Abstract
Balancing of reducing equivalents is a fundamental issue in bacterial metabolism and metabolic engineering. Mutations in the key metabolic genes ldhA and pflB of Escherichia coli are known to stall anaerobic growth and fermentation due to a buildup of intracellular NADH. We observed that the rate of spontaneous mutation in E. coli BW25113 (ΔldhA ΔpflB) was an order of magnitude higher than that in wild-type (WT) E. coli BW25113. We hypothesized that the increased mutation frequency was due to an increased NADH/NAD+ ratio in this strain. Using several redox-impaired strains of E. coli and different redox conditions, we confirmed a significant correlation (P < 0.01) between intracellular-NADH/NAD+ ratio and mutation frequency. To identify the genetic basis for this relationship, whole-genome transcriptional profiles were compared between BW25113 WT and BW25113 (ΔldhA ΔpflB). This analysis revealed that the genes involved in DNA repair were expressed at significantly lower levels in BW25113 (ΔldhA ΔpflB). Direct measurements of the extent of DNA repair in BW25113 (ΔldhA ΔpflB) subjected to UV exposure confirmed that DNA repair was inhibited. To identify a direct link between DNA repair and intracellular-redox ratio, the stringent-response-regulatory gene relA and the global-stress-response-regulatory gene rpoS were deleted. In both cases, the mutation frequencies were restored to BW25113 WT levels.
The genes encoding lactate dehydrogenase (ldhA) and pyruvate-formate lyase (pflB) constitute the primary target for redirecting glucose flux in Escherichia coli growing under anaerobic conditions (3, 4, 29, 32). The pyruvate flux is then diverted toward the formation of desirable bioproducts by overexpression of native (61) and nonnative (62, 71) genes. However, insufficient reduction of pyruvate in such recombinant strains leads to an accumulation of NADH with broad effects on cellular fitness. Intracellular-redox ratios (NADH/NAD+) as high as 3 times that of the wild-type (WT) E. coli strain were reported for an ldhA pflB double knockout strain (60). Previous studies attribute an unusually high redox ratio (NADH/NAD+) in the cytoplasm to the inhibition of the fermentative growth on glucose in minimal or complex medium (8, 44, 61, 71).
Suboptimal growth rates due to various environmental conditions elicit stress responses in bacteria. Bacteria have evolved a battery of mechanisms to cope with the diverse stresses encountered in nature, and the interdependence of these responses is well established (6, 22, 48). In E. coli, the general stress regulator RpoS has been implicated as the primary defense mediator. The level of expression of RpoS is greatest in the stationary phase, although growth rate-dependent control in the exponential phase is also reported (14, 28, 50). This stress-regulatory protein has been implicated in the E. coli responses to nutrient limitation (26), DNA damage (46), osmotic shock (27), oxidative stress (55), ethanol resistance (20), acid stress (38), and biofilm formation (1). RpoS was also shown to regulate the transcription of catalases (encoded by katG and katE) and glutaredoxin, two primary antioxidative cellular defense mechanisms (52, 56). The regulation of the genes involved in DNA repair also provides RpoS a role in adaptive mutagenesis (39). RpoS-dependent downregulation of the mismatch repair (MMR) system and induction of error-prone DNA polymerase IV are believed to be responsible for the increased mutagenesis under stress (39, 69). Adaptive mutagenesis in cells exhibiting the SOS response, the cellular response against oxidative stress, has been studied in extensive detail (19, 21, 34, 36, 49, 63, 72, 74). RecA and RpoS play a central role in protection against damage from superoxide and peroxide radicals generated due to NADH oxidation (45, 66, 73). Downregulation of DNA repair and/or upregulation of error-prone DNA polymerases by stress response regulators leads to an increased rate of mutation of bacteria, a phenomenon known as stress-induced mutagenesis (SIM) (5, 30, 47, 66, 69). It is thought that increased mutagenesis under stress conditions may provide bacterial populations with an avenue for generating beneficial mutations capable of circumventing growth-limiting conditions.
Growth limitation due to nutritional deficiency elicits the stringent response (6, 7). The stringent response in E. coli in response to amino acid starvation is characterized by a rapid inhibition of rRNA and tRNA synthesis and upregulation of the metabolic genes (15, 17, 37, 68). The stringent-response messenger (p)ppGpp, synthesized by the association of the stringent factor relA with the ribosomal protein L11, binds to the β subunit of RNA polymerase to modulate the expression of over a third of all E. coli genes (10, 67, 68, 70). The interdependence of the RpoS-mediated stress response and the stringent response is well documented (35). The expression of RpoS and RecA is increased during the stringent response (17, 43, 68), while several stationary-phase promoters controlled by RpoS also show a requirement for ppGpp (9, 35).
In this study, we sought to investigate the effect of redox imbalance-induced growth defect on genetic stability. It was observed that the frequencies of rifampin resistance in E. coli ldhA pflB double knockout strains BW25113 (ΔldhA ΔpflB) and NZN111 (W1485 Δpfl::cam ldhA::kan) were an order of magnitude higher than those in wild-type E. coli BW25113. These strains, and their derivatives with intermediate redox ratios, were used to demonstrate a statistically significant correlation between cytoplasmic-NADH/NAD+ ratio and rate of spontaneous mutation. Transcriptional profiling revealed that genes involved in DNA repair in BW25113 (ΔldhA ΔpflB) were repressed relative to the levels for the BW25113 WT strain. Thus, we hypothesized that the elevated mutation rate in the double knockouts was a result of decreased DNA repair abilities. This hypothesis was explored by estimating the extent of DNA repair in BW25113 (ΔldhA ΔpflB) by subjecting this strain to direct DNA damage by UV exposure. BW25113 (ΔldhA ΔpflB) showed considerably lower survival rates than the wild-type E. coli strain. BW25113 (ΔldhA ΔpflB) also showed an increased level of expression of energy metabolism genes besides the overexpression of several stress regulators, including rpoS and the stringent-response mediator rplK. These observations, along with the known function of these regulators, led us to speculate that the redox imbalance in BW25113 (ΔldhA ΔpflB) might be eliciting a physiological response similar to that of nutrient limitation, thus causing an increase in spontaneous mutagenesis. The model was verified by deleting the stringent factor (relA) and the stress regulator rpoS in BW25113 (ΔldhA ΔpflB), which restored the frequency of rifampin resistance to BW25113 WT levels.
MATERIALS AND METHODS
Strains used and knockout construction.
Mutation studies were performed on two E. coli strains, BW25113 [Δ(araD-araB)567 ΔlacZ4787(::rrnB3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514] and NZN111 (W1485 Δpfl::cam ldhA::kan). Plasmid clones of NZN111 carried the indicated gene in a medium-copy-number plasmid, pBTL-1 (42), under the control of their native promoter (60). All deletions were constructed in BW25113 by following the method developed earlier (13). The kanamycin resistance cassette was amplified from plasmid pKD13 by PCR using primers with flanking homologous regions for the desired gene. The purified PCR product was electroporated into E. coli BW25113 harboring λ-Red recombinase induced off the plasmid pKD46 by use of 10 mM arabinose. The resulting kanamycin-resistant colonies were screened for the desired gene knockout by PCR amplification and subsequent sequencing. Primers for this confirmation step were designed to bind 300 and 400 bp upstream and downstream, respectively, of the target gene. Plasmid pCP20 carrying the FLP-recombinase was subsequently used to excise the kanamycin selection marker from the knockout strain. All plasmids were cured by propagating the strains at 43°C. Strains and plasmid stocks were obtained from the E. coli Genetic Stock Center at Yale University, New Haven, CT.
Growth conditions and analytical methods.
The fluctuation test developed by Luria and Delbrück (40) was used to test the appearance of rifampin-resistant mutants. The frequency of mutagenesis and the mutation rate were calculated by the method developed by Drake (16). Bacteria were allowed to grow planktonically in MOPS (morpholinepropanesulfonic acid) minimal medium, containing 10 g/liter glucose as the carbon source and supplemented with 1 mg/ml thiamine, from a small population size of ∼103 cells to a final cell count of ∼109. The cultures were grown in 15-ml Nalgene tubes with no headspace to achieve microaerobic conditions, a strategy frequently reported in the literature (11, 12, 41). For the fluctuation test experiments in the presence of an external electron acceptor, 10 mM sodium nitrate was added to the culture medium. Due to the low growth yields, the double mutants and their plasmid derivatives were grown in larger volumes (∼10-fold) to attain the desired cell counts, pelleted, and resuspended in a smaller volume prior to mutagenesis studies. The cells were harvested, diluted, and spread on LB plates supplemented with 100 μg/ml rifampin or not supplemented and were incubated at 37°C in the dark to avoid light-induced degradation of rifampin. Frequency of rifampin resistance was calculated by dividing the number of colonies on LB-rifampin plates by the number of colonies on LB plates.
To measure NADH/NAD+ ratio, cells grown microaerobically were harvested during mid-exponential phase. The cofactors were extracted from the cells lysed by two freeze-thaw cycles and quantified via the NADH cycling assay (NADH quantification kit; Biovision Research Products, Mountain View, CA) in accordance with the manufacturer's procedure.
UV irradiation.
Exponentially growing cells under microaerobic conditions in MOPS minimal medium containing 10 g/liter glucose and supplemented with 1 mg/ml thiamine were harvested and resuspended to an optical density at 600 nm (OD600) of 0.1 in the fresh medium. One hundred microliters of diluted cells was exposed to UV radiation in a flat-bottomed tube (diameter = 1 mm) using an Acticure collimated light source with a 365-nm internal interference filter (EFOS, Inc.). The energy density of UV was set at 5 J/m2/s, and the exposure time was varied to attain ascending levels of UV dose (31, 59). Irradiated cells were washed, diluted in the fresh medium, spread on LB plates, and allowed to grow overnight under aerobic conditions for estimation of viability. The plates were grown in the dark to avoid photorepair of the UV-damaged DNA.
Transcriptional profiling.
For RNA isolation, total RNA was extracted from ∼1 × 109 cells from exponential-phase cultures growing microaerobically in MOPS minimal medium supplemented with 1 mg/ml thiamine, using Qiagen's RNeasy kit in accordance with the manufacturer's protocol. The isolated RNA was quantified by absorbance at 260 nm on a UV-visible-light (UV-Vis) spectrophotometer (Shimadzu Corp.). A SuperScript III reverse transcriptase kit (Invitrogen) was used to synthesize cDNA from the extracted RNA by use of random hexamers (Invitrogen). Following cDNA synthesis, RNA was degraded by adding 1 N NaOH and incubating at 65°C for 30 min. The pH of the solution was adjusted back to neutral by the addition of 1 N HCl. cDNA was subsequently purified using a QIAquick PCR purification kit. Purified cDNA was fragmented with DNase I (Amersham Biosciences) for 10 min at 37°C. DNase I was later heat inactivated at 98°C for 10 min. Fragmented cDNA was then biotin labeled using a terminal labeling kit from Enzo Bioarray in accordance with the manufacturer's protocol. About 3 μg of the labeled fragmented product was hybridized onto a GeneChip E. coli antisense genome array from Affymetrix. Arrays were handled at the University of Colorado DNA Microarray Facility according to the manufacturer's specifications, using a GeneChip hybridization oven, a GeneChip fluidics station, a GeneArray scanner, and GeneChip Operating Software 1.1 (Affymetrix).
Labeled cDNAs corresponding to BW25113 (ΔldhA ΔpflB) and parent strain BW25113 were hybridized onto Affymetrix arrays. The .cel files were processed using the Affymetrix MAS5 normalization routine. Fold changes were calculated from signal log ratios. Identified transcripts were clustered based on the Cluster of Orthologous Group (COG) functional information available in the NCBI and EcoCyc databases.
Microarray data accession number.
The expression profiling data have been deposited in NCBI's Gene Expression Omnibus database and are accessible through GEO Series accession number GSE21995 (18).
RESULTS AND DISCUSSION
Relationship between mutation rate and NADH/NAD+ ratio.
E. coli strains lacking lactate dehydrogenase (ldhA) and pyruvate-formate lyase (pflB) genes are incapable of growing on glucose in rich or minimal medium under anaerobic conditions (8, 61) and are known to accumulate high levels of pyruvate (8, 44, 61, 71). This inability to reduce pyruvate leads to an accumulation of the reduced cofactor NADH in the cytoplasm. In our previous study, we employed a genomic library selection approach for an E. coli strain NZN111 with a similar ΔldhA ΔpflB genotype to identify a set of genetic elements capable of restoring growth by reducing the intracellular-redox ratio (60).
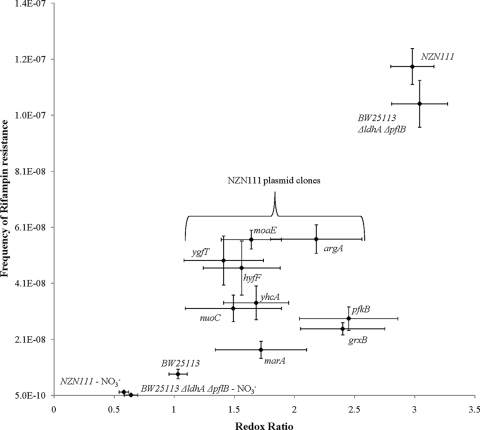
Here, we sought to study the effect of NADH accumulation on genetic stability. We observed that the occurrence of the rifampin-resistant clones was 7-fold higher in BW25113 (ΔldhA ΔpflB) than in wild-type BW25113 during growth under oxygen-limiting conditions (Fig. 1). No such difference was observed in aerobically growing cultures (data not shown), suggesting that an accumulation of NADH under fermentative conditions in the double knockout strain was linked to the observed increase in the frequency of rifampin resistance. The observation was consistent in E. coli strain NZN111 (Fig. 1). In our previous study, we reported that increased copy numbers of several genes affected the intracellular-redox ratio in E. coli NZN111 plasmid clones (60). To further explore the relationship between mutation frequency and redox ratio, fluctuation experiments were extended to E. coli NZN111 plasmid clones with intermediate levels of redox ratio identified in the previous study. As shown in Fig. 1, the mutation rate was found to be a function of the intracellular-redox ratio. Supplementation of the growth medium with electron acceptors (e.g., nitrate salts) was also observed to reduce the NADH/NAD+ ratio as well as the mutagenesis frequency. These results demonstrate that a significant link (P = 0.0008) exists between intracellular-redox ratio and rate of spontaneous mutagenesis in E. coli. While high rates of NADH oxidation associated with oxidative stress have been linked to increased mutagenesis, a relationship between a lack of oxidation, thus NADH accumulation, and increased mutagenesis has not previously been established (23). These results suggest that E. coli has evolved systems to link stress-induced mutagenesis to redox ratios that are either too high or too low.
FIG. 1.
Effect of the intracellular-redox ratio on the frequency of rifampin resistance. Intracellular-redox ratio in the ΔldhA ΔpflB mutant was varied either by 10 mM sodium nitrate supplementation or by transformation with the plasmids carrying the indicated E. coli genes. The increased copy numbers of these genes on the plasmid pBTL-1 vector backbone under the control of their native promoters were previously reported to influence the redox ratio in the ldhA pflB double knockout strain NZN111 (60). Each data point refers to the mean ± standard deviation (SD) of results from 5 independent trials. The mutation rate for all strains was found to be statistically different from that for the wild-type strain BW25113 (P < 0.05). Pearson's product moment correlation coefficient (r) between mutation rate and redox ratio was found to be 0.79 (degrees of freedom= 12; P = 0.0008).
Whole-genome gene expression profiling of BW25113 and BW25113 (ΔldhA ΔpflB).
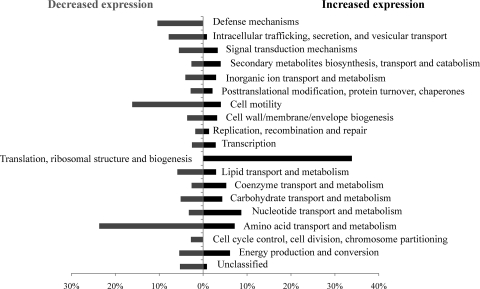
To identify a genetic basis for the observed relationship between redox ratio and mutation frequency, whole-genome transcriptional profiling experiments were performed. Samples were obtained at mid-exponential phase after growth of either BW25113 or BW25113 (ΔldhA ΔpflB) under microaerobic conditions in minimal medium (see Materials and Methods). Transcriptional data were analyzed both by examining the function of the genes with the greatest changes in expression between the two strains and by summarizing the differentially expressed genes according to their respective Cluster of Orthologous Group (COG) assignments and Gene Ontology annotations (2, 64, 65). Figure 2 shows the percentages of genes showing expression change {|log2 [BW25113 (ΔldhA ΔpflB)/BW25113]|} values of >2 in the double mutant compared to the level for the wild-type strain. On the basis of COG classification, energy production, amino acids, lipids, and nucleotide metabolism, genes show both positive and negative expression changes, presumably to counter energy and growth limitation. Interestingly, only translation and ribosomal genes showed uniformly positive expression change, while genes controlling cell cycle and metabolite trafficking and secretion showed uniformly negative changes. It is also interesting to note that the genes belonging to COG-based defense mechanisms were consistently repressed in BW25113 (ΔldhA ΔpflB) compared to the level for the wild type (Fig. 2). Taken together, these data suggest that the response to redox imbalance in the double mutant might involve global stress response functions, which is expected given the central role that NADH/NAD+ play in metabolism. To examine this speculation further, we evaluated the expression of individual genes known to play important roles in stress responses in E. coli that have been implicated in DNA mutagenesis and repair.
FIG. 2.
Extent of gene expression change in BW25113 (ΔldhA ΔpflB) compared to the level for wild-type BW25113. The x axis denotes the percentage of genes showing expression change values {|log2 [BW25113 (ΔldhA ΔpflB)/BW25113]|} of >2 for each COG functional category.
Repression of the defense mechanisms might be caused by the general stress response genes, such as rpoS, hfq, rplK, and crp, each of which showed increased expression in BW25113 (ΔldhA ΔpflB). Table 1 lists cellular stress response genes annotated according to the Gene Ontology classification (2). This observation can be explained by prior reports relating the expression of the stress regulators to growth, which is inhibited in BW25113 (ΔldhA ΔpflB) (14, 26, 28, 50). Consistent with the stringent response, genes coding for tRNA were repressed while the expression of amino acid transport genes was activated. Specifically, proline transport genes (proW) and proline tRNA (proL) showed the greatest positive (log2 ratio = 6.42) and negative (log2 ratio = −5.45) changes in expression in the mutant relative to the level for the wild-type parent. One crucial difference is that contrary to the inhibition of the ribosome synthesis during the stringent response (10, 15, 17, 37, 67, 68, 70), we observed increases in the expression levels of genes involved in translation and ribosomal structure (Fig. 2). However, it should be noted that the inhibition of ribosomal synthesis as part of the stringent response occurs within minutes (15, 17, 37, 68), while we performed transcriptional profiling during the mid-exponential phase after several hours of growth under inhibitory conditions. The stringent response is also characterized by inhibition of peptidoglycan synthesis, leaving the cells insensitive to β-lactam antibiotics. The stringent response can be eliminated (or relaxed) by inhibiting the synthesis of ppGpp, e.g., by introducing a mutation in relA or by treating amino acid-deprived bacteria with certain inhibitors of ribosome function, such as chloramphenicol. Relaxed mutants have normal peptidoglycan synthesis and are thus sensitive to β-lactam-induced lysis (51, 53, 54). Consistent with this property, BW25113 (ΔldhA ΔpflB) was observed to be sensitive to β-lactam-induced lysis (data not shown) in the presence of chloramphenicol.
TABLE 1.
Genes involved in cellular response to stress according to Gene Ontology annotation (GO:0033554)a
| Gene name | Log2 value (mutant/WT) |
|---|---|
| ompT | 5.08 |
| ompC | 4.10 |
| ompA | 3.55 |
| cspA | 3.49 |
| nusA | 2.48 |
| cspG | 2.42 |
| hfq | 2.40 |
| hfq | 2.39 |
| sspA | 1.95 |
| rpoE | 1.92 |
| rfaD | 1.90 |
| iscR | 1.71 |
| uspA | 1.68 |
| cspB | 1.63 |
| pnp | 1.62 |
| rpoS | 1.59 |
| Crp | 1.56 |
| clpP | 1.49 |
| hslV | 1.33 |
| rrmJ | 1.28 |
| hns | 1.24 |
| nudB | 1.11 |
| dps | 1.09 |
| dnaK | 1.08 |
| marC | −1.02 |
| ibpB | −1.12 |
| uspB | −1.14 |
| hslO | −1.15 |
| astD | −1.25 |
| kdpD | −1.25 |
| astB | −1.28 |
| cstA | −1.30 |
| cstA | −1.30 |
| ves | −1.31 |
| pstS | −1.34 |
| ykfG | −1.36 |
| polB | −1.38 |
| phoH | −1.40 |
| yjiY | −1.40 |
| astA | −1.42 |
| pspC | −1.42 |
| hslJ | −1.62 |
| rzpD | −1.66 |
| phoE | −1.76 |
| ydaX | −1.80 |
| ygeG | −1.85 |
| htrE | −1.86 |
| uspC | −1.92 |
| ecpD | −2.20 |
| astE | −2.22 |
Only the genes with |log2 [BW25113 (ΔldhA ΔpflB)/BW25113]| values of >1 are shown.
On the basis of these observations, we hypothesized that the growth inhibition caused by the redox imbalance in BW25113 (ΔldhA ΔpflB) triggers a stress response similar to one faced during nutrient limitation and that this response results in decreased DNA repair. To test this model, we estimated the sensitivity of the double mutant to UV-induced DNA damage.
Attenuated DNA repair functions in BW25113 (ΔldhA ΔpflB).
In E. coli, methyl-directed mismatch repair (MMR) is the primary mechanism for maintaining genetic stability against replication errors caused by DNA polymerases as well as genetic recombination and transposon excision (5, 24, 25, 73). MutH, an endonuclease involved in postreplicative DNA repair, shows decreased expression in BW25113 (ΔldhA ΔpflB), consistent with its reported repression by stress regulators RpoS and Hfq (69). Uracil-DNA-glycosylase, encoded by ung, involved in DNA repair upon misincorporation and cytosine deamination, showed the lowest expression levels among all DNA repair genes. Exonuclease III (XthA) (involved in repair of DNA following removal of damaged bases by DNA glycosylases), uvrA and umuC (encoding proteins involved in DNA repair following UV damage), and recE and recN (general DNA repair and recombination genes) all showed decreased expression (Table 2). The activity of the UvrABC complex is reported to be negatively regulated by the protease OmpT (57, 58), whose expression is greatly upregulated in BW25113 (ΔldhA ΔpflB). Table 2 lists the changes in the expression levels of DNA repair genes annotated according to the Gene Ontology classification (2). Although several genes are not expected to be functional in the absence of any direct DNA damage in BW25113 (ΔldhA ΔpflB), a common trend of decreased expression DNA repair mechanisms is evident.
TABLE 2.
Log2 expression ratio changes for genes involved in DNA repair according to Gene Ontology annotation (GO:0006281)
| Gene name | Log2 value (mutant/WT) |
|---|---|
| recA | 1.16 |
| himA | 0.63 |
| dinI | 0.57 |
| recQ | 0.46 |
| ruvC | 0.37 |
| mutL | 0.34 |
| recC | 0.13 |
| uvrC | 0.08 |
| uvrD | 0.05 |
| dinG | −0.04 |
| ung | −0.05 |
| katG | −0.08 |
| ssb | −0.08 |
| ruvA | −0.2 |
| sodC | −0.31 |
| mutS | −0.34 |
| mutS | −0.34 |
| xthA | −0.36 |
| recJ | −0.38 |
| polA | −0.38 |
| mfd | −0.4 |
| recB | −0.45 |
| sbcB | −0.49 |
| ogt | −0.57 |
| lexA | −0.57 |
| uvrB | −0.58 |
| ada | −0.6 |
| tag | −0.6 |
| ligA | −0.71 |
| phrB | −0.89 |
| uvrA | −0.91 |
| soxs | −0.94 |
| umuD | −0.97 |
| mutM | −0.99 |
| helD | −1.03 |
| nth | −1.04 |
| nth | −1.04 |
| recD | −1.08 |
| vsr | −1.09 |
| vsr | −1.09 |
| dinD | −1.09 |
| ymgE | −1.12 |
| mutT | −1.12 |
| dinF | −1.12 |
| modF | −1.13 |
| yicR | −1.13 |
| yicR | −1.13 |
| sbmC | −1.21 |
| umuC | −1.24 |
| ligB | −1.26 |
| yeeS | −1.27 |
| nfo | −1.34 |
| nfo | −1.34 |
| ykfG | −1.36 |
| polB | −1.38 |
| alkB | −1.42 |
| soxR | −1.46 |
| rusA | −1.57 |
| recX | −1.58 |
| recE | −1.61 |
| ruvB | −1.64 |
| alkA | −1.67 |
| mutH | −1.76 |
| dinP | −1.8 |
| recN | −2.05 |
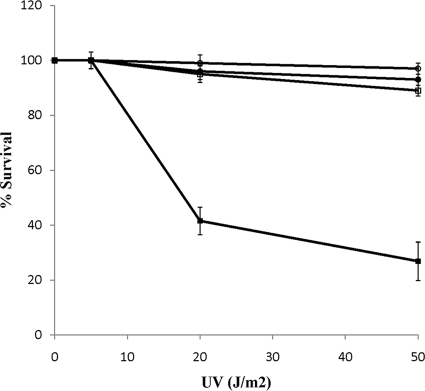
To test the hypothesis that the decreased activity of DNA repair proteins is responsible for the increased mutation frequency, cells were subjected to direct DNA damage by exposure to various doses of UV radiation and their survival rates were measured (31, 59). Wild-type E. coli BW25113 (growing aerobically or microaerobically) showed >90% survival in the UV dose range studied. On the other hand, the survival levels of BW25113 (ΔldhA ΔpflB) growing microaerobically were significantly (P < 0.05) lower than those of the cultures growing aerobically (Fig. 3). This suggests that besides limiting cellular growth, the accumulation of NADH under microaerobic conditions also reduces the ability to repair damaged DNA in E. coli.
FIG. 3.
Survival of the wild-type strains BW25113 (circles) and BW25113 (ΔldhA ΔpflB) (squares) after UV exposure under aerobic (open symbols) and microaerobic (filled symbols) conditions. Each data point refers to the mean ± SD of results from 5 independent trials. The difference between the survival rates for BW25113 under aerobic versus microaerobic conditions was statistically insignificant (P > 0.05), while BW25113 (ΔldhA ΔpflB) showed a significant difference in the survival rates (P < 0.05).
Deletion of relA and rpoS restores the normal mutation rate in BW25113 (ΔldhA ΔpflB).
The RpoS-mediated stress regulon constitutes the primary defense mechanism in E. coli. Deletion of rpoS is known to severely affect the rate of survival against various stress conditions (1, 20, 26, 27, 38, 46, 55). RpoS is also widely implicated in stress-induced mutagenesis in bacteria, plays a role in adaptive evolution (39, 69), and is reported to act in a concerted manner with other stress responses, specifically the stringent response (9, 35). To validate our model indicating that the redox imbalance-induced stress response limits the genetic fidelity function in E. coli, we assessed the effects of deletion of the stringent factor relA and the global stress regulator rpoS on mutation frequency in BW25113 (ΔldhA ΔpflB). The absence of either of these two stress regulators was observed to abolish the increased mutagenesis (SIM) phenotype in the double mutant. The frequencies of rifampin resistance decreased to 1.2E−8 ± 0.08E−8 with rplK deletion and to 1.6E−8 ± 0.01E−8 with deletion of rpoS in BW25113 (ΔldhA ΔpflB). The restoration of the normal mutation levels in BW25113 (ΔldhA ΔpflB) thus provides a link between redox imbalance, growth arrest, and stress-induced mutagenesis in E. coli.
Conclusion.
Deletion of pyruvate-metabolizing lactate dehydrogenase and pyruvate-formate lyase leads to an accumulation of the reduced cofactor NADH in anaerobically growing E. coli (60). Following the observation that the ldhA pflB double knockout strain had elevated mutation rates, we sought to study the effect of NADH accumulation on genetic stability. Fluctuation tests were performed to estimate the appearance of the rifampin-resistant mutants in the population by use of a set of clones with various intracellular-redox ratios (NADH/NAD+). The occurrence of rifampin resistance was observed to be greater in the strains with higher intracellular-redox ratios. Transcriptional profiling revealed repression of the genes involved in cellular defense and increases in the expression levels of stress response genes. The double mutant also showed reduced survival rates following UV exposure, confirming the decreased activity of DNA repair functions. Although this experiment was confounded by the possible photorepair and UV-induced DNA repair, the relatively low survival rate for the double mutant under microaerobic conditions suggests a link between redox imbalance and bacterial survival. Finally, we validated our hypothesis that the repression of DNA repair was due to an increased activity of stress response-regulatory genes rpoS and relA in the ldhA pflB double knockout mutant.
These studies demonstrate that the inhibition of normal redox balancing functions in E. coli can lead to unexpected consequences of direct importance for various basic and applied efforts. While it has been previously recognized that increased oxidative flux can lead to an accumulation of DNA-damaging free radicals, the lack of oxidation and subsequent buildup of NADH has not previously been implicated in increased mutagenesis (33). Given the central role of redox balancing in any effort to manipulate metabolism, we expect that the studies that we have reported here should prove relevant beyond our particular model system. Additional efforts, however, are required to appropriately gauge the importance of these findings. In particular, studies seeking to identify the redox-sensing mechanisms relating NADH buildup to RpoS- and RelA-mediated stress responses are needed. Such sensors, as well as several of the genes described herein, represent attractive potential targets for a broad range of metabolic and strain-engineering efforts where redox balancing and genetic stability are primary concerns.
Acknowledgments
This work was supported by NSF grant BES-0449183. A.K.-F. was supported by NIH grant 5T15LM009451-03.
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Adams, J. L., and R. J. C. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, G. Sherlock, and G. O. Consortium. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atsumi, S., T. Hanai, and J. C. Liao. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86-89. [DOI] [PubMed] [Google Scholar]
- 4.Atsumi, S., and J. C. Liao. 2008. Metabolic engineering for advanced biofuels production from Escherichia coli. Curr. Opin. Biotechnol. 19:414-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Au, K. G., K. Welsh, and P. Modrich. 1992. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 267:12142-12148. [PubMed] [Google Scholar]
- 6.Bonomo, J., and R. T. Gill. 2005. Amino acid content of recombinant proteins influences the metabolic burden response. Biotechnol. Bioeng. 90:116-126. [DOI] [PubMed] [Google Scholar]
- 7.Bonomo, J., M. D. Lynch, T. Warnecke, J. V. Price, and R. T. Gill. 2008. Genome-scale analysis of anti-metabolite directed strain engineering. Metab. Eng. 10:109-120. [DOI] [PubMed] [Google Scholar]
- 8.Bunch, P. K., F. MatJan, N. Lee, and D. P. Clark. 1997. The IdhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143:187-195. [DOI] [PubMed] [Google Scholar]
- 9.Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 10.Chatterji, D., N. Fujita, and A. Ishihama. 1998. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells 3:279-287. [DOI] [PubMed] [Google Scholar]
- 11.Chayabutra, C., and L. K. Ju. 2000. Degradation of n-hexadecane and its metabolites by Pseudomonas aeruginosa under microaerobic and anaerobic denitrifying conditions. Appl. Environ. Microbiol. 66:493-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien, L. J., H. T. Chen, P. F. Yang, and C. K. Lee. 2006. Enhancement of cellulose pellicle production by constitutively expressing Vitreoscilla hemoglobin in Acetobacter xylinum. Biotechnol. Prog. 22:1598-1603. [DOI] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delvigne, F., M. Boxus, S. Ingels, and P. Thonart. 2009. Bioreactor mixing efficiency modulates the activity of a prpoS::GFP reporter gene in E. coli. Microb. Cell Fact. 8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis, P. P., and M. Nomura. 1974. Stringent control of ribosomal protein gene expression in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 71:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drake, J. W. 1970. The molecular basis of mutation. Holden-Day, San Francisco, CA.
- 17.Durfee, T., A. M. Hansen, H. Zhi, F. R. Blattner, and D. J. Jin. 2008. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 190:1084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Sharoud, W. M. 2008. Bacterial physiology: a molecular approach. Springer, Berlin, Germany.
- 20.Farewell, A., K. Kvint, and T. Nystrom. 1998. uspB, a new σS-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J. Bacteriol. 180:6140-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, P. L. 2004. Adaptive mutation in Escherichia coli. J. Bacteriol. 186:4846-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gill, R. T., J. J. Valdes, and W. E. Bentley. 2000. A comparative study of global stress gene regulation in response to overexpression of recombinant proteins in Escherichia coli. Metab. Eng. 2:178-189. [DOI] [PubMed] [Google Scholar]
- 23.Green, J., and M. S. Paget. 2004. Bacterial redox sensors. Nat. Rev. Microbiol. 2:954-966. [DOI] [PubMed] [Google Scholar]
- 24.Grilley, M., J. Griffith, and P. Modrich. 1993. Bidirectional excision in methyl-directed mismatch repair. J. Biol. Chem. 268:11830-11837. [PubMed] [Google Scholar]
- 25.Hall, M. C., and S. W. Matson. 1999. The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem. 274:1306-1312. [DOI] [PubMed] [Google Scholar]
- 26.Hengge-Aronis, R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72:165-168. [DOI] [PubMed] [Google Scholar]
- 27.Hengge-Aronis, R., R. Lange, N. Henneberg, and D. Fischer. 1993. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J. Bacteriol. 175:259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ihssen, J., and T. Egli. 2004. Specific growth rate and not cell density controls the general stress response in Escherichia coli. Microbiology 150:1637-1648. [DOI] [PubMed] [Google Scholar]
- 29.Jantama, K., M. J. Haupt, S. A. Svoronos, X. L. Zhang, J. C. Moore, K. T. Shanmugam, and L. O. Ingram. 2008. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 99:1140-1153. [DOI] [PubMed] [Google Scholar]
- 30.Karp, P. D., I. M. Keseler, A. Shearer, M. Latendresse, M. Krummenacker, S. M. Paley, I. Paulsen, J. Collado-Vides, S. Gama-Castro, M. Peralta-Gil, A. Santos-Zavaleta, M. I. Penaloza-Spinola, C. Bonavides-Martinez, and J. Ingraham. 2007. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 35:7577-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelleher, J. E., and E. A. Raleigh. 1994. Response to UV damage by four Escherichia coli K-12 restriction systems. J. Bacteriol. 176:5888-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, Y., L. O. Ingram, and K. T. Shanmugam. 2007. Construction of an Escherichia coli K-12 mutant for homoethanologenic fermentation of glucose or xylose without foreign genes. Appl. Environ. Microbiol. 73:1766-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohanski, M. A., D. J. Dwyer, B. Hayete, C. A. Lawrence, and J. J. Collins. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797-810. [DOI] [PubMed] [Google Scholar]
- 34.Kuban, W., M. Banach-Orlowska, R. M. Schaaper, P. Jonczyk, and I. J. Fijalkowska. 2006. Role of DNA polymerase IV in Escherichia coli SOS mutator activity. J. Bacteriol. 188:7977-7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kvint, K., A. Farewell, and T. Nystrom. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of sigma(s). J. Biol. Chem. 275:14795-14798. [DOI] [PubMed] [Google Scholar]
- 36.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazzarini, R. A., M. Cashel, and J. Gallant. 1971. On the regulation of guanosine tetraphosphate levels in stringent and relaxed strains of Escherichia coli. J. Biol. Chem. 246:4381-4385. [PubMed] [Google Scholar]
- 38.Lin, J. S., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lombardo, M. J., I. Aponyi, and S. M. Rosenberg. 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166:669-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luria, S. E., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luttersczekalla, S. 1990. Lithoautotrophic growth of the iron bacterium Gallionella ferruginea with thiosulfate or sulfide as energy source. Arch. Microbiol. 154:417-421. [Google Scholar]
- 42.Lynch, M. D., and R. T. Gill. 2006. Broad host range vectors for stable genomic library construction. Biotechnol. Bioeng. 94:151-158. [DOI] [PubMed] [Google Scholar]
- 43.Manganelli, R. 2007. Polyphosphate and stress response in mycobacteria. Mol. Microbiol. 65:258-260. [DOI] [PubMed] [Google Scholar]
- 44.Mat-Jan, F., K. Y. Alam, and D. P. Clark. 1989. Mutants of Escherichia coli deficient in the fermentative lactate dehydrogenase. J. Bacteriol. 171:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. U. S. A. 97:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merrikh, H., A. E. Ferrazzoli, A. Bougdour, A. Olivier-Mason, and S. T. Lovett. 2009. A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc. Natl. Acad. Sci. U. S. A. 106:611-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1998. Mutators in Escherichia coli. Mutat. Res. 409:99-106. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell, A., G. H. Romano, B. Groisman, A. Yona, E. Dekel, M. Kupiec, O. Dahan, and Y. Pilpel. 2009. Adaptive prediction of environmental changes by microorganisms. Nature 460:220-224. [DOI] [PubMed] [Google Scholar]
- 49.Neidhardt, F. C., and R. Curtiss. 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 50.Notley, L., and T. Ferenci. 1996. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J. Bacteriol. 178:1465-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pisabarro, A. G., M. A. Depedro, and E. E. Ishiguro. 1990. Dissociation of the ampicillin-induced lysis of amino acid-deprived Escherichia coli into two stages. J. Bacteriol. 172:2187-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Potamitou, A., P. Neubauer, A. Holmgren, and A. Vlamis-Gardikas. 2002. Expression of Escherichia coli glutaredoxin 2 is mainly regulated by ppGpp and sigma(S). J. Biol. Chem. 277:17775-17780. [DOI] [PubMed] [Google Scholar]
- 53.Rodionov, D. G., and E. E. Ishiguro. 1998. Temperature sensitivity of bacteriolysis induced by beta-lactam antibiotics in amino acid-deprived Escherichia coli. J. Bacteriol. 180:2224-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodionov, D. G., A. G. Pisabarro, M. A. Depedro, W. Kusser, and E. E. Ishiguro. 1995. Beta-lactam-induced bacteriolysis of amino acid-deprived Escherichia coli is dependent on phospholipid synthesis. J. Bacteriol. 177:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schellhorn, H. E. 1995. Regulation of hydroperoxidase (catalase) expression in Escherichia coli. FEMS Microbiol. Lett. 131:113-119. [DOI] [PubMed] [Google Scholar]
- 56.Schellhorn, H. E., and H. M. Hassan. 1988. Transcriptional regulation of katE in Escherichia coli K-12. J. Bacteriol. 170:4286-4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sedliakova, M. 1998. A non-excision uvr-dependent DNA repair pathway of Escherichia coli (involvement of stress proteins). J. Photochem. Photobiol. B 45:75-81. [DOI] [PubMed] [Google Scholar]
- 58.Sedliakova, M., F. Masek, V. Slezarikova, and M. Pirsel. 1997. The effect of the OmpT protease on excision repair in UV-irradiated Escherichia coli. J. Photochem. Photobiol. B 41:245-248. [DOI] [PubMed] [Google Scholar]
- 59.Shibata, T., T. Hishida, Y. Kubota, Y. W. Han, H. Iwasaki, and H. Shinagawa. 2005. Functional overlap between RecA and MgsA (RarA) in the rescue of stalled replication forks in Escherichia coli. Genes Cells 10:181-191. [DOI] [PubMed] [Google Scholar]
- 60.Singh, A., M. D. Lynch, and R. T. Gill. 2009. Genes restoring redox balance in fermentation-deficient E. coli NZN111. Metab. Eng. 11:347-354. [DOI] [PubMed] [Google Scholar]
- 61.Stols, L., and M. I. Donnelly. 1997. Production of succinic acid through overexpression of NAD+-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 63:2695-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stols, L., G. Kulkarni, B. G. Harris, and M. I. Donnelly. 1997. Expression of Ascaris suum malic enzyme in a mutant Escherichia coli allows production of succinic acid from glucose. Appl. Biochem. Biotechnol. 63-65:153-158. [DOI] [PubMed] [Google Scholar]
- 63.Storz, G., and R. Hengge-Aronis. 2000. Bacterial stress responses. ASM Press, Washington, DC.
- 64.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 66.Tenaillon, O., E. Denamur, and I. Matic. 2004. Evolutionary significance of stress-induced mutagenesis in bacteria. Trends Microbiol. 12:264-270. [DOI] [PubMed] [Google Scholar]
- 67.Travers, A. 1976. Modulation of RNA polymerase specificity by ppGpp. Mol. Gen. Genet. 147:225-232. [DOI] [PubMed] [Google Scholar]
- 68.Traxler, M. F., S. M. Summers, H. T. Nguyen, V. M. Zacharia, G. A. Hightower, J. T. Smith, and T. Conway. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68:1128-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsui, H. C. T., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179:7476-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Ooyen, A. J. J., M. Gruber, and P. Jorgensen. 1976. The mechanism of action of ppGpp on rRNA synthesis in vitro. Cell 8:123-128. [DOI] [PubMed] [Google Scholar]
- 71.Vemuri, G. N., M. A. Eiteman, and E. Altman. 2002. Effects of growth mode and pyruvate carboxylase on succinic acid production by metabolically engineered strains of Escherichia coli. Appl. Environ. Microbiol. 68:1715-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Venturi, V. 2003. Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different? Mol. Microbiol. 49:1-9. [DOI] [PubMed] [Google Scholar]
- 73.Wyrzykowski, J., and M. R. Volkert. 2003. The Escherichia coli methyl-directed mismatch repair system repairs base pairs containing oxidative lesions. J. Bacteriol. 185:1701-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. U. S. A. 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]