Abstract
Capping protein (CP) is a ubiquitously expressed, heterodimeric 62-kDa protein that binds the barbed end of the actin filament with high affinity to block further filament elongation. Myotrophin (V-1) is a 13-kDa ankyrin repeat-containing protein that binds CP tightly, sequestering it in a totally inactive complex in vitro. Here, we elucidate the molecular interaction between CP and V-1 by NMR. Specifically, chemical shift mapping and intermolecular paramagnetic relaxation enhancement experiments reveal that the ankyrin loops of V-1, which are essential for V-1/CP interaction, bind the basic patch near the joint of the α tentacle of CP shown previously to drive most of the association of CP with and affinity for the barbed end. Consistently, site-directed mutagenesis of CP shows that V-1 and the strong electrostatic binding site for CP on the barbed end compete for this basic patch on CP. These results can explain how V-1 inactivates barbed end capping by CP and why V-1 is incapable of uncapping CP-capped actin filaments, the two signature biochemical activities of V-1.
Keywords: Actin, Cell Motility, Cytoskeleton, NMR, Protein/Protein Interactions, V-1, Capping Protein
Introduction
Capping protein (CP)3 is a ubiquitously expressed 62-kDa α/β heterodimer that binds the barbed end of the actin filament with high affinity (Kd = 0.1 nm) (1) to prevent further actin monomer association and dissociation, thereby limiting the extent of filament elongation in vivo (2, 3). In actin-based motility, such as that occurring in lamellipodia, new filaments are nucleated by the Arp2/3 complex to create a dendritic actin network at the leading edge. Biochemical, cell biological, and modeling studies all indicate that rapid filament capping by CP is required to sustain a dendritic network that is sufficiently branched to provide the motive force required for leading edge extension (4–7). Consistent with its central role in actin network assembly, CP is one of only five proteins required for the reconstitution of actin-based motility in vitro (4, 5, 8), and cells lacking CP have profound deficiencies in actin cytoskeleton assembly (9–13).
Determination of the CP crystal structure led to the “tentacles” model of barbed end capping by CP (14). The two structurally homologous CP subunits form a central β-sheet that includes the bulk of the protein core, above which there are two antiparallel α-helices, one belonging to each subunit (14). At the end of these helices, each subunit contains a C-terminal “tentacle” which, on CPα, is composed of an unstructured region punctuated in the middle by a short 4-residue helix, and on CPβ, it is composed of a longer amphipathic helix that protrudes from the protein core (Fig. 1). Based on crystallographic evidence, it was proposed that these C-terminal tentacles are flexible in solution, allowing them to bind and cap the barbed end. Extensive mutational studies in yeast (15) and vertebrate (1) CP that focused on the tentacles provided strong support for the tentacles model of capping. Specifically, deletion of the α tentacle decreased the affinity of CP for the barbed end by 6,000-fold and its on-rate by 20-fold, whereas deletion of the β tentacle decreased the affinity of CP by 400-fold, with no effect on its on-rate (1, 15). Moreover, deletion of both tentacles rendered CP unable to cap filaments (15). These results indicated that both the α and β tentacles are important for capping, and they emphasized the greater importance of the α tentacle for fast association with the barbed end and for overall capping activity. That said, peptides corresponding to the β tentacle (C-terminal 28 or 34 residues) exhibited weak capping activity on their own (1). The β tentacle is characterized by acidic residues at its unstructured “joint” (Asp247 and Asp251), followed by the amphipathic α-helix observed in the crystal (14). The β tentacle amphipathic helix was characterized by a hydrophobic side (Tyr255, Leu558, Leu262, Ala265, and Leu266) opposite a side containing mostly polar and charged residues (Asn252, Gln253, Lys254, Lys256, Gln257, Gln259, Arg260, Glu261, and Gln264). The amphipathic helix was followed by an unstructured region with both hydrophobic and charged residues (14).
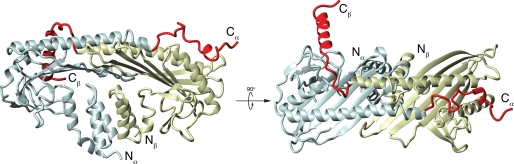
FIGURE 1.
Crystal structure of chicken CPα1β1 (14). The α-subunit is shaded blue; the β-subunit is shaded yellow. N and C termini of each subunit are indicated. Each subunit contains an N-terminal 3-helix bundle followed by a peripheral loop region. The core of the protein is composed of a central β-sheet with each subunit contributing five strands. Above the central β-sheet, there are two anti-parallel α-helices, one from each subunit ending in the C-terminal tentacle. The region of each subunit thought to be flexible, based on crystallographic and mutational analysis, and thus defined as the tentacle, is highlighted in red (Arg259 to the C terminus in CPα and Arg244 to the C terminus in CPβ).
Subsequently, the structure of CP bound to the barbed end was determined at 23 Å resolution by cryo-electron microscopy and was then fitted with the crystal structure of CP and the proposed F-actin structure (16–18). The resulting structure identified additional residues in CP involved in barbed end capping and led to the proposal of a two-step capping mechanism. First, conserved basic residues in the α tentacle, together with additional nearby basic residues from the core of CPβ, present a basic patch that interacts coordinately with a complementary acidic cluster on the barbed end. This electrostatic interaction would drive the initial association of CP with the barbed end, as well as much of the overall binding strength. Second, with CP electrostatically bound to the filament end, the β tentacle undergoes conformational sampling to contact the hydrophobic cleft between subdomains 1 and 3 on the outside of the terminal actin subunit (16). This model requires that the β tentacle be flexible, although this was not directly observed. Overall, this two-step capping mechanism incorporates all available structural and biochemical data to date.
Because actin monomers add mainly onto the barbed end of the filament, the rate of actin-based motility is thought to be largely controlled by the availability of free barbed ends (19). As the primary barbed end capping protein in cells, CP is likely a major focal point for regulation. Indeed, the discrepancy between the half-life of CP bound to the barbed end in vitro (∼30 min) (20, 21) and in vivo (∼1 s) (22) suggests that CP activity is significantly controlled by regulatory molecules in vivo. In fact, several proteins have been found to bind CP to alter its activity (2, 3, 23–26). One possible cellular regulator of CP is CARMIL (26–28), which binds CP in vivo and with high affinity in vitro (Kd = 1 nm) and which drives the removal of CP from the barbed end (26, 29–31). This interaction represents a model for CP “uncapping” in which CARMIL uncaps CP-capped barbed ends, with the CP-CARMIL complex retaining weak capping activity (32).
Another likely cellular regulator of CP is V-1 (myotrophin), a 13-kDa protein that appears to be abundantly expressed in most vertebrate cell types (33, 34). Biochemical studies have shown that V-1 binds CP with high affinity (Kd ∼40 nm) in a 1:1 complex that has no affinity for the barbed end, i.e. V-1 sequesters CP in a totally inactive complex (25, 35). Moreover, V-1 is unable to uncap CP-capped filaments (25, 35). Presumably, just as weak capping and barbed end uncapping are mechanistically coupled in CARMIL, sequestering and the inability to uncap barbed ends are mechanistically coupled in V-1 (32). Although V-1 has been shown to bind tightly to CP in vitro (25, 35) and in vivo (36), its physiological relevance is unknown. However, given the cellular concentration of V-1 of 10 μm (or 10 times that of CP)4 and its high affinity for CP, it could clearly play a major role as a cellular buffer for active, cytosolic CP.
The solution NMR structure of V-1 (37, 38) shows that the protein consists of two central ankyrin repeats flanked by partial repeats at the N and C termini. Ankyrin repeats are present in a large number of proteins and are composed of β-hairpin helix-loop-helix motifs (39). In general, and presumably also in V-1, these variable loops drive specific protein/protein interactions, thereby conferring substrate specificity (40). Consistently, the two V-1 ankyrin loop sequences are highly conserved among vertebrates, and both are crucial for CP binding, as mutations in either dramatically decrease the affinity of V-1 for CP (30–100-fold) (35).
A model for V-1 binding to CP in which the β tentacle is both required and largely sufficient for V-1 interaction has been proposed, based on four main observations (35). First, deletion of the α or β tentacle of CP decreased its affinity for V-1 by 1.6- or 7-fold, respectively, suggesting that the β tentacle was more important for V-1 binding. Second, CPα1β2 bound V-1 4-fold more tightly than did CPα2β2, and these two isoforms differ only at residues close in the structure to the β tentacle. Specifically, the hydrophobic side of the amphipathic helix of the β tentacle was implicated in V-1 binding because V-1 bound equally the β1 and β2 isoforms, which only differ in the hydrophilic side of the amphipathic helix (20). Third, the affinity of V-1 for a peptide corresponding to the CP β tentacle was only slightly weaker (∼1.5-fold) than its affinity for native CP. Finally, changes in simulated molecular dynamics upon hypothetical deletion of the CPα tentacle occurred in a region in CPα close in the structure to the β tentacle, where the α1 and α2 isoforms show greatest sequence divergence. Based on these four observations, a model was proposed in which V-1 binds through its ankyrin loops to β tentacle of CP, although specific molecular details were unknown. Notably, this binding site is distinct from the strong electrostatic actin-binding site on CP, in principle leaving the α tentacle and adjacent basic patch on CP available for barbed end binding even in the V-1/CP complex.
Developments in NMR spectroscopy have allowed studies of larger molecules and protein complexes, and NMR can provide information about the motions of molecules on biologically relevant time scales that are unattainable using other structural methods (41–43). Here, the secondary structure and picosecond-nanosecond dynamics of CP were determined by NMR. We show that the β tentacle is indeed flexible in solution but does not contain an α-helix, as observed in the crystal structure (14). Based on its propensity for assuming helical structure, we propose that a helix could be induced in the β tentacle upon barbed end capping. The flexible portion of the α subunit is limited to the C-terminal 12 residues, which we show are not critical for capping. NMR chemical shift mapping and intermolecular paramagnetic relaxation enhancement experiments were used to determine the structure of the complex of V-1 bound to CP. The complex presented here does not support the previous model of V-1/CP interaction (35) in that it excludes the β tentacle as playing a role in direct binding to V-1. Rather, we demonstrate competitive binding between V-1 and the electrostatic binding site on the barbed end for the basic patch of CP, which includes but is not limited to residues in the α tentacle joint. This structure, which we support by site-directed mutagenesis of CP coupled with measurements of V-1/CP affinity and barbed end capping activity, can explain how V-1 completely inactivates barbed end capping by CP and why V-1 cannot uncap previously capped filaments.
EXPERIMENTAL PROCEDURES
Protein Preparation
Mouse capping protein α1 and β2 subunits were co-expressed on a pET3d plasmid and purified as described previously (29, 31), with minor alterations. Briefly, Escherichia coli BL21 cells expressing CP were grown to A600 of 3–4, at which time they were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Expression was carried out for 3–4 h at 37 °C. For NMR studies, E. coli BL21 DE3 cells expressing CP were always grown in >99.8% D2O. For 15N-labeled samples, cells were grown in M9 minimal media containing 15NH4Cl as the sole nitrogen source. For 13C-,15N-labeled samples, cells were grown as above, except d-[13C]glucose was used as the sole carbon source. 100-ml cultures were grown overnight to A600 > 4.0 and were used to inoculate 1 liter of D2O expression media. Cultures were grown at 37 °C until A600 ∼2.5, at which point 1 mm isopropyl 1-thio-β-d-galactopyranoside was added to induce protein expression, which was carried out at 37 °C for 3–4 h. Cells were harvested by centrifugation at 11,000 × g for 24 min at 4 °C. Cells pellets were resuspended in 3 ml/g of resuspension buffer (1× phosphate-buffered saline, 1 mm DTT, 1 mm EDTA, protease inhibitor mixture (Roche Applied Science)) and lysed by passing through a high pressure cell homogenizer (Emulsi-Flex®-C3, Avestin, Inc, Ottawa, Ontario, Canada) three times at 1500 p.s.i. The insoluble cellular materials were pelleted by centrifugation at 22,000 × g for 45 min at 4 °C. The soluble cell fraction was then applied directly to a CP affinity column, which was composed of CNBR-Sepharose (GE Healthcare) covalently bound to GST-CAH3a/b (29). CAH3a/b was the CP-binding site of the protein CARMIL (31) to which CP bound tightly above pH 5.2 and [NaCl] less than 0.5 m. The column was washed with 5 column volumes of affinity wash buffer (20 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.1% Tween 20, 1 mm DTT, 0.5 mm EDTA), and CP was eluted with elution buffer (100 mm sodium acetate, pH 4.0, 500 mm NaCl, 1 mm DTT, 1 mm EDTA). The affinity column eluate was dialyzed overnight at 4 °C into low salt buffer (10 mm Tris-HCl, pH 8.0, 10 mm KCl, 0.5 mm EDTA, 1 mm DTT, 0.1% NaN3), applied to a column containing Mono Q-Sepharose, and eluted from 0 to 500 mm NaCl. The CP sample was dialyzed into NMR buffer (see below) and concentrated. The sample could not be unfolded and refolded nor could subunits be expressed and labeled separately, followed by refolding the dimer, because of the instability of each of the subunits alone. CP mutants were prepared using the same plasmid as a template, and mutant cDNA was made with the QuikChange® site-directed mutagenesis kit (Stratagene). Mutants were prepared in the same way as wild-type CP, and they were verified by CD spectroscopy to have similar spectra to wild-type CP.
V-1 was purified as described previously (38), and the protein NMR spectrum was confirmed to be identical to that in previous studies by two-dimensional NMR.
Actin was purified from rabbit skeletal muscle by gel filtration chromatography using S-300 resin as described previously (44). Monomeric actin was purified from rabbit skeletal muscle by gel filtration chromatography using S-300 resin to remove oligomers. G-actin was stored in G-buffer (0.2 mm ATP, 1 mm NaN2, 0.1 mm CaCl2, 0.5 mm DTT, and 2 mm Tris-HCl, pH 8.0). Magnesium-actin in KMEI buffer (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole, pH 7.0) was labeled with N-(1-pyrene)iodoacetamide (Invitrogen catalog no. P-29) and purified by centrifugation, dialysis, and gel filtration with G-25 resin. The concentrations of actin and pyrene were estimated using the extinction coefficients A290 = 26,600 m−1 cm−1 and A344 = 22,000 m−1 cm−1 (45). The concentration of labeled actin was determined by subtracting 0.127 times the A344 value from the A290 value.
NMR samples were prepared in 20 mm sodium acetate, pH 6.45, 100 mm KCl, 1 mm DTT, 0.5 mm EDTA, 8% D2O and were prepared to ∼0.4–0.7 mm protein. All NMR experiments were performed at 32 °C. The same conditions were used for V-1, but for samples of the V-1/CP complex, buffers contained 20 mm KCl. The two-dimensional 1H-15N HSQC spectrum was always compared with a reference spectrum before all experiments to ensure sample homogeneity.
NMR Spectroscopy and Chemical Shift Indexing
All experiments, except those involving 13C′ evolution, were recorded on a Bruker 800 MHz spectrometer equipped with a z axis pulsed-field gradient cryoprobe. Experiments involving 13C′ evolution were recorded on a Bruker 600 MHz spectrometer also equipped with a z axis pulsed-field gradient cryoprobe. All pulse sequences contained TROSY selection as described previously (46, 47) and gradient selection (48, 49) during the TROSY component. CP assignments were made using a four-dimensional experiment strategy as described previously (50–52) in which both HNCO(i−1)CA(i−1) and HNCA(i)CO(i) were used to establish sequential connectivities. Both data sets were recorded using (1024X72X24X24) complex data points in the F4 (1H), F3 (15N), F2 (13Cα), and F1 (13C′) with spectral widths of 7183, 1562, 3650, and 1812 Hz, respectively. Each of the four-dimensional experiments was recorded using eight transients, and a recycle delay of 1.3 s. Amino acid identities were obtained using a three-dimensional HN(CO)CACBi−1 and HNCACBi experiments. These data sets were recorded using (1024X96X60) complex data points in the F3(1H), F2(15N), and F1(CACB) dimensions, with spectral widths of 11161, 2083, and 11261 Hz. Chemical shifts and overlaps were resolved using high resolution three-dimensional HNCAi and HNCOi−1 experiments. Data for these experiments were recorded using (1024X96X60(Cα) or 64(C′)) complex data points, with spectral widths of 1161, 2083, and 5000 Hz (Cα) or 2818 Hz (C′). 15N T2 measurements of the 15NH TROSY component were recorded at 800 MHz using a TROSY-selected CPMG experiment (53) using a constant CPMG delay of 200 ms and relaxation delays of 6.2, 9.4, 15.8, 22.2, 31.8, 44.6, 65.4, 84.6, 116.6, and 164.6 ms.
Secondary structure was inferred by comparing the secondary 13Cα chemical shifts to reference values for random coil amino acids as described previously (54, 55). For each region of secondary structure element defined in the crystal structure, the average 13Cα and 13C′ secondary chemical shift was determined for those residues assigned within that region. Regions having average 13Cα secondary shifts greater than 2.0 were considered to be α-helical, although those whose average was less than −1.0 were considered to agree with β-sheet conformation. 13C′ secondary shifts were used qualitatively to verify assignments because no numerical standard is generally used.
Chemical Shift Mapping
Because binding between CP and V-1 is tight (Kd = 14 nm), and the system was in slow chemical exchange on the NMR time scale, free CP and V-1-bound CP were assigned independently. To obtain the chemical shift map on CP, a 2H,15N-labeled sample of CP was bound with 2H-labeled V-1, and the 1H-15N TROSY HSQC was recorded. The V-1 chemical shift map was obtained analogously, using 2H,15N-labeled V-1 titrated with 2H-labeled CP. Resonances whose chemical shift changed by ≥0.15 ppm, calculated as described previously (56), were considered significant. This cutoff represents values >1.5 S.D. from the average calculated Δδ value. These values were used to determine the chemical shift maps.
Paramagnetic Relaxation Enhancement
V-1 was paramagnetically labeled with 3-(2-iodoacetamido)-2,2,5,5-tetramethyl-1-pyrolidinyloxy, free radical (Toronto Research Chemicals Inc., catalog no. I684000), which remained covalently linked to CP in the presence of 1 mm DTT, necessary to prevent disulfide bond formation. Wild-type V-1 contains three cysteine residues, each of which was mutated to serine, whereas a non-native cysteine was introduced as a point mutation at M7C. M7C V-1 gave identical CD and NMR spectra to wild-type V-1 and had normal activity. To produce a sufficient quantity of 2H-labeled M7C V-1, a culture was grown in 500 ml of M9 minimal media in D2O, and the mutant protein was purified in the same way as wild-type V-1. Prior to labeling, the purified protein was soaked in buffer containing 20 mm DTT for 30 min at room temperature to reduce the β-mercaptoethanol adduct and then exchanged into buffer lacking DTT. Spin labeling was carried out in 100 mm Tris-HCl, pH 8.5, 1 mm diethylenetriaminepentaacetic acid overnight at room temperature with 20× molar excess of spin label that had been previously dissolved at 17 mg/ml in DMSO. The reaction was confirmed by liquid chromatography/mass spectrometry (data not shown), and no unlabeled protein was observed. Excess spin label was removed by reversed-phase HPLC using C4 resin. Labeled V-1 was added to 15N,2H-labeled CP at a 1.1:1.0 molar ratio. The diamagnetic sample was made by soaking 2H M7C V-1 with 20 mm ascorbic acid for 6 h at room temperature prior to binding with CP. The buffer was exchanged to remove ascorbic acid, and CP was added to the sample. A 1H-15N HSQC spectrum of each sample was recorded, and peak intensities were directly compared as described previously (57).
Structure Calculation
To determine the structure of the complex of CP and V-1, chemical shift perturbations as well as PRE intensities were used. Chemical shift perturbations were used as ambiguous restraint inputs in XPLOR-NIH (58) calculations with a target distance of 5.0 Å, for a total of 35 restraints from V-1 and 45 (19 from α, 26 from β) from CP. Some residues experienced large chemical shift changes upon V-1 binding, and their resonances could not be confidently assigned. These resonances were all within regions that otherwise experienced chemical shift changes and were thus included as ambiguous restraints in the calculation. PRE restraints were translated into distances from V-1 M7C Sγ. Based on the ratio of peak intensities in the paramagnetic (Ip) and diamagnetic (Id) experiments, distances could be grouped into four bins, where Ip/Id < 0.1, 0.1 < Ip/Id < 0.2, 0.2 < Ip/Id < 0.3, and 0.3 < Ip/Id < 0.5, and given distance restraints of 9, 16, 19, and 26 Å, respectively (57). Residues whose intensity ratio was greater than 0.6 were not used as restraints in the calculation. In total, there were 6, 10, 17, and 16 residues in each bin (49 total PRE restraints). Distance restraints from both datasets were enforced using a soft-square potential with a switching value of 0.5 for both chemical shift and PRE data. The structures were calculated using the simulated annealing protocol with both ambiguous and PRE restraints included and randomized starting structures for both CP and V-1 (59). Both CPαβ and V-1 were treated as rigid bodies in structure refinement, except that the side chains were allowed to be flexible during dynamics calculations. Out of 200 total structures calculated, the 10 lowest energy structures were chosen having no distance violations greater than 0.5 Å from either ambiguous or PRE distance restraints.
CP Mutant Analysis
Various point mutations were introduced into CP and their binding to V-1 was measured using fluorescence anisotropy. Wild-type V-1 labeled with the fluorophore (5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid) bound CP weakly (Kd ∼1 μm), presumably because labeling at multiple sites disrupted the protein structure, and therefore, the same M7C V-1 mutant that had been used for PRE studies was used for fluorescence anisotropy assays. The V-1 sample was labeled with 5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid (catalog no. I14, Molecular Probes, Invitrogen) and purified by reversed-phase HPLC. The same buffer was used as for NMR experiments. Experiments were performed on a double channel T-format quantamaster spectrofluorimeter (Photon Technology International) using built-in Felix32 software. Labeled M7C V-1 (20 nm) was excited at 370 nm, and emission was recorded at 495 nm. Each experimental anisotropy was the average over 300 s with 1 data point recorded every 2 s at 20 °C. Each titration was performed in triplicate, and data represent average values. Data were fitted to a standard binding curve assuming a single binding site (60) using Grace curve-fitting software. Affinities of CP mutants for actin were measured using pyrene-labeled actin polymerization assays at 22 °C as described previously (21). Briefly, pyrene-labeled (100%) and unlabeled Ca-ATP-actin monomers were mixed in G-buffer to obtain a 10 μm stock solution that was 10% pyrene-labeled. To convert Ca-ATP actin monomers in this stock solution to Mg-ATP-actin monomers, a one-tenth volume of ME buffer (10 mm EGTA, 1 mm MgCl2) was gently added, and the mixture was incubated for 2 min at room temperature. Actin polymerization (final actin concentration of 2 μm, diluted with G-buffer) was initiated by adding actin seeds and 10× KMEI buffer. Actin seeds were created by polymerizing 8 μm Mg-ATP G-actin at room temperature for 1 h and then vortexing aliquots for 20 s immediately before addition to the assay. This sheared F-actin was added to a final concentration of 0.8 μm, which should correspond to ∼0.8 nm barbed ends, based on calculations described previously (21). Pyrene fluorescence was monitored with excitation at 365 nm and emission at 407 nm using an LS55 spectrophotometer (PerkinElmer Life Sciences). To measure the barbed end capping activity of CP, the proteins were mixed with actin seeds and incubated for 30 s before adding to the cuvette.
RESULTS
NMR Studies Confirm the Overall Structure of CP and Define the Conformation and Degree of Flexibility of the C-terminal Tentacles
CP samples were prepared at pH 6.5 to make comparisons with the earlier crystal studies. After 1–2 months at 32 °C, some peaks in the 1H-15N-HSQC-TROSY spectrum of CP completely disappeared, accompanied by the appearance of sharp peaks in a region of the spectrum characteristic of unfolded peptides. The disappearing peaks were assigned to the C-terminal portions of CP, namely residues C-terminal to Lys281 from the CPα C terminus and C-terminal to Lys259 in CPβ. Cleavage at these sites was confirmed by liquid chromatography/mass spectrometry. The SDS-PAGE migration pattern of the CPβ subunit was observed to change slightly over time, observed as smearing of the band toward lower molecular weight, presumably because of degradation at its C terminus.5 The affinity of CP for barbed ends was observed to decrease by ∼10-fold after prolonged storage at 4 °C, presumably because of loss of the CPβ tentacle, as the decrease in activity was consistent with that obtained here by mutating β tentacle residues thought to contribute to the interaction with the barbed end.
Of 548 expected peptide backbone N-H resonance peaks for CPα/β dimer, only 435 peaks were observed in the 1H-15N HSQC TROSY spectrum of 15N,2H-labeled CP (supplemental Fig. 1), representing 79% of the peaks. Of the observed peaks, 397 (91%) were assigned. The most probable cause of missing peaks was incomplete exchange of amide deuterium atoms back to hydrogen after protein production in D2O, especially in the case of residues in the hydrophobic core of the protein. In the case of larger proteins, partial unfolding followed by refolding in an H2O environment can increase the amount of re-protonation (52, 61, 62). However, even partial denaturation of CP in the presence of 2 m guanidine hydrochloride rendered the protein insoluble, preventing attempts at partial unfolding and refolding. However, because 72% of the protein backbone could be assigned, reasonable conclusions could be drawn from the data without further optimization.
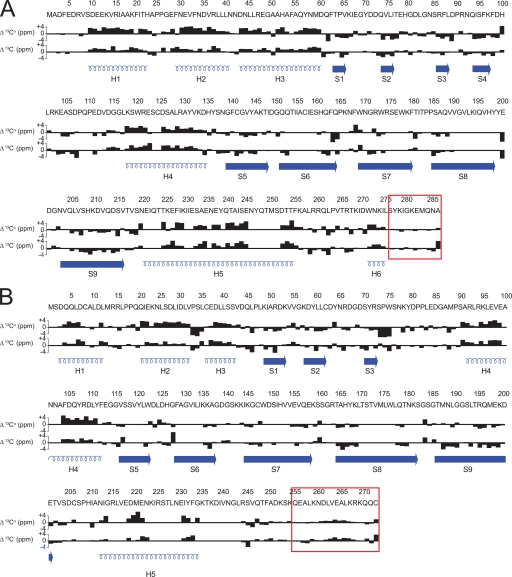
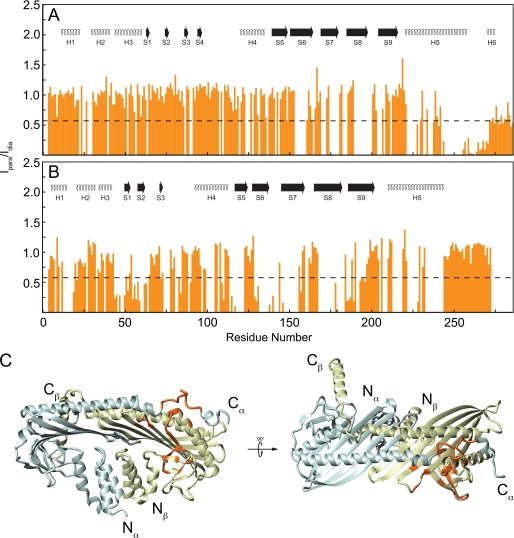
Overall, the secondary structures determined for CP based on backbone chemical shift assignments agreed with those calculated from the crystal structure, with few exceptions (Fig. 2 and supplemental Table 1). 13Cα chemical shifts were used to assign secondary structure from NMR studies, using standards described previously (55); and in all cases, qualitative agreements were obtained from 13C′ secondary shifts. In solution, in both CP subunits, every secondary structure determined was consistent with that from the crystal structure, except as noted below. Helical regions had average secondary shift values greater than 2.0, whereas β-sheets had values less than −1.0, as expected. Interestingly, the region classified as helix 5b of CPα in the crystal structure (Thr254–Leu258) had an average secondary shift value of 0.75, indicating that this region was not a stable helix in solution. The small helix at the extreme C terminus of CPα, defined here as helix 6 (residues Trp271–Leu275), gave an average value of 2.28, consistent with the crystal structure. The side chain of Trp271 is thought to anchor this helix to the core of CP.
FIGURE 2.
Summary of 13Cα, 13C′ secondary chemical shifts for CP. 13Cα and 13C′ chemical shifts were compared with average random coil values for CPα (A) and CPβ (B). Positive bars correspond to the α-helical conformation; negative bars correspond to the β-sheet. Red boxes indicate flexible regions as determined by NMR T2 relaxation studies. Secondary structures are illustrated below the shifts in blue.
The CPβ secondary structures also in general agreed with those from the crystal structure (Fig. 2B), although two significant differences were observed. In the crystal, the long helix on top of the central β-sheet is composed of residues His209–Leu243, but NMR data showed that this helix begins at Asn212 (supplemental Table 1). The most striking result was that for residues comprising the region previously described as the C-terminal β tentacle α-helix (Ser253–Lys267), the average secondary shift was 0.73, which is more characteristic of a random coil than a helix. Although no helical structure was observed for Ser253–Lys259, for Asn260–Lys267, the average secondary shift was 1.09, less than that typical of α-helices, but which nonetheless indicated some helical propensity. This suggested that helical structure may be induced upon barbed-end capping. Overall, assignment results suggested that CP had the same overall structure in solution as under those used for crystallographic studies with the exceptions of residues Thr254–Leu258 from CPα and of the C-terminal tentacle from CPβ which, under solution conditions used for NMR studies, was not helical.
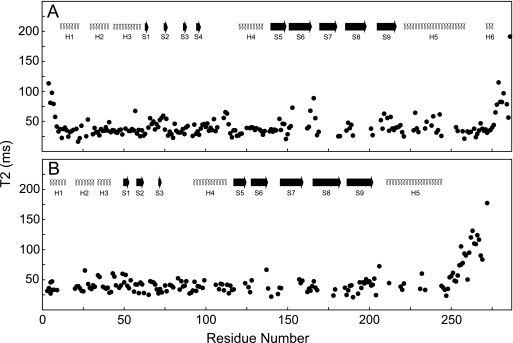
The 15NH-TROSY transverse magnetic relaxation times of CP residues were measured to characterize protein backbone 15NH dynamics on the picosecond-nanosecond time scale. At 32 °C, CP had an overall effective T2 of ∼44 ms, although the C-terminal tentacles of both subunits were considerably more dynamic on this time scale (Fig. 3, A and B). The C-terminal tentacles are defined here as the region for which the effective T2 is longer than 54 ms, or 1.5 S.D. from the average for nonflexible regions. In CPα, the tentacle contains residues Leu275–Ala286 (Fig. 3A), whereas the CPβ tentacle contains residues Phe249–Cys272 (Fig. 3B). CP mutants lacking the C-terminal 28 residues of CPα (Arg259–Ala286), the C-terminal 34 residues of the β subunit (Leu243–Asn276 in chicken CPβ1) (1), or equivalent residues in yeast CP (15) were previously shown to be attenuated for capping. Although extensive regions of the C terminus of each CP subunit may be involved in binding, our NMR relaxation data show that the flexibility of this region in CPα is limited to its C-terminal 12 residues, consistent with B-factors observed in crystallographic studies (14).
FIGURE 3.
15N TROSY effective-T2 relaxation of CP. 15N T2 was acquired at 32 °C at 800 MHz and plotted versus residue number for CPα (A) and CPβ (B). Secondary structures are shown at the top, and numbering corresponds to that in Fig. 1.
The N-terminal nine residues of CPα constitutes a flexible unstructured region not included in the crystal structure (T2avg = 78 ms) (Fig. 3A). The CPβ subunit N-terminal helix began at residue 2, accounting for the lack of flexibility in this region (Fig. 3). The body of CP exhibited generally uniform relaxation rates indicating that CP tumbled in solution as a single unit.
Determination of the Interaction Surfaces of CP and V-1 by Chemical Shift Mapping
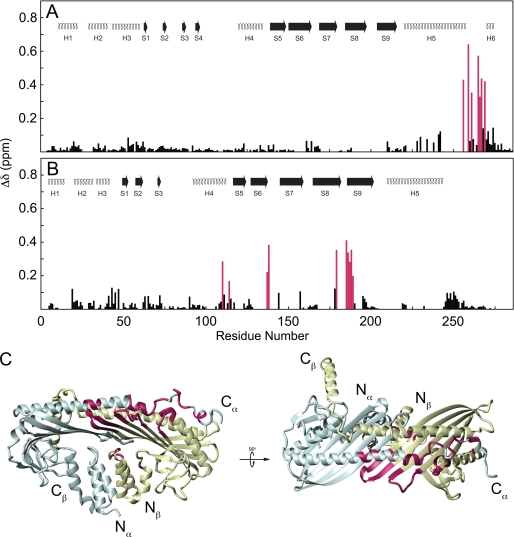
Chemical shift mapping was performed on both CP and V-1 to characterize the binding site between them. Overall, sites on both CPα and CPβ experienced chemical shift changes upon V-1 binding (Fig. 4, A and B). On the α-subunit, significant chemical shift changes (Δδ > 0.15 ppm) were observed only in residues Lys256, Arg259, Leu261, Thr265, Arg266, Thr267, and Ile269, which are immediately N-terminal to the flexible C-terminal 12 residues of CPα (i.e. part of the α tentacle joint). In the crystal structure, this region lacks secondary structure, consistent with the random coil conformation observed in NMR spectra (Fig. 4C). In CPβ, chemical shift changes were extensive. The loops of the central β-sheet experienced the largest chemical shift changes, particularly between strands 6 and 7 and between strands 8 and 9 (Fig. 4B), including residues Ala137–Ile144 and Thr179–Leu189, respectively. These loops are adjacent to each other in the CP structure and contain lysine residues, which we showed (see below) to be involved in electrostatic contacts with V-1. Other significant chemical shift changes were observed surrounding Gly114 in CPβ, which was adjacent to the β-turn between strands 6 and 7 in the central β-sheet. Overall, chemical shift mapping indicated that V-1 binds to a site on CP not previously identified and is composed of the core of CPβ as well as the joint of the CPα tentacle.
FIGURE 4.
Chemical shift map of V-1 binding on CP. Chemical shift change was plotted against residue number for CPα (A) and CPβ (B). Secondary structures are the same as in Fig. 1. Secondary shifts >0.15 ppm are highlighted purple. Residues experiencing large chemical shift changes were plotted onto the CP crystal structure (C). CPα is shaded blue; CPβ is shaded yellow. Residues whose chemical shift changed significantly upon V-1 binding are highlighted magenta. N and C termini of CPα and CPβ are indicated. Molecular structures were rendered using the program MolMol (64).
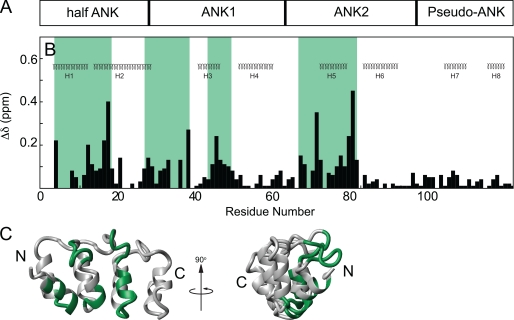
A chemical shift map of the CP binding surface on V-1 was also determined. V-1 is divided into an N-terminal half-ankyrin repeat, two adjacent ankyrin repeats, followed by a C-terminal pseudo-ankyrin repeat (Fig. 5A) (37). Chemical shift changes upon CP binding were observed in the N-terminal three repeats (Fig. 5, B and C). Four regions of V-1, all on its concave side composed of the short inner helices and the ankyrin loops (Fig. 5C), experienced chemical shift changes upon CP binding (Fig. 5B), representing a contiguous surface of V-1 (Fig. 5C). The N-terminal region showed significant chemical shift changes at residues Lys4–Lys19. The second region consisted of residues Glu26–Leu37. This region corresponded to ankyrin loop 1. The next region showing chemical shift changes consisted of residues Ala43–Glu48, which span part of helix 3 and the hairpin turn between helices 3 and 4. The final region undergoing chemical shift changes upon CP binding consisted of residues Asp65–Gly79. This region contains part of ankyrin loop 2 and part of helix 5. The contiguous surface on the V-1 structure showing chemical shift changes (Fig. 5C, bottom) provided a clear map of its interaction surface for CP. The identification of ankyrin loops 1 and 2 in the binding surface was consistent with previous studies (35), but our NMR studies revealed that the total interaction surface also included the inner helices of V-1.
FIGURE 5.
Chemical shift map of CP binding on V-1. The domain arrangement of V-1 is shown (A) aligned on top the secondary structures of V-1, corresponding to the NMR structure (37) (B). Chemical shift changes were plotted against residue number for V-1. Regions affected by binding are highlighted green. Chemical shift changes in V-1 were plotted onto the ribbon diagram of V-1 (C) and its N and C termini are indicated.
Determination of the Orientation of V-1 Binding to CP by PRE
Although the overall binding surface between CP and V-1 could be inferred from chemical shift mapping, the relative orientation of the proteins could not be determined solely based on this technique. Thus, intermolecular PRE experiments were performed to obtain distance restraints that could be used to determine the orientation of V-1 when bound to CP. The addition of a paramagnetic molecule to a protein increases the magnetic relaxation rate of neighboring atoms as a function of distance from the paramagnetic center. Increased relaxation in the presence of a paramagnetic species causes a decrease in NMR peak intensity (Ip) relative to the intensity in the absence of the paramagnetic species (Id), and the intensity ratio (Ip/Id) can be translated into a distance from the affected atom to the PRE center.
2H,15N-CP was mixed with 2H-M7C V-1 paramagnetically labeled at its cysteine, and the NMR spectrum was recorded. Comparing NMR resonance intensities between para- and diamagnetic 15N-1H HSQC spectra proved sufficient to determine the region on CP within ∼26 Å of the paramagnetic center. PRE effects were observed in both CP subunits. On CPα, PRE was strongest at residues Lys256–Trp271 (Fig. 6A), where average Ip/Id = 0.133(±0.089). This region corresponds to the joint of the α tentacle, and it overlaps the chemical shift map. The other regions experiencing enhanced relaxation in the paramagnetic sample were located in CPβ (Fig. 6B). PRE effects on CPβ included areas close to the α tentacle joint, overlapping areas experiencing chemical shift changes, but only a weak effect was observed in residues in the loop between β-strands 8 and 9. Residues Leu47–Leu61, located on β-strands 1 and 2, had average Ip/Id = 0.397(±0.165), although this region did not undergo chemical shift changes, indicating that although close (<26 Å) to V-1 in the structure, it does not directly contact V-1 (Fig. 6C). No PRE effect was observed on the β tentacle.
FIGURE 6.
Intermolecular paramagnetic relaxation enhancement. The ratios of intensity of CP resonance peaks in the paramagnetic sample to those in the diamagnetic sample (orange bars in A and B) were plotted against CPα (A) and CPβ (B) residue number. Secondary structure elements plotted above the intensities are the same as in previous figures. Black dashed line at Ip/Id = 0.6 indicates the upper threshold used for PRE. Residues experiencing significant PRE were highlighted orange on the CP ribbon diagram (C) where the CP subunits are colored as in Fig. 3.
Determination of the Structure of the CP-V-1 Complex Suggests That V-1 Interacts Electrostatically with the Basic Patch on CP
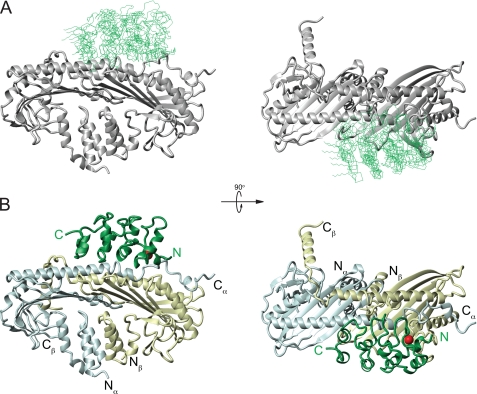
The 10 lowest energy structures of the CP-V-1 complex were chosen from 200 calculated structures as a representative ensemble (Fig. 7A). Each protein backbone was treated as a rigid body during energy minimization and molecular dynamics calculations, although side chains were unrestrained. Structural statistics for the 10 lowest energy structures are presented in Table 1. Because both proteins were treated as rigid during calculation, the protein complexes could be aligned according to the mean structure for the entire complex or by the backbone of CP only, and backbone root mean square deviation values are presented for both cases.
FIGURE 7.
Structure of CP-V-1 complex. The backbone superposition, based on CP coordinates, of the 10 lowest energy structures of the complex of V-1/CP is shown in A. The CPαβ ribbon is shaded gray, and the V-1 backbone is shaded green. The orientation is the same as in B. The ribbon diagram of the lowest energy structure of the V-1/CP complex is shown in B. The location of the PRE center on V-1 at position 7 is shown as a red sphere. V-1 is shaded green, and CP coloring is the same as in Fig. 3. N and C termini are indicated for V-1 (green) and CP (black).
TABLE 1.
Structure statistics of the 10 lowest energy structures of the V-1/CP complex
| Backbone root mean square deviations (Å) of V-1 with CP coordinates fixed | |
| Interfacea backbone | 3.73 ± 2.19 |
| All backbone | 4.20 ± 2.34 |
| Backbone root mean square deviations (Å) of complex with respect to the mean | |
| Interfaceb backbone | 1.76 ± 1.00 |
| All backbone | 1.44 ± 0.80 |
| No. of restraints | |
| Ambiguous from CPα | 19 |
| Ambiguous from CPβ | 26 |
| Ambiguous from V-1 | 35 |
| PRE | 49 |
| Restraint distance root mean square deviations (Å) | |
| Ambiguous | 0.04 ± 0.02 |
| PRE | 0.014 ± 0.01 |
| Interaction surface area (Å2)c | 1338.5 |
a Interface residues were defined as those experiencing significant chemical shift perturbations upon binding, as defined under “Experimental Procedures.” Interface backbone residues included the following: from CPα, Ser244–Arg266; from CPβ, Asp109–Val116, Ala137–Lys145, Gln178–Leu189; and from V-1, Lys4–Leu15, Arg30–Lys37, Ala43–Gly46, Lys66–Ile69, Thr70, and Leu72–His80.
b Backbone residues were defined as NH, Cα, and C′.
c Interaction area was calculated using a sphere of radius 2 Å.
Overall, the binding surface between CP and V-1 includes ∼1300 Å2 (Table 1), and it involves a single patch on the surface of CP and most of one side of V-1 (Fig. 7B). The binding site on V-1 is composed of the ankyrin loops and the short helices. The ankyrin loops are near the CPα tentacle joint (residues Asp252–Asp270), and the short helices of V-1 lie between the CPα long helix and the loops of the central β-sheet. The first ankyrin loop makes closer contact with CPα at the α tentacle joint, although the second ankyrin loop is slightly farther from the long helix on top of CP. This result agrees with previous mutation data showing that ankyrin loop 1 in V-1 is more important for CP inhibition than ankyrin loop 2 (35). The N terminus of V-1 is pointed toward the C terminus of CPα, although the C-terminal pseudo-ankyrin repeat is oriented toward the center of CP but does not contact CP. The β-hairpin loop between strands 6 and 7 in the CPβ central β-sheet protrudes toward the short, inner helices of the two central ankyrin repeats of V-1. This loop on CPβ is part of the “basic patch” previously described as important for actin binding (16).
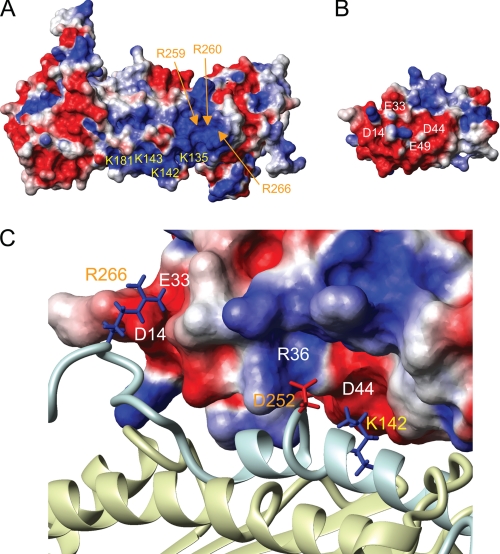
The binding surfaces of both CP and V-1 are composed mainly of charged residues, with those on CP being mostly basic (Fig. 8A) and those on V-1 mostly acidic (Fig. 8B), indicating that their mechanism of binding consists mainly of electrostatic interactions. In fact, in complex with V-1, the basic patch on CP is entirely occluded by V-1, showing that the V-1-binding site is likely competitive with that for the barbed end. On the surface of CPβ, the side chain of Lys142 projects toward an acidic patch on V-1 near Asp44 (Fig. 8C). Also, Asp252 on CPα is located close to a basic patch near Arg36 on V-1. Near the N terminus of V-1, there is another acidic patch that is composed of Asp14 and Glu33 close to which Arg266 on CPα is located.
FIGURE 8.
Characterization of the CP/V-1 interaction surface. Charged residues in the CP basic patch and residues on V-1 thought to interact in the binding surface were labeled on the surface of CPαβ (A) and V-1 (B). Each surface is colored based on charge, with negatively charged residues in red; positively charged residues in blue. An enlargement of the V-1/CP binding interface is shown in C. Residues on CPα, CPβ, and V-1 are labeled orange, yellow, and white, respectively. V-1 is represented as a charged surface, and CP is represented as a ribbon with the same coloring as in previous figures (C). Residues on CP thought to be involved in electrostatic interactions with V-1 are shown as sticks, and the label coloring corresponds to that in A and B.
Mutational Analysis Confirms Competition between V-1 and Barbed Ends for the Basic Patch on CP and Defines the Roles of the CPα Tentacle and Joint in CP Activity
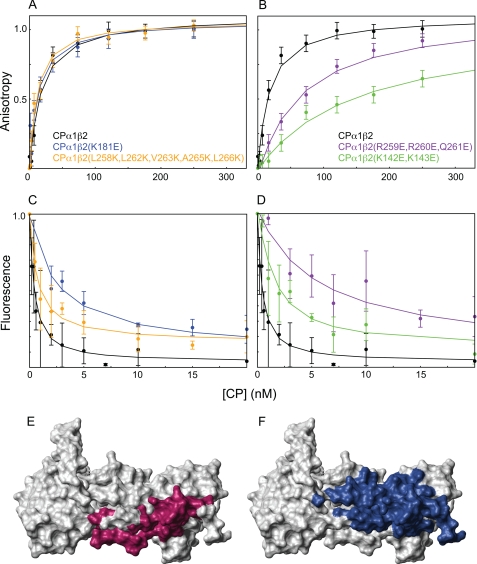
We reasoned that if the binding site on CP for V-1 competes with a significant portion of the binding site on CP for the barbed end, then CP mutations inhibiting V-1 binding should also inhibit barbed end capping. Using pyrene actin polymerization assays (data not shown), we initially found that wild-type V-1 bound wild-type CP with Kd = 13 nm, 3-fold tighter than in a previous study (35). We then used fluorescence anisotropy measurements to determine direct binding of CP to V-1. In controls, fluorescently labeled M7C V-1 was found to bind wild-type CP with Kd = 14 ± 6 nm (Fig. 9, A and B). Similar values were obtained for fluorescently labeled M7C V-1 inhibiting the capping activity of CP by using pyrene actin polymerization assays (data not shown), indicating that M7C-V1 has the same CP-binding affinity to inactivate CP. We then characterized two CP mutants that were attenuated for V-1 binding. V-1 bound to the first mutant, the triple CPα mutant CPα1(R259E,R260E,Q261E)β2, with Kd = 66 ± 12 nm, or 5-fold weaker than wild-type CP (Fig. 9B). This result reveals the importance of the CPα tentacle joint for V-1 binding. V-1 bound to the second mutant, the double CPβ mutant CPα1β2(K142E,K143E), with Kd = 222 ± 40 nm, or 16-fold weaker than to wild-type CP (Fig. 9B). These two residues are adjacent to the CPα tentacle joint in the CP crystal structure, in the turn region between strands 6 and 7 in the central β-sheet. The mutated residues in these two CP samples were all in the interface between CP and V-1. We then measured the affinity of these two CP mutants for the barbed end using actin polymerization assays. In controls, the affinity of wild-type CP for the barbed end agreed with previous measurements (Kd = 0.1 ± 0.08 nm) (Fig. 9, C and D, and Table 2). As anticipated, these two CP mutants (CPα1(R259E,R260E,Q261E)β2 and CPα1β2(K142E,K143E)) exhibited a large decrease in barbed end capping activity (Fig. 9D), having affinities of 6.3 ± 0.8 and 1.9 ± 0.3 nm, corresponding to 63- and 19-fold decreases in affinity, respectively. These results show that some of the same residues in CP's basic patch are critical for binding both V-1 and the barbed end.
FIGURE 9.
CP mutation analysis and mechanism of CP sequestering. V-1 binding to CP mutants was assayed by fluorescence anisotropy of V-1 in A and B. Data points represent the average value of three experiments, and error bars represent the average S.D. CP mutants are indicated according to color. Mutations that had no effect on V-1 binding are as follows: CPα1β2(K181E) (blue), CPα1β2(L258K,L262K,V263K,A265K,L266K) (orange), and wild-type CPα1β2 (black) are shown in A. Mutations that attenuated V-1 binding, CPα1(R259E,R260E,Q261E)β2 (purple), CPα1β2(K142E,K143E) (green), and wild-type CP (black) are shown in B. Equilibrium dissociation constants are presented in Table 2. Binding between CP mutants and actin barbed ends was directly assayed by monitoring pyrene-actin fluorescence in the presence of various concentrations of CP (C and D). The CP mutants used in C and D are the same as in A and B, respectively, and the coloring is the same. The residues on CP implicated in binding V-1 (E) or actin (F), based on chemical shift mapping, or mutational analysis and cryo-EM studies (16), respectively, are highlighted. The binding site for V-1 coincides that for actin in the competitive interaction surface.
TABLE 2.
Summary of equilibrium dissociation constants for CP binding to V-1 and barbed ends
Kd values between V-1/CP were calculated from fluorescence anisotropy data, and those for CP binding to barbed ends were calculated from pyrene actin fluorescence experiments. Data were fitted as described previously (60), and errors represent the S.D. from three independent experiments.
| CP |
Kd |
|
|---|---|---|
| V-1 | Actin | |
| nm | ||
| α1β2 | 14 ± 6 | 0.1 ± 0.08 |
| α1β2(K181E) | 16 ± 6 | 2.4 ± 0.4 |
| α1β2(L258K,L262K,V263K,A265K,L266K) | 14 ± 5 | 1.0 ± 0.2 |
| α1(R259E,R260E Q261E)β2 | 66 ± 12 | 6.3 ± 0.8 |
| α1β2(K142E,K143E) | 222 ± 40 | 1.9 ± 0.3 |
| α1(ΔC28)β2 | 68 ± 12 | 258 ± 168 |
| α1(ΔC10)β2 | 7 ± 2 | 0.91 ± 0.09 |
Two other CP point mutations had no effect on V-1 binding. Mutation of CPα1β2(K181E), located in the turn between strands 8 and 9 in the central β-sheet adjacent to that between strands 6 and 7, had no effect on the affinity of CP for V-1 (Kd = 16 ± 6 nm) (Fig. 9A). This indicates that even though this region is within 8 Å of V-1 in the structure, Lys181 made no electrostatic contacts with V-1. The CPβ tentacle was mutated by changing five residues with hydrophobic side chains to lysines. V-1 bound to this mutant, CPα1β2(L258K, L262K, V263K, A265K, and L266K) with Kd = 14 ± 5 nm (Fig. 9A), which is essentially identical to the affinity of V-1 for wild-type CP. The CPβ tentacle was previously proposed to form an amphipathic α-helix in solution and to interact extensively with V-1 via the hydrophobic face of this amphipathic helix (35). However, our CPβ tentacle mutant shows that these hydrophobic residues are not important for V-1 binding. This result is consistent with our structure, where the CPβ tentacle is located far from the binding site for V-1. These two CP mutant, (CPα1β2(L258K, L262K, V263K, A265K, and L266K) and CPα1β2(K181E)) bound barbed ends with Kd = 1.0 ± 0.2 and 2.4 ± 0.4 nm, respectively, corresponding to 10- and 24-fold decreases in affinity, relative to wild-type CP (Fig. 9C), indicating the β tentacle has an effect on capping activity.
A previous study showed that CPα1(ΔC28)β2 was able to bind V-1 with an affinity similar to wild-type CP (Kd = 65 nm compared with 40 nm, respectively) (35). Our results showed that the same mutant bound V-1 with Kd = 68 ± 12 nm (supplemental Fig. 2B), essentially identical to the affinity reported previously. However, in our study, wild-type CP was observed to bind V-1 with 14 nm affinity, almost three times tighter than previously observed. This mutant bound barbed ends ∼2600-fold weaker than wild-type CP, consistent with previous studies (supplemental Fig. 2D) (1). These results confirm that the CPα tentacle joint is important for binding both V-1 and the barbed end.
Because NMR relaxation experiments showed that only the C-terminal 12 residues of CPα are flexible, a mutant lacking these residues was generated, and its affinities for both V-1 and barbed ends were measured. CPα1(ΔC10)β2 bound V-1 with an affinity similar to that of wild-type CP (Kd = 7 ± 2 nm). Similarly, this mutant bound barbed ends with only slightly reduced affinity relative to wild-type CP (Kd = 0.9 ± 0.1 nm) (supplemental Fig. 2, A and C). These results show that the portion of the “α tentacle” that is actually flexible is much less important in barbed end capping than the α tentacle joint.
DISCUSSION
We characterized the solution structure of CP by NMR and mutational studies, resulting in a precise definition of the C-terminal tentacles. In the crystal structure, the C-terminal region of CPα consists of an unstructured region from Arg259–Asp270, followed by a short helix from Trp271–Ile274, and another unstructured region from Leu275 to the C terminus at Ala286 (14). Deletion of the C-terminal 28 residues of CPα (Arg259–Ala286) severely inhibits capping (supplemental Fig. 2) (1). Although not experimentally supported, this region was initially suggested to be flexible in solution, and it was thus defined as the α tentacle (although speculation about the actual flexibility of this region has persisted). Here, we showed that in solution only residues Leu275–Ala286, C-terminal to the short helix, are flexible on the picosecond-nanosecond time scale and that these residues have little importance for capping activity. Although our results are consistent with the central role that this region of CPα plays in barbed end capping, they clarify the long standing uncertainty about the conformation and dynamics of the α tentacle. Based on our results, we suggest that the α tentacle be defined as including only the flexible residues of the C terminus, Leu275–Ala286. Within the CPα C terminus, Arg259 was described previously as a “pivot” (16), and in analogous fashion we describe the region from Arg259 to Ile274 as the joint of the α tentacle.
Based on the crystal structure, the CPβ tentacle was defined as the C-terminal 34 residues (C-terminal 29 residues in mouse CPβ2) which, based on the high average B-factor and its conformation in the crystal for this region, was thought to be flexible in solution (14). This β tentacle contained an amphipathic helix from Gln253 to Thr267 (Gln253–Lys267 in mouse CPβ2). Deletion of the C-terminal 28 or 34 residues of CPβ1 (1) or equivalent residues in yeast CPβ (Cap2) (15) strongly inhibited the ability of CP to cap barbed ends, and its importance was confirmed as peptides corresponding to these residues showed capping activity (1, 15). Here, we showed that the CPβ tentacle is in fact flexible on the picosecond-nanosecond time scale. Surprisingly, however, the β tentacle is unstructured in solution. In the second step of the two-step mechanism of capping described by Narita et al. (16), the flexible β tentacle, which is an amphipathic helix, undergoes conformational sampling until binding via its hydrophobic face to the barbed end. Although we did not observe helical structure in the β tentacle, it is likely induced upon barbed end binding for several reasons. First, protein structure analysis predicts that this peptide forms an amphipathic helix, and this region was in fact helical in the crystal structure. Second, mutation of residues predicted to lie on the hydrophobic, actin-binding surface of the helix attenuated barbed end capping (1). Finally, this helix is predicted to bind the hydrophobic cleft between subdomains 1 and 3 on the terminal actin protomer at the barbed end, which is known as an important regulatory site on actin (63).
Our experimental determination of the solution structure of CP bound to V-1 offers an explanation for the mechanism of CP sequestration by V-1. Chemical shift mapping revealed the overall binding surfaces involved in the interaction, although paramagnetic relaxation enhancement experiments provided the orientation of V-1 when bound to CP. Analysis of the structure shows that V-1 binds to the basic patch on CP, near the α tentacle joint, using both of its ankyrin loops as well as the short inner helices on the same side of the molecule. Importantly, this binding site on CP for V-1 overlaps almost entirely the strong electrostatic binding site on CP for the barbed end (Fig. 9, E and F). Indeed, we demonstrated that CP mutations that reduce its affinity for V-1 also reduce its affinity for the barbed end, providing additional support for the idea that CP binds V-1 and the barbed end competitively, using many of the same residues in the basic patch. We also showed that point mutations in the β tentacle that reduced affinity of the CPs for the barbed end 10-fold, in agreement with previous studies (1), had no effect on the affinity of CP for V-1.
Our data are consistent with the two-step mechanism of capping described by Narita et al. (16), where residues in the basic patch, consisting of residues in both CPα (Lys256, Arg259, Arg260, Arg266, and Lys268) and CPβ (Lys142, Lys143, Lys145, Lys181, Lys223, and Arg225), make electrostatic interactions with the barbed end to drive the first step in capping. In the second step, the CPβ tentacle undergoes conformational sampling until binding a hydrophobic site on the opposite side of the barbed end. Support for the first step of this model came from point mutations at residues Lys256, Arg259, Arg260, and Arg266 in CPα, all of which inhibited capping (1, 15, 16). In addition to confirming the importance of Arg259 and Arg260 in capping, we showed by site-directed mutagenesis that nearby residues from CPβ (Lys142, Lys143, and Lys181), also within the basic patch, were important for capping.
The structure of CP bound to V-1 determined here is strikingly different from a previously proposed model for V-1 binding to CP (35). Although both studies implicated the ankyrin loops of V-1 in the V-1/CP interaction, the model, based on mutational studies and molecular dynamics simulations, argues that the CPβ tentacle is both necessary and largely sufficient for the interaction of CP with V-1. In contrast, in our structure V-1 binds to the basic patch on CP and does not even contact the CPβ tentacle. Several definitive results obtained in this study argue strongly that the β tentacle region is not involved in binding V-1. First, we observed chemical shift changes and PRE effects only on residues in and close to the basic patch on CP, and no effects were observed in the β tentacle or nearby regions of CP. Second, although the docked model shows V-1 binding to the helical region of the β tentacle, we observed no changes in the peak intensities of residues in the β tentacle upon V-1 binding, indicating no change in their dynamics. Furthermore, no changes in secondary shifts were observed in the β tentacle when CP was bound to V-1, showing that the β tentacle is in fact unstructured in both free CP and V-1 bound CP. Third, the previous model implicated the hydrophobic side of the β tentacle helix in V-1 binding, but we showed that mutating these residues in the β tentacle had no impact on V-1 binding. Finally, although in both studies the CPα tentacle (C-terminal 28 residues) deletion mutants bound V-1 with 66 ± 12 nm affinity, we observed that wild-type CP bound V-1 with 14 ± 6 nm affinity, almost 3-fold stronger than that observed in the previous study. Therefore, the decrease in affinity of CP for V-1 due to deletion of its α tentacle was ∼5-fold rather than ∼1.6-fold, as reported previously.
In the two-step capping mechanism, CP binds the barbed end with two sites, its basic patch first, followed by its β tentacle. In principle, each site can bind and dissociate independently of the other. The electrostatic interaction between the barbed end and the basic patch on CP is responsible for the fast on-rate of capping as well as much of the affinity of CP for the barbed end, although the hydrophobic interaction between the β tentacle and the barbed end affects primarily the off-rate of capping. In our structure, V-1 binds only to the basic patch on CP, and this interaction prevents in a competitive fashion the association of CP with the barbed end, the first step of capping. We note, however, that this interaction appears to leave the β tentacle available for interaction with the barbed end. The converse situation exists with the previous model of V-1/CP interaction, where V-1 ties up just the β tentacle, leaving the basic patch on and around the α tentacle available for binding the barbed end. In that model, the sequestering activity of V-1 was explained using the “wobble” model of barbed end capping where CP wobbles about the barbed end when bound solely by its β tentacle but not when bound solely by its α tentacle. It is thought that sequestering proteins such as V-1 cannot bind CP in the wobble state. We offer an alternative mechanism of sequestering, based on the structure determined here and the two-step mechanism of capping (16). Because the basic patch on CP has been shown repeatedly to be responsible for the large majority of the affinity of CP for the barbed end and for its on-rate, the occlusion of this basic patch by V-1 binding must completely inhibit the contributions made by this basic patch to both the affinity and on-rate for capping. Based on the fact that V-1 completely inactivates CP (35), we hypothesize that V-1 binding to the basic patch prevents for steric reasons the ability of the β tentacle to encounter its hydrophobic binding site on the barbed end.
The results shown here also explain why V-1 cannot uncap previously capped barbed ends. Barbed end uncapping must involve binding to CP and altering its binding site for the barbed end, such as through steric interaction or induction of a conformation change, such that the affinity of CP for the barbed end is greatly reduced. The primary interaction of capping is between the basic patch on CP and a complementary acidic region on the barbed end. Because the basic patch on CP is occluded by binding the barbed end, it is unavailable for V-1 binding. Thus, in much the same way V-1 is able to bind and occlude the high affinity binding site of CP for the barbed end, and the barbed end occludes the V-1-binding site on CP, rendering V-1 unable to bind and uncap the barbed end. Overall, our results explain the two signature biochemical activities of V-1 as follows: its ability to completely sequester CP, and its inability to remove CP from the barbed end. We showed that both V-1 and the barbed end compete for many of the same residues on the basic patch of CP, and binding of either V-1 or the barbed end to CP renders the other unable to bind. The atomic coordinates for the 10 lowest energy structures of the CP-V-1 complex have been deposited in the Protein Data Bank (PDB accession code 2KXP).
Supplementary Material
Acknowledgments
We thank Dr. James Gruschus for help in chemical shift mapping analysis, Dr. Marie-Paule Strub for generating V-1 and CP mutants, and Dr. Motoshi Suzuki and Dr. James Feretti for helpful comments and discussion on this manuscript. The V-1 plasmid was generously provided by Dr. Jun Qin. We acknowledge the professional skills and advice of Dr. Grzegorz Piszczek (Biophysics Core Facility, NHLBI, National Institutes of Health) regarding fluorescence anisotropy and circular dichroism experiments performed in this paper. We thank Dr. Duck-Yeon Lee (Protein Analysis Core Facility, NHLBI, National Institutes of Health) for the expertise and advice regarding mass spectrometry-related experiments performed in this paper.
This work was supported, in whole or in part, by National Institutes of Health grants from Intramural Research Program, NHLBI (to J. A. H. III and N. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and Table 1.
The atomic coordinates and structure factors (code 2KXP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
J. A. Hammer III, unpublished observations.
K. Remmert, unpublished observations.
- CP
- capping protein
- DTT
- dithiothreitol
- HPLC
- high pressure liquid chromatography
- PRE
- paramagnetic relaxation enhancement
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1.Wear M. A., Yamashita A., Kim K., Maéda Y., Cooper J. A. (2003) Curr. Biol. 13, 1531–1537 [DOI] [PubMed] [Google Scholar]
- 2.Wear M. A., Cooper J. A. (2004) Trends Biochem. Sci. 29, 418–428 [DOI] [PubMed] [Google Scholar]
- 3.Cooper J. A., Sept D. (2008) Int. Rev. Cell Mol. Biol. 267, 183–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akin O., Mullins R. D. (2008) Cell 133, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwasa J. H., Mullins R. D. (2007) Curr. Biol. 17, 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mogilner A., Edelstein-Keshet L. (2002) Biophys. J. 83, 1237–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollard T. D., Blanchoin L., Mullins R. D. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 545–576 [DOI] [PubMed] [Google Scholar]
- 8.Loisel T. P., Boujemaa R., Pantaloni D., Carlier M. F. (1999) Nature 401, 613–616 [DOI] [PubMed] [Google Scholar]
- 9.Amatruda J. F., Cannon J. F., Tatchell K., Hug C., Cooper J. A. (1990) Nature 344, 352–354 [DOI] [PubMed] [Google Scholar]
- 10.Amatruda J. F., Gattermeir D. J., Karpova T. S., Cooper J. A. (1992) J. Cell Biol. 119, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sizonenko G. I., Karpova T. S., Gattermeir D. J., Cooper J. A. (1996) Mol. Biol. Cell 7, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mejillano M. R., Kojima S., Applewhite D. A., Gertler F. B., Svitkina T. M., Borisy G. G. (2004) Cell 118, 363–373 [DOI] [PubMed] [Google Scholar]
- 13.Hug C., Jay P. Y., Reddy I., McNally J. G., Bridgman P. C., Elson E. L., Cooper J. A. (1995) Cell 81, 591–600 [DOI] [PubMed] [Google Scholar]
- 14.Yamashita A., Maeda K., Maéda Y. (2003) EMBO J. 22, 1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K., Yamashita A., Wear M. A., Maéda Y., Cooper J. A. (2004) J. Cell Biol. 164, 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narita A., Takeda S., Yamashita A., Maéda Y. (2006) EMBO J. 25, 5626–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narita A., Maéda Y. (2007) J. Mol. Biol. 365, 480–501 [DOI] [PubMed] [Google Scholar]
- 18.Lorenz M., Popp D., Holmes K. C. (1993) J. Mol. Biol. 234, 826–836 [DOI] [PubMed] [Google Scholar]
- 19.Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 20.Schafer D. A., Jennings P. B., Cooper J. A. (1996) J. Cell Biol. 135, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn J. R., Pollard T. D. (2007) J. Biol. Chem. 282, 28014–28024 [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi T., Tsuji T., Higashida C., Hertzog M., Fujita A., Narumiya S., Scita G., Watanabe N. (2006) J. Cell Biol. 175, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruck S., Huber T. B., Ingham R. J., Kim K., Niederstrasser H., Allen P. M., Pawson T., Cooper J. A., Shaw A. S. (2006) J. Biol. Chem. 281, 19196–19203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canton D. A., Olsten M. E., Kim K., Doherty-Kirby A., Lajoie G., Cooper J. A., Litchfield D. W. (2005) Mol. Cell. Biol. 25, 3519–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taoka M., Ichimura T., Wakamiya-Tsuruta A., Kubota Y., Araki T., Obinata T., Isobe T. (2003) J. Biol. Chem. 278, 5864–5870 [DOI] [PubMed] [Google Scholar]
- 26.Jung G., Remmert K., Wu X., Volosky J. M., Hammer J. A., 3rd (2001) J. Cell Biol. 153, 1479–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu P., Zot A. S., Zot H. G. (1995) J. Biol. Chem. 270, 25316–25319 [DOI] [PubMed] [Google Scholar]
- 28.Xu P., Mitchelhill K. I., Kobe B., Kemp B. E., Zot H. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3685–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remmert K., Uruno T., Hammer J. A., 3rd (2009) Protein Expr. Purif. 67, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang C., Pring M., Wear M. A., Huang M., Cooper J. A., Svitkina T. M., Zigmond S. H. (2005) Dev. Cell 9, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uruno T., Remmert K., Hammer J. A., 3rd (2006) J. Biol. Chem. 281, 10635–10650 [DOI] [PubMed] [Google Scholar]
- 32.Fujiwara I., Remmert K., Hammer J. A., 3rd (2010) J. Biol. Chem. 285, 2707–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sivasubramanian N., Adhikary G., Sil P. C., Sen S. (1996) J. Biol. Chem. 271, 2812–2816 [DOI] [PubMed] [Google Scholar]
- 34.Anderson K. M., Berrebi-Bertrand I., Kirkpatrick R. B., McQueney M. S., Underwood D. C., Rouanet S., Chabot-Fletcher M. (1999) J. Mol. Cell. Cardiol. 31, 705–719 [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharya N., Ghosh S., Sept D., Cooper J. A. (2006) J. Biol. Chem. 281, 31021–31030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitazawa M., Yamakuni T., Song S. Y., Kato C., Tsuchiya R., Ishida M., Suzuki N., Adachi E., Iwashita S., Ueno S., Yanagihara N., Taoka M., Isobe T., Ohizumi Y. (2005) Biochem. Biophys. Res. Commun. 331, 181–186 [DOI] [PubMed] [Google Scholar]
- 37.Yang Y., Nanduri S., Sen S., Qin J. (1998) Structure 6, 619–626 [DOI] [PubMed] [Google Scholar]
- 38.Yang Y., Rao N. S., Walker E., Sen S., Qin J. (1997) Protein Sci. 6, 1347–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J., Mahajan A., Tsai M. D. (2006) Biochemistry 45, 15168–15178 [DOI] [PubMed] [Google Scholar]
- 40.Sedgwick S. G., Smerdon S. J. (1999) Trends Biochem. Sci. 24, 311–316 [DOI] [PubMed] [Google Scholar]
- 41.Palmer A. G., 3rd (1997) Curr. Opin. Struct. Biol. 7, 732–737 [DOI] [PubMed] [Google Scholar]
- 42.Kay L. E. (1998) Nat. Struct. Biol. 5, 513–517 [DOI] [PubMed] [Google Scholar]
- 43.Mittermaier A., Kay L. E. (2006) Science 312, 224–228 [DOI] [PubMed] [Google Scholar]
- 44.Kuhn J. R., Pollard T. D. (2005) Biophys. J. 88, 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kouyama T., Mihashi K. (1980) Eur. J. Biochem. 105, 279–287 [DOI] [PubMed] [Google Scholar]
- 46.Pervushin K. V., Wider G., Wuthrich K. (1998) J. Biomol. NMR 12, 345–348 [DOI] [PubMed] [Google Scholar]
- 47.Pervushin K., Riek R., Wider G., Wüthrich K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kay L. E., Keifer P., Saarinen T. (1992) J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 49.Schleucher J., Schwendinger M., Sattler M., Schmidt P., Schedletzky O., Glaser S. J., Sørensen O. W., Griesinger C. (1994) J. Biomol. NMR 4, 301–306 [DOI] [PubMed] [Google Scholar]
- 50.Yang D., Kay L. E. (1999) J. Am. Chem. Soc. 121, 2571–2575 [Google Scholar]
- 51.Kim K., McCully M. E., Bhattacharya N., Butler B., Sept D., Cooper J. A. (2007) J. Biol. Chem. 282, 5871–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tugarinov V., Muhandiram R., Ayed A., Kay L. E. (2002) J. Am. Chem. Soc. 124, 10025–10035 [DOI] [PubMed] [Google Scholar]
- 53.Loria J. P., Rance M., Palmer A. G., 3rd (1999) J. Biomol. NMR 15, 151–155 [DOI] [PubMed] [Google Scholar]
- 54.Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. (1995) J. Biomol. NMR 5, 67–81 [DOI] [PubMed] [Google Scholar]
- 55.Spera S., Bax A. (1991) J. Am. Chem. Soc. 113, 5490–5492 [Google Scholar]
- 56.Garrett D. S., Seok Y. J., Peterkofsky A., Clore G. M., Gronenborn A. M. (1997) Biochemistry 36, 4393–4398 [DOI] [PubMed] [Google Scholar]
- 57.Battiste J. L., Wagner G. (2000) Biochemistry 39, 5355–5365 [DOI] [PubMed] [Google Scholar]
- 58.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 59.Clore G. M., Schwieters C. D. (2003) J. Am. Chem. Soc. 125, 2902–2912 [DOI] [PubMed] [Google Scholar]
- 60.Heyduk T., Lee J. C. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1744–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tugarinov V., Kanelis V., Kay L. E. (2006) Nat. Protoc. 1, 749–754 [DOI] [PubMed] [Google Scholar]
- 62.Gardner K. H., Zhang X., Gehring K., Kay L. E. (1998) J. Am. Chem. Soc. 120, 11738–11748 [Google Scholar]
- 63.Dominguez R. (2004) Trends Biochem. Sci. 29, 572–578 [DOI] [PubMed] [Google Scholar]
- 64.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.