Summary
Acute leukemias induced by MLL chimeric oncoproteins are among the subset of cancers distinguished by a paradoxical dependence on GSK-3 kinase activity for sustained proliferation. We demonstrate here that GSK-3 maintains the MLL leukemia stem cell transcriptional program by promoting the conditional association of CREB and its co-activators TORC and CBP with homedomain protein MEIS1, a critical component of the MLL-subordinate program, which in turn facilitates HOX-mediated transcription and transformation. This mechanism also applies to hematopoietic cells transformed by other HOX genes, including CDX2, which is highly expressed in a majority of acute myeloid leukemias, thus providing a molecular approach based on GSK-3 inhibitory strategies to target HOX-associated transcription in a broad spectrum of leukemias.
Keywords: GSK-3, MLL, Hox, Creb, Meis1, Pbx, c-fos
Introduction
Homeobox (HOX) genes comprise one of the largest groups of annotated oncogenes (Futreal et al., 2004), and are implicated in the pathogenesis of various human cancers (Abate-Shen, 2002; Sitwala et al., 2008; Svingen and Tonissen, 2003). They encode a large and diverse family of transcription factors that share a conserved 60 amino acid homeodomain DNA-binding motif. Originally discovered through their causative roles in homeotic patterning defects, HOX proteins are critical regulators of cell fate, organ and tissue formation, and stem cell functions. In the blood system, HOX proteins regulate hematopoietic stem cell self-renewal, a process that is perturbed in acute leukemias by either activating mutations of HOX genes themselves, or more commonly by mutations or mis-expression of their upstream regulators MLL and CDX2, respectively (Dou and Hess, 2008; Liedtke and Cleary, 2009; Riedt et al., 2009; Scholl et al., 2007). Expression of HOXA9 in particular has been linked with the general prognosis of acute myeloid leukemia (AML). The HOX regulatory pathway, therefore, constitutes a potential target for therapeutic interventions in leukemias and other malignancies.
The DNA-binding and transcriptional properties of HOX proteins are enhanced by interactions with TALE (three amino acid loop extension) class homeodomain proteins of the PBX and MEIS families (Owens and Hawley, 2002; Sitwala et al., 2008). Genetic studies reveal that TALE proteins are required for many HOX-dependent developmental and oncogenic programs. Co-expression of MEIS1 with HOXA9 markedly shortens the latency for myeloid leukemia in mouse models (Kroon et al., 1998), and mutations of HOXA9 that prevent interactions with PBX proteins abrogate its oncogenic properties (Schnabel et al., 2000). MEIS1 is consistently expressed at high levels in MLL and CDX2 leukemias, and serves an essential and rate-limiting role in regulating MLL leukemia stem cell potential (Rawat et al., 2008; Wong et al., 2007). TALE proteins form hetero-oligomeric complexes with HOX proteins to recruit a variety of transcriptional co-regulators with either co-activator or co-repressor properties. PKA signaling has been specifically implicated in the recruitment of co-activators by TALE proteins, and possibly in the inter-conversion of co-regulator recruitment underlying differential transcriptional activity (Goh et al., 2009; Huang et al., 2005). Despite these advances, the signaling pathways that coordinate HOX-TALE transcriptional outputs in normal and neoplastic cells remain largely undefined.
We have previously shown that glycogen synthase kinase 3 (GSK-3) is required for maintenance of leukemias with MLL mutations (Wang et al., 2008). GSK-3 is a serine/threonine kinase that functions on several signaling pathways implicated in various pathological processes including diabetes, inflammation, and neurodegenerative disorders (Cohen and Goedert, 2004; De Ferrari and Inestrosa, 2000; Doble and Woodgett, 2003; Martin et al., 2005). In malignancies, inactivating mutations of GSK-3 underscore its normal tumor suppressor function to down-regulate growth-promoting pathways such as those mediated by WNT, Hedgehog and MYC proteins that are inappropriately activated in cancers (Cohen and Goedert, 2004). However, increasing evidence demonstrates that GSK-3 serves a tumor-promoting role to sustain proliferation in some cancers, thus opening up the possibility of targeting GSK-3 for therapeutic purposes (Luo, 2009). Defining the underlying mechanisms that mediate GSK-3 dependence of specific tumors will provide a molecular rationale for selective application of therapies that target GSK-3. In this report we investigated how GSK-3 facilitates HOX-mediated transcription and oncogenesis.
Results
GSK-3 maintains the MLL leukemia stem cell transcriptional program
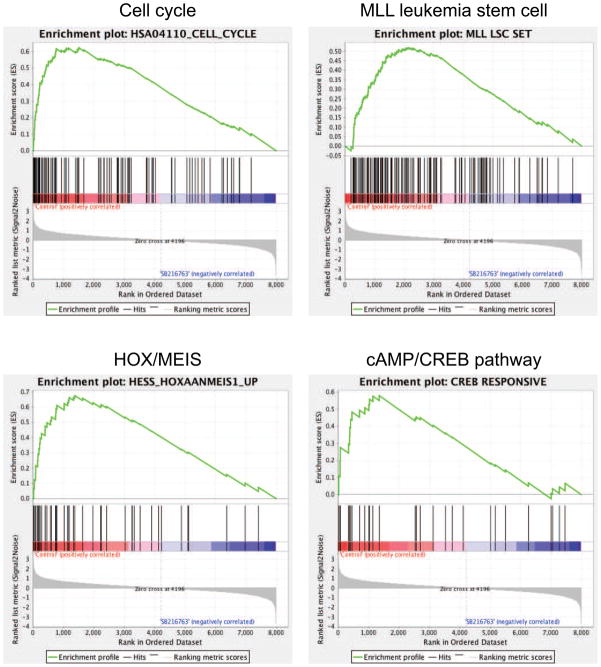
Gene expression profiling was performed to investigate the mechanisms underlying MLL leukemia dependence on GSK-3. Following GSK-3 inhibitor (SB216763) treatment of the RS4;11 human leukemia cell line, which contains an MLL-AF4 chromosomal translocation, 1028 differentially expressed genes were identified of which 554 were up-regulated and 474 down-regulated at least 1.5 fold (Table S1). Comparison of the treatment dataset with curated gene sets derived from diverse published experiments (Subramanian et al., 2005) revealed that down-regulated genes were significantly enriched for gene sets related to cell cycle (Figure 1), as well as MYC-regulated and differentiation-associated genes (Table S2), consistent with growth arrest, decreased MYC expression and differentiation changes in MLL myeloid leukemia cells upon GSK-3 inhibition (Wang et al., 2008).
Figure 1. Global gene expression changes of MLL cells in response to GSK-3 inhibition.
The dataset of gene expression differences resulting from GSK-3 inhibitor treatment (10 μM SB216763 for 20 hours) was used for GSEA. Enrichment plots are shown for selected down-regulated gene sets identified by GSEA (Supplemental Table 2). See also Supplemental Tables S1 and S2.
Genes comprising the MLL leukemia stem cell (LSC) maintenance program, which are shared with embryonic stem cells as well as poor prognosis human cancers (Somervaille et al., 2009), were significantly down regulated (Figure 1 and Table S2), indicating that GSK-3 likely affects MLL LSC potential. Down-regulated genes were also significantly enriched for gene sets associated with HOX over-expression, including those induced by co-expression of HOXA9 and MEIS1 (Figure 1), which are direct MLL transcriptional targets implicated in leukemia pathogenesis. Furthermore, MYB, a downstream mediator of HOXA9/MEIS1 in AML (Hess et al., 2006), and its subordinate transcriptional program, were also down regulated (Table S2). Thus, GSK-3 inhibition appears to target the LSC program at or near the apex of the transcriptional hierarchy initiated by MLL oncoproteins.
The MLL subordinate HOX/MEIS program sensitizes leukemia cells to GSK-3 inhibition
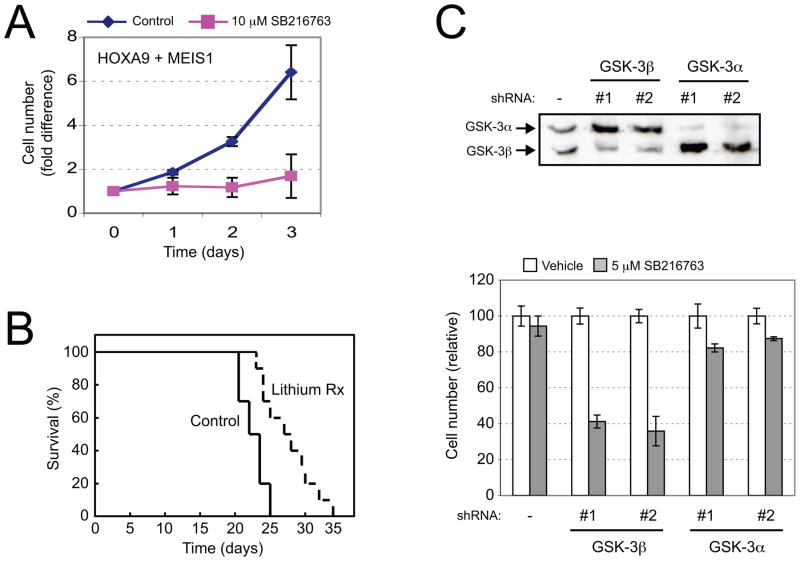
Despite down regulation of the MLL LSC transcriptional program, MLL oncoprotein expression (Wang et al., 2008) and transcriptional activity (Figure S1) were unaffected by GSK-3 inhibition, prompting an assessment of whether GSK-3 may impact at the level of HOX and MEIS, which are immediate downstream effectors of the MLL oncogenic program. Consistent with this possibility, SB216763 inhibited the proliferation of mouse myeloid leukemia cells transformed by the combined actions of HOXA9/MEIS1, which effectively bypass MLL (Figure 2A). Furthermore, treatment with lithium, a GSK-3 inhibitor, modestly prolonged the latency for development of AML in transplanted mice (Figure 2B) similar to the effects of GSK-3 inhibition in murine models of MLL leukemia (Wang et al., 2008). Physiologic GSK-3 inactivation using conditional AKT (CA-AKT-ER), which avoids potential non-specific effects of chemical GSK-3 inhibitors, induced hyper-phosphorylation of GSK-3 and inhibited the growth of HOXA9/MEIS1 cells (Figure S2). Although knockdown of GSK-3α or GSK-3β had no adverse effects on cell proliferation (data not shown), GSK-3β knockdown cells were more sensitive to SB216763 treatment (Figure 2C). These effects phenocopy GSK-3 inhibition in MLL leukemia cells, suggesting that GSK-3 may regulate HOX/MEIS rather than MLL oncoprotein activity.
Figure 2. HOX-induced proliferation is generally sensitive to GSK-3 inhibition.
(A) Mouse myeloid progenitors immortalized by HOXA9 + MEIS1 were cultured in the presence or absence of 10 μM SB216763. Results show mean cell numbers expressed as fold-change compared to day 0 (error bars indicate ±SEM, n = 3).
(B) Survival is shown for mice transplanted with HOXA9/MEIS1 leukemia cells (50,000 cells/mouse) and maintained on regular chow or chow containing 0.4% lithium with saline water (n=10 each cohort; p<0.01). Acute leukemia was confirmed by peripheral blood leukocyte counts and/or necropsy.
(C) Western blot analysis (upper panel) was performed using an anti-GSK-3 antibody on HOXA9/MEIS1 leukemia cells transduced by lentiviral vectors expressing GSK-3 or GSK-3β shRNAs (#1 or #2 for each). Migrations of GSK-3 isoforms are indicated. Bar graph shows the growth of transduced cells in the presence or absence of 5 μM SB216763 for 3 days. Results are shown as relative cell proliferation compared to cell numbers in the absence of inhibitor (error bars indicate ±SEM, n=3). See also Figure S1.
GSK-3 impacts HOX/PBX/MEIS transcriptional activity
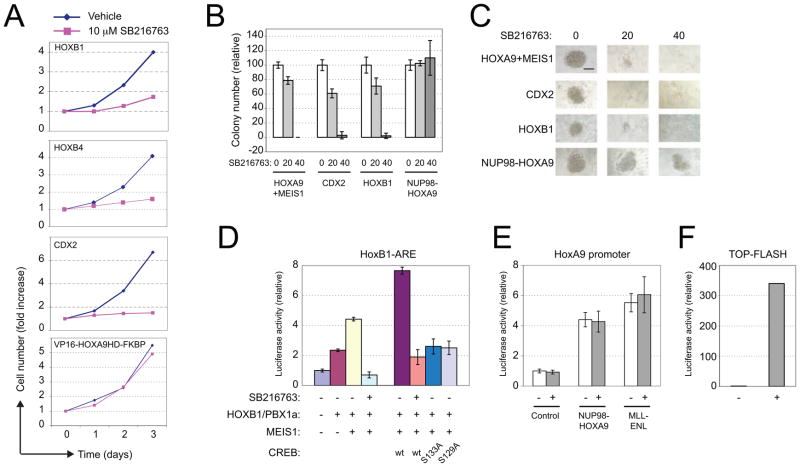
To further assess the correlation of GSK-3 dependence with HOX transformation, we examined its role in mouse myeloid progenitors immortalized by additional homeodomain proteins HOXB1 and HOXB4, or CDX2, an upstream regulator of HOX genes (Rawat et al., 2008) and also MEIS1 (Figure S2D), a HOX DNA-binding cofactor that is necessary (Figure S2E) and rate limiting (Kroon et al., 1998; Wong et al., 2007) for HOX-associated leukemogenesis. When treated with GSK-3 inhibitor, progenitor proliferation was impaired (Figure 3A) and colony-forming abilities were dramatically reduced (Figures 3B and 3C). Activated AKT, which physiologically inhibits GSK-3, also decreased cell proliferation, and GSK-3β knockdown increased sensitivity to GSK-3 inhibitors (Figure S2) phenocopying the responses of MLL-transformed cells. In contrast, cells transformed by NUP98-HOXA9, an oncogenic HOX protein that aberrantly recruits CBP co-activator through the NUP98 moiety, were unaffected by GSK-3 inhibition (Figures 3B and 3C). Furthermore, cells immortalized by an activated HOX mutant (VP16-HD-FKBP, which spontaneously dimerizes to activate transcription) (Figure S2F) were also insensitive to GSK-3 inhibition (Figure 3A), and not dependent on MEIS1 for myeloid transformation (Figure S2E), indicating that mutations conferring constitutive transcriptional effector activity bypass dependence on GSK-3. These data support a broad role for GSK-3 in HOX-associated transformation, and suggest that it may directly impact HOX/MEIS transcriptional function.
Figure 3. GSK-3 regulates HOX/PBX/MEIS transcription complex activity through MEIS1 and CREB association.
(A) Mouse myeloid progenitors immortalized by the indicated genes were cultured in the presence or absence of 10 μM SB216763. Cell numbers are expressed as fold-change compared to day 0. Representative experiments are shown (n = 3 each).
(B) Colony forming ability is shown for myeloid progenitors immortalized by various oncogenes (indicated below) following 5 days culture in the presence or absence of SB216763. Results are shown relative to mean colony numbers without drug set at 100%.
(C) Colony morphologies are shown for the experiment in panel B. Scale bar = 100 μm.
(D) HoxB1 ARE reporter activity was assessed following co-transduction with constructs encoding the proteins indicated below the panel in the presence or absence of 10 μM SB216763. Luciferase activity was normalized to β–galactosidase activity. Results are shown as fold-change compared to control.
(E & F) Activity of the HOXA9 reporter gene (E) or Top-Flash WNT reporter gene (F) was assessed in the presence or absence of 10 μM SB216763. Results of a representative experiment are displayed as fold change compared to control. All error bars indicate ±SEM, n=3. See also Figure S2.
Since HOX-transformed leukemia cells are generally sensitive to GSK-3 inhibition, which down regulates the HOX/MEIS transcriptional program, we tested whether GSK-3 affects HOX/MEIS transcriptional activity. Co-transduction of HOXB1 with its cofactors PBX1 and MEIS1 in transient transcription assays activated expression of a reporter gene under control of the Hoxb1 auto-regulatory element (ARE) as reported previously (Goh et al., 2009; Jacobs et al., 1999). However, transcriptional activation was abrogated in the presence of GSK-3 inhibitor (Figure 3D), which by contrast had no effect on the abilities of NUP98-HOXA9 or MLL-ENL to activate the HOXA9 promoter (Figure 3E). As expected, GSK-3 inhibition induced robust expression of the TOP/Flash WNT pathway reporter gene (Figure 3F). Thus, GSK-3-dependent signaling regulates HOX/MEIS transcriptional activity.
The GSK-3 dependence of MLL and HOX transformed myeloid cells is modulated by CREB
Gene sets linked with cAMP/CREB activity were also enriched among the genes down regulated by GSK-3 inhibition in MLL leukemia cells (Table S2 and Figure 1B). CREB is a transcription factor involved in various biological processes, including cancer, whose activity is promoted by GSK-3 phosphorylation (Fiol et al., 1994; Horike et al., 2008), suggesting that it may play a role in the GSK-3 dependence of HOX-transformed cells.
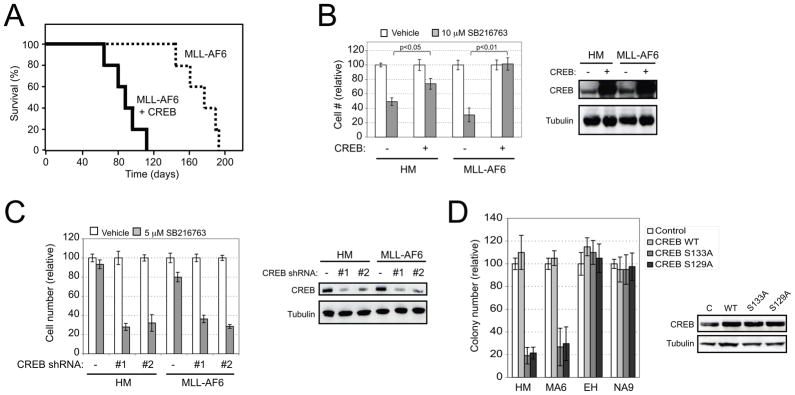
Co-expression of CREB with MLL-AF6 revealed a strong genetic interaction resulting in shortened latency for development of leukemia in transplanted mice (Figure 4A), suggesting that CREB function may be limiting despite mis-regulation of HOX/MEIS expression in MLL leukemia. CREB over-expression also decreased sensitivity to GSK-3 inhibition in cells immortalized by MLL-AF6 or HOXA9/MEIS1 (Figure 4B), whereas CREB knockdown increased their sensitivity (Figure 4C). Similar effects were seen in CDX2 immortalized cells where CREB knockdown reduced proliferation (Figure S3A) and increased sensitivity to GSK-3 inhibition (Figure S3B). Thus, CREB serves an important role to modulate the GSK-3 dependence of cells transformed by mis-regulation of the HOX pathway.
Figure 4. CREB affects MLL and HOX-induced cell proliferation and sensitivity to GSK-3 inhibition.
(A) Mouse myeloid progenitors immortalized by MLL-AF6 were transduced with retrovirus expressing CREB or empty vector. Mice (n=5 each cohort) transplanted with the transduced cells were monitored for leukemia-free survival (p<0.01).
(B) MLL-AF6 or HOXA9/MEIS1 (HM) leukemia cells were stably transduced with CREB (+) or vector (−), and then incubated in the presence or absence of 10 μM SB216763. Cell numbers were enumerated on day 2 and expressed as relative change compared to no SB216763 treatment. Right panel shows protein levels detected by western blot analysis.
(C) MLL-AF6 or HOXA9/MEIS1 leukemia cells were stably transduced with lentiviral vector expressing CREB shRNAs (#1 or #2), and then incubated in the presence or absence of 5 μM SB216763. Cell numbers were enumerated on day 2 and displayed as relative change compared to no SB216763 treatment. Right panel shows CREB protein levels in knockdown cells.
(D) Myeloid progenitors immortalized by the indicated oncogenes were transduced with retroviral vectors expressing the various CREB proteins or empty vector (−). The cells were then plated and colonies enumerated after 5 days culture, and displayed relative to empty vector. Right panel shows CREB protein levels in transduced cells. HM, HOXA9/MEIS1; MA6, MLL-AF6; EH, E2A-HLF; NA9, NUP98-HOXA9. All error bars indicate ± SEM of triplicate analyses. See also Figure S3.
CREB phosphorylation at Ser133 is essential for its transcriptional activity(Carlezon et al., 2005), and primes subsequent phosphorylation by GSK-3 at Ser129 (Fiol et al., 1994), which is required for expression of several genes (Boer et al., 2008; Horike et al., 2008; Tyson et al., 2002). Thus, the effects of CREB phosphorylation on MLL and HOX-mediated proliferation were tested. Forced expression of CREB proteins harboring S129A or S133A mis-sense mutations displayed dominant-negative effects with decreased colony forming abilities of MLL and HOXA9/MEIS1 leukemia cells, but not that of cells transformed by E2A-HLF, which functions independent of the HOX pathway, or NUP98-HOXA9, which is a constitutively active HOX pathway oncoprotein (Figure 4D). In contrast, forced expression of phosphorylation mutants for several other GSK-3 downstream targets (β-catenin, FOXO, and NFAT) did not affect colony-forming abilities (data not shown). These data suggest that CREB phosphorylation by GSK-3 is important for MLL and HOX-mediated transformation.
GSK-3 promotes conditional MEIS1 association with CREB and its co-activators to facilitate HOX-mediated transcription
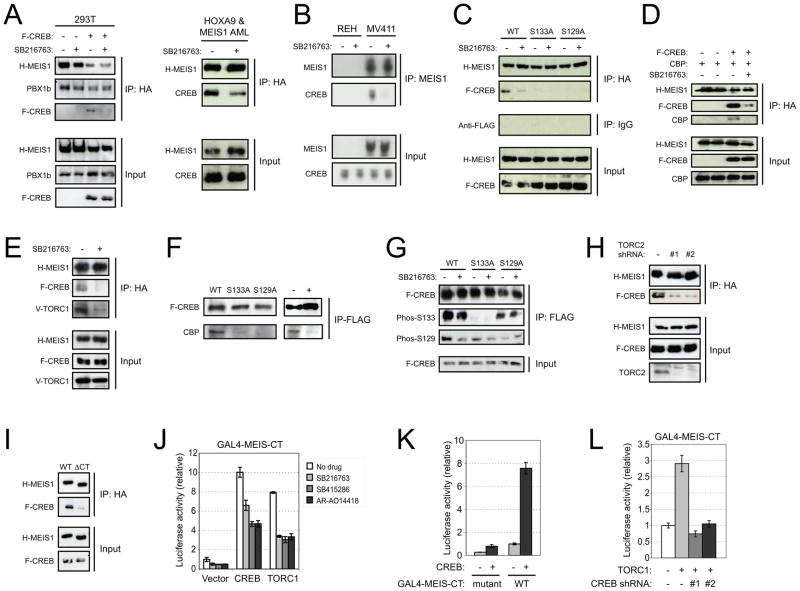
HOX/MEIS transcriptional activity has been linked with the cAMP signaling pathway, which impinges on CREB (Goh, 2009; Carlezon, 2005). Thus, possible physical association of CREB with the HOX/MEIS complex was assessed by IP-western blot analysis. Transfected exogenous CREB in 293T cells and endogenous CREB in AML cells transformed by HOXA9/MEIS1 co-precipitated with MEIS1 (Figure 5A). Notably, GSK-3 inhibitor treatment markedly reduced MEIS1-CREB co-IP, but not interaction of MEIS1 with its dimerization partner PBX1b (Figure 5A). Furthermore, GSK3-dependent association of endogenous CREB and MEIS1 was observed in co-precipitation studies of the human MLL leukemia cell line MV411 (Figure 5B). Association was not the result of DNA tethering since nuclease treatment did not abrogate co-precipitation (data not shown). CREB mutants S129A and S133A did not co-IP with MEIS1 (Figure 5C) further supporting that GSK-3 activity may promote MEIS1-CREB association. MEIS1 also co-precipitated the CREB co-activators CBP and TORC, which was markedly reduced by GSK-3 inhibition (Figures 5D and 5E). CBP co-IP was dependent on the presence of CREB (Figure 5D), and abrogated in the CREB phosphorylation mutants, or by GSK-3 inhibition (Figure 5F), which reduced S129 phosphorylation to background levels (Figure 5G). Conversely, CREB-TORC interaction was not abrogated by GSK-3 inhibition (not shown) consistent with previous studies (Heinrich et al., 2009), but TORC was necessary for CREB-MEIS association (Figure 5H) consistent with previous suggestions that it may directly interact with MEIS1 (Goh et al., 2009; Huang et al., 2005; Jacobs et al., 1999). Taken together, these results support GSK-3 facilitated association of the CBP co-activator complex with MEIS1.
Figure 5. Association of CREB and MEIS1 is phosphorylation dependent.
(A) IP-western blot analysis was performed on 293T cells co-transduced with FLAG-CREB and HA-MEIS1, or AML cells transformed by HOXA9 + HA-MEIS1 in the presence or absence of 20 μM SB216763. Cell lysates were immunoprecipitated with anti-HA antibody conjugated beads, followed by western blot analysis using anti-FLAG (CREB), anti-HA (MEIS1), anti-CREB or anti-PBX1b antibodies.
(B) Human leukemia cell lines (indicated at the top) were cultured overnight in the presence (+) or absence (−) of SB216763 (20 μM), and cell lysates were subjected to immunoprecipitation with anti-MEIS monoclonal antibody conjugated beads, then subjected to western blot analysis with rabbit polyclonal antibodies specific for the indicated endogenous proteins.
(C) HA-MEIS1 was co-expressed with FLAG-tagged CREB proteins (indicated at top) in the presence or absence of 20 μM SB216763. Cell lysates were used for immunoprecipitation with anti-HA or IgG conjugated beads, then subjected to western blot using anti-FLAG (CREB) or anti-HA (MEIS1) antibodies.
(D) HA-MEIS1 was co-expressed with CBP, with or without FLAG-tagged CREB (indicated at top) in the presence or absence of 20 μM SB216763. Cell lysates were used for immunoprecipitation with anti-HA conjugated beads, then subjected to western blot using anti-CBP, anti-FLAG (CREB) or anti-HA (MEIS1) antibodies.
(E) HA-MEIS1 was co-expressed with FLAG-tagged CREB and V5-tagged TORC1 in the presence or absence of 20 μM SB216763. Cell lysates were immuno-precipitated with anti-HA conjugated beads, then subjected to western blot using anti-V5 (TORC1), anti-FLAG (CREB) or anti-HA (MEIS1) antibodies.
(F) Left panel: FLAG-tagged CREB (wt or mutant as indicated at top of panel) proteins were expressed in 293T cells, immunoprecipitated with anti-FLAG antibody conjugated beads, and subjected to western blot to detect the presence of endogenous CBP. Right panel: FLAG-tagged CREB was expressed in 293T cells in the absence (−) or presence (+) of SB216763, immunoprecipitated with anti-FLAG antibody conjugated beads, and subjected to western blot analysis to detect the co-precipitation of endogenous CBP.
(G) FLAG-tagged CREB (or mutant CREB proteins indicated at top of panel) were expressed in 293T cells in the absence (−) or presence (+) of SB216763 and immunoprecipitated with anti-FLAG antibody conjugated beads. The precipitate was washed and subjected to western blot to detect CREB phosphorylation status using phospho-specific antibodies.
(H) HA-MEIS1 was co-expressed with FLAG-tagged CREB in the presence of TORC2 shRNA constructs (#1, #2). Cell lysates were used for immunoprecipitation with anti-HA conjugated beads, then subjected to western blot using anti-TORC2, anti-FLAG (CREB) or anti-HA (MEIS1) antibodies.
(I) HA-MEIS1 (WT) or HA-MEIS1 C-terminal deletion mutant (ΔCT) was co-expressed with FLAG-tagged CREB. Cell lysates were immuno-precipitated with anti-HA conjugated beads, then subjected to western blot using anti-FLAG (CREB) or anti-HA (MEIS1) antibodies.
(J) Gal4-MEIS1 CT activity was assessed following co-transduction with CREB or TORC1 in the presence or absence of different GSK-3 inhibitors.
(K) Gal4-MEIS1 CT or mutant reporter activities were assessed following co-transduction with CREB.
(L) Gal4-MEIS1 CT reporter activity was assessed following co-transduction with TORC1 or CREB shRNA constructs (#1, #2) indicated below the panel. Results are shown as fold-change compared to control. For panels J-L, luciferase activity was normalized to β–galactosidase activity.
Error bars indicate ±SEM.
The potential impact of GSK-3 on CREB and HOX/MEIS cooperative function was assessed in transient transcription assays. Co-expression of wild type CREB significantly enhanced transcriptional activation induced by the HOXB1/PBX1/MEIS1 complex on the Hoxb1 ARE (Figure 3D), an authentic MEIS1 target promoter (Goh et al., 2009; Jacobs et al., 1999). Activation was antagonized by GSK-3 inhibition or co-expression of CREB phosphorylation mutants. In a heterologous GAL4 system, co-expression of CREB or TORC markedly enhanced transcriptional activation mediated by the MEIS1 C-terminus (Figures 5J and 5K), which is implicated in TORC (Goh et al., 2009) and CREB (Figure 5I) interactions, and was sensitive to GSK-3 inhibition (Figure 5J). CREB was necessary for TORC enhancement of MEIS1-mediated transcription (Figure 5L). Thus, GSK-3 activity promotes the physical and functional association of CREB with the HOX/PBX/MEIS transcription complex, and higher-order interactions with CBP and TORC co-activators.
FOS is a critical target gene of HOX/MEIS and CREB responsive to GSK-3 inhibition
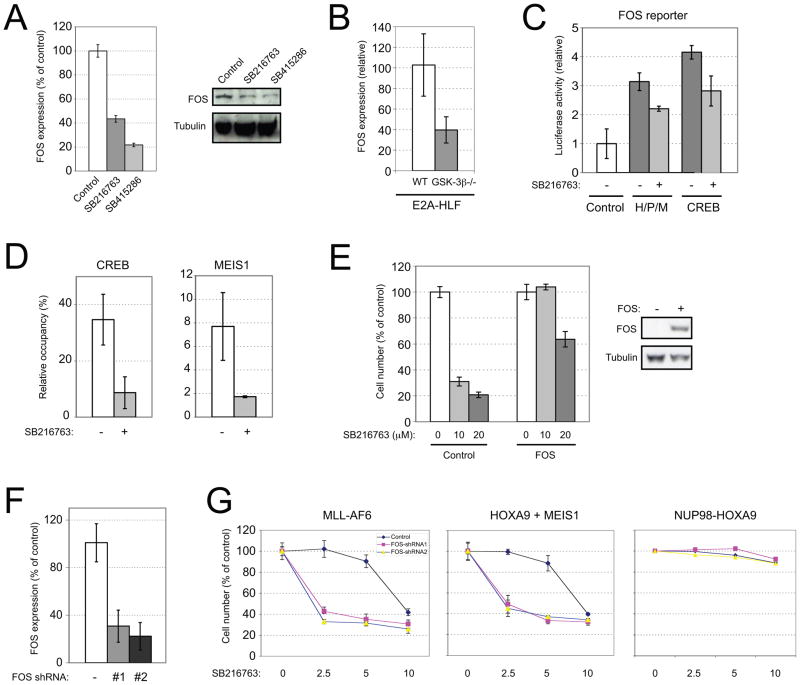
Microarray analysis identified FOS as among the most substantially down-regulated genes in response to GSK-3 inhibition in MLL leukemia cells. Its levels decreased almost 3 fold compared to much more modest or no decreases of other FOS or JUN family genes (Figure S4A), as confirmed by quantitative PCR in cells treated with two different GSK-3 inhibitors (Figure 6A), and consistent with observations that GSK-3β deficiency substantially reduces FOS expression (Figure 6B). Originally identified as a viral oncogene homolog, FOS is involved in many physiological processes (Curran and Teich, 1982; Matthews et al., 2007; Ozanne et al., 2007), and is a known CREB target gene (Gonzales and Bowden, 2002; Ramirez et al., 1997) whose expression levels have been correlated with HOX expression in various cell types (Krosl and Sauvageau, 2000; Potter et al., 2006). In transcription assays, expression of a reporter gene under control of the FOS promoter was induced by CREB or the HOX/PBX/MEIS complex, and was partially sensitive to GSK-3 inhibition (Figure 6C). Furthermore, endogenous FOS expression in human leukemia cells was induced by co-expression of HOXA9 and MEIS1 (Figure S4B). Conversely, FOS levels were reduced in MEIS1-deficient immortalized myeloid progenitors (Figure S4C), consistent with regulation of FOS transcription by the HOX/MEIS/PBX complex. In HOXA9/MEIS1 transformed myeloid progenitors, chromatin immunoprecipitation of the FOS promoter showed MEIS1 and CREB occupancy, which was reduced by GSK-3 inhibition (Figure 6D).
Figure 6. FOS is a critical downstream effector of HOX/PBX/MEIS and CREB to mediate GSK-3 inhibitor sensitivity.
(A) Human MLL-AF4 leukemia cell line RS 4;11 was cultured in the presence or absence of GSK-3 inhibitors (SB216763 or SB415296 for 20 hours). FOS transcripts were then measured by qRT-PCR, and expressed relative to untreated cells.
(B) Expression of FOS in wild type (WT) or GSK-3β null (GSK-3β−/−) cells transformed by E2A-HLF was measured by qRT-PCR, and displayed relative to wild type cells.
(C) FOS reporter was co-transfected with HOXB1, PBX1b and MEIS1a (H/P/M) or CREB in the presence (+) or absence (−) of SB216763. Luciferase activity was normalized with β–galactosidase activity. Results are displayed as fold change compared to control.
(D) Myeloid progenitors immortalized by HOXA/MEIS1 were cultured for 24 hrs in the absence (−) or presence (+) of SB216763. ChIP was performed using antibodies against CREB or HA (MEIS1). Relative occupancy values were normalized against those obtained with IgG.
(E) MLL-AF6 transformed mouse myeloid progenitor cells were stably transduced with a FLAG-FOS construct. Over-expression of FOS is shown by western blot using anti-FOS antibody. Sensitivities of cells to different concentrations of SB216763 are displayed compared to untreated cells.
(F) MLL-AF6 transformed mouse myeloid progenitors stably transduced with vector (−) or FOS shRNAs (#1 or #2) and employed for studies in panel G were assessed for FOS transcript expression by qRT-PCR. Results are displayed relative to vector-transduced cells.
(G) Cells transformed by the indicated oncogenes (top of panels) were transduced with FOS knockdown constructs and cultured in the indicated concentrations of SB216763. Cell numbers were enumerated and displayed as relative change compared to untreated cells. All error bars indicate ± SEM of triplicate analyses. See also Figure S4.
Forced expression of FOS in MLL leukemia cells (MLL-AF6) increased their resistance to GSK-3 inhibition (Figure 6E), consistent with partial bypass of the GSK-3 dependent CREB-MEIS transcription pathway. Alternatively, FOS knockdown enhanced sensitivity to GSK-3 inhibition at least 4 fold (Figures 6F and 6G) but did not affect NUP98-HOXA9 immortalized cells, which are not sensitive to GSK-3 inhibitors. Thus, FOS is a critical target gene that may integrate the role of GSK-3 in promoting conditional CREB function to facilitate HOX-mediated transcription and leukemic transformation.
Discussion
Acute leukemias induced by MLL chimeric oncoproteins are among the subset of cancers distinguished by a paradoxical dependence on GSK-3 kinase activity for sustained proliferation. In this study, we demonstrate that GSK-3 maintains the MLL LSC transcriptional program by promoting the conditional association of CREB and its co-activators with MEIS1, a critical component of the MLL-subordinate HOX/MEIS transcription complex, which in turn facilitates HOX-mediated transcription. This mechanism also applies to cells transformed by a variety of homeodomain proteins, including CDX2, which is highly expressed in a majority of acute myeloid leukemias, suggesting that the spectrum of leukemias susceptible to GSK-3 inhibitory therapies may be much broader than previously anticipated.
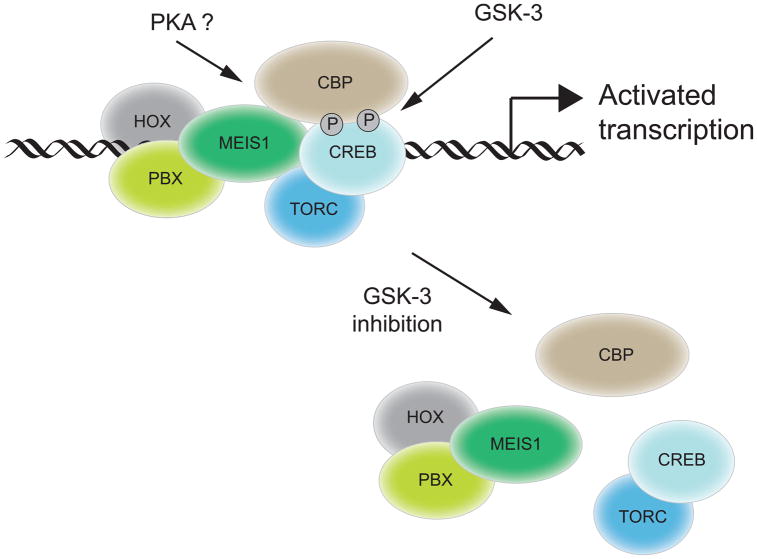
Our conclusions are based on the following key observations: i) HOXA/MEIS leukemia cells are sensitive to GSK-3 inhibition, essentially phenocopying MLL leukemia sensitivity both in vitro and in vivo; ii) CREB along with its co-activators CBP and TORC physically associates with MEIS1, and modulates HOX/MEIS transcriptional activity on an authentic promoter; iii) CREB association with MEIS1 is regulated by phosphorylation at Ser129 of CREB, a known target of GSK-3; and iv) gene programs linked with HOX/MEIS as well as CREB are down-regulated by GSK-3 inhibition in MLL leukemia cells. Together, these observations suggest a model (Figure 7) whereby GSK-3 activity maintains the physical and functional association of CREB with MEIS to promote critical target gene expression responsible for HOX-mediated transformation.
Figure 7. Schematic model depicting role of GSK-3 in promoting HOX-mediated transcription through CREB phosphorylation.
GSK-3 activity promotes conditional association of CREB and its co-activators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis, which are compromised following GSK-3 inhibition in leukemia cells.
In our studies, CREB enhanced the transcriptional activity of a HOX/MEIS/PBX complex on the Hoxb1 ARE, an authentic MEIS1 target promoter, and its role in this context appears to be regulated by GSK-3 phosphorylation. CREB is a multi-functional transcriptional activator that is involved in many physiological pathways under normal and pathologic conditions. CREB activity is regulated by phosphorylation at Ser133, which is a target of various kinases depending on the specific signaling stimulus and cell type. PKA, which is activated by cAMP, is the major kinase that targets Ser133 in many processes. Ser133 phosphorylation primes CREB for phosphorylation by GSK-3 at Ser129. However, unlike Ser133 phosphorylation, which is linked with CREB activation, the physiologic consequences of Ser129 phosphorylation are not well defined, although evidence suggests that it also is linked with CREB activation (Boer et al., 2008; Horike et al., 2008; Tyson et al., 2002). We found that CREB S129A mutation, similar to S133A mutation, antagonized HOX/MEIS activity and decreased colony-forming abilities of MLL or HOX/MEIS transformed cells. Consistent with our study, S129 phosphorylation of CREB by GSK-3 is required for recruitment of CBP and subsequent induction of PEPCK-C gene expression (Horike et al., 2008). Thus, our results support a positive role of S129 phosphorylation by GSK-3 for CREB activation of specific transcriptional programs in MLL-transformed cells.
Our studies demonstrate that MEIS1 is a molecular link that integrates the transcriptional activities of CREB and its co-activators as a higher order complex whose physical and functional integrity is dependent on GSK-3 activity. Previous studies have shown that MEIS1-mediated transcriptional activation is stimulated by PKA and dependent on CBP (Huang et al., 2005). Further evidence in support of a MEIS-CREB nexus is provided by the recent demonstration that MEIS1 interacts with TORC (Goh et al., 2009), a co-activator that also associates with CREB and modulates its activity (Conkright et al., 2003; Siu and Jin, 2007). MEIS1 interaction with TORC is also dependent on PKA signaling, which has been shown to regulate the nuclear translocation of PBX1, an obligate dimerization partner of MEIS1. Despite the prominent role of PKA in CREB signaling pathways, treatment with several PKA inhibitors was equally toxic for leukemia cells transformed by MLL and non-MLL oncogenes (data not shown), unlike the selective anti-proliferative effects of GSK-3 inhibition. This likely reflects the essential role of PKA in various physiological processes that mask any selective role promoting CREB-MEIS interactions in HOX-transformed cells. Thus, it remains to be determined whether GSK-3 may function in concert with PKA to affect a higher order association of CREB with MEIS1 mediated through TORC and CBP.
The underlying biochemical mechanism for how GSK-3 inhibition or CREB S129 phosphorylation affects MEIS association with the TORC/CREB/CBP complex is not entirely clear. Nevertheless, several conclusions can be drawn from our study and others. First, the association is likely mediated through the MEIS1 C-terminus since its deletion abrogates MEIS1 transcriptional activation and association with TORC (Goh et al., 2009) and CREB (Figures 5I and 5K). Second, TORC likely bridges CREB and MEIS1 association since TORC-enhanced activation by MEIS1 depends on CREB (Figure 5L), and MEIS1/CREB association depends on TORC (Figure 5H). Furthermore, CREB S133 and S129 phosphorylation brings CBP/p300 to MEIS1 and this likely further stabilizes MEIS1 and CREB association. Loss of S133 or S129 phosphorylation, which decreases affinity of MEIS1 and CREB, may involve: i) loss of CBP binding that reduces CREB and MEIS1 association, and ii) conformational changes of the TORC/CREB complex that alter TORC/CREB binding (Heinrich et al., 2009) (data not shown). CREB may also directly modulate MEIS1 and PBX expression (Esparza et al., 2008) adding to the complexity of their interrelated function. However, the detailed mechanisms remain to be determined and other unappreciated factors may also be involved.
Multiple genes are down-regulated in MLL-transformed cells following GSK-3 inhibition. Our studies focused on FOS as a prototype since it was one of the most differentially expressed genes in MLL leukemia cells. It encodes a member of the AP-1 transcription factor family, and as an early response gene induced by CREB activation, its role in cancer has been widely studied and shown to enhance the proliferation of transformed cells. In addition to increased proliferation, forced expression of FOS in our studies rendered MLL leukemia cells more resistant to GSK-3 inhibition whereas its knockdown rendered cells more sensitive, suggesting a downstream role for FOS in mediating the response to GSK-3 inhibitors. FOS expression levels are likely to be regulated by HOX transcription complexes as originally suggested by enforced expression of HOXB4 or HOXC13, which induced FOS expression in fibroblasts or epithelial cells, respectively (Krosl and Sauvageau, 2000; Potter et al., 2006). Moreover, FOS levels are decreased following deletion of MEIS1 (Figure S4C). In our studies, FOS transcript levels were increased in human leukemia cells following forced expression of HOXA9 and MEIS1, but not HOXA9 alone, indicating a regulatory role for the HOX/MEIS complex. However, reduced FOS levels alone are unlikely to account for the cell cycle arrest associated with GSK-3 inhibition since knockdown of FOS to similar low levels achieved by GSK-3 inhibition did not result in complete cell cycle arrest. Consistent with our expression profiling studies, multiple downstream genes are likely to mediate the adverse effects of GSK-3 inhibition on MLL leukemia cells.
In conclusion, GSK-3 promotes conditional association of CREB and its co-activators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. This provides a mechanistic basis for the paradoxical dependence of MLL-associated leukemias on GSK-3 activity, which critically maintains the MLL LSC transcriptional program, and suggests a therapeutic approach to molecularly target HOX-associated transcription. In addition to MLL leukemias, accumulating evidence indicates that GSK-3 inhibition blocks proliferation or induces apoptosis in a variety of cancers including melanoma, myeloma, glioblastoma and pancreatic cancer among others (Korur et al., 2009; Miyashita et al., 2009; Smalley et al., 2007; Wilson and Baldwin, 2008; Zhou et al., 2008). It will be of interest to determine how much of a role, if any, CREB-MEIS interactions contribute to the GSK-3 dependence of these malignancies, which are candidates for GSK-3 inhibitor therapies.
Experimental Procedures
Mice
Mice were maintained on an inbred C57BL/6 background. All experiments on mice in this study were performed with the approval of and in accordance with Stanford University’s Administrative Panel on Laboratory Animal Care.
GSK-3 inhibitors
GSK-3 inhibitors SB216763 (Cat. No. S3442), SB415286 (Cat. No. S3567), AR-A014418 (Cat. No. A3230) and TDZD-8 (Cat. No. T8325) were purchased from Sigma and have been described previously (Bhat et al., 2003; Smith et al., 2001) (Martinez et al., 2002). They were dissolved in DMSO and used at the indicated concentrations.
Cell culture
Human leukemia cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). Immortalized mouse myeloid cells or leukemia cells were cultured in RPMI 1640 supplemented with 20% FCS and 20% WEHI conditioned medium, or in methylcellulose-containing medium (Methocult M3231, Stem Cell Technologies) with cytokines, as described previously (Lavau et al., 1997). 293T and Phoenix cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS.
Microarray and bioinformatics analyses
Human leukemia cell line RS4;11 was cultured in 10 μM SB216763 prior to total RNA isolation. cDNA was synthesized, fragmented, and hybridized to Affymetrix Human Gene (Hu 1.0 ST) microarrays (Affymetrix, Santa Clara, CA) according to the manufacturer’s instructions. Microarray data were normalized with Expression Console software (Affymetrix), using RMA algorithms. Low signals (below 50) were filtered out using the PreprocessDataset module in GenePattern (http://www.broad.mit.edu/cancer/software/genepattern/). Hierarchical clustering of microarray samples (Eisen et al., 1998) was based on pair-wise Pearson correlation across preprocessed probe sets. Differentially expressed genes in SB216763 treated cells versus control were identified using Significance Analysis of Microarrays software (Tusher et al., 2001). Genes with a false-discovery rate below 5% and fold-change over 1.5 were considered significant. Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) was used to compare the SB216763-treated dataset with gene sets in Molecular Signatures Database (MSigDB) and gene lists derived from previous publications. GO analysis was performed using MGI Gene Ontology Term Finder (http://proto.informatics.jax.org/prototypes/GOTools/web-docs/MGI_Term_Finder.html).
DNA constructs and virus production
Retroviral constructs encoding MLL-ENL(C), MLL-GAS7, E2A-PBX1, NUP98-HOXA9 and E2A-HLF were reported previously (Kasper et al., 1999; Smith et al., 1997; Smith et al., 2002; So et al., 2004). VP16-HD-FKBP was constructed by fusing the HOXA9 homeodomain with the VP16 activation domain and FKBP dimerization domain, respectively. The expression constructs Gal-MEIS1A (335–390), mutant Gal-MEIS1A(GQWHYM), and FLAG-TORC1/2 were gifts from Mark Featherstone (Nanyang Technological University, Singapore) (Goh et al., 2009). V5-His-TORC1 was a gift from Dong-Yan Jin (The University of Hong Kong, Hong Kong)(Siu et al., 2006). HOXA9 reporter gene was a gift from Xu Cao (University of Alabama, Birmingham, AL) (Wang et al., 2006). Retroviral vectors encoding c-FOS, CREB and CREB mutants were constructed using standard cloning techniques. shRNA sequences for CREB knockdown were designed using pSicoOligomaker v1.5 (http://web.mit.edu/ccr/labs/jacks/protocols_table.html) (see Supplemental Experimental Procedures for sequences). shRNA oligos were cloned into pSicoR-puro. Lentiviruses encoding murine FOS, human CREB, and TORC2 shRNAs were purchased from Open Biosystems. Retrovirus and lentivirus production was performed as described previously (Wong et al., 2007).
Transduction of immortalized cells or leukemia cells
Myeloid progenitor transformation assays were performed as described previously (Lavau et al., 1997). For colony forming assays, mouse immortalized or leukemia cells (20,000 cells) were transduced with retrovirus or lentivirus by spinoculation at 2500 g at 37°C for 2 hours. The cells were then incubated in medium with cytokines and seeded into methylcellulose-containing medium (Methocult) with cytokines in the absence or presence of indicated concentrations of SB216763. Colonies were counted after 5 days and morphology was captured with digital photography. The human leukemia cell line RCH-ACV stably expressing the ecotropic retroviral receptor was transduced with retrovirus encoding HOXA9 or MEIS1. Cells were selected in the presence of 1 μg/ml puromycin or 250 μg/ml hygromycin.
In vivo leukemogenesis assay
MLL-AF6 immortalized mouse myeloid cells secondarily transduced with empty vector or CREB were cultured in Methocult and selected with 1 μg/ml of puromycin. Cells (106) harvested from methylcellulose were transplanted intravenously into sub-lethally irradiated (400 rads) C57BL/6 mice. For lithium treatment, mice transplanted with leukemia cells (50,000 cells) were maintained on chow (Tekada) containing 0.4% lithium and 0.9% saline water. Development of acute leukemia was confirmed by blood smear, peripheral blood leukocyte counts, and/or histology.
Transcriptional assay
Transient transcriptional assays were performed in 293T cells as described previously (Jacobs et al., 1999) using the Top-Flash reporter or reporter constructs under control of the HOXB1 ARE (Jacobs et al., 1999), Gal-Meis1 (Goh et al., 2009), HOXA9 promoter (Wang et al., 2006), or FOS promoter (Addgene, plasmid 11983). CMV-LacZ was used as internal control for transfection efficiency.
Immunoprecipitation and western blot
Cells were lysed in buffer A [20 mM sodium phosphate pH 7.0, 250 mM NaCl, 30 mM sodium pyrophosphate, 0.1% NP-40, 5 mM EDTA, 10 mM NaF, 0.1 mM Na3VO4 and 1 mM phenylmethylsulfonyl fluoride (PMSF)] supplemented with protease inhibitors (Complete, Roche). The lysates were cleared by centrifugation at 30,000 g for 10 min at 4°C and pre-cleared with Anti-IgG agarose beads, then incubated with anti-FLAG (Sigma) or anti-HA (Roche) conjugated agarose beads with rotation at 4°C for 4 h. The beads were washed five times with buffer A and then washed four times with cold PBS. Immunoblotting was performed using antibodies specific for GSK-3 (Upstate), anti-HA-HRP (Roche), anti-FLAG-HRP (Sigma), phosph-GSK3 (Cell Signaling), AKT (Cell Signaling), tubulin (Sigma), CREB and CBP (Santa Cruz Biotechnology), or CDX2 (Santa Cruz Biotechnology). For co-precipitation of endogenous proteins, cell lysates prepared from human leukemia cell lines (REH and MV411) were incubated with CNBr-activated sepharose beads conjugated with anti-MEIS monoclonal antibody (9.2.7) for 4 hrs at 4°C. Beads were washed six times with lysis buffer, and immunoblotting was performed with antisera specific for CREB or MEIS1 (Abcam).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as previously described (Yokoyama et al., 2005) using primary antibodies specific for CREB (sc-186, Santa Cruz Biotechnology) and HA (ab9110, Abcam), or IgG antibody. Immuno-complexes were precipitated using protein G beads (Dynabeads, Invitrogen). Quantitative real-time PCR was performed on the precipitated DNA using primers flanking the consensus CREB site (AGACGTCA) in the murine FOS gene (mFOS forward: cggcagcctggagcacggagg; mFOS reverse: cagtgcctgtctcttccatcc). The relative values to input were determined using SYBR green (Applied Biosystems).
Quantitative RT-PCR
Total RNA was isolated using Trizol (Invitrogen) from cells growing in log phase, and converted to cDNA using Superscriptase III (Invitrogen). Real-time PCR was performed with TaqMan probes (Applied Biosystems) using β-ACTIN as an internal control.
Significance
Increasing evidence indicates that inhibition of the GSK-3 multi-functional serine/threonine kinase impairs the proliferation and induces the differentiation of a variety of cancers including leukemias induced by MLL oncogenes. Conversely, GSK-3 inhibition also stimulates the activities of several oncogenic proteins, therefore it is critical to determine the underlying mechanisms that dictate its biphasic oncogenic properties. In this report, we demonstrate that GSK-3 activity maintains the physical and functional association of CREB and its co-activators with MEIS1, a HOX DNA-binding co-factor and critical downstream mediator of the MLL oncogenic program. This in turn promotes critical target gene expression responsible for HOX-mediated transformation. These findings provide a molecular rationale for targeting HOX-associated transcription through GSK-3 inhibition in a subset of leukemias.
Supplementary Material
Acknowledgments
We thank M. Ambrus and C. Nicolas for technical assistance; members of the Cleary lab for helpful discussions, and K. Sakamoto for comments on the manuscript. F.F. was supported by a fellowship from the American-Italian Cancer Foundation and an ASH Fellow Scholar Award in basic research. C.J.M. was supported by PHS grant T32 CA09151 awarded by the National Cancer Institute, DHHS. We acknowledge support from the Children’s Health Initiative of the Packard Foundation, PHS grant CA116606, the Leukemia and Lymphoma Society, the Williams Lawrence Foundation, and a Developmental Research Award from the Stanford Cancer Center.
Footnotes
Accession number
Microarray raw data are available for download at Gene Expression Omnibus (http://ncbi.nlm.nih.gov/geo, Accession Number GSE 19736).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- Bhat R, Xue Y, Berg S, Hellberg S, Ormö M, Nilsson Y, Radesäter AC, Jerning E, Markgren PO, Borgegård T, et al. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J Biol Chem. 2003;278:45937–45945. doi: 10.1074/jbc.M306268200. [DOI] [PubMed] [Google Scholar]
- Boer U, Cierny I, Krause D, Heinrich A, Lin H, Mayr G, Hiemke C, Knepel W. Chronic lithium salt treatment reduces CRE/CREB-directed gene transcription and reverses its upregulation by chronic psychosocial stress in transgenic reporter gene mice. Neuropsychopharmacology. 2008;33:2407–2415. doi: 10.1038/sj.npp.1301640. [DOI] [PubMed] [Google Scholar]
- Carlezon WAJ, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Cohen P, Goedert M. GSK3 inhibitors: development and therapeutic potential. Nat Rev Drug Discov. 2004;3:479–487. doi: 10.1038/nrd1415. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Canettier iG, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Curran T, Teich NM. Identification of a 39,000-dalton protein in cells transformed by the FBJ murine osteosarcoma virus. Virology. 1982;116:221–235. doi: 10.1016/0042-6822(82)90415-9. [DOI] [PubMed] [Google Scholar]
- De Ferrari GV, Inestrosa NC. Wnt signaling function in Alzheimer’s disease. Brain Res Brain Res Rev. 2000;33:1–12. doi: 10.1016/s0165-0173(00)00021-7. [DOI] [PubMed] [Google Scholar]
- Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Hess JL. Mechanisms of transcriptional regulation by MLL and its disruption in acute leukemia. Int J Hematol. 2008;87:10–18. doi: 10.1007/s12185-007-0009-8. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esparza SD, Chang J, Shankar DB, Zhang B, Nelson SF, Sakamoto KM. CREB regulates Meis1 expression in normal and malignant hematopoietic cells. Leukemia. 2008;22:665–667. doi: 10.1038/sj.leu.2404933. [DOI] [PubMed] [Google Scholar]
- Fiol CJ, Williams JS, Chou CH, Wang QM, Roach PJ, Andrisani OM. A secondary phosphorylation of CREB341 at Ser129 is required for the cAMP-mediated control of gene expression. A role for glycogen synthase kinase-3 in the control of gene expression. J Biol Chem. 1994;269:32187–32193. [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh SL, Looi Y, Shen H, Fang J, Bodner C, Houle M, Ng AC, Screaton RA, Featherstone M. Transcriptional activation by MEIS1A in response to protein kinase A signaling requires the transducers of regulated CREB family of CREB co-activators. J Biol Chem. 2009;284:18904–18912. doi: 10.1074/jbc.M109.005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales M, Bowden GT. Ultraviolet B (UVB) induction of the c-fos promoter is mediated by phospho-cAMP response element binding protein (CREB) binding to CRE and c-fos activator protein 1 site (FAP1) cis elements. Gene. 2002;293:169–179. doi: 10.1016/s0378-1119(02)00723-0. [DOI] [PubMed] [Google Scholar]
- Heinrich A, Böer U, Tzvetkov M, Oetjen E, Knepel W. Stimulation by lithium of the interaction between the transcription factor CREB and its co-activator TORC. Biosci Rep. 2009;29:77–87. doi: 10.1042/BSR20080116. [DOI] [PubMed] [Google Scholar]
- Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, Frampton J, Slany RK. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike N, Sakoda H, Kushiyama A, Ono H, Fujishiro M, Kamata H, Nishiyama K, Uchijima Y, Kurihara Y, Kurihara H, Asano T. AMP-activated protein kinase activation increases phosphorylation of glycogen synthase kinase 3beta and thereby reduces cAMP-responsive element transcriptional activity and phosphoenolpyruvate carboxykinase C gene expression in the liver. J Biol Chem. 2008;283:33902–33910. doi: 10.1074/jbc.M802537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Rastegar M, Bodner C, Goh SL, Rambaldi I, Featherstone M. MEIS C termini harbor transcriptional activation domains that respond to cell signaling. J Biol Chem. 2005;280:10119–10127. doi: 10.1074/jbc.M413963200. [DOI] [PubMed] [Google Scholar]
- Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korur S, Huber RM, Sivasankaran B, Petrich M, Morin PJ, Hemmings BA, Merlo A, Lino MM. GSK3beta regulates differentiation and growth arrest in glioblastoma. PLoS One. 2009;4:e7743. doi: 10.1371/journal.pone.0007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. Embo J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosl J, Sauvageau G. AP-1 complex is effector of Hox-induced cellular proliferation and transformation. Oncogene. 2000;19:5134–5141. doi: 10.1038/sj.onc.1203897. [DOI] [PubMed] [Google Scholar]
- Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. Embo J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke M, Cleary ML. Therapeutic targeting of MLL. Blood. 2009;113:6061–6068. doi: 10.1182/blood-2008-12-197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J. Glycogen synthase kinase 3beta (GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer Lett. 2009;273:194–200. doi: 10.1016/j.canlet.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Rehani K, Jope RS, Michalek SM. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol. 2005;6:777–784. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Alonso M, Castro A, Pérez C, Moreno FJ. First non-ATP competitive glycogen synthase kinase 3 beta (GSK-3beta) inhibitors: thiadiazolidinones (TDZD) as potential drugs for the treatment of Alzheimer’s disease. J Med Chem. 2002;45:1292–1299. doi: 10.1021/jm011020u. [DOI] [PubMed] [Google Scholar]
- Matthews CP, Colburn NH, Young MR. AP-1 a target for cancer prevention. Curr Cancer Drug Targets. 2007;7:317–324. doi: 10.2174/156800907780809723. [DOI] [PubMed] [Google Scholar]
- Miyashita K, Kawakami K, Nakada M, Mai W, Shakoori A, Fujisawa H, Hayashi Y, Hamada J, Minamoto T. Potential therapeutic effect of glycogen synthase kinase 3beta inhibition against human glioblastoma. Clin Cancer Res. 2009;15:887–897. doi: 10.1158/1078-0432.CCR-08-0760. [DOI] [PubMed] [Google Scholar]
- Owens BM, Hawley RG. HOX and non-HOX homeobox genes in leukemic hematopoiesis. Stem Cells. 2002;20:364–379. doi: 10.1634/stemcells.20-5-364. [DOI] [PubMed] [Google Scholar]
- Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26:1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- Potter CS, Peterson RL, Barth JL, Pruett ND, Jacobs DF, Kern MJ, Argraves WS, Sundberg JP, Awgulewitsch A. Evidence that the satin hair mutant gene Foxq1 is among multiple and functionally diverse regulatory targets for Hoxc13 during hair follicle differentiation. J Biol Chem. 2006;281:29245–29255. doi: 10.1074/jbc.M603646200. [DOI] [PubMed] [Google Scholar]
- Ramirez S, Ait-Si-Ali S, Robin P, Trouche D, Harel-Bellan A. The CREB-binding protein (CBP) cooperates with the serum response factor for transactivation of the c-fos serum response element. J Biol Chem. 1997;272:31016–31021. doi: 10.1074/jbc.272.49.31016. [DOI] [PubMed] [Google Scholar]
- Rawat VP, Thoene S, Naidu VM, Arseni N, Heilmeier B, Metzeler K, Petropoulos K, Deshpande A, Quintanilla-Martinez L, Bohlander SK, et al. Overexpression of CDX2 perturbs HOX gene expression in murine progenitors depending on its N-terminal domain and is closely correlated with deregulated HOX gene expression in human acute myeloid leukemia. Blood. 2008;111:309–319. doi: 10.1182/blood-2007-04-085407. [DOI] [PubMed] [Google Scholar]
- Riedt T, Ebinger M, Salih HR, Tomiuk J, Handgretinger R, Kanz L, Grunebach F, Lengerke C. Aberrant expression of the homeobox gene CDX2 in pediatric acute lymphoblastic leukemia. Blood. 2009;113:4049–4051. doi: 10.1182/blood-2008-12-196634. [DOI] [PubMed] [Google Scholar]
- Schnabel CA, Jacobs Y, Cleary ML. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19:608–616. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- Scholl C, Bansal D, Dohner K, Eiwen K, Huntly BJ, Lee BH, Rucker FG, Schlenk RF, Bullinger L, Dohner H, et al. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J Clin Invest. 2007;117:1037–1048. doi: 10.1172/JCI30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitwala KV, Dandekar MN, Hess JL. HOX Proteins and Leukemia. Int J Clin Exp Pathol. 2008;1:461–474. [PMC free article] [PubMed] [Google Scholar]
- Siu YT, Chin KT, Siu KL, Yee WCE, Jeang KT, Jin DY. TORC1 and TORC2 coactivators are required for tax activation of the human T-cell leukemia virus type 1 long terminal repeats. J Virol. 2006;80:7052–7059. doi: 10.1128/JVI.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu YT, Jin DY. CREB--a real culprit in oncogenesis. FEBS J. 2007;274:3224–3232. doi: 10.1111/j.1742-4658.2007.05884.x. [DOI] [PubMed] [Google Scholar]
- Smalley KS, Contractor R, Haass NK, Kulp AN, Atilla-Gokcumen GE, Williams DS, Bregman H, Flaherty KT, Soengas MS, Meggers E, Herlyn M. An organometallic protein kinase inhibitor pharmacologically activates p53 and induces apoptosis in human melanoma cells. Cancer Res. 2007;67:209–217. doi: 10.1158/0008-5472.CAN-06-1538. [DOI] [PubMed] [Google Scholar]
- Smith DG, Buffet M, Fenwick AE, Haigh D, Ife RJ, Saunders M, Slingsby BP, Stacey R, Ward RW. 3-Anilino-4-arylmaleimides: potent and selective inhibitors of glycogen synthase kinase-3 (GSK-3) Bioorg Med Chem Lett. 2001;12:635–639. doi: 10.1016/s0960-894x(00)00721-6. [DOI] [PubMed] [Google Scholar]
- Smith KS, Jacobs Y, Chang CP, Cleary ML. Chimeric oncoprotein E2a-Pbx1 induces apoptosis of hematopoietic cells by a p53-independent mechanism that is suppressed by Bcl-2. Oncogene. 1997;14:2917–2926. doi: 10.1038/sj.onc.1201249. [DOI] [PubMed] [Google Scholar]
- Smith KS, Rhee JW, Cleary ML. Transformation of bone marrow B-cell progenitors by E2a-Hlf requires coexpression of Bcl-2. Mol Cell Biol. 2002;22:7678–7687. doi: 10.1128/MCB.22.21.7678-7687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svingen T, Tonissen KF. Altered HOX gene expression in human skin and breast cancer cells. Cancer Biol Ther. 2003;2:518–523. doi: 10.4161/cbt.2.5.441. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson DR, Swarthout JT, Jefcoat SC, Partridge NC. PTH induction of transcriptional activity of the cAMP response element-binding protein requires the serine 129 site and glycogen synthase kinase-3 activity, but not casein kinase II sites. Endocrinology. 2002;143:674–682. doi: 10.1210/endo.143.2.8626. [DOI] [PubMed] [Google Scholar]
- Wang N, Kim HG, Cotta CV, Wan M, Tang Y, Klug CA, Cao X. TGFbeta/BMP inhibits the bone marrow transformation capability of Hoxa9 by repressing its DNA-binding ability. Embo J. 2006;25:1469–1480. doi: 10.1038/sj.emboj.7601037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Wr, Baldwin AS. Maintenance of constitutive IkappaB kinase activity by glycogen synthase kinase-3alpha/beta in pancreatic cancer. Cancer Res. 2008;68:8156–8163. doi: 10.1158/0008-5472.CAN-08-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Uddin S, Zimmerman T, Kang JA, Ulaszek J, Wickrema A. Growth control of multiple myeloma cells through inhibition of glycogen synthase kinase-3. Leuk Lymphoma. 2008;49:1945–1953. doi: 10.1080/10428190802304966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.