Abstract
Pyridoxal 5′-phosphate (PLP)-dependent basic amino acid decarboxylases from the β/α-barrel-fold class (group IV) exist in most organisms and catalyze the decarboxylation of diverse substrates, essential for polyamine and lysine biosynthesis. Herein we describe the first x-ray structure determination of bacterial biosynthetic arginine decarboxylase (ADC) and carboxynorspermidine decarboxylase (CANSDC) to 2.3- and 2.0-Å resolution, solved as product complexes with agmatine and norspermidine. Despite low overall sequence identity, the monomeric and dimeric structures are similar to other enzymes in the family, with the active sites formed between the β/α-barrel domain of one subunit and the β-barrel of the other. ADC contains both a unique interdomain insertion (4-helical bundle) and a C-terminal extension (3-helical bundle) and it packs as a tetramer in the asymmetric unit with the insertions forming part of the dimer and tetramer interfaces. Analytical ultracentrifugation studies confirmed that the ADC solution structure is a tetramer. Specificity for different basic amino acids appears to arise primarily from changes in the position of, and amino acid replacements in, a helix in the β-barrel domain we refer to as the “specificity helix.” Additionally, in CANSDC a key acidic residue that interacts with the distal amino group of other substrates is replaced by Leu314, which interacts with the aliphatic portion of norspermidine. Neither product, agmatine in ADC nor norspermidine in CANSDC, form a Schiff base to pyridoxal 5′-phosphate, suggesting that the product complexes may promote product release by slowing the back reaction. These studies provide insight into the structural basis for the evolution of novel function within a common structural-fold.
Keywords: Crystallography, Decarboxylase, Enzyme Structure, Metabolism, Polyamines, Arginine Decarboxylase, Carboxynorspermidine Decarboxylase, Protein Structure/Function, Pyridoxal Phosphate
Introduction
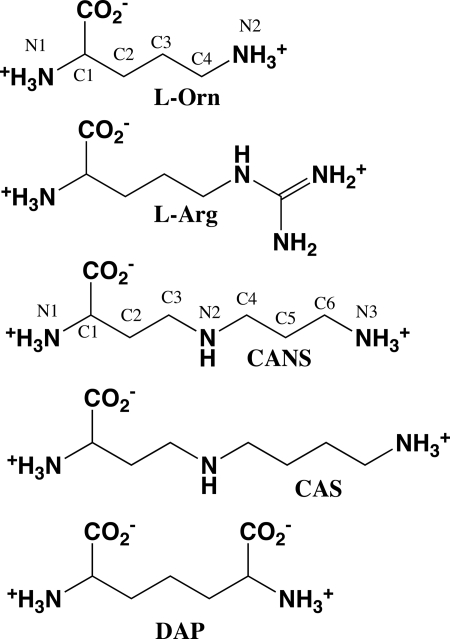
Studies of large protein families with variable function provide a mechanism to gain insight into structure/activity relationships in proteins. Approaches to define functionally important residues rely on both three-dimensional structure determination and on sequence-based methods that utilize the evolutionary blueprint as a guide (1, 2). The β/α-barrel-fold basic amino acid decarboxylase family (group IV pyridoxal 5′-phosphate (PLP)2 enzymes (3)) provides an excellent model to study the evolution of substrate specificity within a conserved structural fold-type. This family of PLP-dependent enzymes functions on a broad range of basic amino acid substrates required for polyamine biosynthesis (l-ornithine, l-arginine, l-lysine, carboxynorspermidine, and carboxyspermidine) in both eukaryotes and bacteria (ODC, ADC, L/ODC, CANSDC), and lysine biosynthesis (meso-diaminopimelate) (DAPDC) in bacteria and archaea (4) (Table 1 and Fig. 1). These enzymes play key roles in the primary metabolism in both eukaryotic and bacterial cells.
TABLE 1.
β/α-Barrel-fold basic amino acid decarboxylases
| Enzyme | Species range |
|---|---|
| Ornithine decarboxylase (ODC) | Eukaryotes and mainly α-proteobacteria |
| Arginine decarboxylase (ADC) | Plants, Gram negative bacteria, P. bursaria chlorella virus |
| Dual-specificity Lysine/ornithine decarboxylase (L/ODC) | Limited and sporadic in bacteria |
| Diaminopimelate decarboxylase (DAPDC) | Bacteria, euryarchaeotes, plants |
| Carboxynorspermidine decarboxylase (CANSDC) | Bacteria |
FIGURE 1.
Chemical structures of basic amino acid decarboxylase substrates. CANS, carboxynorspermidine; CAS, carboxyspermidine; DAP, diaminopimelate.
Phylogenetic analysis shows that eukaryotic and bacterial ODCs and L/ODCs are the most closely related, whereas bacterial/plant ADCs, DAPDCs, and CANSDCs have significantly diverged, and share only ∼15% overall sequence identity with each other and with ODC (5). Paramecium bursaria chlorella virus ADC (PBCVADC) is an exception sharing high sequence similarity (∼40%) with the eukaryotic ODCs but preferring l-Arg as substrate (6, 7). Some bacteria also contain aspartate aminotransferase fold-type decarboxylases that are involved in polyamine biosynthesis, but these enzymes are structurally unrelated to the β/α-barrel-fold basic amino acid decarboxlyase family and are restricted to activity on l-Orn, l-Lys, and l-Arg (8, 9).
Polyamines are necessary for cell growth, leading to therapeutic utility of pathway inhibitors; dl-α-difluoromethylornithine, an irreversible inhibitor of ODC, cures African sleeping sickness (10). Polyamines play important roles in the cell cycle and cancer (11) and in bacteria they have been implicated in biofilm formation and motility (12–14). Norspermidine (NSpd), a polyamine analog absent in most eukaryotic cells, is required for vibriobactin biosynthesis, a peptide iron-chelator needed for growth and virulence in Vibrio species (15). CANSDC provides the only route to spermidine and NSpd biosynthesis in Vibrio (16) and deletion of the gene abolishes spermidine and NSpd intracellular pools leading to defects in biofilm formation (13).
The x-ray structures of several eukaryotic ODCs (17–22), of PBCVADC (6), Vibrio vulnificus L/ODC (VvL/ODC) (5), and several bacterial DAPDCs have been reported (23, 24). Comparison of the available structures identified the helix (termed “specificity helix”) (Fig. 2), which sits at the back of the substrate-binding site at the 2-fold axis of the dimer, as a key specificity element (5, 6). Substrates of different size are accommodated by changes in the distance from this helix to PLP, whereas variation in its amino acid composition provides specificity of interaction with the range of substrates.
Two functional classes within the family, however, remain structurally uncharacterized, including bacterial/plant ADC and CANSDC. The bacterial ADCs contain a long insertion between the N-terminal β/α-barrel domain and a C-terminal β-barrel domain, both of which are novel within the family, and the function of which was unknown. Furthermore, the structural basis for substrate specificity in the bacterial ADCs cannot be deduced by sequence analysis alone. Determination of the x-ray structure of a bacterial ADC has been of long standing interest as evidenced by the number of crystallization reports that have been published (25–28). Despite this significant effort, solution of the structure remained elusive until now. CANSDC catalyzes the decarboxylation of carboxynorspermidine and carboxyspermidine, both substrates are considerably larger than the substrates utilized by other enzymes in the family, and structural data are needed to elucidate how the enzyme accommodates these larger substrates.
Herein we report the first x-ray structure determination of the two remaining functional enzyme types in the β/α-barrel-fold basic amino acid decarboxlyase family: bacterial ADC (V. vulnificus; VvADC) and bacterial CANSDC (Campylobacter jejuni; CjCANSDC). These studies round out the structural analysis of the five characterized substrate specificity types (Table 1) in the β/α-barrel decarboxylase-fold. Taken together with previous structural analysis of enzymes from this family, these studies provide a comprehensive example of how enzymes evolve to generate novel function through gene duplication and divergence.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification of CjCANSDC and VvADC
The gene for CjCANSDC was cloned from C. jejuni 81116 by PCR using genomic DNA as the template followed by cloning into the pET100 expression vector. The original clone of CjCANSDC contained a spontaneous single nucleotide mutation at amino acid residue 184 resulting in Glu (GAA) to Lys (AAA) mutation. This mutation was not observed in any other sequenced strain of CjCANSDC and led to loss of activity. Lys184 was mutated to Glu by QuikChangeTM mutagenesis (5′-gctgttttaaaggtctttgaagagaaatttggtaaatgg-3′) and the corrected clone was used for further study including kinetic analysis and protein crystallization. The pET-22b expression vector for V. vulnificus CMCP6 arginine decarboxylase (VvADC) was previously described (5).
Native proteins of CjCANSDC and VvADC were overexpressed in Escherichia coli BL21(DE3) and purified using Ni2+-affinity and gel filtration chromatography as previously described (5). Selenomethionine (SeMet) derivatives of CjCANSDC and VvADC were expressed in E. coli BL21(DE3) using the Met pathway inhibition method as previously described (29). Cells were grown in M9 minimal medium containing 100 μg/ml of ampicillin at 37 °C until A600 nm reached 0.6–0.7. Cells were induced with 200 μm isopropyl 1-thio-β-d-galactopyranoside at room temperature for 4 h after the addition of SeMet stock solution (Athena Enzyme Systems, Baltimore, MD) and the feedback inhibition amino acids (100 mg/liter each of l-Thr, l-Lys, and l-Phe, and 50 mg/liter each of l-Leu, l-Ile, and l-Val). Harvested cells were lysed and proteins were purified as above in the presence of 20 mm dithiothreitol. ESI-MS analysis of the purified proteins (Protein Technology Center, UT Southwestern Medical Center) revealed that all Met sites in each protein (14 sites in VvADC and 13 sites in CjCANSDC) were replaced with SeMet (data not shown).
Synthesis of Carboxynorspermidine and Carboxyspermidine
Substrates were synthesized previously as described (13).
Steady-state Kinetic Analysis of CjCANSDC
Steady-state kinetic analysis was performed utilizing a coupled enzyme assay that links decarboxylation to the oxidation of NADH through the activity of phosphoenolpyruvate carboxylase and malate dehydrogenase as previously described (13). Data were fitted to the Michaelis-Menten equation to determine kcat and Km using Prism (GraphPad).
Crystallization and X-ray Diffraction Data Collection
SeMet-substituted crystals of CjCANSDC were co-crystallized with 10 mm NSpd in hanging drops containing 1.5 μl of protein (20 mg/ml in 50 mm HEPES, pH 8.0, 300 mm NaCl, 10% glycerol, 20 mm dithiothreitol, 0.03% Brij, and 0.5 mm EDTA) and 1.5 μl of reservoir solution (2.75 m AmSO4, 0.1 m Bicine, pH 8.5). Rod shape crystals appeared after 4–5 days at 20 °C and grew to 80 μm thick × 600 μm long by 2 weeks. Crystals were cryoprotected in 2.7 m AmSO4, 0.1 m Bicine, pH 8.5, 0.3 m NaCl, and 17% glycerol and flash frozen in liquid nitrogen. SAD data were collected at beamline 19ID of Advanced Photon Source. Crystals of SeMet-substituted CjCANSDC exhibited the symmetry of space group P43212 with unit cell parameters of a = b = 144.2 Å and c = 79.9 Å. They contained two molecules per asymmetric unit and diffracted isotropically to a dmin of 1.9 Å when exposed to synchrotron radiation.
SeMet-derivative crystals of VvADC were co-crystallized with 5 mm l-Arg in sitting drops containing 2 μl of protein (2.5 mg/ml in 50 mm HEPES, pH 7.5, 300 mm NaCl, 10% glycerol, 20 mm dithiothreitol, 0.03% Brij, and 0.5 mm EDTA), 2 μl of reservoir solution (0.1 m HEPES, pH 7.0, 0.2 m MgCl2, 12% PEG-4000), and 0.4 μl of 1 m NaI. Crystals were cryoprotected in 0.1 m HEPES, pH 7.5, 0.2 m MgCl2, 15% PEG-4000, 0.1 m NaI, 0.3 m NaCl, and 25% glycerol. Crystals of SeMet-substituted VvADC exhibited the symmetry of space group P21 with unit cell parameters of a = 101.6 Å, b = 119.4 Å, c = 121.8 Å, and β = 96.3°, contained four molecules of VvADC per asymmetric unit and diffracted isotropically to a dmin of 2.30 Å when exposed to synchrotron radiation. Data were indexed, integrated, and scaled using the HKL-3000 program package (30). Data collection statistics are provided in Table 2.
Phase Determination and Structure Refinement
Phases for SeMet-substituted CANSDC were obtained from a single wavelength anomalous dispersion experiment with data to a resolution of 1.9 Å, and 17 selenium sites were located using the program SHELXD (31). Phases were refined with the program MLPHARE (32), resulting in an overall figure-of-merit of 0.24 for data between 44.7 and 1.9 Å. Phases were further improved by density modification and 2-fold non-crystallographic averaging with the program DM (33). An initial model was automatically generated by ARP/wARP (34) and additional residues were manually modeled in Coot (35). Refinement was performed to a resolution of 1.9 Å using the program Refmac (36) with a random 5% of all data set aside for an Rfree calculation. The structure was refined to a Rwork of 0.179 and a Rfree of 0.218 (Table 2). The final model contains two CjCANSDC monomers in the asymmetric unit; molecule A includes residues 2–129, 140–382, PLP, and NSpd, and whereas molecule B contains residues 6–129, 140–382, and PLP. Both subunits also contain a bound glycerol. The final refined structure contains 385 waters. A Ramachandran plot generated with Molprobity indicated that 97.5% of all protein residues are in the most favored regions with the remaining 2.5% in allowed regions.
TABLE 2.
Data collection, phasing, and refinement statistics for CjCANSDC and VvADC structures
Data for the outermost shell are given in parentheses.
| Data collection | ||
| Crystal | CANSDC SeMeta peak | VvADC SeMeta peak |
| Energy (eV) | 12,659.6 | 12,659.9 |
| Space group | P43212 | P21 |
| Unit cell dimensions | a = b = 144.5Å, c = 79.9Å | a = 101.6 Å, b = 119.4 Å, c = 121.8 Å |
| α = β = γ = 90° | α = 90°, β = 96.3°, γ = 90° | |
| Resolution (Å) | 41.2-2.00 (2.03-2.00) | 49.7-2.30 (2.34-2.30) |
| Unique reflections | 57,401 (2,800) | 127,349 (6.302) |
| Redundancy | 7.1 (7.0) | 4.6 (4.6) |
| Data completeness (%) | 100.0 (100.0) | 100.0 (100.0) |
| Rmerge (%)b | 13.6 (98.0) | 12.7 (88.8) |
| I/σ(I) | 21.8 (2.4) | 17.4 (2.1) |
| Phase determination | ||
| Anomalous scatterers (Se) | 17 out of 18 possible sites | 52 out of 56 possible sites |
| Figure of merit | 0.24 | 0.13 |
| Refinement statistics | ||
| Resolution range (Å) | 102.0–1.9 (1.95–1.90) | 121.1–2.30 (2.36–2.30) |
| Unique reflections | 66,670 (4,761) | 128,245 (7,954) |
| Data completeness (%) | 99.8 (98.2) | 98.8 (98.0) |
| Atoms (protein/PLP/water/other) | 5,890/30/385/21 | 20,053/60/443/36 |
| Rwork (%) | 17.9 (26.3) | 17.8 (24.9) |
| Rfree (%) | 21.8 (34.1) | 23.9 (32.5) |
| R.m.s.d. bond length (Å) | 0.02 | 0.02 |
| R.m.s.d. bond angle (°) | 1.82 | 1.83 |
| Ramachandran plot (%) (favored/additional)c | 97.5/2.5 | 97.6/2.4 |
a Bijvoet-pairs were kept separate for data processing.
b Rmerge = 100 ΣhΣi|Ih,i — 〈Ih|/ΣhΣiIh,i, where the outer sum (h) is over the unique reflections and the inner sum (i) is over the set of independent observations of each unique reflection.
c As defined by the validation suite MolProbity (51).
Phases for SeMet-substituted VvADC were obtained from a single-wavelength anomalous dispersion experiment with data to a resolution of 2.3 Å, and 52 selenium sites were located using the program SHELXD. Phases were refined with the program MLPHARE, resulting in an overall figure-of-merit of 0.13 for data between 49.7 and 2.3 Å. Phases were further improved by density modification and 4-fold non-crystallographic averaging with the program DM. An initial model containing ∼91% of all residues was automatically generated by alternating cycles of the programs ARP/wARP. Refinement to a resolution of 2.3 Å was performed as described for CjCANSDC, except that the 2-fold non-crystallographic restraints on the protein main chain were applied between the A to C subunit and B to D subunit. The final Rwork is 0.178, and the Rfree is 0.239 (Table 2). The final model contains four VvADC monomers in the asymmetric unit and a PLP and the product of l-Arg decarboxylation, agmatine, bound to each monomer; including residues 10–639, in molecule A; residues 11–639, molecule B and D; residues 12–639 in molecule C; and 440 waters. A Ramachandran plot generated with Molprobity indicated that 97.6% of all protein residues are in the most favored regions with the remaining 2.4% in allowed regions. Phasing and model refinement statistics are provided in Table 2.
Molecular Modeling
Structures were displayed using the graphics program PyMol (52). Buried surface area was calculated by “Define secondary structure of proteins” analysis (37). Structures were superimposed by alignment of 4 β-strands and 2 α-helixes located in the most conserved part of the β/α-barrel domain using LSQKab (38) as follows: CjCANSDC (subunit A and B) residues 233–239, 199–202, and 340–342; VvADC (subunit A and B) residues 343–349, 292–295, and 549–551; Trypanosoma brucei ODC (TbODC) (PDB code 1F3T) (subunit C and D), residues 274–280, 234–237, and 387–389; and Methanocaldococcus jannaschii DAPDC (MjDAPDC) (PDB code 1TWI) (subunit A and B) residues 305–311, 261–264, and 399–401. The r.m.s. deviation was calculated over the monomer for the backbone atoms only. R.m.s. deviation values were calculated for the superimposed structures based on the Cα positions using LSQMAN (39, 40) and Moleman2 (41) was used to manipulate the PDB files before the analysis. Software was obtained from the Uppsala Software Factory. Values are as follows: VvADC to TbODC, 3.1 Å; CjCANSDC to TbODC, 2.7 Å; VvADC to CjCANSDC, 3.0 Å; VvADC to MjDAPDC, 3.2 Å; CjCANSDC to MjDAPDC, 2.4 Å.
Multisequence Alignment
The amino acid sequences of VvADC and CjCANSDC were aligned with other sequences in the family for which x-ray structural data are available, including a representative of each substrate specificity type: TbODC (PDB code 1F3T), PBCVADC (PDB code 2NVA), VvL/ODC (PDB code 2PLK), MjDAPDC (PDB code 1TWI). The sequence alignment was generated from the x-ray structure alignment using the program PROMALS3D (42, 43). The secondary structure elements were then annotated using PDBsum (44).
Analytical Ultracentrifugation
Sedimentation velocity experiments were performed using a Beckman Optima XL-1 with An-50 Ti rotor, charcoal-filled dual sector centerpiece, and sapphire windows. The wavelength of the absorbance optics was set at 280. Experiments were performed at 45,000 rpm and at 20 °C. Three data set were collected using 3 VvADC concentrations (0.1, 0.4, and 0.8 OD280, respectively), 1 OD280 = 0.93 mg/ml in buffer (50 mm HEPES, pH 7.8, 150 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine), and a total volume per cell of 0.4 ml. Data were analyzed using SEDFIT software to determine the oligomeric structure as described (45). Solvent density (1.0083 g/ml), partial specific volume (0.7324 ml/g), and viscosity (0.010473 g/s/cm) were calculated using the SEDNTERP program.
RESULTS
Purification and Kinetic Analysis of VvADC and CjCANSDC
VvADC and CjCANSDC were expressed and purified from E. coli as described under “Experimental Procedures.” Steady-state kinetic analysis of VvADC was described previously (5). Steady-state kinetic analysis for CjCANSDC was performed using both carboxyspermidine (Km = 4.1 ± 0.36 mm and kcat = 0.24 ± 0.0077 s−1) and carboxynorspermidine (CANS) (Km = 2.1 ± 0.13 mm and kcat = 0.58 ± 0.013 s−1) as substrates. CjCANDC has similar activity on both CANS and carboxyspermidine, whereas VvCANSDC has evolved as higher activity on CANS (13). This is consistent with the cellular requirement for both norspermidine and spermidine in Vibrio species (13).
Crystallization and Structural Refinement of VvADC and CjCANSD
VvADC was crystallized in the presence of the substrate l-Arg, and the structure was refined to 2.30-Å resolution (Table 2). A tetramer was observed in the asymmetric unit and the final refined structure contains 1 molecule of PLP and 1 molecule of the product agmatine per subunit (Figs. 3–5). Non-crystallographic symmetry was constrained along the 2-fold dimers for the main chain during the refinement. Small differences in the subunits are observed in the position of the bound agmatine.
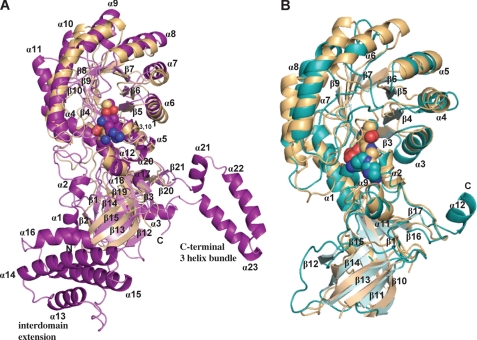
FIGURE 3.
Ribbon diagram of VvADC and CjCANSDC monomers. A, VvADC monomer (purple) superimposed with TbODC (tan). B, CjCANSDC monomer (teal) superimposed with TbODC (tan). The α-helices and β-strands are labeled for VvADC and CjCANSDC and correspond to the numbers shown in supplemental Fig. S2. The T. brucei ODC structure is complexed with putrescine (Put).
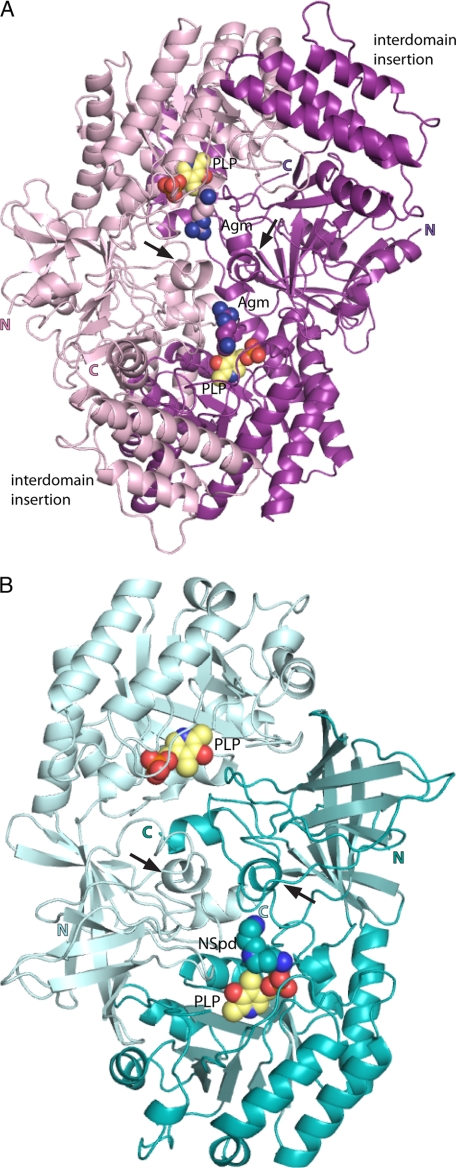
FIGURE 4.
Ribbon diagram of VvADC and CjCANSDC dimers. The two VvADC monomers are colored pink and purple, respectively, whereas the CjCANSDC monomers are colored in light teal and teal. PLP (yellow), Agm (agmatine) (pink and purple), and NSpd (teal) are shown as spheres. The N (N) and C (C) terminus of each chain is labeled. Arrows indicate the position of the specificity helix.
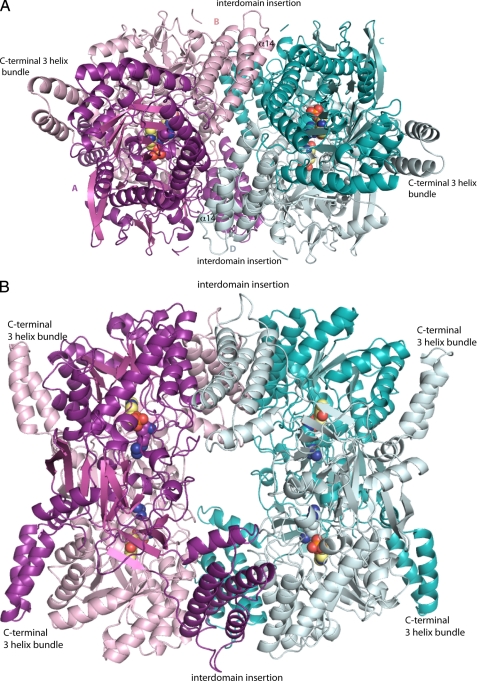
FIGURE 5.
Ribbon diagram of VvADC tetramer. A, side view showing the β/α-barrel of subunits A and C and the interdomain region of subunits B and D. The active dimer pairs are A/B (purple and pink) and C/D (teal and light teal). B, top view showing the central hole around the entrance to the four active sites. PLP (yellow), agmatine (pink and purple), and NSpd (light teal and teal) are shown as spheres.
CjCANSDC was co-crystallized in the presence of the product NSpd, and the structure was refined to 1.9-Å resolution (Table 2). A dimer was observed in the asymmetric unit and the final refined structure contained 1 molecule of PLP per subunit, and a molecule of NSpd was found in one of the two active sites (Figs. 3 and 4). Additionally, a molecule of glycerol used in the crystallization buffer was observed in both subunits.
Overall Fold and Oligomeric Structure of VvADC
The structure of the VvADC monomer is similar to that observed for other members of the β/α-barrel-fold decarboxylase family, and it contains both the β/α-barrel N-terminal domain, and the C-terminal β-barrel domain observed in the other structures (Fig. 3A). Structural alignment with T. brucei ODC shows a r.m.s. deviation of 3.1 Å for the monomer. No significant domain rotations are observed. However, in addition to the core conserved domains, VvADC contains three unique insertions: 1) the N-terminal residues (Val14–Gln49) form a broken helix, followed by a 2-stranded anti-parallel β-sheet, 2) the interdomain extension (residues Lys366–Glu462) forms a right-handed superhelix composed of a 4-helix bundle, and 3) the C terminus (Val589–Glu638), which extends beyond that observed in T. brucei ODC, forms a 3-helical bundle, with all helices in a single plane (Fig. 3A).
The oligomeric unit required for activity is the dimer (AB or CD dimer in Fig. 4A) and the active site is formed as previously observed for ODC at the subunit and domain boundaries. Packing of the active dimer unit is similar to what has been previously observed for this fold type. The dimer interface of the A/B or C/D subunits sits between the N-terminal β/α-barrel domain of one subunit, the C-terminal β-barrel domain of the second subunit, and the α17-α20 helices and loop region composed of residues 555–560 from the β-barrel domains of the two subunits. Two additional interfaces are observed in the ADC dimer that have not been previously observed in enzymes from this fold type: 1) the 4-helical bundle interdomain extension interacts (helices α14 and α15) with the β/α-barrel domain of the opposite subunit (helices α9 and α11), and the C-terminal extension 3-helical bundle (helices α22 and α23) interacts with the β/α-barrel domain (helices α6 and α7) of the opposite subunit, but on the opposite face from the 4-helical bundle. These two unique helical bundles form a cradle around the β/α-barrel of the opposite subunit. This results in a large buried surface area for the dimer of 12,300 Å2 per dimer (6,180 Å2 per monomer).
Within the asymmetric unit VvADC packs as a tetramer of two active dimer units (Fig. 5). The interface between the two dimers contains residues from all 4 subunits. They are arranged such that the 4 helical bundles from the interdomain extension of monomers B and D (or A and C for the symmetry related pair) are in direct contact with each other over a short region of helix α14. They in turn sit between the two β/α-barrel domains from monomers A and C (or B and D) (interacting with helices α9-α11) allowing the formation of an extensive packing interaction. The tetramer is packed as a donut with a large central cavity. The four active sites point into that cavity with the 4-helical bundles packed around the cavity. The additional buried surface area that occurs by formation of the dimer-dimer interface (tetramer) is 7,370 Å2 (3,680 Å2 buried surface area per dimer).
Sedimentation velocity analysis of VvADC was performed at three concentrations of enzyme. The calculated molecular mass of the VvADC monomer is 73,558 Da. In solution, the molecular mass determined by the sedimentation velocity analysis was on average 280 ± 27 kDa for data collected at three protein concentrations in the range of 0.1, 0.4, and 0.8 OD (supplemental Fig. 3S). These data are consistent with the tetramer being the dominant solution form for VvADC. A slight amount of dissociation to the dimer (<10%) was evident from the data, but this species was not present in sufficient amounts for quantitation.
Active Site of VvADC
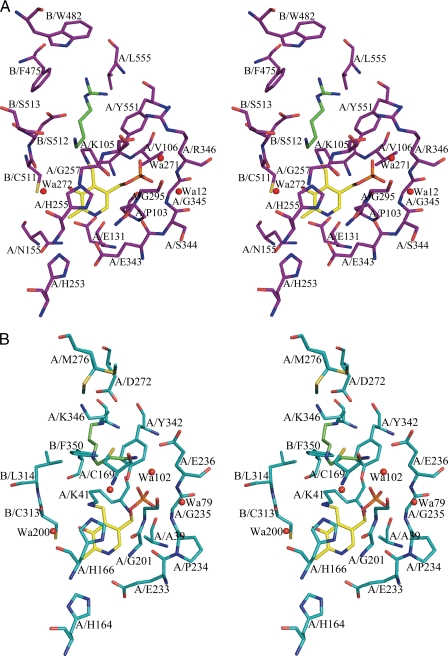
The VvADC crystals were grown in the presence of the substrate l-Arg, and good density (Fo − Fc map) was observed for the decarboxylated product agmatine before refinement (supplemental Fig. 1S). Strong electron density was also observed for PLP in all four subunits.
PLP-binding Site of VvADC
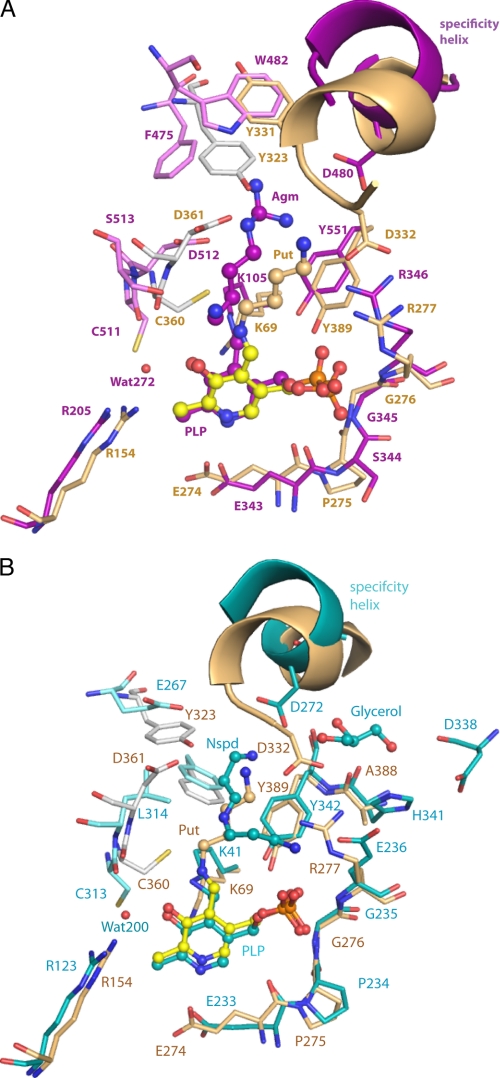
The PLP-binding site is formed between the end of β-strands β10 and β11 and helix α12 of the β/α-barrel (Figs. 6A, 7A, and 8), and interactions between PLP and the protein are largely conserved in comparison with other enzymes from the family (Fig. 2 and supplemental Fig. S2). VvADC-Lys105 (TbODC-Lys69) is the catalytic Lys that forms a Schiff base with PLP, and which has been implicated in accelerating the rates of substrate binding, decarboxylation, and product release in TbODC (46). VvADC-His255 (TbODC-His196) stacks against the PLP ring, VvADC-Arg205 (TbODC-Arg154) forms an interaction with the hydroxyl of PLP through an ordered water molecule (water 272), and VvADC-Glu343 (TbODC-Glu270) forms a short H-bond with the pyridine nitrogen of PLP (Glu343). The phosphate oxygens of PLP form interactions with VvADC-Arg346 (TbODC-Arg277), VvADC-Tyr551 (TbODC-Tyr389), and the backbone of VvADC-Gly295 (TbODC-Gly237). In the ODC structures TbODC-Arg277 forms a salt bridge with TbODC-Asp332, a key substrate specificity determinant. Because of the change in the position of the specificity helix (α18) in VvADC this interaction is not preserved. Instead the equivalent Arg residue (VvADC-Arg346) forms an H-bond with Tyr300 (2.9 Å). TbODC-Asp88, which forms an interaction with TbODC-Lys69 in the ligand bound structure of TbODC is replaced with VvADC-Glu131, which presumably functions in the same role. The mobile surface loop that was observed to cover the substrate-binding site in PBCVADC (residues 135–152) (6) is in the open conformation in VvADC (residues 211–225).
FIGURE 6.
Stereo diagram of the active sites of VvADC and CjCANSDC. The figures show the 4-Å shell around the PLP and bound product ligands. A, VvADC (purple) bound to PLP (yellow) and agmatine (green). B, CjCANSDC (teal) bound to PLP (yellow) and NSpd (green). Residues are labeled to show the contribution from the two monomers (A or B).
FIGURE 7.
Active site comparison of VvADC and CjCANSDC to TbODC. A limited set of residues within the 4-Å shell are displayed. A, comparison of VvADC (monomer A (purple); monomer B (pink)) to TbODC (monomer A (tan); monomer B (gray)). B, comparison of CjCANSDC (monomer A (teal); monomer B (light teal)) to TbODC (monomer A (tan); monomer B (gray)). PLP (yellow for ODC, purple for VvADC, and teal for CjCANSDC), putrescine (Put) (tan), agmatine (Agm) (purple), and NSpd (teal) are shown as ball and stick.
FIGURE 8.
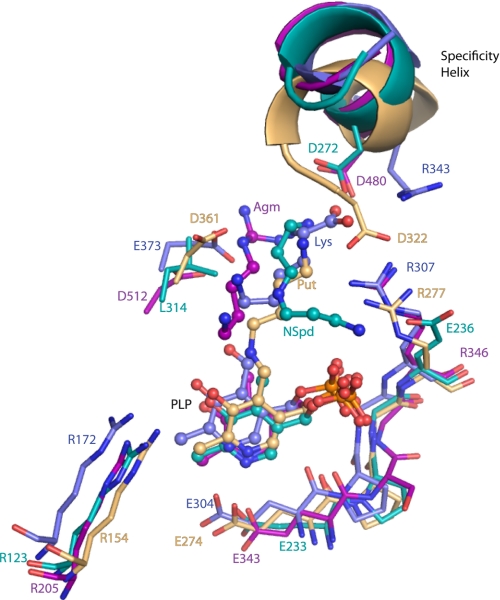
Specificity determinants in the active sites of enzyme from the four major specificity classes. The substrate and PLP binding sites including the specificity helix and key residues from the other subunit are shown, VvADC (pink-purple), CjCANSDC (teal), TbODC (tan), and DAPDC (blue-purple). PLP, agmatine (Agm), NSpd, putrescine (Put), and Lys are shown as ball and stick.
FIGURE 2.
Structure based sequence alignment of the β/α-barrel-fold basic amino acid decarboxylases in the region of the specificity helix (α18 in VvADC and α11 in CjCANSDC). VvADC, V. vulnificus BAC94750; TbODC, T. brucei AAA30218; PBCVADC, chlorella virus PBCV-1 ADC; NP_048554; VvL/ODC, V. vulnificus NP_762948; CjCANSDC, C. jejuni AAW36121; MjDAPDC, M. jannaschii NP_248090. A more complete alignment of representative sequences from the family has been previously published as supplemental data in Ref. 5. β-Sheets are highlighted in blue, α-helices are highlighted in red, and 310-helices are highlighted in green. The position of the specificity helix is indicated by the black bar. The key amino acids involved in substrate binding that project from this element into the active site are underlined.
VvADC Agmatine-binding Site
The substrate-binding site lies between the β/α-barrel domain on one subunit, which encompasses the PLP-binding site, the interdomain region of this same subunit, which contains helix α18 (found in the same position as the previously described 310-helix or “specificity element” (5, 6)), and the C-terminal β-barrel from the opposite subunit (Figs. 6A, 7A, and 8). Agmatine is observed in the substrate-binding site but it does not form a Schiff base with PLP, and represents a structure of the enzyme product Michaelis complex. The substrate N1 is not within bonding distance of the PLP C4′ atom and thus the position of the substrate when bound to PLP in the productive reaction complex will minimally require bond rotations in the ligand to bring the N1 atom into bonding distance of the PLP cofactor. Within the β/α-barrel subunit the guanidinium group of agmatine (NH1 and NH2) forms a salt bridge interaction with VvADC-Asp480 in 2 of the 4 active sites (distance 3.5–4.5 Å depending on subunit), and the wall of the binding pocket that runs down the length of the aliphatic portion of the substrate is formed by VvADC-Tyr551 (Tyr389 in TbODC). Additional interactions are contributed from across the subunit boundary: VvADC-Asp512 forms a salt bridge with NE of the substrate in 3 of 4 active sites (3.2–4.1 Å depending on the subunit), N1 of agmatine forms an H-bond with Ser513, and VvADC-Asp514, whereas just outside of van der Waals range, likely provides additional charge stabilization. All three residues are invariant within the ADC enzymes that contain the insertions (5). An acidic residue at position VvADC-Asp512 is also conserved in ODC (TbODC-Asp361). The catalytic base VvADC-Cys511 (TbODC-Cys360) is observed in the down position pointing away from the ligand, but its position preserves a similar potential to play a role in catalysis as observed for TbODC (20). TbODC-Phe397, conserved in ODC type enzymes and implicated in decarboxylation (19), is replaced by VvADC-His559 in the ADCs and TbODC-Tyr323 is replaced by VvADC-Phe475, which sits further away from the active site.
Overall Fold and Oligomeric Structure of CjCANSDC
The monomer of CjCANSDC superimposes with TbODC with a r.m.s. deviation of 2.7 Å, and again no significant domain rotations were observed (Fig. 3B). CjCANSDC is a dimer (Fig. 4B). The CjCANSDC sequence contains additional C-terminal residues (residues Tyr375–Asn382) that extend beyond the C-terminal domain of TbODC. This extension forms a short α-helix (α12) that interacts to form part of the dimer interface on the back side of the dimer (opposite face from the active sites) (supplemental Fig. 4S). These helices in turn interact with helix α3 of the β/α-barrel of the opposite subunit to form a 4-helix stack, with helices alternating between monomers. This generates an additional interface between the monomeric subunits not observed in ODC or ADC. The buried surface area upon dimerization is 7950 Å2 (3970 per monomer).
Active Site of CjCANSDC
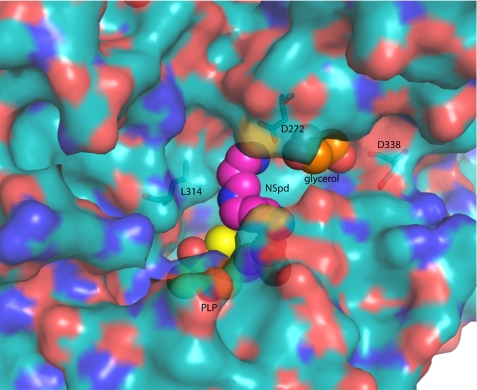
Strong electron density (Fo − Fc map) for the PLP cofactor was observed for both subunits of the CjCANSDC structure and density for the co-crystallized product (NSpd) was present but only in subunit A (supplemental Fig. 1S). The NSpd density is discontinuous but clearly indicates the presence of the ligand. In addition, density for glycerol, a component of the crystallization buffer, was observed in both subunits in a solvent accessible channel adjacent to the distal end (relative to PLP) of the ligand binding site.
PLP-binding Site of CjCANSDC
The PLP-binding site retains most of the key contacts described for VvADC (Figs. 6B and 7B): Lys41 forms a Schiff base with PLP, His166 stacks against the PLP ring, Glu66 (Glu94 in TbODC) forms an H-bond with the α-amino group of Lys41, Arg123 forms an H-bond with the hydroxyl of PLP through a bound water molecule, and Glu233 interacts with the pyridine nitrogen of PLP. The mobile surface loop (residues 130–139) is disordered in the CjCANSDC structure.
Several unusual differences in the CjCANSDC PLP-binding site are present. First, TbODC-Arg277 (VvADC-Arg346) is replaced with CjCANSDC-Glu236, which removes the charge stabilization of the PLP-phosphate that has been shown to be important for PLP binding in TbODC (47). Glu236 is mostly conserved in CANSDC members of the family, although in some species this residue is replaced with Ser (5). CjCANSDC-Glu236 is involved in a H-bond network with His341 and Asp338. Although Asp338 is conserved among the CANSDCs, His341 is not (5). Second, Asp88 in TbODC is replaced with Thr60 in CjCANSDC, which retains the potential to form an H-bond to the catalytic Lys41 in the ligand bound structure, although in this structure the hydroxyl group of Thr60 is not oriented toward Lys41. Additionally because the ligand in CjCANSDC is not observed in a Schiff base with PLP, Lys41 has not rotated to its substrate bound position, leaving open the question of what additional residues may form interactions with Lys41 when substrate is bound to PLP as a Schiff base.
CjCANSDC NSpd-binding Site
NSpd is bound in the typical substrate-binding site at the subunit interface between PLP and helix α11, as described above for VvADC (Figs. 6B, 7B, and 8). NSpd does not form a Schiff base with PLP and instead the α-amino group is turned away from C4′ of PLP and forms an ion pair with Glu236. Thus as with the VvADC structure, the Michaelis complex is observed. A rotation of the bound C2-C3 would bring the amino group into position to form a Schiff base with PLP. The N3-amino group forms an ion pair with Asp272 from the specificity helix α11, with the bond distances being short and consistent with a strong interaction (OD2 to N3 = 2.6 Å and OD1 to N3 = 3.3 Å). However, unlike all other substrate specificity types within the family, CANSDC does not position an acidic residue across the subunit boundary to form a salt bridge with the ligand (e.g. VvADC-Asp512 and TbODCA-Asp361). The equivalent residue in CjCANSDC is Leu314 and this residue is conserved among the CANSDCs (13). Both carboxynorspermidine and carboxyspermidine have a 3-carbon linker between the α-amino group and N2, which is 1 carbon shorter than observed for other substrates in the family. The positioning of the ligand within the binding site places the aliphatic portion of NSpd near Leu314, providing structural insight into why an aliphatic residue in this position is required instead of the acidic residue typical of other enzymes in the family. The adjacent catalytic base from the same loop, Cys313 (TbODC-Cys360), is conserved and is observed in the down position.
CjCANSDC Glycerol-binding site
A glycerol molecule from the crystallization solvent was observed bound in a solvent accessible channel adjacent to the specificity helix α11 (Figs. 2, 7B, and 9). The hydroxyl residues form H-bond interactions with Asp272, Asp338, and His341. This channel provides the potential for a more extended substrate-binding site than observed for other enzymes in the family. The second substrate of the enzyme, carboxyspermidine, is longer by 1 carbon than NSpd suggesting that the extra chain length could be accommodated by turning into the channel occupied by glycerol in the NSpd CANSDC structure.
FIGURE 9.
Surface representation of CjCANSDC showing the ligand-binding sites. NSpd (pink), PLP (yellow), and glycerol (orange) are displayed as spheres. The surface was generated against the full protein minus the ligands (PLP and NSpd). The side chain of Lys41 was also removed before the surface was generated to allow the PLP to be more visible.
Comparison of VvADC and CjCANSDC to Other Members of the Family
Alignment of the VvADC and CjCANSDC structures with prior x-ray structures for enzymes in the family shows that the actives sites are highly conserved, both in structure and composition (Figs. 7 and 8), and indeed key residues in the active site overlay closely on each other with PLP bound in near identical position in all structures, despite the very low overall sequence identity between members of the family. Within this context the structural basis for specificity differences was evaluated to identify differences between the enzymes from the family. The primary structural change is observed in the specificity helix.
In the VvADC and CjCANSDC structures the specificity helix is shifted further back in the pocket when compared with TbODC, which allows a larger distance between PLP and the key acidic residue that contacts ligand (Figs. 6–8). The acidic residues (VvADC-Asp480; helix α18) in VvADC and CjCANSDC (CjCANSDC-Asp272; helix α11) form salt bridges with the ligand project from the start of the specificity helix. TbODC-Asp332 also projects from the equivalent helix but originates from the C-terminal end of the helix (Figs. 2 and 8). The change in register allows equivalent salt bond interactions to be formed despite the larger distance between the helix and PLP and accommodates ligands of different size. DAPDC is the only enzyme that catalyzes decarboxylation of a dicarboxylate substrate and the binding site of this enzyme has evolved to stabilize the carboxylate on C5. However, the same strategy is in play, and an arginine residue (Arg343) projects from the specificity helix from the N terminus of the helix to interact with the C5-carboxylate, and Glu348 from the C terminus of the helix interacts with substrate N2.
Across the domain boundary, differences that provide insight into the substrate specificity spectrum in the family are also observed. VvADC-Asp512 forms an equivalent interaction with ligand to TbODC-Asp361. The position of VvADC-Asp512 has shifted away from the ligand-binding site allowing additional room to accommodate the larger agmatine. The equivalent residue in CjCANSDC (Leu314) interacts with the aliphatic portion of the NSpd ligand, accommodating the shorter carbon skeleton between N1 and N3 for substrates that bind CANSDC. In VvADC TbODC-Tyr331 is replaced with VvADC-Trp482 and NE1 is within H-bond distance of the NH2 of agmatine. In both cases this residue projects from the specificity helix and forms an interaction with its symmetry related partner from the other subunit. However, the relationship to the active site structure is reversed. In TbODC-Tyr331 from the same subunit the β/α-barrel domain caps the active site, although it is not within van der Waals contact with putrescine, and in VvADC-Trp482 is contributed from the opposite subunit. This residue is replaced with Ile275 in CANSDC, and like TbODC the residue from the same subunit as the β/α-barrel is closest to the active site, but its position on the helix does not allow for direct interaction with ligand. Furthermore, this residue is not conserved within the CANSDCs (5) and is unlikely to play a role in ligand binding.
DISCUSSION
The basic amino acid decarboxylases from the β/α-barrel-fold family encompass enzymes with diverse substrate specificity ranging from the smallest ligand l-Orn to the largest, carboxyspermidine. Members of the family are found in all three domains of life, and play essential roles in amino acid metabolism and polyamine biosynthesis (5, 13). Completion of the ADC and CANSDC structures allows for the first time a comprehensive structural analysis of how substrate specificity has evolved within the five major specificity types within the family. These data provide significant insight into the structural basis for the observed specificity differences and provide a powerful example of how change of function evolves within the context of a conserved structural domain.
Structural comparison of the major specificity classes in the family (ODC, ADC, DAPDC, and CANSDC) shows that the overall fold and active site structure is strongly conserved, despite low overall sequence identity (in the range of 15%). This allows the active site structural elements controlling specificity to be clearly identified. At the overall structural level ODC, DAPDC, and CANSDC appear to be most similar to each other with all three sharing the same overall monomer organization and dimeric structure. ADC diverges most significantly and has acquired significant additional structural complexity augmenting the basic monomeric structure. Both the interdomain insertion (4-helical bundle) and the extended C terminus (3-helixal bundle) are unique to ADC and these participate in interactions at the dimer interface and in the formation of the tetrameric structure observed in the asymmetric unit. The additional interactions, and significantly greater buried surface area, formed at the dimer interface as the result of these insertions suggests the dimer may be more stable than observed for shorter members of the family that do not contain these features. The tetrameric structure was shown to be the relevant solution species by sedimentation velocity analysis, suggesting that the tetramer may play a unique role in the biology of the bacterial ADCs. All four active sites are oriented within the central donut of the tetramer, providing a potential mechanism for allosteric regulation. These data provide the first example of a tetrameric structure within the fold class. A tetrameric donut-like structure of Mycobacterium tuberculosis DAPDC was recently reported, however, this tetramer displays a small surface area of interaction (900 Å2 per dimer) that only involves 2 monomers (48) in comparison to the VvADC tetramer that involves all 4 monomers (3,680 Å2 buried surface area per dimer) in a more extensive interaction. Furthermore, solution studies with mtDAPDC were consistent with a dimeric structure.
The substrate specificity differences in the family are reflected in two key differences in the active site structures. The first is the position and amino acid composition of the specificity helix, which sits at the back of the active site. The distance between the PLP cofactor and the key residues on this helix that interact with substrate serves as a molecular ruler to restrict catalysis to the basic amino acid ligand of the correct size. For ODC, which has the shortest ligand, the helix is positioned closest to PLP, whereas for ADC, CANSDC, and DAPDC the helix has shifted away from PLP allowing accommodation of the larger ligand (Fig. 8). The functional importance of the amino acid residues that project from the helix to interact with the substrate has been shown for both ODC and chlorella virus ADC, where mutation of TbODC-Asp332 to Glu increases the Km for l-Orn by 20-fold, and mutation of the equivalent residue in chlorella virus ADC increases the Km by 10-fold, whereas decreasing kcat by 100-fold (7).
The second specificity determinant is contributed from residues across the domain boundary on the loop that also hosts the key catalytic base (e.g. TbODC-Cys360) (20). For ODC, ADC, and DAPDC, an acidic residue is positioned from this loop to interact with the distal amino group of the ligand. In ODC, mutation of TbODC-Asp332 to Glu or Ala increases the Km for l-Orn by 100–1000-fold, respectively, demonstrating the functional importance of the interaction (49). However, the CANSDC structure reveals the first example in the family where this residue is replaced by a hydrophobic amino acid (CjCANSDC-Leu314), and this change allows interaction with the aliphatic portion of the substrate because of the change in register due to the shorter carbon backbone found between N1 and N2 of the carboxyspermidine and carboxynorspermidine substrates, in comparison to other enzymes in the family.
Although the overall structure diverges most for the large ADCs, the active site composition is most different for CANSDC. ODC and ADC retain almost all of the same key catalytic residues, whereas for CANSDC, in addition to the substitution of Leu314 for an acidic residue, the residue interacting with the phosphate of PLP has also diverged (Glu or Ser at position 236 replaces TbODC-Arg277, and equivalent in ADC and DAPDC). This is at first a surprising change in active site structure given the demonstration that TbODC-Arg277 is necessary for high affinity PLP binding (47). However, CjCANSDC-Glu236 is involved in an extensive H-bonding network that may mitigate the charge replacement and provide an alternative mechanism to stabilize the PLP phosphate moiety.
An open question remains as to how CANSDC accommodates the larger carboxyspermidine substrate. The observation of a solvent accessible channel that binds glycerol in the CjCANSDC-NSpd structure suggests that the longer substrate could be accommodated by extending into this channel. The interaction between N3 and Asp272 could potentially be preserved, and additionally Asp338, which interacts with glycerol, is conserved throughout the CANSDC enzymes (5) and could potentially form another interaction point for N3 of the longer carboxyspermidine substrate.
The ligands bound to the VvADC and CjCANSDC structures are not found in the typical Schiff base configuration with PLP that has been observed in most of the structures of other enzymes in the family. Both appear to be representatives of the Michaelis complex between enzyme and product. The N1 group of both ligands forms an H-bond interaction in the active site: N1 of agmatine interacts with Ser513 in VvADC, whereas the NSpd N1 of CjCANSDC interacts with Glu236. These data suggest that after decarboxylation these interactions may help to facilitate product release and dissociation away from PLP.
As all polyamine biosynthetic pathways have evolved from amino acid metabolism, it is likely that ADC, ODC, and CANSDC evolved from the lysine biosynthetic enzyme DAPDC by gene duplication and functional divergence. This idea is also supported by the fact that DAPDC is found in most bacteria and euryarcheaota, whereas ADC, although widespread in bacteria, is not found in single-membrane bacteria (Firmicutes and Actinobacteria) or archaea (5). An important aspect of the evolution of ADC was the four-helical bundle interdomain insertion, raising the possibility of an ancestral form of ADC lacking the insertion. Within bacteria and archaea, ODC is found mainly in the α-proteobacteria, which suggests that the eukaryotic ODC may have originated from the α-proteobacterial ancestor of the mitochondrion. Gene duplication not only led to diversification of polyamine biosynthesis, from l-Arg and l-Orn by ADC and ODC but also led to elongation of the pathway, in the case of CANSDC. The viral PBCVADC evolved relatively recently from ODC (6, 7), and in vertebrates, antizyme inhibitor, a polyamine regulatory protein, evolved by gene duplication of ODC followed by loss of catalytic activity, which was accompanied by loss of dimer formation (50). This family of decarboxylases exemplifies the evolutionary processes sculpting metabolism by elaboration of a single structural fold through gene duplication and functional divergence to produce biosynthetic diversification, pathway elongation, and the formation of regulatory proteins.
Supplementary Material
Acknowledgments
We thank Bruce Pearson of the Institute of Food Research, Norwich, for the C. jejuni CANSDC clone. The structures described in this report were derived from work performed on beamline 19-ID at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by the University of Chicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under contract DE-AC02–06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI34432 and R37AI34432 (to M. A. P.) and Welch Foundation Grant I-1257 (to M. A. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 3N2O and 3N29) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PLP
- pyridoxal 5′-phosphate
- VvADC
- V. vulnificus arginine decarboxylase
- CjCANSDC
- C. jejuni carboxynorspermidine decarboxylase
- TbODC
- T. brucei ornithine decarboxylase
- MjDAPDC
- M. jannaschii diaminopimelate decarboxylase
- PBCVADC
- P. bursaria chlorella virus ADC
- L/ODC
- dual specificity lysine/ornithine decarboxylase
- NSpd
- norspermidine
- L-Orn
- L-ornithine
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- ADC
- arginine decarboxylase
- CANSDC
- carboxynorspermidine decarboxylase
- r.m.s.
- root mean square
- PDB
- Protein Data Bank
- SeMet
- selenomethionine.
REFERENCES
- 1.Poole A. M., Ranganathan R. (2006) Curr. Opin. Struct. Biol. 16, 508–513 [DOI] [PubMed] [Google Scholar]
- 2.Kinch L. N., Grishin N. V. (2002) Curr. Opin. Struct. Biol. 12, 400–408 [DOI] [PubMed] [Google Scholar]
- 3.Sandmeier E., Hale T. I., Christen P. (1994) Eur. J. Biochem. 221, 997–1002 [DOI] [PubMed] [Google Scholar]
- 4.Grishin N. V., Phillips M. A., Goldsmith E. J. (1995) Protein Sci. 4, 1291–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J., Michael A. J., Martynowski D., Goldsmith E. J., Phillips M. A. (2007) J. Biol. Chem. 282, 27115–27125 [DOI] [PubMed] [Google Scholar]
- 6.Shah R., Akella R., Goldsmith E. J., Phillips M. A. (2007) Biochemistry 46, 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah R., Coleman C. S., Mir K., Baldwin J., Van Etten J. L., Grishin N. V., Pegg A. E., Stanley B. A., Phillips M. A. (2004) J. Biol. Chem. 279, 35760–35767 [DOI] [PubMed] [Google Scholar]
- 8.Momany C., Ghosh R., Hackert M. L. (1995) Protein Sci. 4, 849–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andréll J., Hicks M. G., Palmer T., Carpenter E. P., Iwata S., Maher M. J. (2009) Biochemistry 48, 3915–3927 [DOI] [PubMed] [Google Scholar]
- 10.Fries D. S., Fairlamb A. H. (2003) in Burger's Medicinal Chemistry and Drug Discovery (Abraham D. ed) 6th Ed., pp. 1033–1087, John Wiley & Sons, Inc., New York [Google Scholar]
- 11.Pegg A. E. (2009) IUBMB Life 61, 880–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sturgill G., Rather P. N. (2004) Mol. Microbiol. 51, 437–446 [DOI] [PubMed] [Google Scholar]
- 13.Lee J., Sperandio V., Frantz D. E., Longgood J., Camilli A., Phillips M. A., Michael A. J. (2009) J. Biol. Chem. 284, 9899–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel C. N., Wortham B. W., Lines J. L., Fetherston J. D., Perry R. D., Oliveira M. A. (2006) J. Bacteriol. 188, 2355–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keating T. A., Marshall C. G., Walsh C. T. (2000) Biochemistry 39, 15513–15521 [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto S., Sugahara T., Tougou K., Shinoda S. (1994) Microbiology 140, 3117–3124 [DOI] [PubMed] [Google Scholar]
- 17.Almrud J. J., Oliveira M. A., Kern A. D., Grishin N. V., Phillips M. A., Hackert M. L. (2000) J. Mol. Biol. 295, 7–16 [DOI] [PubMed] [Google Scholar]
- 18.Grishin N. V., Osterman A. L., Brooks H. B., Phillips M. A., Goldsmith E. J. (1999) Biochemistry 38, 15174–15184 [DOI] [PubMed] [Google Scholar]
- 19.Jackson L. K., Brooks H. B., Myers D. P., Phillips M. A. (2003) Biochemistry 42, 2933–2940 [DOI] [PubMed] [Google Scholar]
- 20.Jackson L. K., Brooks H. B., Osterman A. L., Goldsmith E. J., Phillips M. A. (2000) Biochemistry 39, 11247–11257 [DOI] [PubMed] [Google Scholar]
- 21.Jackson L. K., Goldsmith E. J., Phillips M. A. (2003) J. Biol. Chem. 278, 22037–22043 [DOI] [PubMed] [Google Scholar]
- 22.Jackson L. K., Baldwin J., Akella R., Goldsmith E. J., Phillips M. A. (2004) Biochemistry 43, 12990–12999 [DOI] [PubMed] [Google Scholar]
- 23.Gokulan K., Rupp B., Pavelka M. S., Jr., Jacobs W. R., Jr., Sacchettini J. C. (2003) J. Biol. Chem. 278, 18588–18596 [DOI] [PubMed] [Google Scholar]
- 24.Ray S. S., Bonanno J. B., Rajashankar K. R., Pinho M. G., He G., De Lencastre H., Tomasz A., Burley S. K. (2002) Structure 10, 1499–1508 [DOI] [PubMed] [Google Scholar]
- 25.Liu X. Y., Lei J., Liu X., Su X. D., Li L. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 282–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song J., Zhou C., Liu R., Wu X., Wu D., Hu X., Ding Y. (2010) Mol. Biol. Rep. 37, 1823–1829 [DOI] [PubMed] [Google Scholar]
- 27.Patel C. N., Adcock R. S., Sell K. G., Oliveira M. A. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2396–2398 [DOI] [PubMed] [Google Scholar]
- 28.Rodrigez B. R., Carroll D. W., Mitchell D., Momany C., Hackert M. L. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 175–177 [DOI] [PubMed] [Google Scholar]
- 29.Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31.Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z. (1991) in Isomorphous Replacement and Anomalous Scattering (Wolf W., Evans P. R., Leslie A. G. W. eds) pp. 80–86, Science & Engineering Research Council, Cambridge, UK [Google Scholar]
- 33.Cowtan K. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 750–756 [DOI] [PubMed] [Google Scholar]
- 34.Morris R. J., Zwart P. H., Cohen S., Fernandez F. J., Kakaris M., Kirillova O., Vonrhein C., Perrakis A., Lamzin V. S. (2004) J. Synchrotron Radiat. 11, 56–59 [DOI] [PubMed] [Google Scholar]
- 35.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W., Kabsch H., Eisenberg D. (1976) J. Mol. Biol. 100, 283–291 [DOI] [PubMed] [Google Scholar]
- 39.Kleywegt G. J., Jones T. A. (1997) Methods Enzymol. 277, 525–545 [DOI] [PubMed] [Google Scholar]
- 40.Kleywegt G. J., Jones T. A. (1994) CCP4/ESF-EACBM Newsletter on Protein Crystallogr. 31, 9–14 [Google Scholar]
- 41.Kleywegt G. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1878–1884 [DOI] [PubMed] [Google Scholar]
- 42.Pei J., Tang M., Grishin N. V. (2008) Nucleic Acids Res. 36, W30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei J., Kim B. H., Grishin N. V. (2008) Nucleic Acids Res. 36, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskowski R. A. (2007) Bioinformatics 23, 1824–1827 [DOI] [PubMed] [Google Scholar]
- 45.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osterman A. L., Brooks H. B., Jackson L., Abbott J. J., Phillips M. A. (1999) Biochemistry 38, 11814–11826 [DOI] [PubMed] [Google Scholar]
- 47.Osterman A. L., Brooks H. B., Rizo J., Phillips M. A. (1997) Biochemistry 36, 4558–4567 [DOI] [PubMed] [Google Scholar]
- 48.Weyand S., Kefala G., Svergun D. I., Weiss M. S. (2009) J. Struct. Funct. Genomics 10, 209–217 [DOI] [PubMed] [Google Scholar]
- 49.Osterman A. L., Kinch L. N., Grishin N. V., Phillips M. A. (1995) J. Biol. Chem. 270, 11797–11802 [DOI] [PubMed] [Google Scholar]
- 50.Albeck S., Dym O., Unger T., Snapir Z., Bercovich Z., Kahana C. (2008) Protein Sci. 17, 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.