelt (D12S and D13S) back-slopped sourdoughs were prepared through spontaneous fermentation. Therefore, fresh 8-kg sourdough fermentations with a dough yield of 400 [(dough mass/flour mass) × 100] were started up in Biostat C fermentors (Sartorius AG/B. Braun Biotech International, Melsungen, Germany) and incubated at 30°C for 24 h. The mixture was kept homogeneous by stirring at 300 rpm. The sourdoughs were daily back-slopped over a period of 10 days by inoculating a fresh water-flour mixture with ripe sourdough (10% [wt/wt]), after which the fresh dough was incubated under the same conditions as mentioned above. At each refreshment step, samples of the ripe sourdough were collected for culture-dependent and culture-independent identifications.
For culture-dependent identification, a 10-fold dilution series of sourdough samples was plated on MRS-5 agar (de Man-Rogosa-Sharpe agar supplemented with a vitamin solution) (24) and incubated at 30°C. Per sample, on average 10 to 15 purified Gram-positive and catalase-negative isolates were subjected to (GTG)5-PCR fingerprinting, and the resulting fingerprints were numerically analyzed by means of the BioNumerics version v4.0 software package (Applied Maths, Sint-Martens-Latem, Belgium) using an extensive in-house reference database (29, 33). To verify the identifications tentatively assigned to a (GTG)5-PCR cluster and to identify the remaining unknown clusters and/or single isolates, representatives of each cluster and the single isolates were subjected to a polyphasic taxonomic approach, including amplified fragment length polymorphism, sodium dodecyl sulfate-polyacrylamide gel electrophoresis of whole-cell proteins, 16S rRNA gene sequence analysis, pheS gene sequence analysis, and/or DNA-DNA hybridizations, as described previously (29, 33). In addition, the composition of the microbiota of the sourdough samples was investigated through a culture-independent approach by means of denaturing gradient gel electrophoresis (DGGE) of PCR amplicons of the V3 region of the 16S rRNA gene (16S rRNA-PCR-DGGE), followed by purification and sequencing of the DGGE bands, as described previously (29, 33).
RNA isolation.
RNA was isolated to represent the metatranscriptome of the sourdough fermentation samples. Therefore, additional samples were taken 3 h after the refreshment step (i.e., at 27, 51, 75, 99, 123, 147, 171, 195, and 219 h), as approximately half of the acidification had taken place at these time points, indicating that the bacteria were in their exponential growth phase (33). Ten grams of sourdough was mixed with 40 ml of RNAprotect (Qiagen, Hilden, Germany) that was 2:1 diluted with 1× phosphate-buffered saline (Invitrogen, Carlsbad, CA). This mixture was kept at room temperature for 5 min, followed by centrifugation to remove solids (1,000 × g for 5 min). The supernatant was centrifuged for a second time (5,000 × g for 15 min), and the metatranscriptome RNA was isolated from the resulting cell pellet by applying an enzymatic cell lysis using mutanolysin and lysozyme, after which the RNA was extracted from the resulting mixture by using an RNeasy minikit (Qiagen) following standard instructions and including mechanical disruption of the cells using glass beads, as described previously (35).
Microarray hybridization and data analysis.
An LAB functional gene microarray was used, containing 2,269 oligonucleotides that target in total 406 key genes (35). The isolated RNA was linearly amplified (aRNA) using a Genisphere SensAmp kit (Genisphere, Hatfield, PA), labeled with Cy3 and Cy5 dyes in a reverse transcription reaction, and hybridized for 16 h using a HS 4800 Pro automated hybridization station (Tecan Systems, Inc., San Jose, CA), as described previously (35), except that 60 pmol instead of 50 pmol of labeled aRNA was used to prepare the hybridization mixtures. The labeled aRNA of the different sourdough fermentation samples was hybridized to the microarray, using a loop design over the different time points: i.e., two consecutive samples (e.g., 27 h and 51 h, 51 h and 75 h, etc.) were hybridized on the same microarray slide, each labeled with another fluorescent dye (Cy3 and Cy5), and the loop was closed by hybridizing the last sample (219 h) together with the first (27 h) (Fig. 1).
FIG. 1.
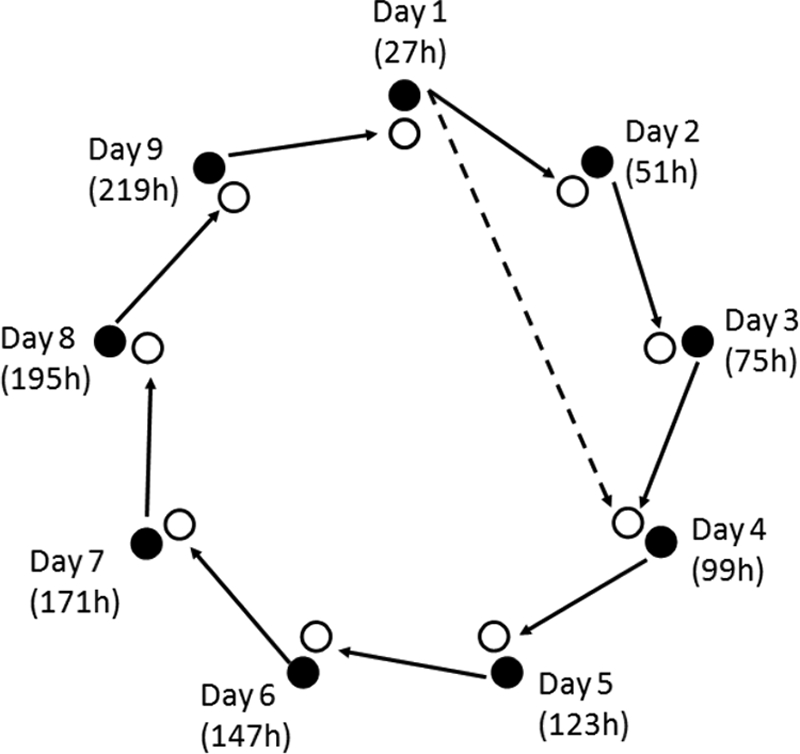
Schematic representation of the loop design applied to hybridize labeled aRNA to the microarray. Open circles represent Cy3-labeled aRNA, and closed circles represent Cy5-labeled aRNA. Solid lines represent the hybridization of the two connected aRNA samples on the same microarray slide. The dashed line represents the hybridization of the Cy5-labeled aRNA at 27 h with the Cy3-labeled aRNA at 99 h for fermentation D13S, since no labeled aRNA samples were available for day 2 (51 h) and day 3 (75 h) in this particular case.
Given that each oligonucleotide was spotted four times on the array and each sample was hybridized twice (i.e., once labeled with Cy3 and once labeled with Cy5), the intensity of each oligonucleotide was measured eight times. The intensity of an oligonucleotide was considered above background level if the intensities of at least six out of eight spots were above the background level, as described previously (35). Furthermore, intensity values were normalized for array and dye effects, and only those oligonucleotides with a significant change in their hybridization intensity profile over time were retained for clustering. The hybridization intensity profiles were transformed into Z-score profiles by subtracting the average hybridization intensity of all oligonucleotides from the hybridization intensity of the oligonucleotide considered and dividing that result by the standard deviation of all hybridization intensities (8). Subsequently, these Z-score profiles were hierarchically clustered with the complete linkage option with a distance measure of one minus the Pearson correlation (16, 18).
To indicate the species present in the sourdough ecosystems, a significance per species of at least 10% had to be reached: i.e., at least 10% of the species-specific oligonucleotides (with a minimum of two oligonucleotides) had to have an intensity above background. This percentage was based on the outcome of validation hybridizations using DNA and RNA of 18 LAB strains, covering 86% of all oligonucleotides on the microarray, whereby the highest number of false-positive signals appeared to be 6% (35). In addition, only species that were represented by at least 10 oligonucleotides were considered for further analysis.
Microarray data accession numbers.
The microarray data were deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GPL5459 (microarray including detailed annotation), GSE15686 (D12W), GSE15693 (D13W), GSE15692 (D12S), and GSE15691 (D13S).
RESULTS
Bacterial community dynamics analyzed through a combined culture-dependent and culture-independent approach.
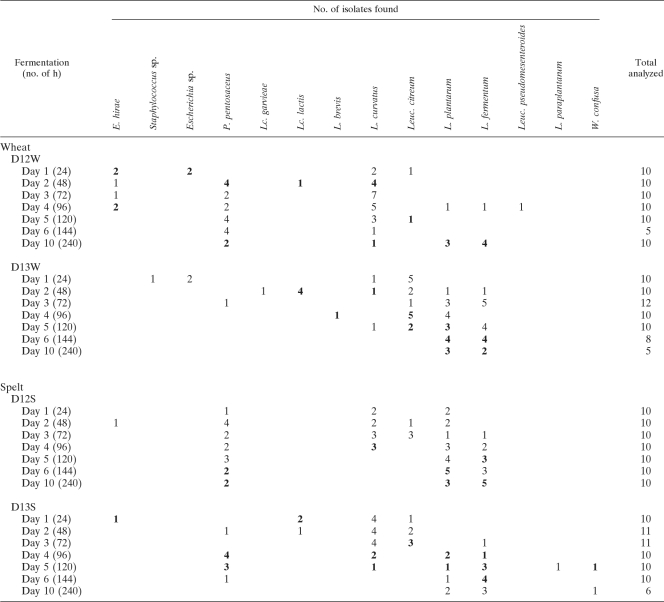
The combination of the culture-dependent and culture-independent data from the four 10-day back-slopped sourdough fermentations studied (D12W, D13W, D12S, and D13S) showed that the LAB compositions and hence the community dynamics of the sourdough ecosystem followed a time-related evolution, whereby most changes occurred during the first 5 days of fermentation (Table 1). In general, species such as Lactococcus lactis, Enterococcus hirae, Leuconostoc citreum, and Lactobacillus curvatus predominated during the first 2 days of all fermentations. From day 3 to 5, Lc. lactis and E. hirae disappeared, while Leuc. citreum and L. curvatus remained present. In addition, Pediococcus pentosaceus, L. plantarum, and L. fermentum appeared. From day 6 on, a more uniform ecosystem was found, consisting basically of L. plantarum and L. fermentum in all fermentations. In addition, P. pentosaceus remained in fermentations D12S, D13S, and D12W from day 2 on. Furthermore, also L. curvatus (D12W) and Weissella confusa (D13S) were found from days 2 and 5 on, respectively.
TABLE 1.
Bacterial community dynamics of LAB during back-slopped sourdough fermentationsa
Shown are results from four 10-day back-slopped sourdough fermentations carried out with wheat or spelt at 30°C, as revealed by a combined culture-dependent and culture-independent identification approach. The numbers correspond to the number of isolates found for the given species at the given time point by culture-dependent identification. The numbers in boldface indicate that the species was also found by culture-independent identification. No species were found by culture-independent identification only.
Bacterial community dynamics analyzed through metatranscriptome hybridizations.
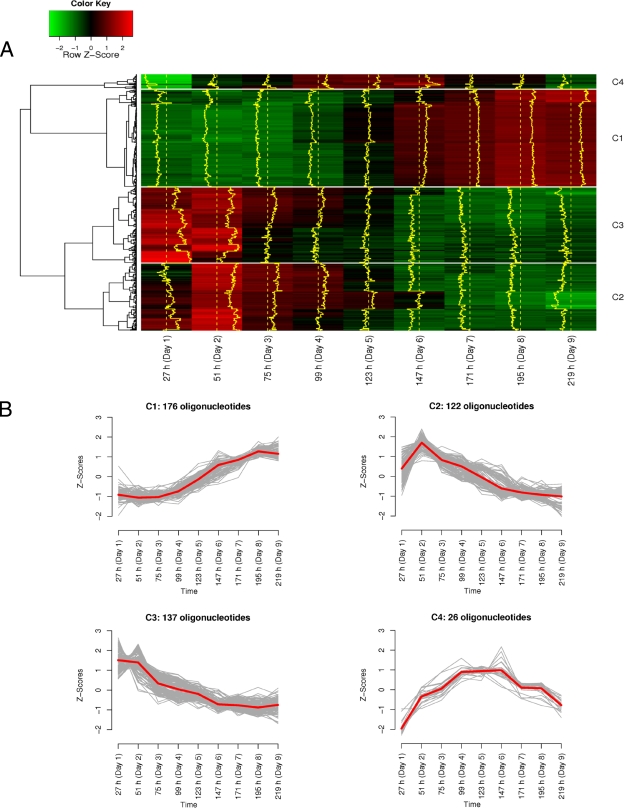
For wheat fermentation D12W, 461 oligonucleotides displayed a Z-score profile that showed a significant change over time. The Z-score profiles were grouped into four clusters (Fig. 2). Cluster 1 comprised 176 oligonucleotides with increasing Z-scores over time. The oligonucleotides that met the requirements as set out above were related to L. plantarum and L. fermentum (Table 2). Clusters 2 and 3 showed decreasing Z-scores over time, although cluster 2 had a peak at day 2 and a slower decrease over time compared to cluster 3. Cluster 2 comprised 122 oligonucleotides, mainly related to L. curvatus, Lactobacillus reuteri, Lactobacillus sakei, and Lc. lactis (Table 2). Cluster 3 comprised 137 oligonucleotides, mainly related to Enterococcus faecalis, Enterococcus faecium, E. hirae, and Lc. lactis (Table 2). Cluster 4 showed increasing Z-scores, followed by a decrease, with maximum values at days 4 to 6. This cluster comprised 26 oligonucleotides, mainly related to P. pentosaceus (Table 2).
FIG. 2.
Intensity profiles obtained through hybridization of metatranscriptome samples of wheat fermentation D12W. (A) For a total of 461 out of 2,269 oligonucleotides, the intensity was above the background level for at least one time point of the 10-day back-slopped sourdough fermentation process. The 461 oligonucleotides whose intensities were above the background level could be clustered into four groups (C1 to C4) according to their Z-score profiles. The Z-scores varied from green (−2; i.e., below the average response of that oligonucleotide) to red (+2; i.e., above the average response of that oligonucleotide). (B) Representation of the hybridization intensity profiles of the four clusters of oligonucleotides. x axis, sampling times; y axis, Z-scores of the different oligonucleotides.
TABLE 2.
Species present in the four ecosystems as revealed by metatranscriptome hybridizations
| Fermentation and cluster (no. of oligonucleotides) | % of significant oligonucleotides of speciesa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. hirae (11) | E. faecium (113) | P. pentosaceus (72) | Lc. lactis (551) | L. brevis (17) | L. curvatus (10) | L. plantarum (302) | L. fermentum (14) | L. reuteri (31) | L. sakei (187) | |
| Wheat | ||||||||||
| D12W | ||||||||||
| Cluster 1 (176) | 50 | 29 | ||||||||
| Cluster 2 (122) | 15 | 30 | 11 | |||||||
| Cluster 3 (137) | 82 | 27 | 13 | |||||||
| Cluster 4 (26) | 29 | |||||||||
| D13W | ||||||||||
| Cluster 1 (242) | 65 | 21 | ||||||||
| Cluster 2 (73) | 27 | 12 | 20 | |||||||
| Cluster 3 (259) | 27 | 41 | 13 | |||||||
| Cluster 4 (86) | 13 | |||||||||
| Cluster 5 (8) | 10 | |||||||||
| Spelt | ||||||||||
| D12S | ||||||||||
| Cluster 1 (203) | 58 | 29 | ||||||||
| Cluster 2 (49) | 18 | 30 | 13 | |||||||
| Cluster 3 (54) | 28 | |||||||||
| Cluster 4 (24) | 55 | 23 | ||||||||
| D13S | ||||||||||
| Cluster 1 (150) | 46 | |||||||||
| Cluster 2 (58) | 12 | 30 | 14 | |||||||
| Cluster 3 (256) | 46 | 12 | 39 | 16 | ||||||
| Cluster 4 (16) | 21 | |||||||||
The total number of oligonucleotides present on the microarray is given in parentheses after the species name.
The Z-score profiles of the fermentations D13W, D12S, and D13S were clustered likewise (Table 2; see the supplemental material). For these fermentations, clusters with increasing Z-scores over time comprised mainly oligonucleotides related to L. plantarum and L. fermentum, although the latter LAB species was not found in spelt fermentation D13S. Clusters with a fast decrease in Z-scores over time comprised mainly oligonucleotides related to E. hirae, E. faecium, L. reuteri, and Lc. lactis, although the latter two LAB species were not found in spelt fermentation D12S. Clusters with less-steep decreasing Z-scores over time comprised mainly oligonucleotides related to L. sakei, L. curvatus, L. brevis, and E. hirae. For fermentation D13W, one additional cluster was obtained with decreasing Z-scores over time. This cluster comprised only oligonucleotides related to Lc. lactis and showed a faster decrease of the Z-scores over time compared to the other two clusters. Clusters with increasing Z-scores, followed by a decrease over time, comprised mainly oligonucleotides related to P. pentosaceus. In spelt fermentation D13S, for which no samples were available for day 2 (51 h) and day 3 (75 h), also oligonucleotides related to L. fermentum were found in the cluster with increasing Z-scores followed by a decrease over time.
DISCUSSION
In the present study, a LAB functional gene microarray, comprised of 2,269 oligonucleotides that target 406 key genes (35), was applied for the first time to follow up the bacterial community dynamics of two wheat and two spelt sourdough fermentations that were back-slopped for a period of 10 days. Time-related metatranscriptome samples were hybridized to this LAB functional gene microarray, thereby relying on the species annotation of oligonucleotides whose intensities were above the background level. The identification as revealed by the microarray hybridization data was in agreement with the outcome of a combined culture-dependent and culture-independent identification approach. In addition, the differences revealed interesting views on the bacterial community dynamics of a sourdough ecosystem.
The bacterial community dynamics followed a three-phase evolution. During the first 2 days, a variety of LAB, atypical for mature sourdough ecosystems, as well as some Enterococcus species were found in the fermenting ecosystems. During a transition phase from day 2 until day 5 witnessed in all four fermentations, the Enterococcus species fully disappeared from the ecosystem. Some of the atypical LAB lasted for some more days, such as L. curvatus, L. sakei, and Lc. lactis. The latter species was not able to survive in a mature sourdough ecosystem, although it is an autochthonous species of the cereal microbiota (12). This indicates that factors such as refreshment steps, fermentation times, and/or pH influence the prevalence of certain species (13). Nevertheless, Lc. lactis seemed to play a functional role in the ecosystem during the first days of fermentation, given the Z-scores of the oligonucleotides related to this species in three of the four fermentations. This role may be ascribed to its ability to hydrolyze starch through amylase activity (3, 14). The occurrence of L. sakei in the early sourdough ecosystem of fermentations D12W, D12S, and D13S is unclear, as earlier findings indicated that L. sakei is very well adapted to its natural meat environment (6). However, that L. sakei can be found and proliferate in other environments too has not been excluded (7).
Already during the transition phase from day 2 until day 5, sourdough-specific LAB species such as L. plantarum and L. fermentum were present. Based on increasing Z-scores, these species increased in number and/or activity over time, to dominate the ecosystem after day 5. Interestingly, in three of the four sourdough fermentations (i.e., D12W, D13W, and D12S), P. pentosaceus was a prominent member of the ecosystem, besides L. plantarum and L. fermentum, although it followed a different Z-score profile. Indeed, oligonucleotides related to P. pentosaceus reached a higher Z-score much earlier in the fermentations compared to the oligonucleotides related to L. plantarum and L. fermentum. They decreased again from days 6 to 7, leaving L. plantarum and L. fermentum as the sole dominating species after day 7. This was in contrast with the combined outcome of the culture-dependent and culture-independent identification approaches, where P. pentosaceus was found during the whole fermentation period. However, the DNA-based identification techniques indicated that P. pentosaceus was still present, probably as resting cells, explaining why this species was not detected with the RNA-based microarray hybridizations. Although information on the identity of metabolically active cells is valuable and can be considered as a strength of the microarray approach, the apparent impossibility to detect resting cells is a point of attention. However, this can be circumvented by hybridizing metagenome DNA, isolated from the same samples, in addition to metatranscriptome RNA. Furthermore, it should be noted that only species that are represented on the microarray can be detected, making it impossible to discover new species.
In general, species of Lactobacillus, in particular, L. plantarum and L. fermentum, dominate sourdough fermentation processes (13). Lactococcus, Enterococcus, Leuconostoc, and Weissella are typically associated with cereal kernels and flour, but do not survive a long-term acidification process (10, 11, 33). Lactobacillus fermentum and L. plantarum are more acid tolerant and often dominate fermentation processes of vegetables and cereals, in particular because of their ability to transport and metabolize different carbohydrates (2, 4, 5). LAB species that are much less frequently encountered in mature sourdoughs, such as P. pentosaceus (11, 28, 31), may dominate some sourdoughs as well, for instance depending on back-slopping practices, temperature of incubation, and pH of the dough (13).
To conclude, the LAB functional gene microarray used in the present study was successfully applied to follow-up LAB community dynamics of two wheat and two spelt 10-day sourdough fermentations with daily back-slopping, through hybridization of time-related metatranscriptome samples. The overall hybridization data indicated rapid growth of multiple LAB during the first days of fermentation, after which sourdough-specific LAB (i.e., L. plantarum and L. fermentum) contributed to the maturation of the sourdough. Compared with the combined outcome of the culture-dependent and culture-independent identification techniques used, the hybridization data indicated that Lc. lactis seemed to play a functional role during the first 2 days of some fermentations. Also, P. pentosaceus was dominant for some days as well, suggesting a certain impact on the course of the concomitant sourdough ecosystems. Consequently, the LAB functional gene microarray hybridization data analysis showed the method to be an interesting alternative to microbiological analysis of the bacterial community dynamics of complex microbial ecosystems.
Supplementary Material
Acknowledgments
This work was financed by SBO project IWT-030263 of the Institute for the Promotion of Innovation by Science and Technology in Flanders, Belgium (IWT-Vlaanderen). S.W. and L.D.V. acknowledge financing from the Research Council of the Vrije Universiteit Brussel, in particular their BOF and IOF projects. G.H. is a postdoctoral fellow of the Fund for Scientific Research Flanders, Belgium (FWO-Vlaanderen).
We are grateful to Kizi Coeck, Ruth Maes, Ilse Scheirlinck, and Ann Van Schoor for their contributions.
Footnotes
Published ahead of print on 25 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bae, J. W., S. K. Rhee, J. R. Park, W. H. Chung, Y. D. Nam, I. Lee, H. Kim, and Y. H. Park. 2005. Development and evaluation of genome-probing microarrays for monitoring lactic acid bacteria. Appl. Environ. Microbiol. 71:8825-8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boekhorst, J., R. J. Siezen, M. C. Zwahlen, D. Vilanova, R. D. Pridmore, A. Mercenier, M. Kleerebezem, W. M. de Vos, H. Brussow, and F. Desiere. 2004. The complete genomes of Lactobacillus plantarum and Lactobacillus johnsonii reveal extensive differences in chromosome organization and gene content. Microbiology 150:3601-3611. [DOI] [PubMed] [Google Scholar]
- 3.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis spp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderon-Santoyo, M., G. Loiseau, and J. P. Guyot. 2003. Fermentation by Lactobacillus fermentum Ogi E1 of different combinations of carbohydrates occurring naturally in cereals: consequences on growth energetics and α-amylase production. Int. J. Food Microbiol. 80:161-169. [DOI] [PubMed] [Google Scholar]
- 5.Calderon-Santoyo, M., G. Loiseau, R. R. Sanoja, and J. P. Guyot. 2003. Study of starch fermentation at low pH by Lactobacillus fermentum Ogi E1 reveals uncoupling between growth and α-amylase production at pH 4.0. Int. J. Food Microbiol. 80:77-87. [DOI] [PubMed] [Google Scholar]
- 6.Chaillou, S., M. C. Champomier-Verges, M. Cornet, A. M. Crutz-Le Coq, A. M. Dudez, V. Martin, S. Beaufils, E. Darbon-Rongere, R. Bossy, V. Loux, and M. Zagorec. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527-1533. [DOI] [PubMed] [Google Scholar]
- 7.Chaillou, S., M. Daty, F. Baraige, A.-M. Dudez, P. Anglade, R. Jones, C.-A. Alpert, M.-C. Champomier-Verges, and M. Zagorec. 2009. Intraspecies genomic diversity and natural population structure of the meat-borne lactic acid bacterium Lactobacillus sakei. Appl. Environ. Microbiol. 75:970-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheadle, C., M. P. Vawter, W. J. Freed, and K. G. Becker. 2003. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 5:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2000. Development of a rapid method for the identification of Lactobacillus spp. isolated from naturally fermented Italian sausages using a polymerase chain reaction-temperature gradient gel electrophoresis. Lett. Appl. Microbiol. 30:126-129. [DOI] [PubMed] [Google Scholar]
- 10.Corsetti, A., L. Settanni, C. C. Lopez, G. E. Felis, M. Mastrangelo, and G. Suzzi. 2007. A taxonomic survey of lactic acid bacteria isolated from wheat (Triticum durum) kernels and non-conventional flours. Syst. Appl. Microbiol. 30:561-571. [DOI] [PubMed] [Google Scholar]
- 11.Corsetti, A., L. Settanni, S. Valmorri, M. Mastrangelo, and G. Suzzi. 2007. Identification of subdominant sourdough lactic acid bacteria and their evolution during laboratory-scale fermentations. Food Microbiol. 24:592-600. [DOI] [PubMed] [Google Scholar]
- 12.De Vuyst, L., and P. Neyssens. 2005. Biodiversity of sourdough lactic acid bacteria. Trends Food Sci. Technol. 16:43-56. [Google Scholar]
- 13.De Vuyst, L., G. Vrancken, F. Ravyts, T. Rimaux, and S. Weckx. 2009. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 26:666-675. [DOI] [PubMed] [Google Scholar]
- 14.Diaz-Ruiz, G., J. P. Guyot, F. Ruiz-Terna, J. Morlon-Guyot, and C. Wacher. 2003. Microbial and physiological characterization of weakly amylolytic but fast-growing lactic acid bacteria: a functional role in supporting microbial diversity in pozol, a Mexican fermented maize beverage. Appl. Environ. Microbiol. 69:4367-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes—characterization of a gene coding for 16S-ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ercolini, D. 2004. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56:297-314. [DOI] [PubMed] [Google Scholar]
- 18.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giraffa, G., and D. Carminati. 2008. Molecular techniques in food fermentation: principles and applications, p. 1-30. In L. Cocolin and D. Ercolini (ed.), Molecular techniques in the microbial ecology of fermented foods. Springer, Heidelberg, Germany.
- 20.Gobbetti, M. 1998. The sourdough microflora: interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 9:267-274. [Google Scholar]
- 21.Harris, L. J. 1998. The microbiology of vegetable fermentation, p. 45-72. In B. J. B. Wood (ed.), Microbiology of fermented foods. Blackie Academic and Professional, London, United Kingdom.
- 22.Huyghe, A., P. Francois, Y. Charbonnier, M. Tangomo-Bento, E. J. Bonetti, B. J. Paster, I. Bolivar, D. Baratti-Mayer, D. Pittet, and J. Schrenzel. 2008. Novel microarray design strategy to study complex bacterial communities. Appl. Environ. Microbiol. 74:1876-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 24.Meroth, C. B., J. Walter, C. Hertel, M. J. Brandt, and W. P. Hammes. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam, Y.-D., H.-W. Chang, K.-H. Kim, S. W. Roh, and J.-W. Bae. 2009. Metatranscriptome analysis of lactic acid bacteria during kimchi fermentation with genome-probing microarrays. Int. J. Food Microbiol. 130:140-146. [DOI] [PubMed] [Google Scholar]
- 26.Peplies, J., C. Lachmund, F. O. Glockner, and W. Manz. 2006. A DNA microarray platform based on direct detection of rRNA for characterization of freshwater sediment-related prokaryotic communities. Appl. Environ. Microbiol. 72:4829-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rademaker, J. L. W., M. Peinhopf, L. Rijnen, W. Bockelmann, and W. H. Noordman. 2005. The surface microflora dynamics of bacterial smear-ripened Tilsit cheese determined by T-RFLP DNA population fingerprint analysis. Int. Dairy J. 15:785-794. [Google Scholar]
- 28.Robert, H., V. Gabriel, and C. Fontagné-Faucher. 2009. Biodiversity of lactic acid bacteria in French wheat sourdough as determined by molecular characterization using species-specific PCR. Int. J. Food Microbiol. 135:53-59. [DOI] [PubMed] [Google Scholar]
- 29.Scheirlinck, I., R. Van der Meulen, A. Van Schoor, M. Vancanneyt, L. De Vuyst, P. Vandamme, and G. Huys. 2007. Influence of the geographical origin and flour type on the diversity in lactic acid bacteria. Appl. Environ. Microbiol. 73:6262-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwan, R. F., and A. E. Wheals. 2004. The microbiology of cocoa fermentation and its role in chocolate quality. Crit. Rev. Food Sci. Nutr. 44:205-221. [DOI] [PubMed] [Google Scholar]
- 31.Siragusa, S., R. Di Cagno, D. Ercolini, F. Minervini, M. Gobbetti, and M. De Angelis. 2009. Taxonomic structure and monitoring of the dominant population of lactic acid bacteria during wheat flour sourdough type I propagation using Lactobacillus sanfranciscensis starters. Appl. Environ. Microbiol. 75:1099-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solieri, L., and P. Giudici. 2008. Yeasts associated to traditional balsamic vinegar: ecological and technological features. Int. J. Food Microbiol. 125:36-45. [DOI] [PubMed] [Google Scholar]
- 33.Van der Meulen, R., I. Scheirlinck, A. Van Schoor, G. Huys, M. Vancanneyt, P. Vandamme, and L. De Vuyst. 2007. Population dynamics and metabolite target analysis of lactic acid bacteria during laboratory fermentations of wheat and spelt sourdoughs. Appl. Environ. Microbiol. 73:4741-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber, D. G., K. Sahm, T. Polen, V. F. Wendisch, and G. Antranikian. 2008. Oligonucleotide microarrays for the detection and identification of viable beer spoilage bacteria. J. Appl. Microbiol. 105:951-962. [DOI] [PubMed] [Google Scholar]
- 35.Weckx, S., J. Allemeersch, R. Van der Meulen, G. Vrancken, I. Scheirlinck, G. Huys, P. Vandamme, P. Van Hummelen, and L. De Vuyst. 2009. Development and validation of a species-independent functional gene microarray that targets lactic acid bacteria. Appl. Environ. Microbiol. 75:6488-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood, B. J. B. 1998. Microbiology of fermented foods. Blackie Academic & Professional, London, United Kingdom.
- 37.Wu, L., L. Kellogg, A. H. Devol, J. M. Tiedje, and J. Zhou. 2008. Microarray-based characterization of microbial community functional structure and heterogeneity in marine sediments from the Gulf of Mexico. Appl. Environ. Microbiol. 74:4516-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.