Abstract
β-Ketothiolases catalyze the first step of poly(3-hydroxybutyrate) [poly(3HB)] synthesis in bacteria by condensing two molecules of acetyl coenzyme A (acetyl-CoA) to acetoacetyl-CoA. Analyses of the genome sequence of Ralstonia eutropha H16 revealed 15 isoenzymes of PhaA in this bacterium. In this study, we generated knockout mutants of various phaA homologues to investigate their role in and contributions to poly(3HB) metabolism and to suppress biosynthesis of 3HB-CoA for obtaining enhanced molar 3-mercaptopriopionate (3MP) contents in poly(3HB-co-3MP) copolymers when cells were grown on gluconate plus 3-mercaptopropionate or 3,3′-dithiodipropionate. In silico sequence analysis of PhaA homologues, transcriptome data, and other aspects recommended the homologues phaA, bktB, H16_A1713/H16_B1771, H16_A1528, H16_B1369, H16_B0381, and H16_A0170 for further analysis. Single- and multiple-deletion mutants were generated to investigate the influence of these β-ketothiolases on growth and polymer accumulation. The deletion of single genes resulted in no significant differences from the wild type regarding growth and polymer accumulation during cultivation on gluconate or gluconate plus 3MP. Deletion of phaA plus bktB (H16Δ2 mutant) resulted in approximately 30% less polymer accumulation than in the wild type. Deletion of H16_A1713/H16_B1771, H16_A1528, H16_B0381, and H16_B1369 in addition to phaA and bktB gave no differences in comparison to the H16Δ2 mutant. In contrast, deletion of H16_A0170 additionally to phaA and bktB yielded a mutant which accumulated about 30% poly(3HB) (wt/wt of the cell dry weight [CDW]). Although we were not able to suppress poly(3HB) biosynthesis completely, the copolymer compositions could be altered significantly with a lowered percentage ratio of 3HB constituents (from 85 to 52 mol%) and an increased percentage ratio of 3MP constituents (from 15 to 48 mol%), respectively. In this study, we demonstrated that PhaA, BktB, and H16_A0170 are majorly involved in poly(3HB) synthesis in R. eutropha H16. A fourth β-ketothiolase or a combination of several of the other β-ketothiolases contributed to a maximum of only 30% (wt/wt of CDW) of the remaining (co)polymer.
Polyhydroxyalkanoates (PHAs) are naturally occurring polyoxoesters that are synthesized and accumulated as cytoplasmic inclusions by diverse bacteria. Poly(3-hydroxybutyrate) [poly(3HB)] was detected in 1926 by Maurice Lemoigne as an intracellular compound of Bacillus megaterium (16). Generally the accumulation of PHAs proceeds under unbalanced cultivation conditions when a carbon source is available in excess and if another macroelement like nitrogen, phosphorus, or oxygen is limiting growth at the same time (36, 44). Ralstonia eutropha strain H16, a Gram-negative facultative chemolithoautotrophic hydrogen-oxidizing bacterium, accumulates poly(3HB) as insoluble granules as a storage compound for carbon and energy in the cytoplasm. The genome of R. eutropha H16 harbors the PHA operon, which comprises three genes encoding a β-ketothiolase (phaA), an acetoacetyl-CoA-reductase (phaB), and a PHA synthase (phaC) (38). The β-ketothiolase (PhaA) condenses two acetyl coenzyme A (acetyl-CoA) molecules to acetoacetyl-CoA, and a stereospecific acetoacetyl-CoA-reductase (PhaB) reduces the latter to R-(−)-3-hydroxybutyryl-CoA (25). Finally, the PHA synthase (PhaC) polymerizes the 3-hydroxybutyryl moieties of 3HB-CoA to poly(3HB). Besides 3HB, more than 150 different PHA constituents are currently known as components of microbial polyesters (45).
Since 2001, polythioesters (PTEs) consisting of various 3-mercaptoalkanoic acids have been identified which constitute a new class of biopolymers with thioester linkages in the backbone (19). PTEs exhibit interesting physical and biological properties, and their thermal properties, such as melting point temperature or glass transition temperature, deviate significantly from those of the corresponding polyoxoester analogues containing oxoester linkages (13, 21, 22, 46). A recombinant strain of Escherichia coli expressing the artificial BPEC pathway, relying on the enzymes butyrate kinase (Buk) and phosphotransbutyrylase (Ptb) from Clostridium acetobutylicum and a two-component PHA synthase (PhaEC) from Thiococcus pfennigii, synthesizes PTE homopolymers when cultivated in the presence of the respective 3-mercaptoalkanoic acids (17, 18). Thus, E. coli cells harboring a plasmid containing the BPEC genes synthesized poly(3-mercaptopropionate) [poly(3MP)], poly(3-mercaptobutyrate) [poly(3MB)], or poly(3-mercaptovalerate) [poly(3MV)] (21). Additionally, it was discovered that poly(3MP) is practically nonbiodegradable (8, 14). R. eutropha H16 synthesizes copolymers of 3HB and 3MP, 3-mercaptobutyrate (3MB) or 3-mercaptovalerate (3MV), when cells are cultivated in the presence of various thiochemicals (20, 23). Since R. eutropha H16 accumulates poly(3HB) up to 90% of the cell dry weight (CDW) (1), it is an interesting microorganism to produce also other homopolyoxoesters and homopolythioesters like poly(3MP).
The genome of R. eutropha H16 consists of three replicons. The megaplasmid pHG1 and the two chromosomes of R. eutropha H16 were entirely sequenced and the genes annotated, and DNA microarrays were employed (28, 29, 39). The knowledge of the complete genome sequence and transcriptome analyses provided the first complex, in-depth insights into the remarkable metabolic versatility of this bacterium. Genome-wide transcriptome analyses of R. eutropha H16 detected genes that are differentially transcribed during PHB biosynthesis (28). Here, the obtained data were taken into consideration to evaluate the role of different β-ketothiolases in poly(3HB) biosynthesis. In general, the β-ketothiolases/acetyl-CoA acetyltransferases belong to the class of transferases or acyltransferases which transfer groups other than aminoacyl groups. A β-ketothiolase catalyzes the acetylation of acetyl-CoA to acetoacetyl-CoA while one molecule of CoA is released. In addition to the fatty acid and PHA metabolism, β-ketothiolases participate in ketogenesis, sterol synthesis, and propanoate and butanoate metabolism as well as in the degradation of some amino acids like valine, leucine, and isoleucine.
The genome sequence of R. eutropha H16 revealed 14 homologues in addition to phaA (29, 33). Until this study, it was known that, besides PhaA, at least two other β-ketothiolases (BktB, BktC) ought to be active in R. eutropha H16 (11, 43). The condensation of acetyl coenzyme A and propionyl-CoA is required to form β-ketovaleryl-CoA and to synthesize the copolymer poly(3HB-co-3HV). It was assumed that β-ketothiolase PhaA accomplishes this condensation reaction; however, surprisingly, recombinant Escherichia coli harboring the R. eutropha H16 phaCAB operon synthesized only poly(3HB) homopolymer when fed with propionate and acetate (43). Production of poly(3HB-co-3HV) became possible only after the induction of BktB, a β-ketothiolase with a broad substrate specificity, in addition to the phaCAB operon. PhaA seems to be restricted to synthesis of acetoacetyl-CoA, whereas BktB is also capable of synthesizing β-ketovaleryl-CoA (41, 42, 43).
When analyzing other PHA-accumulating bacteria, it is conspicuous that many of them harbor more than one β-ketothiolase homologue in their genomes. Other Ralstonia species, like R. eutropha JMP134, also possess 14 acetyl-CoA acetyltransferases, nearly all identical to those of R. eutropha H16. In phylogenetically closely related bacteria, like species of the genus Burkholderia, multiple β-ketothiolase isoenzymes were also detected. Burkholderia cenocepacia AU 1054 possesses eight β-ketothiolases with high levels of homology to the homologues of R. eutropha H16. Pseudomonas putida KT 2440 possesses altogether nine β-ketothiolase homologues, and Rhodospirillum rubrum ATCC 11170 possesses six different acetyl-CoA acetyltransferases.
In this study, the role of different β-ketothiolases in poly(3HB) metabolism was investigated. It was assumed that by suppressing 3HB-CoA synthesis copolymers with reduced 3HB contents are produced by R. eutropha H16, thereby enhancing the fraction of 3MP in the copolymers. Therefore, the results of an in silico analysis of phaA and of the 14 homologues were related to recently generated transcriptome data (28), to identify those β-ketothiolases which are probably most important for poly(3HB) synthesis in this bacterium. According to these transcriptome data and other information, different β-ketothiolase single- and multiple-deletion mutants of R. eutropha H16 were generated. These mutants were analyzed with regard to growth and accumulation of poly(3HB) homopolymer and poly(3HB-co-3MP) copolymers.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Cells of R. eutropha H16 were cultivated at 30°C in 300 ml mineral salt medium (MSM) which was supplemented with 1% (wt/vol) sodium gluconate as a carbon source in 2-liter Erlenmeyer flasks with baffles (37). The concentration of ammonium chloride was reduced to 0.05% (wt/vol) to provide conditions permissive for PHA accumulation. As a precursor substrate for PTE synthesis, 3-mercaptopropionic acid (99%; Acros Organics) or 3,3′-dithiodipropionic acid (DTDP) (99%; Fluka) was added to the medium to a concentration of 0.05% (vol/vol) or 1% (wt/vol), respectively. Tetracycline was used at a final concentration of 25 μg ml−1. Cells of Escherichia coli were cultivated at 37°C in Luria-Bertani (LB) medium (35). Solid media contained 1.5% (wt/vol) agar-agar. Growth of cells was measured photometrically in a Klett-Summerson photometer (Manostat) using filter no. 54 (520 to 580 nm).
TABLE 1.
Bacterial strains used in this study
| Straina | Description | Reference or source |
|---|---|---|
| Ralstonia eutropha | ||
| H16 | Wild type | DSM 428 |
| ΔphaA | phaA precise-deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔbktB | bktB precise-deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔH16_A1528 | H16_A1528 precise-deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔH16_B1369 | H16_B1369 precise-deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔH16_A0170 | H16_A0170 precise-deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔH16_B0381 | H16_B0381 precise-deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔH16_A1713ΔH16_B1771 | H16_A1713/H16_B1771 precise-deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔphaAΔbktB (Δ2) | phaA and bktB deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔphaAΔbktBΔH16_A0170 (Δ3) | phaA, bktB, and H16_A0170 deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔphaAΔbktBΔH16_A1713ΔH16_B1771 (Δ4) | phaA, bktB, H16_A1713, and H16_B1771 deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔphaAΔbktBΔH16_A1713 ΔH16_B1771ΔH16_B1369 (Δ5) | phaA, bktB, H16_A1713, H16_B1771, and H16_B1369 deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔphaAΔbktBΔH16_A1713 ΔH16_B1771ΔH16_B1369ΔH16_A1528 (Δ6) | phaA, bktB, H16_A1713, H16_B1771, H16_B1369 and H16_A1528 deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔphaAΔbktBΔH16_A1713ΔH16_B1771 ΔH16_B1369ΔH16_A1528ΔH16_A0170 (Δ7) | phaA, bktB, H16_A1713, H16_B1771, H16_B1369, H16_A1528, and H16_A0170 deletion gene replacement strain derived from R. eutropha H16 | This study |
| ΔphaAΔbktBΔH16_A1713ΔH16_B1771 ΔH16_B1369ΔH16_A1528ΔH16_A0170Δ H16_B0381(Δ8) | phaA, bktB, H16_A1713, H16_B1771, H16_B1369, H16_A1528, H16_A0170, and H16_B0381 deletion gene replacement strain derived from R. eutropha H16 | This study |
| PHB−4 | Poly(3HB)-negative mutant | DSM 541 |
| Escherichia coli | ||
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) f80lacZ ΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| S17-1 | thi proA hsdR17 hasdM+ recA RP4 tra function | 40 |
The short-form designations of multiple mutants are shown in parentheses.
Isolation and manipulation of DNA.
Genomic DNA of R. eutropha H16 was isolated by the method of Marmur (24). Plasmid DNA was isolated by the protocol of Birnboim and Doly (3). DNA restriction fragments were purified from agarose gels using the peqGOLD gel extraction kit (Peqlab) by following the instructions of the manufacturer. Ligase and restriction enzymes (Fermentas) were used according to the manufacturer's instructions.
Transfer of DNA.
Plasmids used in this study are listed in Table 2. Competent cells of E. coli were prepared and transformed by the CaCl2 procedure as described by Hanahan (10). Spot agar mating of R. eutropha H16 or mutant derivatives with E. coli S17-1 as a donor was carried out on nutrient broth (NB) agar plates at 30°C. The sacB gene selection was performed on NB agar plates supplemented with 10% sucrose at 30°C.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pJQ200mp18Tc | sacB oriV oriT traJ Tcr | 31 |
| pJQ200mp18Tc::ΔphaA | ΔphaA gene replacement plasmid; Tcr | This study |
| pJQ200mp18Tc::ΔbktB | ΔbktB gene replacement plasmid; Tcr | This study |
| pJQ200mp18Tc::ΔH16_A1713/H16_B1771 | ΔH16_A1713/H16_B1771 gene replacement plasmid; Tcr | This study |
| pJQ200mp18Tc::ΔH16_B1369 | ΔH16_B1369 gene replacement plasmid; Tcr | This study |
| pJQ200mp18Tc::ΔH16_B0381 | ΔH16_B0381 gene replacement plasmid; Tcr | This study |
| pJQ200mp18Tc::ΔH16_A0170 | ΔH16_A0170 gene replacement plasmid; Tcr | This study |
| pJQ200mp18Tc::ΔH16_A1528 | ΔH16_A1528 gene replacement plasmid; Tcr | This study |
PCR amplification.
Amplification of DNA by PCR was done according to the method of Sambrook et al., using Taq DNA polymerase (Invitrogen) in an Omnigene HBTR3CM DNA thermocycler (Hybaid) (35). The oligonucleotides employed for amplification are listed in Table S1 in the supplemental material.
DNA sequencing.
Sequencing reactions of DNA fragments were carried out according to standard procedure in the Institut für Klinische Chemie und Laboratoriumsmedizin (Münster, Germany).
Generation of phaA, bktB, H16_A1713, H16_B1771, H16_A0170, H16_B0381, H16_A1528, and H16_B1369 gene replacement strains employing the sacB system.
The plasmids and oligonucleotides used in this study are listed in Table 2 and Table S1 in the supplemental material, respectively. The flanking regions upstream and downstream (each 400 to 800 bp) of each target gene were amplified by PCR with concomitant introduction of a BamHI or EcoRI restriction site and an XbaI or SacI restriction site. The resulting fragments were digested with BamHI or EcoRI and ligated to yield an approximately 1,000-bp fragment. This fragment was then digested with XbaI or SacI and cloned into the corresponding site of plasmid pJQ200mp18Tc. Correspondingly, all gene replacement plasmids were constructed: pJQ200mp18Tc::ΔphaA, pJQ200mp18Tc::ΔbktB, pJQ200mp18Tc:: ΔH16_A0170, pJQ200mp18Tc::ΔH16_A1528, pJQ200mp18Tc::ΔH16_B0381, pJQ200mp18Tc::ΔH16_B1369, and pJQ200mp18Tc::ΔH16_A1713/H16_B1771. These precise-deletion gene replacement plasmids were then used to generate the corresponding single- or multiple-deletion mutants R. eutropha H16ΔphaA, R. eutropha H16ΔbktB, R. eutropha H16ΔH16_A0170, R. eutropha H16ΔH16_A1528, R. eutropha H16ΔH16_B0381, R. eutropha H16ΔH16_B1369, R. eutropha H16ΔH16_A1713/H16_B1771, R. eutropha H16Δ2, R. eutropha H16Δ3, R. eutropha H16Δ4, R. eutropha H16Δ5, R. eutropha H16Δ6, R. eutropha H16Δ7, and R. eutropha H16Δ8 (see Table 1 for nomenclature of mutants). The generation of deletion mutants was performed by adaptation of standard protocols by using plasmid pJQ200mp18Tc (30, 31). The plasmids were transformed into the donor strain E. coli S17-1 and were from there mobilized into the corresponding R. eutropha H16 recipient strains (12). The identification of mutants was carried out on NB agar plates supplemented with 10% (wt/vol) sucrose and mineral salts medium agar plates containing 25 μg ml−1 tetracycline (30, 31). Correct gene replacement strains were confirmed by PCR analyses and DNA sequencing employing primers which bind beyond the primers used for constructing the deletion gene replacement plasmids.
RNA isolation and cDNA synthesis.
Cells of R. eutropha H16 were harvested after 4 (exponential growth phase), 7 (early stationary phase), and 12 (late stationary phase) hours of cultivation in MSM under storage conditions containing 1% (wt/vol) sodium gluconate and 0.05% (wt/vol) ammonium chloride by centrifugation (15 min, 4,000 U/min, 4°C). The harvested cells were directly shock-frozen in liquid nitrogen and stored at −70°C. Isolation of total RNA was performed as previously described (28) using the RNeasy minikit (Qiagen, Hilden, Germany) and zirconia-silica beads (Roth, Karlsruhe, Germany).
Subsequently, 25 μg total RNA was used for random hexamer-primed synthesis of fluorescence-labeled cDNA using the CyScribe first-strand cDNA labeling kit and the fluorescent nucleotide analogues FluoroLink Cy3-dUTP and Cy5-dUTP (GE Healthcare).
Microarray hybridization and scanning.
Full genomic R. eutropha H16 oligonucleotide microarrays were used for transcription analyses (28). To achieve reliable data, each hybridization was repeated on a second slide using a reverse combination of dye labels (7). Portions of 80 pmol of each incorporated fluorescent dye were used for microarray hybridization on CodeLink activated slides (SurModics Inc., Eden Prairie, MN). The labeled cDNA was denatured at 98°C for 5 min, hybridization buffer was added up to 220 μl, and the hybridization was carried out for 15 h at 58°C using an automatic slide processor (Lucidea SlidePro hybridization chamber; GE Healthcare) (28). Microarray images were acquired for both channels (Cy3/Cy5) using a GenePix 4000B array scanner, and the image files were analyzed using GenePix Pro software (Axon Instruments).
Data normalization and filtering.
Data were normalized by multiplication of a constant factor so that the mean of the ratio of medians of all features became equal to 1. Arrays with normalization factors larger than 2 were excluded from further analysis. After the fluorescence intensities of both channels were determined, a background correction was made by subtracting the local background value from the foreground intensity (2). Data with an intensity value smaller than the background in both channels were excluded from further analyses. To exclude irregular spots, dye precipitates, or misplaced feature indicators, the ratio of medians, the ratio of means, and the regression ratios were calculated for each feature. When these ratios deviated more than 30% from each other, the corresponding feature was excluded from further analyses.
Analysis of PHA content.
Cells of R. eutropha were harvested by centrifugation (15 min, 6,000 × g, 4°C), washed in 0.9% (wt/vol) sodium chloride, and then lyophilized for 24 h. The PHA contents of the cells were determined upon methanolysis of 5 to 10 mg lyophilized cells in the presence of 85% (vol/vol) methanol and 15% (vol/vol) sulfuric acid. The resulting methyl esters of 3HB and 3MP were analyzed by gas chromatography as described previously (4, 48).
Sequence data analysis.
The sequences of the homologues were extracted from the National Center for Biotechnology Information (NCBI) nucleotide sequence database (http://www.ncbi.nlm.nih.gov/GenBank/index.html). Protein and nucleotide sequences were aligned using CLUSTAL W program with PhaA/phaA as the reference.
Microarray data accession number.
Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-2440.
RESULTS
In silico analysis of the β-ketothiolase homologues of R. eutropha H16.
Before the genome sequence of R. eutropha H16 was published in 2006 (29), it was supposed that at least four β-ketothiolases exist in this bacterium: PhaA, BktB, BktC, and a fourth β-ketothiolase which is expressed only if the cells are cultivated in the presence of fatty acids as a carbon source (43). Surprisingly, the analysis of the genome sequence revealed 15 homologues in total. In Table 3, all 15 homologues present in R. eutropha H16 are listed.
TABLE 3.
Overview of β-ketothiolases in R. eutropha H16
| Homologue | Gene | Protein | e-valuea | No. of nucleotides/no. of amino acids | Mol wt | % identitya at: |
Gene expressionb | |
|---|---|---|---|---|---|---|---|---|
| Amino acid level | Nucleotide level | |||||||
| 1 | H16_A1438 (phaA) | Acetyl-CoA acetyltransferase | 0.0 | 1,182/393 | 40,548.8 | 100 | 100 | + |
| 2 | H16_A1445 (bktB) | β-Ketothiolase | 3e−95 | 1,185/394 | 40,903.8 | 52 | 65 | + |
| 3/4 | H16_A1713/H16_B1771 | Acetyl-CoA acetyltransferase | 8e−87 | 1,188/395 | 40,827.5 | 47 | 58 | + |
| 5 | H16_A0170 | Acetyl-CoA acetyltransferase | 1e−85 | 1,179/392 | 41,031.1 | 47 | 62 | + |
| 6 | H16_B0381 | Acetyl-CoA acetyltransferase | 9e−81 | 1,179/392 | 40,713.6 | 45 | 60 | − |
| 7 | H16_B1369 | Acetyl-CoA acetyltransferase | 3e−75 | 1,209/402 | 43,081.1 | 44 | 60 | − |
| 8 | H16_B0200 (pcaF) | β-Ketoadipyl CoA thiolase | 3e−71 | 1,203/400 | 41,889.0 | 44 | 59 | − |
| 9 | H16_A1528 | Acetyl-CoA acetyltransferase | 2e−53 | 1,179/392 | 41,057.1 | 39 | 57 | + |
| 10 | H16_A0462 | Acetyl-CoA acetyltransferase | 4e−55 | 1,197/398 | 41,776.1 | 38 | 58 | − |
| 11 | H16_B0668 | Acetyl-CoA acetyltransferase | 3e−55 | 1,203/400 | 41,992.9 | 40 | 56 | − |
| 12 | H16_A1720 | Acetyl-CoA acetyltransferase | 2e−52 | 1,182/393 | 41,500.4 | 41 | 55 | − |
| 13 | H16_B0759 | Acetyl-CoA acetyltransferase | 8e−52 | 1,176/391 | 40,515.5 | 38 | 56 | − |
| 14 | H16_B0662 | Acetyl-CoA acetyltransferase | 5e−50 | 1,152/383 | 40,436.2 | 37 | 54 | − |
| 15 | H16_A1887 | Acetyl-CoA acetyltransferase | 2e−60 | 1,179/392 | 41,525.3 | 37 | 54 | − |
The e-value (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and amino acid and nucleotide acid identities are with respect to phaA.
Gene expression in R. eutropha H16 as taken from the raw data set obtained by Peplinski et al. (28). +, gene expression was detected; −, gene expression was not detected.
With regard to their substrate specificity, two groups of β-ketothiolases are distinguished. Members of the first group are involved in biosynthetic pathways like those for PHAs or isoprenoids and are specific for acetoacetyl-CoA. In contrast, members of the second group exhibit a broader carbon chain length specificity and are involved in fatty acid β-oxidation (27). During growth on fructose and propionate, PhaA is essentially restricted to the synthesis of acetoacetyl-CoA, whereas BktB produces primarily β-ketovaleryl-CoA; the latter enzyme is, however, able to complement PhaA for poly(3HB) synthesis in E. coli (43). An alignment of the 15 homologues revealed two highly conserved cysteine residues at positions 88 and 378 which were already previously described (26, 47). In addition to these two cysteine residues, a histidine residue at position 348 is located in a conserved region (Fig. 1). These three amino acids take part in formation of an active site cavity.
FIG. 1.
Multiple-amino-acid alignment of PhaA and of its homologues in R. eutropha H16. Identical amino acids are highlighted in black, while conserved and similar amino acids are highlighted in dark or light gray, respectively. Labeled amino acids putatively form a catalytic cavity.
The aims of this study were to see whether and to what extent poly(3HB) biosynthesis can be eliminated in R. eutropha H16 and to learn about the contribution of the different β-ketothiolases to poly(3HB) synthesis. Therefore, different single- and multiple-deletion mutants of selected β-ketothiolases were constructed to identify the isoenzymes involved in PHA metabolism. The target genes were selected by amino acid sequence homology in combination with the available transcriptome data of R. eutropha H16 (28).
Identification of β-ketothiolases relevant for PHA metabolism by available transcriptome data.
To experimentally evaluate the in silico predictions and to obtain hints which β-ketothiolase may be involved in PHA metabolism and which not, recently obtained transcriptome data were taken into consideration. The data obtained during genome-wide transcriptome analyses of R. eutropha H16 applying DNA microarrays detected genes that are differentially transcribed during PHB biosynthesis (28). The detection of only one (PhaA) of the 15 putative β-ketothiolases is due to a 3-fold cutoff value, which was set to identify only genes with significant regulation. In the present study, all data obtained for the putative β-ketothiolases were taken into consideration, regardless of a cutoff value, because our interest was not the extent of the transcription level but whether a particular β-ketothiolase was expressed at all. Thus, the transcriptome data revealed an expression for 6 of the 15 genes encoding putative β-ketothiolases: phaA, bktB, H16_A1713/H16_B1771, H16_A0170, and H16_A1528 (Table 3). For PhaA, the capability of synthesizing acetoacetyl-CoA has already been shown (43). H16_A1713 and H16_B1771 represent completely identical phaA homologues, as mentioned above, whose products exhibit 46.7% amino acid identity with respect to PhaA (Table 3). bktB is known to encode a β-ketothiolase with a broad substrate specificity (43), and H16_A0170 and H16_A1528 represent homologues of phaAy with products sharing 47 and 38.6% amino acid identity (Table 3).
As a conclusion of the results of the transcriptome analyses, we decided to delete the genes phaA, bktB, H16_A1713/H16_B1771, H16_A0170, and H16_A1528 in this bacterium. The other nine β-ketothiolases were not detected.
In addition to these six β-ketothiolases, which were selected for further analysis, Raberg et al. determined during proteome analysis of similar cell samples of the wild type that the amount of homologue H16_B1369 increased significantly from the exponential to the stationary growth phase after 21 h of cultivation (32). Therefore, the gene of this homologue was also included in the deletion procedure. After the deletion of the homologue H16_A0170 in the multiple mutants H16Δ3 and H16Δ7 and determination of the resultant effect on poly(3HB) synthesis, we also decided to delete the closest homologue to H16_A0170. For that reason H16_B0381, the fifth homologue in Table 3, was also chosen for deletion.
Generation of β-ketothiolase single- and multiple-deletion mutants.
All mutants generated in this study are precise-deletion mutants established by gene replacements, which employed recombinant pJQ200mp18Tc suicide vectors harboring the upstream and downstream regions of the respective target gene. This vector comprises the sacB system from Bacillus subtilis, which is induced by adding sucrose to the medium and which is lethal when expressed in Gram-negative bacteria. This sacB system can be applied for the generation of multiple R. eutropha H16 deletion mutants and to avoid the introduction of antibiotic resistance markers (31). Tetracycline resistance is used for the selection of homogenotes by revealing those R. eutropha H16 cells that still harbor the suicide vector. All mutants were generated as described in Materials and Methods. The first set of mutants established in this study comprised seven mutants: six mutants were defective in only one gene, phaA, bktB, H16_A0170, H16_A1528, H16_B0381, or H16_B1369, and one mutant was defective in two genes. The latter was the H16_A1713/H16_B1771 double mutant, which because of the deletion of two identical homologues resembled a single β-ketothiolase mutant. All other mutants comprised multiple ß-ketothiolase homologue gene deletions, i.e., mutants which were defective in at least two and up to eight β-ketothiolase genes: H16Δ2, H16Δ3, H16Δ4, H16Δ5, H16Δ6, H16Δ7, and H16Δ8. All mutants and their nomenclature are shown in Table 1.
Growth and PHA accumulation of β-ketothiolase mutants.
To analyze the growth behavior and the ability of the mutants to synthesize poly(3HB) or the sulfur-containing copolymer poly(3HB-co-3MP), the various mutants defective in different β-ketothiolases and the wild type were cultivated under storage conditions permitting polymer accumulation in liquid MSM containing 1.0% (wt/vol) sodium gluconate and 0.05% (wt/vol) ammonium chloride. To enable biosynthesis of the copolymer, poly(3HB-co-3MP), 0.05% 3MP (vol/vol) or 1% DTDP (wt/vol) was added to the medium. In R. eutropha H16, DTDP is cleaved into two molecules of 3MP and is then further metabolized (23). Samples were taken after 12 h, 24 h, 36 h, and 48 h from each culture to analyze the polymer contents. All experiments were done in duplicate as described in Materials and Methods.
Cultivation on gluconate.
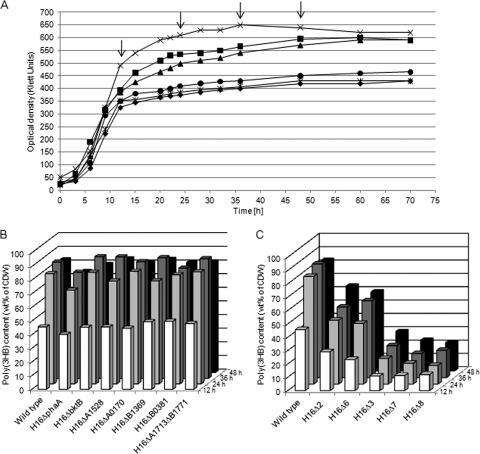
Cells of the wild-type strain H16 of R. eutropha and of all single mutants showed similar growth behavior when cultivated in MSM containing gluconate as the sole carbon source. After 12 h of incubation, the increase of optical density of the H16ΔphaA culture became slightly slower than those of the wild type and of the other single mutants. The poly(3HB) contents in the single-deletion strains were nearly identical to that of the wild type in all four samples withdrawn during the time course of the experiment (Fig. 2B). Cultures of the wild type and of the single mutants H16ΔbktB, H16ΔH16_A1528, H16ΔH16_A0170, H16ΔH16_B0381, H16ΔH16_B1369, and H16ΔH16_A1713/ΔH16_B1771 reached after 30 h the same cell densities of about 600 Klett units, and the poly(3HB) contents of the cells were about 80% of their cell dry weight after 36 h. In contrast, mutant H16ΔphaA accumulated approximately 10% less poly(3HB) than the others (Fig. 2B).
FIG. 2.
Growth behavior (A) and PHB accumulation (B and C) of R. eutropha H16 and various mutants in MSM under storage conditions containing 1.0% (wt/vol) sodium gluconate. After 12, 24, 36, and 48 h poly(3HB) contents were analyzed by gas chromatography. (A) Growth of the wild type (×) and of the mutants H16Δ2 (▪), H16Δ3 (•), H16Δ6 (▴), H16Δ7 (⧫), and H16Δ8 (*). The arrows indicate the times of sample drawing. (B and C) Poly(3HB) contents of cells of strains as indicated and as shown in panels B (single mutants) or C (multiple mutants), respectively. Samples were withdrawn after 12 h (white bars), 24 h (light gray bars), 36 h (dark gray bars) or 48 h (black bars).
Cultures of the double mutant H16Δ2 lacking phaA plus bktB reached cell densities of only about 550 Klett units after 36 h, and the gas chromatographic analysis revealed about 30% less poly(3HB) in the cells than in any single mutant and in the wild type (Fig. 2C). Mutants H16Δ2, H16Δ4, H16Δ5, and H16Δ6 showed growth and poly(3HB) accumulation behaviors similar to those of H16Δ2, which were already mentioned above.
A deletion of H16_A0170 in addition to phaA and bktB yielded an optical density of only about 400 to 450 Klett units of the respective culture and a poly(3HB) content of the cells of only approximately 30% (wt/wt) of CDW after 48 h of incubation (Fig. 2A and C). The deletion of seven homologues in H16Δ7 yielded a much smaller amount of accumulated poly(3HB) after 48 h (20% [wt/wt] of CDW). The additional deletion of the homologue H16_B0381 in H16Δ8 showed in comparison to H16Δ7 no significant difference in the amount of accumulated poly(3HB) (Fig. 2C). The phenotypes of the mutants cultivated on solid MSM agar plates containing 1.5% (wt/vol) sodium gluconate and 0.05% (wt/vol) ammonium chloride differed in opacity (see Fig. S1 in the supplemental material). These differences in the phenotypes are due to the different poly(3HB) contents of the cells.
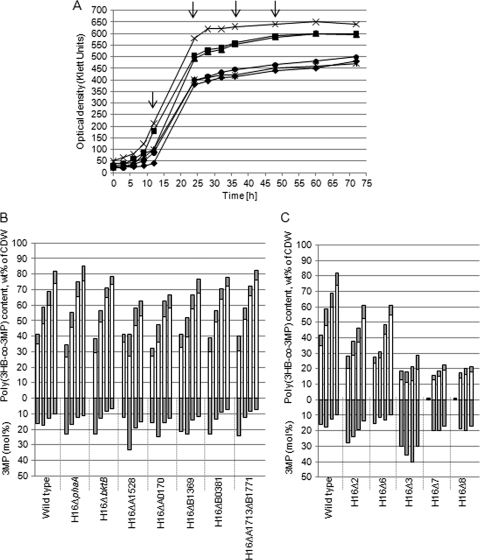
Cultivation on gluconate plus 3MP or DTDP as a precursor substrate.
All strains grew more slowly in MSM containing 1% (wt/vol) sodium gluconate and 0.05% (vol/vol) 3MP as the precursor substrate under storage conditions than in MSM containing only sodium gluconate (Fig. 3A). While after 36 h the highest poly(3HB) contents were obtained in cells of the gluconate cultures, the highest poly(3HB-co-3MP) contents of cells were obtained after 48 h in cultures containing gluconate plus 3MP (Fig. 3B and C). The wild type and the single mutants H16ΔphaA, H16ΔbktB, H16ΔH16_B1369, H16ΔH16_B0381, and H16ΔH16_A1713ΔH16_B1771 exhibited similar growth and storage behaviors (Fig. 3B). In contrast to cells cultivated in the presence of gluconate alone, cells of the single mutants H16ΔH16_A1528 and H16ΔH16_A0170 accumulated after 48 h only about 60% copolymer, whereas in cells of the other single mutants and of the wild type poly(3HB-co-3MP) was accumulated up to 80% (wt/wt) of the CDW. Deletion of the homologues phaA and bktB resulted in a different polymer composition with a significantly lower molar fraction of 3HB constituents. After 36 to 48 h, the copolyester in the cells of the multiple β-ketothiolase mutants H16Δ2 (Fig. 3C), H16Δ4, and H16Δ5 (data are not shown) consisted of 15 to 20 mol% 3MP (wt/wt) constituent, whereas the copolyester in the wild type consisted of only about 10 mol% 3MP. H16Δ6 stored altogether 60% (wt/wt of CDW) copolymer but incorporated a smaller amount of 3MP constituents (10 mol%) than the above-mentioned mutants H16Δ2, H16Δ4, and H16Δ5. The additional deletion of the homologue H16_A0170 in H16Δ7 led to slower growth, lower optical density, and less polymer accumulation. The highest optical density reached was about 430 Klett units, and the highest yield of copolymer was approximately 20% (wt/wt) of CDW after 48 h (Fig. 3A and C). The incorporation of 3MP (20 mol%) into the copolymer after 36 h to 48 h of in H16Δ7 was again higher than in the wild type, whereas the total amount of the produced copolymer decreased. Also here, the additional deletion of H16_B0381 in mutant H16Δ8 led to no significant difference in the amount of accumulated copolymer in comparison to H16Δ7 (Fig. 3C).
FIG. 3.
Growth behavior (A) and poly(3HB-co-3MP) accumulation (B and C) of the wild type and of the mutants of R. eutropha H16 in MSM under storage conditions containing 1.0% (wt/vol) sodium gluconate and 0.05% (vol/vol) 3MP. After 12, 24, 36, and 48 h, poly(3HB-co-3MP) contents of the cells of strains were analyzed by gas chromatography. (A) Growth of the wild type (×) and of the mutants H16Δ2 (▪), H16Δ3 (•), H16Δ6 (▴), H16Δ7 (⧫), and H16Δ8 (*). The arrows indicate the times of sample drawing. (B [single mutants] and C [multiple mutants]) Upper y axis: poly(3HB-co-3MP) content of cells. Bars for each strain from the left to the right follow the times at which samples were withdrawn from the cultivation vessel for analysis (12, 24, 36, and 48 h). White bars indicate the 3HB contents (wt% of CDW) in the copolymers, whereas gray bars indicate the 3MP contents (wt% of CDW) in the copolymers. The lower y axis shows the molar 3MP contents of the corresponding copolymer bars above.
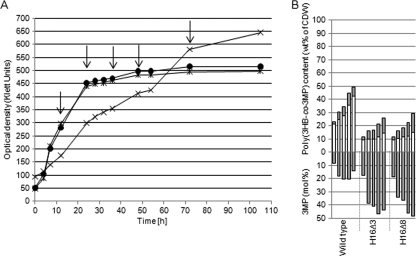
Mutant H16Δ3 lacking the genes phaA, bktB, and H16_A0170 showed a growth behavior similar to that of the 7-fold mutant, but the fraction of 3MP in the copolymer was, in contrast to H16Δ7, higher (30 to 40 mol%) after 36 h to 48 h of cultivation (Fig. 3C). Selected mutants were also tested on DTDP as a precursor substrate at a concentration of 1% (wt/vol). As shown in Fig. 4A, growth of the mutants H16Δ3 and H16Δ8, in contrast to that of the wild type, was not inhibited. The wild type accumulated 49% (wt/wt of CDW), approximately half as much copolymer as cultivated on 3MP as a precursor, whereas the mutants synthesized the same amount but with higher molar fractions of 3MP (Fig. 4B). After 48 h, the copolymer of the wild type contained approximately 15 mol% of 3MP while the mutant's copolymer consisted of almost 50% (mol/mol) of each constituent.
FIG. 4.
Growth behavior (A) and poly(3HB-co-3MP) accumulation (B) of the wild type and of the mutants of R. eutropha H16 in MSM under storage conditions containing 1.0% (wt/vol) sodium gluconate and 1% (wt/vol) DTDP. After 12, 24, 36, 48, and 72 h, the poly(3HB-co-3MP) contents of the cells were analyzed by gas chromatography. (A) Growth of the wild type (×) and of the mutants H16Δ3 (•) and H16Δ8 (*). The arrows indicate the times of sample drawing. (B) Upper y axis: poly(3HB-co-3MP) content of cells. Bars for each strain from the left to the right follow the times at which samples were withdrawn from the cultivation vessel for analysis (12, 24, 36, 48, and 72 h). White bars indicate the 3HB contents (wt% of CDW) in the copolymers, whereas gray bars indicate the 3MP contents (wt% of CDW) in the copolymers. The lower y axis shows the molar 3MP contents of the corresponding copolymer bars above.
Residual β-ketothiolase activity in H16Δ7 was additionally studied, and a protein exhibiting the capability to catalyze a conversion of acetoacetyl-CoA to CoA was purified and sequenced (matrix-assisted laser desorption ionization-time of flight mass spectrometry [MALDI-TOF]). H16_B0200 (pcaF) appeared to be active in this mutant (unpublished data).
DISCUSSION
Recent studies and knowledge of the R. eutropha H16 genome sequence revealed that the poly(3HB) metabolism in this bacterium is much more complex than previously assumed (29). Overall, 15 homologues can be classified as acetyl-CoA acetyltransferases (Table 3). Numerous homologues of several other proteins relevant for the metabolism of poly(3HB) also exist in this bacterium.
The detailed in silico analysis revealed that the identity at the amino acid level in comparison to PhaA varies from 37% to 52% while at the nucleotide level it varies from 54% to 65%. This shows that homologues might have accumulated nonsynonymous substitutions after gene duplications. The theory of gene duplication states that the functionally redundant gene will be released from selective constraints (15). Moreover, the homologues H16_A1713 and H16_B1771 (homologues 3 and 4 in Table 3) are completely identical. Interestingly, the corresponding genes are located on different chromosomes. Each of these homologues has one putative transcriptional regulator gene upstream (H16_A1712/H16_B1772) and two genes downstream, encoding an acyl dehydratase (H16_A1714/H16_B1770) and a ketopantoate reductase (apbA1/apbA3), that are also identical. The genes H16_A1713 and H16_B1771 and the adjacent genes might be paralogues resulting from a very recent gene duplication. It has been described that gene duplications occur as an evolutionary progression in microorganisms that have been exposed to different selection pressures such as starvation and stress conditions (5, 34). By duplication of genes, the adaptation to changing environmental conditions can be facilitated. Gevers et al. observed that in bacteria the products of most duplicated genes take part in transcription, metabolism, or defense mechanisms and that small-scale duplications, such as tandem or operon duplications, predominate (9).
In previous studies PhaA and BktB were already isolated and characterized in detail, and a third homologue referred to as BktC was detected (11, 43). Due to the putative presence of 15 β-ketothiolase isoenzymes in crude extracts it may be assumed that at least some previously isolated β-ketothiolase samples consisted not of only one single β-ketothiolase but of a mixture of several β-ketothiolases. For instance, PhaA appears to be essentially limited to biosynthesis of acetoacetyl-CoA, while BktB also synthesizes β-ketovaleryl-CoA during growth on fructose and propionate (43). Therefore, the existence of 15 β-ketothiolase candidates might point to a potential for synthesis of several other polyesters containing various constituents other than 3-hydroxybutyric acid, e.g., 3-hydroxypropionic acid, 3-hydroxyvaleric acid, or 3-mercaptopropionic acid.
PTEs are being discussed for medical devices and applications because of their high thermal stability and putative antibacterial properties and could be therefore of biotechnological interest. One aim of this study was also to suppress synthesis of 3HB-CoA in R. eutropha H16 by inactivating β-ketothiolases to synthesize poly(3HB-co-3MP) copolymers with an elevated 3MP ratio or even 3MP homopolymers, both containing more sulfur in the backbone. A complete suppression of 3HB biosynthesis was not possible, but it could be drastically reduced from 80% (wt/wt of CDW) in the wild type to approximately 30% (wt/wt of CDW) in H16Δ3 and even to 20% (wt/wt of CDW) in the mutants H16Δ7 and H16Δ8 (Fig. 2C). Therefore, the molar composition of the synthesized copolymer was modified in the multiple mutants H16Δ3 and H16Δ8 to nearly 50 mol% 3HB and 50 mol% 3MP instead of 85 mol% 3HB and 15 mol% 3MP in the wild type (Fig. 4B).
Based on recently obtained transcriptome data, putative β-ketothiolases, which were expressed under conditions permissive for PHA accumulation, were determined. For six homologues in total, the expression was detected: phaA, bktB, H16_A1713/H16_B1771, H16_A0170, and H16_A1528. For all other nine putative β-ketothiolases, no mRNA could be detected. Obviously, the genes are not expressed under the conditions investigated. Whether they are silent genes or whether they are expressed only under other cultivation conditions, for example, during growth on a particular carbon source, is not known.
To gain more information about the functions of the expressed isoenzymes and their contributions to PHA biosynthesis in R. eutropha, deletion mutants were generated, and the impacts of the presence or absence of these β-ketothiolases on growth and polymer content were investigated. The deletion of the homologues phaA plus bktB in the double mutant H16Δ2 led to approximately 30% less polymer accumulation than in the wild type, whereas the single-deletion mutants H16ΔphaA and H16ΔbktB exhibited no significant effect. Slater et al. showed that bktB is able to complement phaA for PHA production in Escherichia coli (43). This is also true in R. eutropha: phaA and bktB are capable of complementing each other in synthesis of acetoacetyl-CoA. The deletions in the multiple mutants H16Δ4, H16Δ5, and H16Δ6 did not affect the ability to synthesize acetoacetyl-CoA to a greater extent than in the double mutant H16Δ2. Possibly the homologues H16_A1713/H16_B1771, H16_1528, and H16_B1369, which are defective in these mutants, have a function in fatty acid degradation or they are expressed under other cultivation conditions during growth on different carbon sources. This might also be true for the homologue H16_B0381: the multiple mutant H16Δ8 did not accumulate less polymer than its parent strain H16Δ7.
R. eutropha H16 accumulates poly(3HB-co-3HHx) and poly(3HB-co-3HV) when even- and odd-numbered fatty acids, respectively, are provided as the carbon source (6). The deletion of the homologue H16_A0170 in H16Δ7 and in H16Δ3, in which phaA and bktB were already deleted, resulted in less polymer accumulation, with poly(3HB) only 20 to 30% of the cell dry weight. Thus, we can conclude that the acetyl-CoA acetyltransferases PhaA, BktB, and H16_A0170 are functionally active enzymes during synthesis of 3HB-CoA. Isoenzyme H16_A0170 might be the third enzyme found by Slater and coworkers (BktC) in 1998 (43). The deletion of only one of these three homologues did not result in less poly(3HB) accumulation because the missing homologue could then be completely complemented by one or more of the other β-ketothiolases.
Despite the circumstances that several (up to eight) homologues were deleted, poly(3HB) synthesis was still not fully suppressed; even the multiple mutant in which eight β-ketothiolase homologues were deleted could accumulate poly(3HB) up to about 20% (wt/wt) of the CDW. Whereas the homologues phaA, bktB, and H16_A0170 were identified as encoding major β-ketothiolases contributing to PHA synthesis in this study, at least one additional acetyl-CoA acetyltransferase must be involved in poly(3HB) synthesis in R. eutropha H16. Which of the other multiple homologues contributes to acetoacetyl-CoA synthesis in R. eutropha under conditions permissive for poly(3HB) accumulation remained unknown, though one possible candidate may be pcaF, which seems to be still active in H16Δ7. Very interestingly, many PHA-accumulating bacteria, like R. solanacearum, R. rubrum ATCC 11170, and P. putida KT2440, possess six or even nine β-ketothiolase isoenzymes. It is obvious that synthesis of acetoacetyl-CoA for poly(3HB) biosynthesis not only relies on one β-ketothiolase but is mediated by several of the isoenzymes. Continuative further studies would be necessary to fully unravel the functions of the different β-ketothiolase isoenzymes in R. eutropha and other bacteria. This study demonstrated how this question can be addressed and how a combined approach of isolating defined single and multiple mutants, of transcriptome analyses, of physiological investigations, and of other methods was able to identify the few most relevant β-ketothiolases that are available for this bacterium.
Supplementary Material
Acknowledgments
We thank Silke Özer and Christina Döring for technical assistance with DNA microarrays.
This study was supported by a grant provided by the Bundesministerium für Bildung und Forschung (BMBF, Förderkennzeichen 0313751E).
Footnotes
Published ahead of print on 2 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benes, V., and M. Muckenthaler. 2003. Standardization of protocols in cDNA microarray analysis. Trends Biochem. Sci. 28:244-249. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 277:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandl, H., R. A. Gross, R. W. Lenz, and R. C. Fuller. 1988. Pseudomonas oleovorans as a source of poly(ß-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caporale, L. H. 2003. Natural selection and the emergence of a mutation phenotype: an update of the evolutionary synthesis considering mechanisms that affect genome variation. Annu. Rev. Microbiol. 57:467-485. [DOI] [PubMed] [Google Scholar]
- 6.Dennis, D., M. McCoy, A. Stangl, H. E. Valentin, and Z. Wu. 1998. Formation of poly(3-hydroxybutyrate-co-hydroxyhexanoate) by PHA synthase from Ralstonia eutropha. J. Biotechnol. 64:177-186. [DOI] [PubMed] [Google Scholar]
- 7.Ehrenreich, A. 2006. DNA microarray technology for the microbiologist: an overview. Appl. Microbiol. Biotechnol. 73:255-273. [DOI] [PubMed] [Google Scholar]
- 8.Elbanna, K., T. Lütke-Eversloh, D. Jendrossek, H. Luftmann, and A. Steinbüchel. 2004. Studies on the biodegradability of polythioester copolymers and homopolymers by polyhydroxyalkanoate (PHA)-degrading bacteria and PHA depolymerases. Arch. Microbiol. 182:212-225. [DOI] [PubMed] [Google Scholar]
- 9.Gevers, D., K. Vandepoele, C. Simillion, and Y. Van de Peer. 2004. Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends Microbiol. 44:148-154. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Haywood, G. W., A. J. Anderson, L. Chu, and E. A. Dawes. 1988. Characterization of two 3-ketothiolases possessing differing substrate specificities in the polyhydroxyalkanoate synthesizing organism Alcaligenes eutrophus. FEMS Microbiol. Lett. 52:91-96. [Google Scholar]
- 12.Hogrefe, C., D. Römermann, and B. Friedrich. 1981. Alcaligenes eutrophus hydrogenase genes (Hox). J. Bacteriol. 158:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawada, J., T. Lütke-Eversloh, A. Steinbüchel, and R. H. Marchessault. 2003. Physical properties of microbial polythioesters: characterization of poly(3-mercaptoalkanoates) synthesized by engineered Escherichia coli. Biomacromolecules 4:1698-1702. [DOI] [PubMed] [Google Scholar]
- 14.Kim, D. Y., T. Lütke-Eversloh, K. Elbanna, N. Thakor, and A. Steinbüchel. 2005. Poly(3-mercaptopropionate): a nonbiodegradable biopolymer? Biomacromolecules 6:897-901. [DOI] [PubMed] [Google Scholar]
- 15.Kondrashov, F. A., I. B. Rogozin, Y. I. Wolf, and E. V. Koonin. 2003. Selection in the evolution of gene duplications. Genome Biol. 3:0008.1-0008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemoigne, M. 1926. Produits de deshydration et de polymerization de l'ácide β-oxybutyrique. Bull. Soc. Chim. Biol. 8:770-782. [Google Scholar]
- 17.Liu, S. J., and A. Steinbüchel. 2000. Exploitation of butyrate kinase and phosphotrans-butyrylase from Clostridium acetobutylicum for the in vitro biosynthesis of poly(hydroxyalkanoic acid). Appl. Microbiol. Biotechnol. 53:545-552. [DOI] [PubMed] [Google Scholar]
- 18.Liu, S. J., and A. Steinbüchel. 2000. A novel genetically engineered pathway for synthesis of poly(hydroxyalkanoic acids) in Escherichia coli. Appl. Environ. Microbiol. 66:739-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lütke-Eversloh, T., K. Bergander, H. Luftmann, and A. Steinbüchel. 2001. Identification of a new class of biopolymer: bacterial synthesis of a sulfur-containing polymer with thioester linkages. Microbiology 147:11-19. [DOI] [PubMed] [Google Scholar]
- 20.Lütke-Eversloh, T., K. Bergander, H. Luftmann, and A. Steinbüchel. 2001. Biosynthesis of poly(3-hydroxybutyrate-co-3-mercaptobutyrate) as a sulfur analogue to poly(3-hydroxybutyrate) (PHB). Biomacromolecules 2:1061-1065. [DOI] [PubMed] [Google Scholar]
- 21.Lütke-Eversloh, T., A. Fischer, U. Remminghorst, J. Kawada, R. H. Marchessault, A. Bögershausen, M. Kalwei, H. Eckert, R. Reichelt, S. J. Liu, and A. Steinbüchel. 2002. Biosynthesis of novel thermoplastic polythioesters by engineered Escherichia coli. Nat. Mater. 1:236-240. [DOI] [PubMed] [Google Scholar]
- 22.Lütke-Eversloh, T., J. Kawada, H. Marchessault, and A. Steinbüchel. 2002. Characterization of microbial polythioesters: physical properties of novel copolymers synthesized by Ralstonia eutropha. Biomacromolecules 3:159-166. [DOI] [PubMed] [Google Scholar]
- 23.Lütke-Eversloh, T., and A. Steinbüchel. 2003. Novel precursor substrates for polythioesters (PTE) and limits of PTE synthesis in Ralstonia eutropha. FEMS Microbiol. Lett. 221:191-196. [DOI] [PubMed] [Google Scholar]
- 24.Marmur, J. 1961. A procedure for the isolation of desoxyribonucleic acids from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 25.Oeding, V., and H. G. Schlegel. 1973. β-Ketothiolase from Hydrogenomonas eutropha H16 and its significance in the regulation of poly-β-hydroxybutyrate metabolism. Biochem. J. 134:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer, M. A. J., E. Differding, R. Gamboni, S. F. Williams, O. P. Peoples, C. T. Walsh, A. J. Sinskey, and S. Masamune. 1991. Biosynthetic thiolase from Zoogloea ramigera: evidence for a mechanism involving CYS-378 as the active site base. J. Biol. Chem. 266:8369-8375. [PubMed] [Google Scholar]
- 27.Peoples, O. P., and A. J. Sinskey. 1989. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Characterization of the genes encoding β-ketothiolase and acetoacetyl-CoA reductase. J. Biol. Chem. 264:15293-15297. [PubMed] [Google Scholar]
- 28.Peplinski, K., A. Ehrenreich, C. Döring, M. Bömeke, F. Reinecke, C. Hutmacher, and A. Steinbüchel. 2010. Genome-wide transcriptome analyses of the “Knallgas” bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology 156:2136-2152. [DOI] [PubMed] [Google Scholar]
- 29.Pohlmann, A., W. F. Fricke, F. Reinecke, B. Kusian, H. Liesegang, R. Cramm, T. Eitinger, C. Ewering, M. Pötter, E. Schwartz, A. Strittmatter, I. Voß, G. Gottschalk, A. Steinbüchel, B. Friedrich, and B. Bowien. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24:1257-1262. [DOI] [PubMed] [Google Scholar]
- 30.Pötter, M., H. Müller, and A. Steinbüchel. 2005. Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology 151:825-833. [DOI] [PubMed] [Google Scholar]
- 31.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 32.Raberg, M., F. Reinecke, R. Reichelt, U. Malkus, S. König, M. Pötter, W. F. Fricke, A. Pohlmann, B. Voigt, M. Hecker, B. Friedrich, B. Bowien, and A. Steinbüchel. 2008. Ralstonia eutropha H16 flagellation changes according to nutrient supply and state of poly(3-hydroxybutyrate) accumulation. Appl. Environ. Microbiol. 74:4477-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinecke, F., and A. Steinbüchel. 2009. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J. Mol. Microbiol. Biotechnol. 16:91-108. [DOI] [PubMed] [Google Scholar]
- 34.Riehle, M. M., A. F. Bennett, and A. D. Long. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 98:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Schlegel, H. G., G. Gottschalk, and V. Bartha. 1961. Formation and utilization of poly-β-hydroxybutyric acid by knallgas bacteria (Hydrogenomonas). Nature 29:463-465. [DOI] [PubMed] [Google Scholar]
- 37.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209-222. [PubMed] [Google Scholar]
- 38.Schubert, P. A., A. Steinbüchel, and H. G. Schlegel. 1988. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-β-hydroxybutyrate. J. Bacteriol. 170:5837-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz, E., A. Henne, R. Cramm, T. Eitinger, B. Friedrich, and G. Gottschalk. 2003. Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H2-based lithoautotrophy and anaerobiosis. J. Mol. Biol. 332:369-383. [DOI] [PubMed] [Google Scholar]
- 40.Simon, R., U. Priefer, and A. Pühler. 1983. Vector plasmids for in vivo and in vitro manipulations of Gram-negative bacteria, p. 98-106. In A. Pühler (ed.), Molecular genetics of the bacteria-plant interaction. Springer, Berlin, Germany.
- 41.Slater, S. C., W. H. Voige, and D. E. Dennis. 1988. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-β-hydroxybutyrate biosynthesis pathway. J. Bacteriol. 170:4431-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slater, S., T. Gallaher, and D. Dennis. 1992. Production of poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) in a recombinant Escherichia coli strain. Appl. Environ. Microbiol. 58:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slater, S., K. L. Houmiel, M. Tran, T. A. Mitsky, N. B. Taylor, S. R. Padgette, and K. J. Gruys. 1998. Multiple β-ketothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J. Bacteriol. 180:1979-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinbüchel, A., and H. G. Schlegel. 1989. Excretion of pyruvate by mutants of Alcaligenes eutrophus, which are impaired in the accumulation of poly(β-hydroxybutyric acid) (PHB), under conditions permissive for synthesis of PHB. Appl. Microbiol. Biotechnol. 31:168-175. [Google Scholar]
- 45.Steinbüchel, A., and H. E. Valentin. 1995. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol. Lett. 128:219-228. [Google Scholar]
- 46.Tanaka, M., M. Takebayashi, M. Miyama, J. Nishada, and M. Shimomura. 2004. Design of novel biointerfaces (II). Fabrication of self-organized porous polymer film with highly uniform pores. Biomed. Mater. Eng. 14:439-446. [PubMed] [Google Scholar]
- 47.Thompson, S., F. Mayer, O. P. Peoples, S. Masamune, A. J. Sinskey, and C. T. Walsh. 1989. Mechanistic studies on β-ketoacyl thiolase from Zoogloea ramigera: identification of the active-site nucleophile as Cys89, its mutation to Ser89, and kinetic and thermodynamic characterization of wild type and mutant enzymes. Biochemistry 28:5735-5742. [DOI] [PubMed] [Google Scholar]
- 48.Timm, A., and A. Steinbüchel. 1990. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl. Environ. Microbiol. 56:3360-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.