Abstract
The foliar pathogen Pseudomonas syringae pv. syringae exhibits an exceptional ability to survive on asymptomatic plants as an epiphyte. Intermittent wetting events on plants lead to osmotic and matric stresses which must be tolerated for survival as an epiphyte. In this study, we have applied bioinformatic, genetic, and biochemical approaches to address water stress tolerance in P. syringae pv. syringae strain B728a, for which a complete genome sequence is available. P. syringae pv. syringae B728a is able to produce the compatible solutes betaine, ectoine, N-acetylglutaminylglutamine amide (NAGGN), and trehalose. Analysis of osmolyte profiles of P. syringae pv. syringae B728a under a variety of in vitro and in planta conditions reveals that the osmolytes differentially contribute to water stress tolerance in this species and that they interact at the level of transcription to yield a hierarchy of expression. While the interruption of a putative gene cluster coding for NAGGN biosynthesis provided the first experimental evidence of the NAGGN biosynthetic pathway, application of this knockout strain and also a gfp reporter gene fusion strain demonstrated the small contribution of NAGGN to cell survival and desiccation tolerance of P. syringae pv. syringae B728a under in planta conditions. Additionally, detailed investigation of ectC, an orphan of the ectoine cluster (lacking the ectA and ectB homologs), revealed its functionality and that ectoine production could be detected in NaCl-amended cultures of P. syringae pv. syringae B728a to which sterilized leaves of Syringa vulgaris had been added.
Any bacterium growing in an environment with variable external wetness conditions will need a method of adjusting its own intracellular osmolarity. In order to counteract the deleterious effects of high osmolarity and a lack of water itself on cell physiology and the resulting loss of cytosolic water (dehydration), many bacteria rapidly synthesize or take up compatible solutes. These osmotically active small molecules are compatible with cellular function even at high concentrations and serve to maintain cell turgor by balancing the osmotic pressure across the cellular membrane without compromising protein folding or other processes which could be inhibited by high levels of cytosolic salts (26, 38, 42, 65, 66, 74).
Because they are necessarily inert, there are only a limited variety of compounds that can serve as compatible solutes, including certain polyols, amino acids, amino acid derivatives, and peptides. Typical prokaryotic osmolytes include glycine betaine, carnitine, proline, l-α-glutamate, mannitol, trehalose (α-d-glucopyranosyl-α-d-glucopyranoside), N-acetylglutaminylglutamine amide (NAGGN), glucosylglycerol, ectoine, and hydroxyectoine (65). These osmolytes can be produced de novo but are often accumulated by uptake from the environment to reduce the energetic cost to the cell. It is unclear what proportion of its compatible solutes an epiphyte must produce de novo, although it has been hypothesized that choline is widely available in plants for uptake and conversion to betaine (also known as glycine betaine or N,N,N-trimethylglycine) (14). Regardless of the mode of accumulation, bacteria must be able to respond quickly, especially in harsh environments where water potentials can change dramatically in a short period of time.
Waxy leaf surfaces are a particularly hostile bacterial habitat due to their rapidly and frequently changing environment. In addition to coping with a limited heterogeneous nutrient supply and daily fluxes in solar UV irradiation and temperature, epiphytes must also respond to frequent fluctuations in moisture availability from factors such as dew, rainfall, and atmospheric water content (33, 43). The bacterial communities on the plant surface regularly experience water stresses in the forms of both osmotic and matric stresses. Osmotic stress can result from the concentration of leaf surface solutes as water films evaporate, while matric stress can occur even in the absence of solutes during periodic desiccation events (78). Therefore, water stress tolerance likely is very robust in epiphytes, although its comprehensive analysis in such bacteria has not been undertaken previously.
The Gram-negative gammaproteobacterium Pseudomonas syringae pv. syringae is a common epiphyte on a variety of plants and the causative agent of bacterial brown spot disease in Phaseolus vulgaris L. (snap bean). P. syringae pv. syringae establishes a large epiphytic population on healthy plants, serving as a source of inoculum for the organism to invade the plant tissue and cause infections under permissive conditions. Due to its importance as a plant pathogen and the fact that adequate chemical or biological control of such bacterial diseases is not available, the epidemiology, epiphytic lifestyle (33), and recently, also the genome of P. syringae pv. syringae strain B728a (27) have been intensively studied. P. syringae pv. syringae B728a is thus a good model bacterium for studying osmoprotection in an epiphyte.
In a previous study by Feil et al. in which they first analyzed the P. syringae pv. syringae B728a genome sequence, it was hypothesized that the strain is able to produce betaine and possibly ectoine as osmolytes (24). However, biochemical studies have not been undertaken to confirm these bioinformatic predictions, and other possible responses of P. syringae pv. syringae to water stress have not been addressed. In order to gain novel insight into the osmotic stress responses of B728a, this study investigated the strain's biosynthetic capability to produce osmolytes using a genome-driven strategy, complemented by genetic and biochemical experiments.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. For identification of compatible solutes, P. syringae pv. syringae B728a and Pseudomonas syringae pv. tomato DC3000 were grown in Davis minimal broth without dextrose (41) but containing 20 mM glycerol (DMBgly), supplemented with 1% to 3% (wt/vol) NaCl for 72 h with shaking (160 rpm) at 25°C. For simulated in planta ectoine production, P. syringae pv. syringae B728a was cultivated for 8 days at 10°C in modified minimal medium MM63 (half of the glucose was replaced by fructose, and 1% NaCl was added) (40). In some experiments, fresh shredded surface-sterilized leaves of lilac (Syringa vulgaris) were added to the medium at the beginning of the experiment. Shredded leaves, incubated under identical conditions without added bacteria, and the extract of fresh shredded leaves served as negative controls. Restoration of the Halomonas elongata ΔectC mutant WUB01 was tested in DMBgly with 5% added NaCl. Production of ectoine and betaine from precursors by P. syringae pv. syringae B728a and P. syringae pv. tomato DC3000 was tested in DMB containing 1% NaCl and amended with 2 mM N-γ-acetyldiaminobutyric acid or choline, respectively. Betaine uptake was tested in both Luria-Bertani (LB) medium (6) and antibiotic assay medium (assay broth) 3 (30) containing 1% NaCl. Production of solutes by P. syringae pv. syringae B728a and P. syringae pv. tomato DC3000 under pseudo in planta conditions was checked by cultivation in SRM medium (55) for 7 days at 25°C without shaking. Escherichia coli strain DH5α was maintained on LB agar and cultured at 37°C. The antibiotics kanamycin, rifampin, and tetracycline were used at final concentrations of 50, 100, and 15 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Relevant characteristics or purposea | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Multipurpose cloning | 69 |
| S17-1 | Conjugal transfer of mobilizable plasmids | 73 |
| P. syringae pv. syringae | ||
| B728a | Rifampin-resistant mutant of a wild-type strain originally collected from bean leaves | 45 |
| B728a ΔPsyr_3747 | Tetr | This study |
| B728a (pPnaggn:gfp) | Isolate B728a harboring NAGGN reporter plasmids containing transcriptional fusions of the promoter for Psyr_3747 with a GFP-encoding gene | This study |
| B728a (p519gfp) | Isolate B728a harboring constitutive p519gfp | 52 |
| P. syringae pv. tomato DC3000 | Rifampin derivative of DC52 (wild-type P. syringae pv. tomato race 0 strain), pathogen of tomato and Arabidopsis spp. | 20 |
| P. stutzeri DSM 5190T | Wild type, contains the full functional ectABC gene cluster | 67 |
| H. elongata | ||
| DSM 2581T | Wild type, contains the full functional ectABC gene cluster | 79 |
| WUB01 | Isolate DSM2581 ΔectC (in-frame deletion) | E. Witt and A. Ures, Galinski group, University of Bonn, Germany |
| WUB pPESyC | Isolate DSM2581 ΔectC plus ectC gene of P. syringae pv. syringae B728a; Chlr | This study |
| WUB pPEStC | Isolate DSM2581 ΔectC plus ectC gene of P. stutzeri DSM5190T; Chlr | This study |
| Plasmids | ||
| pENTR/D-Topo | Entry vector | Invitrogen |
| pLVC-D | Destination vector | 47 |
| pPE | Cloning vector; pBBR1MCS derivative equipped with the salt-dependent H. elongata promoter region upstream of H. elongata ectC | 12, 35, 36 |
| pPESyC | ectC gene of P. syringae pv. syringae B728a subcloned in frame into pPE under the control of the H. elongata promoter | This study |
| pPEStC | ectC gene of P. stutzeri DSM5190T subcloned in frame into pPE under the control of the H. elongata promoter | This study |
| pTOPO-Pnaggn | Vector equipped with the upstream promoter region of the NAGGN biosynthetic gene cluster | This study |
| pPROBE-OT | Vector containing a promoterless gfp gene | 54 |
| pPnaggn:gfp | Cloning vector containing the upstream promoter region of the NAGGN cluster fused with the gfp gene | This study |
Chlr, chloramphenicol resistant; Tetr, tetracycline resistant.
Construction of mutants.
To generate the NAGGN synthesis-defective (ΔPsyr_3747) mutant, a DNA fragment of approximately 500 bases in length harboring an internal portion of Psyr_3747 was amplified from P. syringae pv. syringae B728a genomic DNA using the primers 3747-KO-F (5′-CACCTACGGCCCGCACGAAGACGAG-3′) and 3747-KO-R (5′-CCAGTGGTAACCGGCGAACAGTTCG-3′). This fragment was placed in the Gateway vector pENTR/D-Topo (Invitrogen, Carlsbad, CA), and then transferred to the destination vector pLVC-D, as described by Marco et al. (47). The resulting construct was transformed into P. syringae pv. syringae B728a, and tetracycline was used to select for single-crossover events, as in other studies (47). The resulting interruptions in Psyr_3747 were confirmed using PCR with primers specific to the pLVC-D vector and genomic DNA.
For the construction of a Pnaggn::gfp transcriptional fusion, the upstream promoter region of the P. syringae pv. syringae B728a Psyr_3747 NAGGN biosynthetic gene was amplified from genomic DNA by PCR with primers naggn-HindIII, 5′-TAAGCTTTGGAGACCGATCGGGGCTC-3′, and naggn-EcoRI, 5′-TGAATTCAAGTCCTTACCGGGTTTCGTTGGGG-3′, to generate a 400-bp promoter region (underlining shows corresponding restriction sites). PCR conditions used were as follows: 28 cycles of 95°C, 61°C, and 72°C for 1 min each, with a final extension time of 10 min at 72°C. The PCR product was first cloned into pTOPO Blunt (Invitrogen) to generate pTOPO-Pnaggn, and then transformed into E. coli DH5α. The insert was sequenced to verify its identity. pTOPO-Pnaggn was digested with HindIII and EcoRI, and the resulting fragment was cloned into pPROBE-OT (54), which contains a promoterless gfp gene, in order to generate pPnaggn:gfp. pPnaggn:gfp was electroporated into P. syringae pv. syringae B728a to yield the transformed reporter strain B728a (pPnaggn:gfp).
In order to confirm the activity of the ectoine synthase EctC of P. syringae pv. syringae B728a, the ectC gene from B728a was cloned into the vector pPE. Plasmids were then transformed into E. coli S17-1 for conjugal transfer into the ectC-deficient strain H. elongata WUB01, as described elsewhere (28). The ectC gene of ectoine-producing Pseudomonas stutzeri was included as a positive control.
Analysis of intracellular osmolyte contents.
Extraction of freeze-dried cell material from an exponentially growing shaking culture followed a modified protocol of the technique of Bligh and Dyer (7) as described previously (27). Solute content was analyzed by isocratic high-performance liquid chromatography (HPLC) on a Macherey-Nagel Nucleosil NH2 100-3 column (125 by 4 mm, 3 μ) and an 80:20 (vol/vol) acetonitrile/water ratio at a flow rate of 1 ml min−1 as a mobile phase, with simultaneous refractive index and UV detection (220 nm).
Bioinformatics and phylogenetic tree construction.
In order to detect homologous protein sequences from each biosynthetic pathway, previously reported sequences, preferably from pseudomonads or bacteria related to Pseudomonas, were used. Searches for related proteins were performed with the BLASTP program at the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/BLAST) (2), and sequences with minimum E values of 0.0001 without filtering were identified.
To estimate the phylogenetic relationships of EctC, a BLAST search for sequences with similarity either to that of EctC of P. stutzeri (YP_001170728), representing a full functional EctC derived from a classic three-component gene ectABC operon, or to that of EctC of P. syringae pv. syringae B728a (YP_233444), originating from a single and isolated ectC gene, was conducted. Sequence alignments were performed with ClustalW2 software using default values (39). The tree was visualized using the software PhyloDraw (16).
Isolation and purification of NAGGN.
P. syringae pv. syringae B728a was grown for 72 h at 25°C with shaking (160 rpm). Two cultures in 1 liter of DMBgly supplemented with 1% (wt/vol) NaCl yielded 708.4 mg of crude extract. Chromatographic separation of the crude extract employing isocratic HPLC on a Macherey-Nagel Nucleosil NH2 100-5 column (250 by 4.6 mm, 5 μ) and an 80:20 (vol/vol) acetonitrile/water ratio at a flow rate of 1 ml min−1 as a mobile phase yielded 4.7 mg of pure NAGGN.
NMR spectroscopy.
1H and 13C nuclear magnetic resonance (NMR) spectra were recorded on a Bruker Avance 300 DPX spectrometer. Spectra were calibrated to the solvent signal (13C, d6-dimethyl sulfoxide [d6-DMSO], δ 39.5 ppm) and the signal contributed by the nondeuterated portion of the solvent (1H, D2O, δ 4.68 ppm; DSMO, δ 2.50 ppm).
Absolute configuration of the peptide moiety of NAGGN.
NAGGN (0.9 mg) was dissolved in 600 μl 6 N HCl and hydrolyzed in a sealed vial at 105°C for 16 h. The solvent was removed by using a stream of dry N2, and the sample was analyzed by chiral HPLC with a Phenomenex Chirex 3126 phase (4.6 by 250 mm, 2 mM CuSO4-MeCN [95:5], 1.0 ml/min, UV detection at 254 nm). The retention times of the standards were 42.8 min for l-Glu and 45.6 min for d-Glu. The hydrolysate showed a peak coincident with that of the l-Glu standard (see the supplemental material).
Plant assays.
Cells of P. syringae pv. syringae B728a strains were grown overnight on KB medium with appropriate antibiotic selection, harvested by scraping them from the plate surface, washed, and resuspended to a final cell concentration of 106 cells ml−1 in 1 mM potassium phosphate buffer. The cell suspension was then spray inoculated onto fully expanded trifoliate leaves of bean plants (Phaseolus vulgaris cv. Bush Blue Lake 274; approximately 2 weeks old). Plants were placed in a humid chamber at room temperature (approximately 24°C) and periodically exposed to a fine mist of water to maintain high relative humidity and to maintain thin films of water on the leaf surfaces. Artificial light was maintained for 16-h periods within the 24-h cycle. For epiphytic growth analysis, plants were taken out of the humid chamber after 48 h and placed on a greenhouse bench for an additional 24 h of drying. Cells were washed from individual trifoliate leaves with 5 min of sonication in 1 mM potassium phosphate buffer (pH 7.0), and epiphytic population sizes were determined by plating of appropriate dilutions, as previously described (63, 64). Alternatively, for the fluorescence analysis, after an initial 24 h of humidity the plants were exposed to the following conditions for 24 h: continued humidity for “wet plants,” turning off the humidifier for “slow-dry plants” and moving the inoculated plants to a dry chamber with 40% humidity and at 28°C for “fast-dry plants.”
Fluorescence assays.
For analysis of expression of NAGGN in vitro, cells of the reporter P. syringae pv. syringae B728a (pPnaggn:gfp) and a constitutively fluorescent strain B728a (p519gfp) (52) were grown with shaking overnight in Davis minimal medium broth and then spiked with NaCl (final concentration, 1%); half of the NaCl cultures were also spiked with choline (2 mM), and all were incubated for an additional 4 h. Cells were then recovered by centrifugation, washed, and resuspended in phosphate buffer (10 mM, pH 7.5) to a concentration of approximately 3 × 108 cell ml−1 (optical density at 600 nm [OD600] = 0.2). The green fluorescent protein (GFP) fluorescence intensity was determined using a TD-700 fluorometer (Turner Designs, Sunnyvale, CA) with a 486-nm band-pass excitation filter and a 510- to 700-nm combination emission filter. A relative fluorescence unit (RFU) was defined as the fluorescence of the suspensions normalized for the suspension turbidity, measured as OD600.
For in vivo applications, P. syringae pv. syringae B728a (pPnaggn:gfp) and B728a (p519gfp) were grown and inoculated as described above in plant assays. Cells were washed from individual leaves and recovered by successive rounds of centrifugation at 8,000 × g. Cells were mixed with a DAPI (4′,6-diamidino-2-phenylindol) solution (final concentration, 2 μg ml−1) on a microscope slide and then flash frozen on dry ice to permeabilize the cells. Epifluorescent microscopy was then done using a Zeiss Axio Imager M1 fluorescence microscope using a 20× lens objective and a xenon light source. The GFP was excited with a band-pass filter at 470/40 nm. Emissions were collected using a band-pass filter at 525/50 nm. DAPI was excited with a band-pass filter at 350/50 nm, and the emissions were collected using a 400-nm long-pass filter for 0.4 s. Images were collected with a Hamamatsu Orca camera, using exposure times of 6 s for GFP and 0.4 s for DAPI. Images were captured, and relative GFP fluorescence was quantified using iVision software (BioVision Technologies).
RESULTS
Identification of compatible solute biosynthetic pathways in the P. syringae pv. syringae B728a genome.
The complete genome sequence of B728a was investigated for the presence of open reading frames (ORFs) with similarity to genes known to code for compatible solutes typical for prokaryotes.
Trehalose.
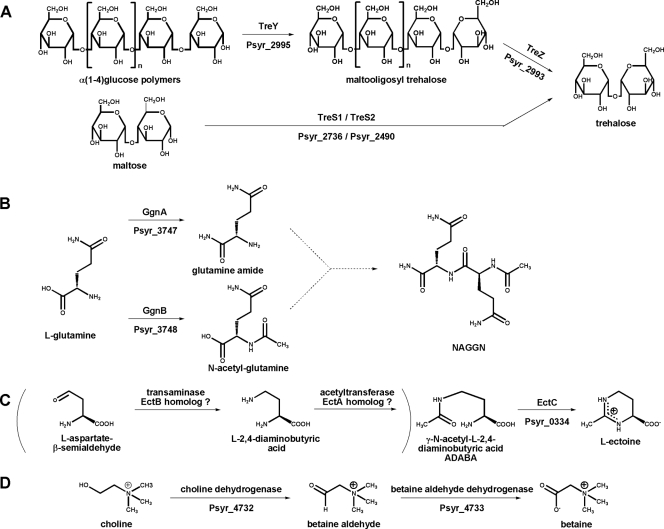
Using a BLAST search against proteins carried by the three typical eubacterial trehalose biosynthesis pathways TreXYZ, TS, and TPS/TPP (4, 17, 48-51), we identified a TreXYZ pathway (Psyr_2997, Psyr_2995, and Psyr_2993) and two potential TS pathways (Psyr_2736 and Psyr_2490) in P. syringae pv. syringae B728a (Fig. 1). The predicted Psyr_2997 protein shares 48% sequence identity with TreX of Arthrobacter sp. Q36, whereas the Psyr_2995 and Psyr_2993 proteins showed 65% and 66% sequence identity with Pseudomonas fluorescens Pf-5 maltooligosyl-trehalose synthase TreY and maltooligosyl-trehalose trehalohydrolase TreZ, respectively. Psyr_2736 and Psyr_2490 were significantly related (73% sequence identity each) to the two trehalose synthases of Pseudomonas putida W619 (PputW619_1821 and PputW619_1816) and are here referred to as TreS-1 and TreS-2, respectively. Genes encoding the TPS/TPP pathway (as found in Pseudomonas savastanoi and P. stutzeri A1501) were not found in the genome of P. syringae pv. syringae B728a.
FIG. 1.
Compatible solute biosynthesis pathways found in P. syringae pv. syringae B728a. (A) Three pathways leading from either maltose or glucose polymers to trehalose. (B) Proposed biosynthesis of N-acetylglutaminylglutamine amide (NAGGN). (C) Formation of the osmolyte ectoine from its precursor ADABA. The enzymatic steps in parentheses are speculative. (D) Conversion of choline into betaine by P. syringae pv. syringae B728a.
NAGGN.
An early study on the biosynthesis of the compatible solute NAGGN in Rhizobium meliloti reported that NAGGN is produced nonribosomally by an ATP-dependent enzyme, although no candidate loci were identified in R. meliloti (76). This prediction aided our search for candidate genes in P. syringae pv. syringae B728a. As a working hypothesis, it was envisioned that two glutamines serve as precursors which are fused directly before N-acetylation and amidation occurs, or in turn, the free amino acids are first derivatized and subsequently connected to yield a dipeptide. Hence, a clustered set of at least three genes was hypothesized to be involved in the biosynthesis, i.e., an acetyltransferase, an amidotransferase, and a dipeptide synthase, possibly accompanied by regulatory genes. A BLAST search for annotated acetyltransferases in the genome of P. syringae pv. syringae B728a yielded numerous potential candidate genes. Exclusion of apparently specialized acetyltransferases halved the number of candidate genes. Closer inspection of flanking regions of the remaining candidate genes identified the acetyltransferase Psyr_3748 to be of particular interest due to the adjacent amidotransferase (Pysr_3747) and aminopeptidase (Psyr_3749). This hypothesis was further corroborated by a microarray study by Aspedon and coworkers which demonstrated that the expression of orthologs of Psyr_3747 to Psyr_3749 in Pseudomonas aeruginosa (PA3459 to PA3461) was upregulated by 5- to 6-fold in osmotically shocked cells (3).
Ectoine.
P. syringae pv. syringae B728a possesses a homolog of ectC (Psyr_0334), although ectoine biosynthesis is uncertain because there are no homologs of ectA and ectB, which synthesize the ectoine precursor N-γ-acetyldiaminobutyric acid (ADABA) (Fig. 1) and are usually spatially linked with ectC (24).
Betaine.
Putative choline and betaine dehydrogenase enzymes (Psyr_4732 and Psyr_4733, respectively) were already annotated in the first analysis of the P. syringae pv. syringae B728a genome sequence (24). Like many other bacteria, this indicates that B728a can synthesize betaine only when supplied with its precursor choline (Fig. 1). Additionally, orthologs of osmoprotectant transporters BetT (Psyr_3758), OpuC (Psyr_4249), and CbcXWV (Psyr_4709 to Psyr_4711), responsible for the transport of the essential precursor choline, have already been identified by Beattie and coworkers (13-15).
Other compatible solutes.
Searches of the P. syringae pv. syringae B728a genome for homologs of GgpPS from Pseudomonas mendocina YMP (32) (accession no. YP_001188605) that encode glucosylglycerol biosynthesis did not yield any significant matches, suggesting that glucosylglycerol is not produced by B728a. Due to their dual role in primary metabolism, it was not possible to attribute genes for amino acid-based osmolytes (e.g., proline or glutamate) or mannitol exclusively to water stress tolerance.
Identification of compatible solutes of salt-stressed P. syringae pv. syringae B728a.
In order to test the bioinformatic hypotheses, B728a was subjected to salt stress (1% NaCl) and analyzed for compatible solute production. First, natural-abundance 1H and 13C NMR spectroscopy of the crude polar extract was employed to identify the osmolyte composition of P. syringae pv. syringae B728a. Inspection of the 1H NMR spectrum and particularly the 13C NMR spectrum (see the supplemental material) indicated the presence of NAGGN (δH D2O: 1.85 m + 1.98 m, 1.89 s, 2.23 t, J = 7.3 Hz, 4.14 m; δC D2O: 179.3, 179.2, 177.2, 175.9, 175.1, 54.7, 54.3, 2 × 32.4, 28.1, 28.0, and 23.0) and trehalose (δH D2O: 3.21 to 3.74 m, 5.03 d 3.7; δC D2O: 94.6, 73.8, 73.5, 72.4, 71.0 and 61.9) as the two principal osmolytes. The identity of these compounds was readily established by direct comparison with literature data (22, 75) and the spectra of reference substances. Additionally, by two-dimensional NMR (2D-NMR), high-resolution mass spectrometry (HR-MS), and chiral amino acid analysis, NAGGN was semipreparatively purified, and its identity was further confirmed (see the supplemental material).
Inactivation of the P. syringae pv. syringae B728a NAGGN biosynthesis pathway.
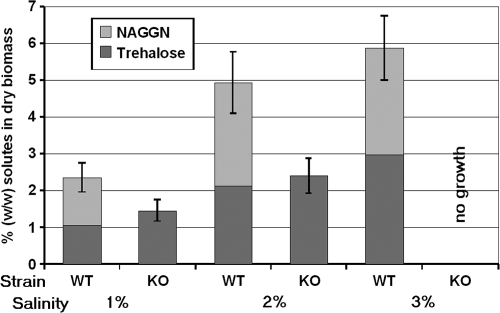
In order to confirm that the pathway proposed from the genome data analysis (Psyr_3747 to Psyr_3749) was involved in the production of NAGGN, a mutant was constructed by chromosomal inactivation of the first gene (Psyr_3747) of the pathway, encoding a putative amidotransferase. While the mutant still produced trehalose under conditions of 1% and 2% salinity, it was unable to accumulate NAGGN (Fig. 2; see also Fig. S10 in the supplemental material).
FIG. 2.
Osmolytes accumulated by the P. syringae pv. syringae B728a wild type (WT) and NAGGN-deficient mutants (knockouts [KO]) grown at different salinities (wt/vol).
Osmotic regulation of NAGGN and trehalose in salt-stressed cells.
The dependence of osmolyte accumulation on medium osmolality was examined further by determining the intracellular levels of NAGGN and trehalose as a function of salinity. The levels of both of the osmolytes increased at a 1:1 dilution ratio as the concentration of NaCl was increased (Fig. 2). In the absence of NaCl, NAGGN and trehalose were undetectable, indicating that both compounds are osmoregulated. The mutant of P. syringae pv. syringae B728a deficient in NAGGN production responded to 1% and 2% salinity exclusively with the synthesis of trehalose, and although the amount of trehalose produced was always slightly higher than that produced by the wild type at any given level of NaCl, this increase did not fully compensate for the loss of NAGGN. The limited compensation for increased trehalose production is also evident in the lack of growth of the NAGGN mutant in 3% NaCl. While no difference in growth was observed between the wild type and the NAGGN mutant without salt stress, the mutant strain was severely impaired in growth compared to that of the wild-type strain when the osmolarity of the medium was raised to 3% NaCl (see Fig. S13 in the supplemental material). As further proof of osmotic regulation, the transcription of the NAGGN biosynthetic locus was estimated by measuring the GFP fluorescence of cells harboring a fusion of the promoter for Psyr_3747 to a promoterless gfp reporter gene. As with accumulation of NAGGN, the expression of Psyr_3747 increased directly with the increasing NaCl concentration of the culture media (see Fig. S14 in the supplemental material).
Betaine as a compatible solute in P. syringae pv. syringae B728a.
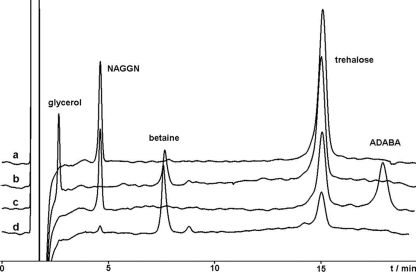
P. syringae pv. syringae B728a is not expected to be able to perform de novo biosynthesis of betaine due to its predicted inability to produce the precursor choline, which was further confirmed when no betaine was detected in osmotically stressed cells. Therefore, choline was supplied in order to probe the functionality of the predicted betaine biosynthetic pathway. HPLC analysis of cellular extracts revealed that betaine was produced when choline was added to the growth medium (Fig. 3). Moreover, the accumulation of betaine was accompanied by a decrease of the other endogenously synthesized osmoprotectants. The production of NAGGN was nearly abolished upon addition of choline to culture media, while the accumulation of trehalose appeared to be reduced to only 20% of that seen in media without choline (Fig. 3). A direct uptake of betaine was also demonstrated by growing P. syringae pv. syringae B728a in a betaine-containing media; after 48 h of growth in the presence of betaine and 1% NaCl, HPLC analysis of cellular extracts revealed a prominent betaine peak (Fig. 3), thus confirming that B728a possesses efficient transport mechanisms for this compatible solute, as noted by Beattie et al. (13-15). Similar to the addition of choline, there was a near-complete elimination of NAGGN biosynthesis but only a slight decrease in trehalose production upon addition of betaine to culture media in the presence of NaCl (Fig. 3). Furthermore, transcription of Psyr_3747 involved in NAGGN production was strongly downregulated in the presence of choline and NaCl compared to that in the presence of NaCl alone (see Fig. S14 in the supplemental material).
FIG. 3.
Compatible solute profile of the P. syringae pv. syringae B728a wild type grown under different conditions at 1% salinity (amounts of solutes are given in mg [dry biomass] per g). (a) DMBgly (trehalose, 10.7; NAGGN, 2.3); (b) betaine-containing AB1 medium (trehalose, 7.7; NAGGN, 0.0; betaine, 0.9); (c) DMBgly supplemented with the ectoine precursor ADABA (trehalose, 4.9; NAGGN, 1.9; ADABA, 2.3); (d) DMBgly supplemented with choline (trehalose, 2.3; NAGGN, 0.1; betaine, 2.0).
Verification of the functionality of P. syringae pv. syringae B728a EctC.
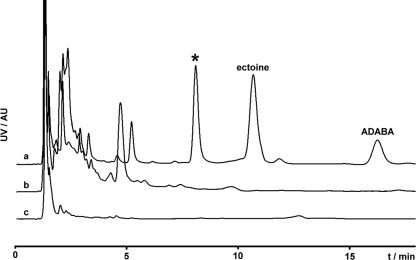
To test if P. syringae pv. syringae B728a ectC encodes a functional cyclase, the gene was transferred into the ectC-deficient strain Halomonas elongata WUB01, which does not produce ectoine, and ectoine production was quantified. This strain retains functional ectA and ectB genes, so it can produce the ectoine precursor ADABA. Ectoine production was restored in the ectC-deficient H. elongata strain complemented with pPESyC to produce half the level of ectoine compared with that produced by the wild-type strain harboring this plasmid, indicating that P. syringae pv. syringae ectC encodes a fully functional but less efficient ectoine synthase. Although no ectoine was detected in cells exposed to NaCl, it was hypothesized that B728a might be able to produce ectoine de novo but that appropriate conditions for its synthesis were not provided under these culture conditions. Since the production of secondary metabolites depends on culture conditions (9) and since many genes in P. syringae pv. syringae are modulated by plant signal molecules (47, 55), simulated in planta conditions were explored to determine if the ectC gene and, hence, ectoine production might be conditional in its expression. Surprisingly, of the many conditions tested, only the addition of surface-sterilized leaves of lilac (the organism from which P. syringae pv. syringae was first isolated), which were devoid of other microbes, was shown to induce ectoine production in wild-type B728a (Fig. 4). While such leaves would be expected to contain arbutin, a compound previously shown to induce toxin production in P. syringae pv. syringae (55), arbutin-containing medium had no influence on the osmolyte production profile, suggesting that arbutin itself is not a signal for ectoine production (data not shown). Additionally, an osmolyte profile of P. syringae pv. syringae B728a grown in media containing the ectoine precursor ADABA revealed that ADABA was readily taken up (Fig. 3), demonstrating the presence of the respective transport capacity in B728a. ADABA uptake was accompanied by a decrease in the production of NAGGN and trehalose, since it may also be used as a compatible solute (58). However, feeding of ADABA alone was not sufficient to yield ectoine production in P. syringae pv. syringae B728a.
FIG. 4.
(a) UV-HPLC osmolyte profile of P. syringae pv. syringae B728a grown in the presence of shredded lilac leaves. The asterisk indicates an unknown component of the extract. In the negative controls, which comprised shredded leaves incubated under identical conditions but without bacteria added (b) and fresh shredded leaves (c), peaks for the ectoine precursor ADABA or for ectoine itself were not observed. AU, arbitrary units.
Role of NAGGN in colonization of plant surfaces by P. syringae pv. syringae B728a.
In order to assess the role of NAGGN in the osmoadaption of P. syringae pv. syringae B728a in planta, wild-type and NAGGN-knockout strains of B728a were inoculated onto the surface of bean plants (Phaseolus vulgaris L.) that were subsequently exposed to various environmental conditions. The NAGGN-deficient mutant grew to population sizes equal to those of the wild-type strain under humid conditions, exhibiting about 100-fold epiphytic growth after 2 days. Given that moist leaves probably confer little water stress to epiphytic bacteria, the lack of difference in growth between the NAGGN mutant and wild-type strains under humid conditions was not surprising. However, little difference in survival of the NAGGN mutant compared to that of the wild-type strain was also seen on dry leaves (see Fig. S15 in the supplemental material). Water stress conditions were imposed by a slight increase in temperature and a great reduction in leaf surface moisture after the initial moist growth conditions. Both strains exhibited about a 10-fold decrease in population size on the leaf surface in response to the drying of the leaf environment, and the presence or absence of NAGGN biosynthesis had no effect on this response.
In order to clarify if NAGGN is actually produced by P. syringae pv. syringae B728a on leaves, an expression analysis of the NAGGN biosynthetic genes using gfp reporter gene fusion strains of wild-type B728a (pPnaggn:gfp) and its corresponding NAGGN mutant (p519gfp) was performed. In agreement with the epiphytic growth experiments, only small differences in the levels of GFP fluorescence under all conditions tested on leaves were observed, indicating that there is no evidence of NAGGN biosynthesis induction on leaves, even under desiccation conditions.
DISCUSSION
Genomic mining for compatible solutes in P. syringae pv. syringae B728a revealed that this plant pathogen has the capacity to produce a wide repertoire of cell-protecting osmolytes to counteract water stress. The predicted compatible solutes of P. syringae pv. syringae B728a appear to be typical of the osmolytes produced by other pseudomonads (Table 2). While we demonstrated that all four predicted osmoprotectants could actually be produced by P. syringae pv. syringae, their production in many cases was very context dependent; while trehalose and NAGGN were readily biosynthesized in NaCl-containing growth medium, betaine and ectoine were produced only when provided with medium conditions mimicking in planta conditions, and the supplementation of exogenous osmolytes such as betaine suppressed the production of many of these compounds, suggesting that osmolyte production itself is generally inhibited when osmolyte uptake is possible.
TABLE 2.
Compatible solutes of pseudomonads
| Pseudomonas strain |
De novo biosynthesized compatible solute results |
Uptake/conversion of betainec | Reference(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Alanine | Glutamateb | NAGGN | Trehalose | Mannitol | Glucosyl- glycerol | Ectoine | OH- ectoine | |||
| P. aeruginosa ATCC 17933 | + | ND/ND | 34 | |||||||
| P. aeruginosa PAO1 | + | + | + | U/C | 22 | |||||
| P. chlororaphis L10-2-1a | + | + | ND/ND | 53 | ||||||
| P. fluorescens | + | + | + | ND/ND | 75 | |||||
| P. fluorescens ATCC 11250 | + | ND/ND | 34 | |||||||
| P. fluorescens DSM 8569a | + | + | ND/ND | 53 | ||||||
| P. fluorescens DSM 50090T | + | + | −/C | This study | ||||||
| P. fluorescens EPS62e | + | + | + | U/ND | 10 | |||||
| P. fluorescens Pf-5 | + | + | U/C | This study | ||||||
| P. fluorescens ST | + | ND/ND | 34 | |||||||
| P. halophila DSM 3050 | + | + | ND/C | 72 | ||||||
| P. halosaccharolytica CCM 2851 | + | + | + | U/C | 72 | |||||
| P. mendocina ATCC 25411 | + | + | + | ND/ND | 62 | |||||
| P. mendocina SKB70 | + | + | + | U/ND | 62 | |||||
| P. pseudoalcaligenes ATCC 14440 | + | + | + | ND/ND | 62 | |||||
| P. psychrotolerans BO001 | + | + | + | + | −/− | 31; this study | ||||
| P. putida Idaho NRRL B-18435 | + | + | + | ND/ND | 34 | |||||
| P. putida L30-3-8a | + | + | ND/ND | 53 | ||||||
| P. putida PpG1 ATCC 17453 | + | + | + | ND/ND | 34 | |||||
| P. putida S12 | + | + | U/ND | 34 | ||||||
| P. stutzeri DSM 5190T | + | + | + | + | U/C | 37, 71; this study | ||||
| P. syringae L20-3-1a | + | + | ND/ND | 53 | ||||||
| P. syringae pv. syringae B728a | + | + | (+)d | U/C | This study | |||||
| P. syringae pv. tomato DC3000 | + | + | + | U/C | 25; this study | |||||
Amino acids and other charged osmolytes were not determined in this study.
The role of glutamate as a compatible solute is under debate (70).
U, uptake occurred; C, conversion occurred; ND, not determined in the corresponding study.
(+), production occurs only when medium conditions mimic in planta conditions.
Compatible solutes in P. syringae pv. syringae B728a.
Under the moderate-salt stress conditions of 1% NaCl, P. syringae pv. syringae B728a produces trehalose and NAGGN in equivalent amounts. With increasing salinity, the strain increases the biosynthesis of both compatible solutes. Although we have determined only one biosynthetic cluster for NAGGN and demonstrated its sufficiency for NAGGN biosynthesis, the production of trehalose may be conferred by several biosynthetic pathways. In fact, not only are there multiple biosynthetic pathways for trehalose production in B728a, but there are even multiple copies of some of these pathways. Such redundancy is a commonly observed phenomenon in halotolerant and halophilic bacteria (4), which may be due to a need for rapid accumulation of trehalose under changeable environmental conditions and substrate availability. Trehalose appears to be a particularly effective osmolyte; production of trehalose enables P. syringae pv. syringae B728a to tolerate up to 2% NaCl, while trehalose in combination with NAGGN raises the halotolerance to 3% NaCl (Fig. 2). At salinities of 3% and higher, P. syringae pv. syringae B728a was hardly cultivable. Overall, the in vitro osmolyte profile and the mechanisms of osmotic stress adaption of P. syringae pv. syringae B728a were largely similar to those of other pseudomonads (22).
Choline participates in the osmotolerance of P. syringae pv. syringae B728a in complex ways. A large number of bacteria are not able to synthesize betaine de novo (57) but require the presence of the precursor choline or glycine betaine aldehyde in the growth medium for betaine formation (8). This study showed that P. syringae pv. syringae B728a actually produces betaine as bioinformatically predicted but only if its precursor choline is added to the growth medium. Under these conditions, betaine becomes the dominant osmoprotectant, replacing the endogenously synthesized osmolytes trehalose and NAGGN. Such shifts in osmolyte preference by exogenously added compatible solutes or their precursors are well known (18, 19, 22) and can be explained by the fact that the production of the small 5-carbon-containing betaine is energetically less costly than the larger 12-carbon-containing trehalose or NAGGN molecules (60). P. syringae pv. syringae B728a is somewhat more flexible in osmolyte accumulation than some other bacteria, because even in the presence of exogenous betaine, it still produces trehalose (Fig. 3). With its ability to derive better osmoprotection from choline than from betaine, P. syringae pv. syringae B728a is distinct from most other characterized bacterial strains. However, this behavior may be explained by the higher capacity for uptake of choline than that for uptake of betaine by the concerned transporter systems OpuC, BetT, and CbcXWV (13-15). Since choline is ubiquitous in plants, whereas betaine is produced by only some plants (77), it has been speculated that P. syringae, as a plant-associated organism living in the phyllosphere, particularly evolved the preference of choline rather than betaine in order to enhance its survival rate in or on plants. Based on these facts, we hypothesize that the low efficacy of the bacterial transporter systems for betaine uptake may restrict the ability of P. syringae pv. syringae B728a to accumulate enough betaine for optimal osmotolerance, and it thus retains the capacity for de novo synthesis of trehalose for maximal osmoprotection.
Gene ectC of P. syringae pv. syringae B728a.
Ectoine biosynthetic enzymes are usually encoded by the gene cluster ectABC (46, 59) (Fig. 1C). Intriguingly, under simulated in planta conditions, the wild-type strain of P. syringae pv. syringae B728a is able to produce the osmolyte ectoine despite the absence of the acetyltransferase EctA and the transaminase EctB. Two possible explanations for this observation can be envisioned, as follows: (i) the precursor ADABA is supplied by the host plant (11, 23, 44), or (ii) ADABA may be produced by a nonorthologous set of enzymes which do not reside in an operon with ectC (Psyr_0334). Several genes coding for N-acetyltransferases, as well as Psyr_1925 encoding a diaminobutyrate aminotransferase, were suggested to fulfill the functions of ectA and ectB, respectively (24). From some ectoine-producing microorganisms, it was already known that EctB is not essential because its respective ΔectB mutants were still capable of biosynthesizing ectoine (29; D. Stumpfe and M. Kurz, unpublished data). However, there is no precedence in the literature showing that EctA can also be replaced. This observation raises the question of the origin and the development of the ectC fragment in the genome of P. syringae pv. syringae B728a. A phylogenetic analysis (see Fig. S12 in the supplemental material) which compared EctC of P. syringae pv. syringae B728a with all other currently known bacterial ectoine synthases revealed (i) that several other bacteria, particularly plant-associated bacteria, possess a solitary ectC fragment that might be functional and (ii) that the products of solitary ectC genes differ significantly from those of clustered ectC genes, hence forming a distinctive subgroup. It can therefore be envisioned that the solitary ectC gene fragments represent a remnant of originally complete ectABC clusters, which evolved in their ecological niche toward a minimalistic cluster by evolution. It is possible that the corresponding regulatory mechanisms also coevolved and depend on a signal factor from the host plant, since simple addition of ADABA did not yield ectoine production.
NAGGN-biosynthetic pathway of P. syringae pv. syringae B728a.
The analysis of the available P. syringae pv. syringae B728a genome sequence data led to the proposal of a 4.7-kb biosynthesis gene cluster for the osmolyte NAGGN. The pathway consists of the three contiguous genes, Psyr_3747 to Psyr_3749 (see Fig. S11 in the supplemental material), for which the designation ggnABC is proposed. The product of ggnA (Psyr_3747) is predicted to code for an amidotransferase and is hypothesized to perform an amidation at the C-terminal end of its substrate, glutamine. Chromosomal disruption of ggnA abolished NAGGN production and proved ggnA to be involved and essential for NAGGN biosynthesis. The adjacent ggnB gene (Psyr_3748) encodes an acetyltransferase acetylating the N terminus of another glutamine. The reaction by which the two amino acids are fused is unknown at this time. Since no dipeptide synthase was detected in addition to their regular predicted function, we assume that the corresponding catalytic activity is embedded in either GgnA or GgnB, or elsewhere, e.g., by an unspecific peptide synthase. GgnC is predicted to be an aminopeptidase, with similarity to deblocking aminopeptidases. Aminopeptidases, particularly deblocking aminopeptidases, act at the amino terminals of peptides to release amino acids from regular peptides or peptides modified with amino-terminal acyl-type blocking groups, such as N-terminal acylated amino acids. Hence, GgnC might be necessary to cleave the dipeptide product from either GgnA or GgnB upon assembly. Alternatively, it could be hypothesized that GgnC cleaves NAGGN into glutamine amide and N-acetyl-glutamine and thereby possesses negative regulatory properties. A more detailed understanding of the roles of GgnA, GgnB, and GgnC requires more in-depth biochemical studies and is currently being investigated in our laboratories. A survey of all available Pseudomonas sp. genomes revealed that the NAGGN biosynthetic gene cluster is both widespread and highly conserved across the genus (see Fig. S11 in the supplemental material).
Contribution of the compatible solutes of P. syringae pv. syringae B728a to its epiphytic fitness.
Depending on the bioavailability of precursors and on the given water availability conditions, P. syringae pv. syringae B728a can choose from a variety of osmolytes to assemble an optimal balance of compatible solutes. This flexibility in its stress response allows B728a to accomplish osmoregulation under a wide variety of conditions and reflects an evolutionary necessity for this bacterium to counteract desiccation stress in the harsh leaf environment. It can therefore be hypothesized that compatible solute production in P. syringae pv. syringae B728a contributes to the epiphytic fitness of the strain. The osmolyte NAGGN plays, in this context, a minor role, as demonstrated by reporter gene expression assays. In planta, the NAGGN biosynthetic genes were found to be weakly expressed in P. syringae pv. syringae B728a, and a deficient mutant of B728a exhibited the same survival on dry leaves as the wild-type strain. A possible explanation for this observation could be that the surviving bacteria are protected from desiccation by other osmoprotectants (i.e., betaine, trehalose, or ectoine), other mechanisms (i.e., aggregation [56], polysaccharides, or biofilms), or a combination of both, which renders NAGGN biosynthesis superfluous. In particular, the first hypothesis is in agreement with the finding that exogenous betaine or choline acts to suppress production of NAGGN. Although betaine is most likely not present in the phyllosphere, the betaine precursor choline is hypothesized to be ubiquitous and relatively abundant (77, 80) because it represents a component of the major membrane lipid phosphatidylcholine. Choline bound to phospholipids could be made disposable for epiphytic microorganisms through the action of phospholipases (1, 5), which in turn are activated in response to wounding (68) and microbial infection (21, 61). Rapid uptake of choline has been demonstrated in Pseudomonas syringae pv. tomato DC3000 (14), and B728a is predicted to have a homologous choline transport system. This transport is so efficient that DC3000 derives more osmoprotection from choline (which it must convert to betaine) than betaine itself, unlike bacteria from other environments where betaine is more abundant. Indeed, our studies found that addition of choline to the medium resulted in more intracellular betaine accumulation than addition of betaine itself (Fig. 3). Aside the choline/betaine system, trehalose can be further envisioned as a major compatible solute of B728a, particularly since its role in survival and population maintenance in the phyllosphere was very recently demonstrated for P. syringae pv. tomato DC3000 (25). However, the osmoadaption profile of P. syringae pv. syringae B728a in the presence of lilac strongly suggests that ectoine is the major compatible solute utilized by the bacterium in planta, particularly since no production of betaine or trehalose could be observed under these culture conditions (Fig. 4). This fact would also explain the better epiphytic performance exhibited by P. syringae pv. syringae B728a compared with that exhibited by P. syringae pv. tomato DC3000, because both strains exhibit comparable osmolyte profiles (Fig. 3; see also Fig. S9 in the supplemental material) and the only biosynthetic difference identified between the strains was the capacity to produce the compatible solute ectoine.
In summary, with this study we obtained a comprehensive and detailed overview on genetic and metabolic levels of the strategies used by P. syringae pv. syringae B728a to confer with osmotic stress. The results presented here thus serve as an important step toward a better understanding of the ecological roles of osmolytes in P. syringae biology. In addition to these findings, we identified during our explorative study the biosynthetic gene cluster coding for NAGGN and gained insights into a new minimalistic biosynthetic locus for ectoine production.
Supplementary Material
Acknowledgments
We gratefully acknowledge E. A. Galinski and U. Habermann for interesting discussions and critical reading of the manuscript. We also thank E. Witt and A. Ures for providing the H. elongata ΔectC mutant WUB01 and A. N. S. Brünig for providing the vector pPE. We are grateful to S. Höfs for helping with the reconstitution of H. elongata WUB01 and with the in planta experiments and to S. Ruzin and D. Schichnes for support with microscope imaging and processing. We greatly appreciate the assistance of M. Al-Kaddah and R. Sherzada for semipreparative HPLC isolation work.
Footnotes
Published ahead of print on 25 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abousalham, A., M. Teissere, A. M. Gardies, R. Verger, and G. Noat. 1995. Phospholipase D from soybean (Glycine max L.) suspension-cultured cells: purification, structural and enzymatic properties. Plant Cell Physiol. 36:989-996. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aspedon, A., K. Palmer, and M. Whiteley. 2006. Microarray analysis of the osmotic stress response in Pseudomonas aeruginosa. J. Bacteriol. 188:2721-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avonce, N., A. Mendoza-Vargas, E. Morett, and G. Iturriaga. 2006. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 6:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bargmann, B. O., and T. Munnik. 2006. The role of phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 9:515-522. [DOI] [PubMed] [Google Scholar]
- 6.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 8.Boch, J., B. Kempf, and E. Bremer. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bode, H. B., R. Höfs, B. Bethe, and A. Zeeck. 2002. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3:619-627. [DOI] [PubMed] [Google Scholar]
- 10.Bonaterra, A., J. Cabrefiga, J. Camps, and E. Montesinos. 2007. Increasing survival and efficacy of a bacterial biocontrol agent of fire blight of rosaceous plants by means of osmoadaption. FEMS Microbiol. Ecol. 61:185-195. [DOI] [PubMed] [Google Scholar]
- 11.Brown, D. H., and L. Fowden. 1966. Characterization of δ-acetyl-l-ornithine isolated from Onobrychis viciifolia SCOP. Phytochemistry 5:881-886. [Google Scholar]
- 12.Brünig, A. N. S. 2005. Diploma thesis. University of Bonn, Bonn, Germany.
- 13.Chen, C., and G. A. Beattie. 2007. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-β-synthase domains are required for its osmoregulatory function. J. Bacteriol. 189:6901-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, C., and G. A. Beattie. 2008. Pseudomonas syringae BetT is a low-affinity choline transporter that is responsible for superior osmoprotection by choline over glycine betaine. J. Bacteriol. 190:2717-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, C., A. A. Malek, M. J. Wargo, D. A. Hogan, and G. A. Beattie. 2010. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75:29-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi, J. H., H. Y. Jung, H. S. Kim, and H. G. Cho. 2000. PhyloDraw: a phylogenetic tree drawing system. Bioinformatics 16:1056-1058. [DOI] [PubMed] [Google Scholar]
- 17.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Conner, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 18.Csonka, L. N. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Csonka, L. N., and A. D. Hanson. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569-606. [DOI] [PubMed] [Google Scholar]
- 20.Cuppels, D. A. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jong, C. F., A. M. Laxalt, B. O. Bargmann, P. J. de Wit, M. H. Joosten, and T. Munnik. 2004. Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J. 39:1-12. [DOI] [PubMed] [Google Scholar]
- 22.D'Souza-Ault, M. R., L. T. Smith, and G. M. Smith. 1993. Roles of N-acetylglutaminylglutamine amide and glycine betaine in adaptation of Pseudomonas aeruginosa to osmotic stress. Appl. Environ. Microbiol. 59:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans, C. S., J. Clardy, P. F. Hughes, and E. A. Bell. 1985. 2-Amino-4-acetylaminobutyric acid, 2,4-diaminobutyric acid and 2-amino-6N-oxalylureidopropionic acid (oxalylalbizziine) in seeds of Acacia angustissima. Phytochemistry 24:2273-2275. [Google Scholar]
- 24.Feil, H., W. S. Feil, P. Chain, F. Larimer, G. DiBartolo, A. Copeland, A. Lykidis, S. Trong, M. Nolan, E. Goltsman, J. Thiel, S. Malfatti, J. E. Loper, A. Lapidus, J. C. Detter, M. Land, P. M. Richardson, N. C. Kyrpides, N. Ivanova, and S. E. Lindow. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 102:11064-11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman, B. C., C. Chen, and G. A. Beattie. 2010. Identification of the trehalose biosynthetic loci of Pseudomonas syringae and their contribution to fitness in the phyllosphere. Environ. Microbiol. 12:1486-1497. [DOI] [PubMed] [Google Scholar]
- 26.Galinski, E. A. 1995. Osmoadaption in bacteria. Adv. Microb. Physiol. 37:272-328. [PubMed] [Google Scholar]
- 27.Galinski, E. A., and R. M. Herzog. 1990. The role of trehalose as a substitute for nitrogen-containing compatible solutes (Ectothiorhodospira halochloris). Arch. Microbiol. 153:607-613. [Google Scholar]
- 28.Göller, K., A. Ofer, and E. A. Galinski. 1998. Construction and characterization of an NaCl-sensitive mutant of Halomonas elongata impaired in ectoine biosynthesis. 1998. FEMS Microbiol. Lett. 161:293-300. [DOI] [PubMed] [Google Scholar]
- 29.Göller, K. 1999. Ph.D. thesis. University of Bonn, Bonn, Germany.
- 30.Grove, D. C., and W. A. Randall. 1955. Assay methods of antibiotics: a laboratory manual. Antibiotics monograph no. 2. Medical Encyclopedia Inc., New York, NY.
- 31.Grunwald, K. R. 2009. Diploma thesis. University of Bonn, Bonn, Germany.
- 32.Hagemann, M., K. Ribbeck-Busch, S. Klähn, D. Hasse, R. Steinbruch, and G. Berg. 2008. The plant-associated bacterium Stenotrophomonas rhizophila expresses a new enzyme for the synthesis of the compatible solute glucosylglycerol. J. Bacteriol. 190:5898-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kets, E. P. W., E. A. Galinski, M. De Wit, J. A. M. De Bont, and H. J. Heipieper. 1996. Mannitol, a novel bacterial compatible solute in Pseudomonas putida S12. J. Bacteriol. 178:6665-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 36.Kovach, M. E., P. H. Elzer, S. D. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS1, carrying different antibiotic resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 37.Kurz, M., C. Montermann, and E. A. Galinski. 2007. A Pseudomonas stutzeri surprise: production of the compatible osmolyte hydroxyectoine, abstr. PT015, p. 195. Abstr. Annu. Conf. Assoc. Gen. Appl. Microbiol. VAAM, Osnabrück, Germany.
- 38.Kurz, M. 2008. Compatible solute influence on nucleic acids: many questions but few answers. Saline Systems 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. ClustalW and ClustalX version 2. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 40.Larsen, P. I., L. K. Sydnes, B. Landfald, and A. R. Strøm. 1987. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: betaines, glutamic acid and trehalose. Arch. Microbiol. 147:1-7. [DOI] [PubMed] [Google Scholar]
- 41.Lederberg, J. 1950. Isolation and characterization of biochemical mutants of bacteria, p. 5-22. In R. W. Gerard (ed.), Methods in medical research, vol. 3. Year Book Publishers Inc., Chicago, IL. [Google Scholar]
- 42.Lentzen, G., and T. Schwarz. 2006. Extremolytes: natural compounds from extremophiles for versatile applications. Appl. Microbiol. Biotechnol. 72:623-634. [DOI] [PubMed] [Google Scholar]
- 43.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liss, I. 1962. N-Acetyldiaminobuttersäure, eine neue Aminosäure aus dem Latex von Euphorbia pulcherrima Willd ex Klotzsch. Phytochemistry 1:87-88. [Google Scholar]
- 45.Loper, J. E., and S. E. Lindow. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449-1454. [Google Scholar]
- 46.Louis, P., and E. A. Galinski. 1997. Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in E. coli. Microbiology 143:1141-1149. [DOI] [PubMed] [Google Scholar]
- 47.Marco, M. L., J. Legac, and S. E. Lindow. 2005. Pseudomonas syringae genes induced during colonization of leaf surfaces. Environ. Microbiol. 7:1379-1391. [DOI] [PubMed] [Google Scholar]
- 48.Maruta, K., K. Hattori, T. Nakada, M. Kubota, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of trehalose biosynthesis genes from Arthrobacter sp. Q36. Biochim. Biophys. Acta 1289:10-13. [DOI] [PubMed] [Google Scholar]
- 49.Maruta, K., K. Hattori, T. Nakada, M. Kubota, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of trehalose biosynthesis genes from Rhizobium sp. M-11. Biosci. Biotechnol. Biochem. 60:717-720. [DOI] [PubMed] [Google Scholar]
- 50.Maruta, K., H. Mitsuzumi, T. Nakada, M. Kubota, H. Chaen, S. Fukuda, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of a cluster of genes encoding novel enzymes of trehalose biosynthesis from thermophilic archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 1291:177-181. [DOI] [PubMed] [Google Scholar]
- 51.Maruta, K., M. Kubota, S. Fukuda, and M. Kurimoto. 2000. Cloning and nucleotide sequence of a gene encoding a glycogen debranching enzyme in the trehalose operon from Arthrobacter sp. Q36. Biochim. Biophys. Acta 1476:377-381. [DOI] [PubMed] [Google Scholar]
- 52.Matthysse, A. G., S. Stretton, C. Dandie, N. C. McClure, and A. E. Goodman. 1996. Construction of GFP vectors for use in Gram-negative bacteria other than Escherichia coli. FEMS Microbiol. Lett. 145:87-94. [DOI] [PubMed] [Google Scholar]
- 53.Mikkat, S., E. A. Galinski, G. Berg, A. Minkwitz, and A. Schoor. 2000. Salt adaption in pseudomonads: characterization of glucosylglycerol-synthesizing isolates from brackish coastal waters and the rhizosphere. Syst. Appl. Microbiol. 23:31-40. [DOI] [PubMed] [Google Scholar]
- 54.Miller, W. G., J. H. J. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 55.Mo, Y.-Y., and D. C. Gross. 1991. Plant signal molecules activate the syrB gene, which is required for syringomycin production by Pseudomonas syringae pv. syringae. J. Bacteriol. 173:5784-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monier, J. M., and S. E. Lindow. 2003. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. U. S. A. 100:15977-15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nyyssölä, A., J. Kerovuo, P. Kaukinen, N. von Weymarn, and T. Reinikainen. 2000. Extreme halophiles synthesize betaine from glycine by methylation. J. Biol. Chem. 275:22196-22201. [DOI] [PubMed] [Google Scholar]
- 58.Ono, H., M. Okuda, S. Tongpim, K. Imai, A. Shinmyo, S. Sakuda, Y. Kaneko, Y. Murooka, and M. Takano. 1998. Accumulation of compatible solutes, ectoine and hydroxyectoine, in a moderate halophile, Halomonas elongata KS3 isolated from dry salty land in Thailand. J. Ferment. Bioeng. 85:362-368. [Google Scholar]
- 59.Ono, H., K. Sawada, N. Khunajakr, T. Tao, M. Yamamoto, M. Hiramoto, A. Shinmyo, M. Takano, and Y. Murooka. 1999. Characterization of biosynthetic enzymes for ectoine as a compatible solute in a moderately halophilic eubacterium Halomonas elongata. J. Bacteriol. 181:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oren, A. 1999. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63:334-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peleg-Grossman, S., H. Volpin, and A. Levine. 2007. Root hair curling and Rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. J. Exp. Bot. 58:1637-1649. [DOI] [PubMed] [Google Scholar]
- 62.Pocard, J.-A., L. T. Smith, G. M. Smith, and D. Le Rudulier. 1994. A prominent role for glucosylglycerol in the adaptation of Pseudomonas mendocina SKB70 to osmotic stress. J. Bacteriol. 176:6877-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quiñones, B., C. J. Pujol, and S. E. Lindow. 2004. Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant Microbe Interact. 17:521-531. [DOI] [PubMed] [Google Scholar]
- 64.Quiñones, B., G. Dulla, and S. E. Lindow. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 18:682-693. [DOI] [PubMed] [Google Scholar]
- 65.Roberts, M. F. 2005. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Systems 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roessler, M., and V. Müller. 2001. Osmoadaption in bacteria and archaea: common principles and differences. Environ. Microbiol. 3:743-754. [DOI] [PubMed] [Google Scholar]
- 67.Rosello, R., E. Garcia-Valdes, J. Lalucat, and J. Ursing. 1991. Genotypic and phenotypic diversity of Pseudomonas stutzeri. Syst. Appl. Microbiol. 14:150-157. [Google Scholar]
- 68.Ryu, S. B., and X. Wang. 1996. Activation of phospholipase D and the possible mechanism of activation in wound-induced lipid hydrolysis in castor bean leaves. Biochim. Biophys. Acta 1303:243-250. [DOI] [PubMed] [Google Scholar]
- 69.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 70.Saum, S. H., and V. Müller. 2008. Regulation of osmoadaption in the moderate halophile Halobacillus halophilus: chloride, glutamate and switching osmolyte strategies. Saline Systems 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seip, B., M. Stein, E. A. Galinski, and M. Kurz. 2009. Pseudomonas stutzeri DSM5190T as a potential production strain for the compatible osmolyte hydroxyectoine, abstr. PX23, p. 200. Abstr. Annu. Conf. Assoc. Gen. Appl. Microbiol. VAAM, Bochum, Germany.
- 72.Severin, J., A. Wohlfarth, and E. A. Galinski. 1992. The predominant role of recently discovered tetrahydrpyrimidines for the osmoadaption of halophilic eubacteria. J. Gen. Microbiol. 138:1629-1638. [Google Scholar]
- 73.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 74.Sleator, R. D., and C. Hill. 2001. Bacterial osmoadaption: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49-71. [DOI] [PubMed] [Google Scholar]
- 75.Smith, L. T., and G. M. Smith. 1989. An osmoregulated dipeptide in stressed Rhizobium meliloti. J. Bacteriol. 171:4714-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith, L. T., A. A. Allaith, and G. M. Smith. 1994. Mechanism of osmotically regulated N-acetylglutaminylglutamine amide production in Rhizobium meliloti. Plant Soil 161:103-108. [Google Scholar]
- 77.Storey, R., and R. G. Wyn Jones. 1975. Betaine and choline levels in plants and their relationship to NaCl stress. Plant Sci. Lett. 4:161-168. [Google Scholar]
- 78.van de Mortel, M., and L. J. Halverson. 2004. Cell envelope components contributing to biofilm growth and survival of Pseudomonas putida in low-water-content habitats. Mol. Microbiol. 52:735-750. [DOI] [PubMed] [Google Scholar]
- 79.Vreeland, R. H. 1980. The family Halomonadaceae. In M. P. Starr et al. (ed.), The prokaryotes, 2nd ed. Springer Press, Berlin, Germany.
- 80.Zeisel, S. H., M. H. Mar, J. C. Howe, and J. M. Holden. 2003. Concentrations of choline-containing compounds and betaine in common foods. J. Nutr. 133:1302-1307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.