Abstract
The Hippo signaling pathway is a key regulator of growth during animal development, while loss of normal Hippo pathway activity is associated with a wide range of cancers. Hippo signaling represses growth by inhibiting the activity of a transcriptional co-activator protein, known as Yorkie in Drosophila and Yap in vertebrates. In the five years since the first report linking Yorkie to Hippo signaling, intense interest in this pathway has led to rapid increases in our understanding of the action and regulation of Yorkie/Yap, which we review here. These studies have also emphasized the complexity of Yorkie/Yap regulation, including multiple, distinct mechanisms for repressing its transcriptional activity, and multiple DNA-binding partner proteins that can direct Yorkie to distinct downstream target genes.
Yorkie/Yap, the key effector of growth control by Hippo signaling
The control of growth is among the most fundamental aspects of development. A comprehensive understanding of what dictates the size of a particular organ (how does the liver “know” how large it should be), or why a mouse is small and an elephant is large, remains elusive. Nonetheless, insights into how growth is controlled have come from the identification and characterization of intercellular signaling pathways that are required for the normal control of organ growth. Many of these signaling pathways are highly conserved among different phyla. Consequently, studies in simple model systems like Drosophila have had a profound influence on our understanding of how organ growth is controlled in humans. Moreover, since in both Drosophila and mammals dysregulation of signaling pathways that control growth can result in tumor formation, characterization of these pathways is of fundamental importance to oncology.
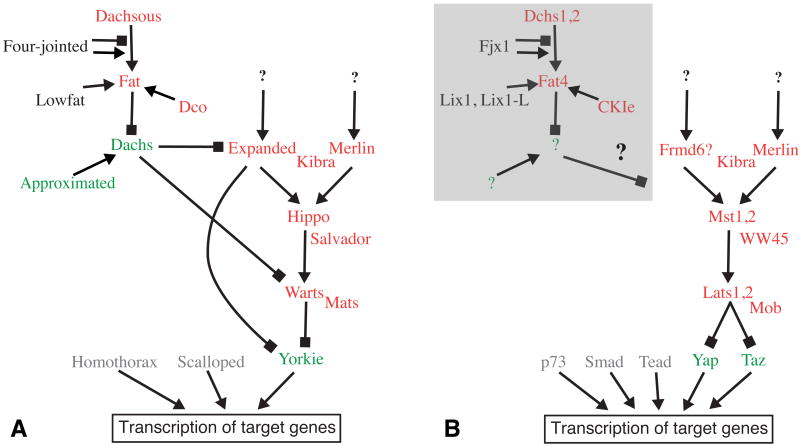
Over the past several years, studies of growth control in Drosophila have led to the identification of a new signaling pathway that controls growth 1, 2. This pathway has been most commonly referred to as “Hippo signaling”, although it is also referred to by a variety of other names, including Fat signaling, Warts signaling, and Salvador-Warts-Hippo (SWH) signaling. The identification of this pathway came about largely through identification and linkage into a common pathway of a series of Drosophila tumor suppressor genes. To date, ten tumor suppressors (warts (wts), hippo (hpo), salvador, mob-as-tumor suppressor (mats), expanded (ex), Merlin (Mer), kibra, discs overgrown (dco), fat, and dachsous) have been linked to Hippo signaling. Upstream regulation of this pathway is complex, as it encompasses three interconnected branches (Fig. 1). However, these upstream branches all converge and act through a common downstream component, the transcriptional co-activator protein Yorkie (Yki). Yki can act as an oncogene, and its potent growth-promoting activity is normally kept in check by the upstream tumor suppressor proteins. Initial studies of Yki identified a simple mechanism for this negative regulation, in which the Warts kinase blocked Yki activity by phosphorylating it 3, and other upstream components influenced the levels and activity of Warts 1, 2. However, recent studies have begun to uncover increasing complexity in the regulation of Yki and its mammalian homologues, Yap and Taz. This complexity includes distinct roles for multiple phosphorylation sites, phosphorylation-independent regulation, effects on expression and stability, and the identification of additional DNA-binding partner proteins (Fig. 2). In this review, we discuss recent developments in our understanding of Hippo signaling, focusing on those advances that impinge on our understanding of the regulation and function of Yki/Yap.
Figure 1. The Hippo signaling pathway.
Schematic depictions of the regulatory interactions among genes linked to Hippo signaling. Pointed arrows indicate a positive regulatory connection, blocks indicate an inhibitory regulatory connection. Genes depicted in red can function as tumor suppressors (loss of function mutations can cause overgrowth), genes depicted in green can function as oncogenes (over expression and/or activation can cause overgrowth), genes depicted in black have not been shown to function as tmor suppressors or oncogenes. At the botom, genes depicted in gray encode DNA binding proteins that can partner with Yki/Yap to influence transcription of downstream target genes. Hippo signaling influences growth by influencing the transcription of target genes, but transcriptional targets that are not directly related to growth control have also been identified. A) Depicts genes and regulatory links identified in Drosophila. B) Depicts some of the genes identified in mammals. The Fat regulatory branch is shaded by a grey box to indicate that it is not yet known whether these genes actually influence Hpo signaling in mammals.
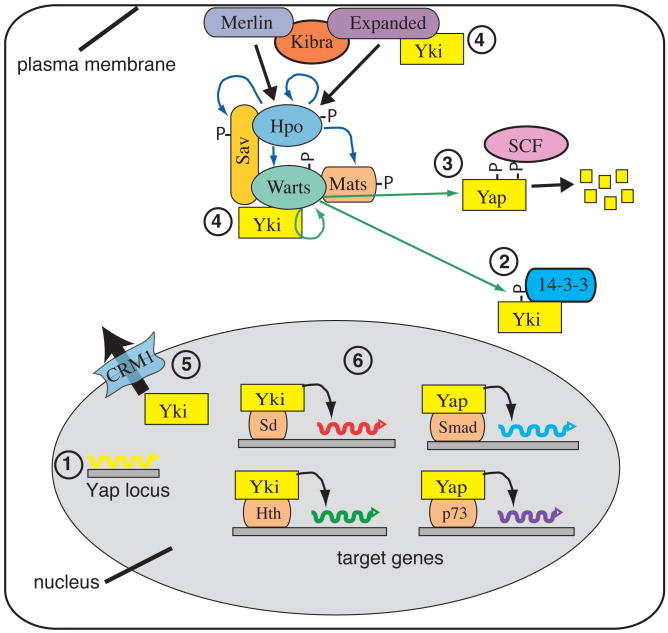
Figure 2. Multiple modes of Yki/Yap regulation.
This schematic depicts some of the different ways in which Yki/Yap is known to be regulated within the cell, gray oval represents the nucleus. 1) Yki/Yap can be transcriptionally regulated; increased transcription has been implicated in medulloblastoma. 2) Phosphorylation of Yki/Yap by Warts/Lats (green arrows) creates a 14-3-3 binding site, which promotes cytoplasmic localization of Yki/Yap. Wts activity is regulated by Hpo-dependent phosphorylation (blue arrows). 3) Phosphorylation of Yap by Lats makes it a substrate for CKI phosphorylation, which recruits SCF, leading to Yap ubiquitination and degradation (small yellow boxes). 4) Direct binding of Yki to Wts and Ex can retain Yki in the cytoplasm. 5) Yki/Yap can be exported from the nucleus in a CRM1-dependent fashion. 6) Nuclear Yki/Yap can associate with distinct DNA-binding transcription factors to regulate distinct downstream target genes.
Regulation of Warts/Lats activity
Crucial phosphorylation-dependant regulation of Yki is mediated by Warts 3, a member of Nuclear Dbf2-related (NDR) family of Ser/Thr kinases 4, 5. The activity of Warts is controlled through a series of phosphorylation events. Warts is directly phosphorylated by Hippo (Hpo), a member of the Sterile-20 family of Ser/Thr kinases, in a reaction that is facilitated by the Salvador protein 6. Hpo also phosphorylates the Warts co-factor Mob-as-tumor suppressor (Mats), which promotes Warts-Mats interaction, and thereby Warts activity, and Warts activation also involves autophosphorylation 7.
Two related proteins, Lats1 and Lats2, have been identified as vertebrate homologues of Warts 8, 9, whereas Mst1 and Mst2 have been identified as homologues of Hpo 6, 10-14. However, a recent study of Yap regulation in mice indicates that additional kinases can influence Yap phosphorylation at its major regulatory site (Ser127). In liver cells mutant for both Mst1 and Mst2, Yap phosphorylation was impaired, but Lats1 and Lats2 phosphorylation were not visibly affected, and instead a distinct kinase appeared to be involved 15. This alternative kinase was not identified, but other members of the NDR kinase family are possible candidates. The same authors also found that even though Mst1 and Mst2 were required for Yap phosphorylation in liver cells, they were not required for Yap or Lats phosphorylation in embryonic fibroblasts, which implies that other kinases provide Mst function in these cells 15. There are additional Ste-20 kinase family members in mice, but whether or not any of them are responsible for Lats activation in fibroblasts remains to be determined. Intriguingly, hpo mutant phenotypes are weaker than wts mutant phenotypes in some Drosophila tissues 6, 16, which is consistent with the possibility that additional Hpo-like kinases might also have a role in Drosophila. These observations emphasize two themes that are emerging as studies of Hpo signaling intensify – the increasing complexity of the pathway, and the existence of cell-type specific requirements for different components.
Hpo signaling can be regulated by cell contact, and is important for contact-dependent inhibition of proliferation in cultured mammalian cells and regulation of cell fate in the early embryo 17-19. Although the mechanism of contact-dependent signaling remains poorly understood, insights into regulatory steps at the membrane are beginning to emerge. Three interconnected, upstream regulatory branches of Hippo signaling have been identified: Fat-dependent, ex-dependent, and Mer-dependent (Fig. 1) 2. Fat is a transmembrane receptor protein, and both Fat and its ligand Dachsous are large, atypical cadherins. Dachsous promotes a phosphorylation of the Fat cytoplasmic domain, which is mediated by the CKIε protein Dco 20, 21. Fat influences downstream events in Hippo signaling by antagonizing the activity of the myosin Dachs, which accumulates at the membrane in the absence of Fat activity 16, 22, 23.
Ex and Mer are members of the FERM-domain containing protein family, which associate with both the membrane and the actin cytoskeleton 24. Ex and Mer function in a partially redundant manner to regulate Hippo signaling 25, 26. Recently, a new player in the Ex- and Mer-dependent branches has been identified, Kibra 27-29. Kibra interacts physically and genetically with both Mer and Ex. Kibra binds to Mer and Ex through distinct protein motifs, and the three proteins can form a trimeric complex. Mer and Ex can also bind directly to Sav and Hpo, respectively, consistent with the notion that a regulatory complex among these tumor suppressors forms at the membrane 27. Studies of Mats have also emphasized the importance of membrane association for regulation of Wts. A fraction of Mats normally localizes to the membrane, and forced localization of Mats to the membrane induces Wts activation, even in the absence of Hpo 30. Similarly, forced membrane localization of human MOB proteins can activate NDR/Lats kinases in cultured mammalian cells 31, 32. Thus, although the biochemical mechanisms by which Mer or Ex regulate Wts activation requires further investigation, it seems that activation occurs in a protein complex at the cell membrane.
Wts/Lats-mediated regulation of Yki/Yap localization
Investigations of the regulation of Yki/Yap by Wts/Lats identified Ser168 of Yki (Ser127 of Yap) as a key site 18, 33-35. Phosphorylation of this Ser enables interaction with 14-3-3 proteins, which promote cytoplasmic retention and nuclear export, and phosphorylation of Yki/Yap decreases its nuclear localization. The expectation that 14-3-3 proteins should thus be functionally involved in Yki regulation has recently been confirmed by knock-down experiments in Drosophila 36. These authors also showed that both wild-type and phosphorylation site mutant isoforms of Yki are subject to CRM1-mediated nuclear export, which implies that normal pathway regulation may involve additional, as yet undefined, nuclear export processes.
Although Ser168/127 of Yki/Yap is a crucial Wts/Lats phosphorylation site, there are two additional Wts sites within Yki (Ser111 and Ser250) 36, 37, and four additional sites within Yap (Ser61, Ser109, Ser164 and Ser381) 18, 38. Mutations of Yki Ser111 and Ser250 give mild overgrowth phenotypes on their own, and reduce Ser168 phosphorylation, which presumably contributes to their influence on Yki activity, and implies that Yki phosphorylation is cooperative. However, since combinations of mutations in the different sites result in additive phenotypes, each site also makes independent contributions to Yki regulation 36, 37. In the case of Yap, mutation of all five sites results in stronger activation than just mutation of Ser127 18, 39, but most of this can be accounted for by the site at Ser381 39, 40.
Unlike Ser168/127, the other Wts/Lats phosphorylation sites lack consensus 14-3-3 binding motifs, and biochemical and genetic experiments argue against a role for 14-3-3 proteins in regulating Yki/Yap through these sites 18, 33, 34, 36, 37. They do influence Yki/Yap localization, which might be mediated through interactions with additional binding partners. Alternatively, they might act through distinct mechanisms, such as the effect of Ser381 of Yap on protein stability 40.
Phosphorylation-independent regulation of Yki localization by upstream tumor suppressors
Protein-protein interactions mediated by binding between WW-domains and Pro-rich motifs are found in a range of biological processes (see Box 1), but appear to be especially common in Hpo signaling. Recent studies have implicated direct binding of Yki to Ex, Wts or Hpo, mediated by WW domains of Yki and PPxY motifs of Ex, Wts or Hpo, in a phosphorylation-independent mechanism of Yki repression 41, 42. Over-expressed Ex or Wts can repress Yki mediated transcriptional activation, in vivo and S2 cell experiments, even when all of the Wts phosphorylation sites on Yki are mutant 36, 37, 41. Moreover, this repression requires PPxY motifs of Ex and Wts 41, 42. A fraction of Yki localizes to the sub-apical membrane in an Ex-dependent fashion 41, 42, and this apical localization of Yki requires its WW domains 43.
BOX 1. WW domain – PPxY interactions in Hpo signaling.
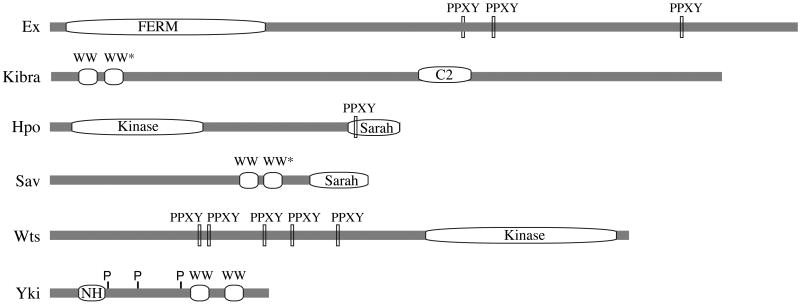
WW domains are 35-40 amino acid protein-protein interaction domains, which are named for their pair of conserved Trp residues 82, and which generally interact with Pro-rich sequence motifs. WW domain-Pro motif interactions are found among proteins that function in a wide range of biological processes, including cytoskeletal regulation, signal transduction, protein trafficking, protein degradation, and transcriptional regulation 83, 84. They appear to be especially common in Hpo signaling. WW domains have been classified into different subgroups based on either on differences in sequence composition of the WW domain, or in the sequence of the Pro-rich motif to which they bind 83, 84. Most of the WW domains found in Hpo pathway components are of the group I type, which bind to PPxY sequence motifs. Three core components of Hpo signaling (Yki, Kibra, and Sav) contain WW domains, whereas three other components (Wts, Ex, and Hpo) contain PPxY motifs (see Figure). Moreover, several DNA binding transcription factors have been identified that contain PPxY motifs and interact with WW domains of Yap or Taz 56, 57. The existence of binding between many of the possible WW-PPxY pairs within the pathway has been confirmed, at least in cultured cells. Recently it has been reported that certain atypical WW domains, found in Sav and Kibra, can dimerize, which can mediate homodimerization of Sav or heterdimerization between Kibra and Sav 27, 85. The prevalance of this protein interaction module among pathway components suggests that sequential or competive interactions amongst proteins with these motifs might be important. Indeed, competitive binding between Ex and Wts for Yki was demonstrated by titration experiments in S2 cells 42. Moreover, interaction between Mer and Kibra does not involve WW-PPxY interactions, but was enhanced when the WW-domains of Kibra were deleted 29, which suggests that interactions between Mer and Kibra are impeded by other WW-domain interacting proteins. Figure I depicts linear schematics of Drosophila components of the Hpo pathway with WW domains and PPxY motifs, and also indicates the location of other key motifs in these proteins (WW* = atypical WW domain that can dimerize, C2 = C2 domain, Sarah = Sarah domain, NH = Sd binding domain, P = Wts phosphorylation sites).
Assessing the respective contributions of phosphorylation-dependent versus phosphorylation-independent repression in vivo is complicated by the fact that both modes are mediated by the same upstream tumor suppressors, and by the dual positive and negative roles of the WW domains in Yki regulation (discussed below). Although phosphorylation-independent repression has not yet been demonstrated in vertebrates, its discovery in Drosophila suggests a dual repression model, with the same upstream tumor suppressors repressing Yki activity by two distinct mechanisms, and emphasizes the importance of the relative levels of Yki/Yap to its upstream tumor suppressors. As discussed below, these two themes also apply to the regulation of Yap in vertebrates.
Additional mechanisms of Yki/Yap Regulation
In addition to Wts-mediated phosphorylation of Yki and Yap, there is also evidence for additional forms of phosphorylation-mediated regulation. Two Ser residues near Ser168 of Yki (Ser169 and Ser172) are highly conserved in Yap, and mutating these to Ala in Yki elevated phosphorylation of Ser168 in vivo, whereas phosphomimetic mutations (Ser to Asp) inhibited Ser168 phosphorylation 37. Although the functional significance of phosphorylation at these sites has not been directly demonstrated, a phosphoproteomic analysis of Drosophila embryos established that at least one, and possibly both, of these sites can be phosphorylated in vivo 44. Interestingly, this study also identified several additional amino acids of Yki that can be phosphorylated, the significance of which remains to be determined.
Mass-spec analysis of Yap expressed in cultured cells identified five phosphorylation sites (Thr63, Ser138, Ser289, Ser351, and Ser384) in addition to the five Lats sites 40. Notably, Ser384 is phosphorylated by Casein kinase I in a reaction that is dependent upon a priming phosphorylation of Ser381 by Lats 40. Phosphorylation of Ser384 then recruits an E3 ubiquitin ligase complex to promote Yap degradation. This study thus identified an additional mechanism by which Wts-mediated phosphorylation negatively regulates Yap. The existence of this mechanism in vivo is supported by the observation that Yap levels are elevated in the livers of Mst mutant mice 15, 45.
The frequency with which the Yap locus is amplified and/or Yap levels are elevated in human cancers emphasizes the importance of Yap levels to tumorigenesis 34, 46-50. Although transcriptional regulation of Yap has not received much attention so far, a recent study identified Yap as a key mediator of Shh-associated medulloblastoma 46. In addition to increased nuclear localization, the authors found that Yap expression is transcriptionally upregulated in Shh-driven medulloblastoma, which should stimulate a greater focus on transcriptional regulation of Yki/Yap in other contexts.
Mammals have a single Yap locus, but through alternative splicing it can generate three different polypeptides, Yap1 and Yap2, and Yap2L 51. These isoforms are structurally related, and can be regulated by Hpo signaling 18, 34, 38, 52, 53, but Yap2 isoforms differ in the inclusion of a C-terminal PDZ binding domain, which is also found on the Yap-related gene Taz 54. The PDZ domains are important for Yap2 and Taz activity and regulate their nuclear localization 54, 55, which thus identifies another mode of Yap/Taz regulation. Drosophila Yki does not have a C-terminal PDZ domain, so this form of regulation may be specific to vertebrates.
Role of the WW domains in regulating Yki/Yap activity
Yki was identified as an interactor and substrate of Wts 3. Although it was initially assumed that this binding would be required for Yki/Yap phosphorylation by Wts/Lats, subsequent studies have revealed that this is not the case 37, 39, raising the possibility that it may serve other functions. Indeed, as noted above, the PPxY motifs of Ex and Wts are required for phosphorylation-independent repression 41, 42. However, if the WW domains of Yki were only required for Yki repression, then mutation of these domains should result in elevation of Yki activity, but instead the opposite occurs–mutation of the WW domains abolishes the activity of Yki 34, 37, 43. Since the WW domains are not required for nuclear localization of Yki, yet they are required for its transcriptional activity, it appears that the WW domains may recruit an as yet unidentified PPxY-motif containing transcriptional co-factor to Yki. Indeed, even when Yki was bound to the promoter of a synthetic reporter gene, using a Gal4-DNA binding domain-Yki fusion protein, the WW domains were required 37. WW domain-PPxY interactions play multiple roles in Hippo signaling (see Box 1), and the dual role of the WW domains, facilitating repression of Yki in the cytoplasm, but participating in its activity in the nucleus, raises the possibility that competitive binding between PPxY-containing proteins for Yki might contribute to Yki regulation.
In the case of Yap, the requirement for the WW domains appears to be more complex. They are required for transcriptional activation by Yap with some DNA binding partner proteins, and on some target genes, but not others 39, 43, 53. For example in they are required for transformation of NIH3T3 cells, but in breast epithelial cells, Yap without WW-domains was even more potent at promoting cellular transformation than wild-type Yap 43, which implies that in this context, the major function of the WW domains is to contribute to Yap repression. Such cell-type specific responses presumably reflect the differential expression and regulation of Yap partner proteins in different cells.
DNA binding co-factors and targets of Yki/Yap
Yki/Yap function as transcriptional co-activators, and lack their own DNA binding domain. Before Yap was linked to the Hpo pathway, several different DNA binding partners for it were identified 56, 57. However, the biological significance of many of these interactions was unclear, and it was not known which might function as partners for Yki/Yap's role in Hpo signaling. This began to be addressed with the discovery that Sd could function as partner for Yki in Hpo signaling in Drosophila, and its mammalian homologues the Tead/Tef proteins could function as partners for Yap in Hpo signaling in vertebrates 19, 35, 58-61. The significance of Sd/Tead as Yki/Yap co-factors was further solidified by the demonstration that they bound directly to regulatory sequences of a known Yki target gene, Diap1 (thread) 35, 58, and a known Yap target gene, Connective Tissue Growth Factor (CTGF) 60.
The identification of Sd as a partner for Yki in Drosophila was unexpected, because most Drosophila tissues that require yki for normal growth don't require sd 62, 63. Recently, a new DNA-binding partner for Yki, has been identified, Homothorax (Hth) 64. Together with two other DNA-binding proteins (Extradenticle and Teashirt), Hth regulates the proliferation of uncommitted progenitor cells in the eye imaginal disc. Genetic experiments revealed that Yki and Hth require each other to promote growth in the eye, and also interact physically. Hth does not detectably influence expression of Diap1, but instead the authors identified another Yki target, the micro RNA gene bantam 65, 66, as a direct target of the Yki:Hth complex 64. The identification of both Sd and Hth as DNA binding partners of Yki thus reveals a surprising complexity in Hpo signaling. On the one hand, since they are expressed in different patterns, it suggests that one resolution to the limited requirements for sd may be that Yki interacts with multiple, distinct partner proteins in different tissues. However, this raises the question of how Yki executes a common developmental fuction, promoting tissue growth, in combination with distinct DNA binding proteins that appear to regulate distinct sets of genes.
Another class of Yki/Yap partners that have been further characterized recently are the Smad proteins, which function as transcription factors for TGFβ signaling pathways 67. Smad7 was identified as a Yap-binding protein several years ago 68. More recent studies have identified Smad1 as a Yap partner 69, Mad as Yki partner 69, and the Smad2/3-4 complex as a Taz partner 70. These recent studies focussed on the ability of Yap or Taz to potentiate transcriptional activation by Smads to regulate Smad target genes. The significance of these interactions to growth control by Hippo signaling remains to be determined, but their detection identifies the potential for cross-talk between TGFβ and Hippo signaling pathways.
The interaction between Yki/Yap/Taz and Smads is partially dependent upon WW-domains in Yap-related proteins and PPxY motif ins SMADs 68-70. In addition to the Smads, several other transcription factors have been reported to interact with Yap through WW-domain-PPxY interactions 56, 57. These partners appear to utilize Yap to execute diverse functions. For example, while activation of Yki/Yap in Hpo signaling inhibits apoptosis, Yap can have a pro-apoptotic role in conjunction with p73 under some conditions 53, 71, and this pro-apoptotic activity of Yap can also be regulated by Hpo signaling 53.
The Yki/Yap partners that have been linked to its role in Hpo pathway-regulated growth control, namely Sd/Tead and Hth, lack PPxY motifs. Sd/Tead interacts with an N terminal region of Yki/Yap that is well separated from its WW domains (see Box 1 Figure) 35, 58, 59, 72. Thus, different regions of Yki/Yap associate with different DNA binding partners. One question that needs further study is how Yki or Yap interact with distinct partners to mediate transcriptional activation. For example, since Yki/Yap appear to employ their WW domains to recruit factors required for transcription when they are bound to Sd/Tead, when complexed with DNA binding partners that contain PPxY motifs, do they still employ one of their WW domains to recruit other transcription factors, or do they employ a distinct mechanism of transcriptional activation? The observation that at least in some contexts the WW domains of Yap are not required for transcriptional activation suggests that Yki/Yap may be able to act as a versatile scaffold to assemble distinct transcriptional regulatory complexes.
Figure I.
Linear schematics of Drosophila components of the Hpo pathway
Another key question is how the association of Yki/Yap amongst distinct DNA-binding partners within the same cell is modulated. Related to this is the identification of downstream target genes regulated by Yki/Yap. A comprehensive discussion of target genes is beyond the scope of this review, but we note that even in Drosophila, which seems to have fewer DNA-binding partners for Yki/Yap than vertebrates, not all downstream targets are related to growth control, as exemplified by the recent discovery that Yki activity influences the apical membrane of epithelial cells 73, 74. Moreover, even in the context of its growth control functions, we can consider distinct types of downstream target genes, as both autonomous and non-autonomous effects are important. Autonomous growth promoting targets of Yki/Yap include genes that promote cellular growth (Myc, bantam), genes involved in cell cycle progression (CyclinB, CyclinD, CyclinE, E2F1), and inhibitors of apoptosis (Diap1, cIap1) 6, 34, 59, 65, 66, 75-79. Non-autonomous growth promoting targets include genes that encode ligands for other signaling pathways. In Drosophila, the Wnt pathway ligand Wg is regulated by Hippo signaling in the proximal wing, where it is a major contributor to growth regulation 16, 23. The Notch pathway ligand Ser, and the EGF-R pathway ligand vein, can also be regulated by Yki in some contexts 16, 22, 80. Additionally, the glypicans Dally and Dally-like, which influence the movement of secreted signals through tissues, can be regulated by Yki 81. Studies in mammalian cells identified Connective Tissue Growth Factor (CTGF) as a Yap target 52, 60, and the importance of non-autonomous growth promoting targets in vertebrates has been emphasized recently by the discovery that the ability of Yap to transform breast epithelial cells is dependent upon its ability to upregulate expression of the EGFR ligand amphiregulin 80. Thus, the biological effects of Yki/Yap on growth control in vivo appear to reflect a combination of autonomous and non-autonomous effects, which may vary in different cell types.
Conclusions and future directions
Hpo signaling is an evolutionarily conserved signaling pathway that controls organ size from flies to humans. The novelty of the pathway, and its fundamental importance to growth control and oncogenesis, has attracted many researchers to the field, and the pace of discovery has been rapid. These discoveries have identified sophisticated, multi-layered mechanisms for controlling Yki/Yap activity. Nonetheless, many steps in Hippo signaling remain poorly understood, and it is likely that core components are still missing. For example, despite the fundamental importance of kinases and protein phosphorylation at multiple steps of Hippo signaling, phosphatases that act on pathway components have not yet been identified.
As emphasized by the existence of multiple DNA binding partners for Yki and Yap, a particularly important challenge for the future will be to elucidate the logic of the transcriptional regulatory network relevant to Hpo signaling. What is the full complement of Yki/Yap partner proteins for transcriptional activation? How is the availability or activity of the partner proteins controlled, and what modulates the association of Yki among different possible partners within the same cell? How are different partner proteins employed to effect common or distinct transcriptional responses to Hpo signaling in different cell types? The answers to these questions should provide new insights into growth control during development, and could, in the future, help to guide diagnoses and therapies and for cancers or other diseases involving dysregulation of Hpo signaling.
Acknowledgments
Research in K.D.I.'s lab is supported by the HHMI and NIH grant GM078620; H.O. was supported by NIH post-doctoral fellowship 1F32GM079817. We thank C Rauskolb for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Badouel C, et al. Herding Hippos: regulating growth in flies and man. Curr Opin Cell Biol. 2009;21:837–843. doi: 10.1016/j.ceb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- 3.Huang J, et al. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Justice RW, et al. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 5.Xu T, et al. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 6.Wu S, et al. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 7.Wei X, et al. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. Embo J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yabuta N, et al. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics. 2000;63:263–270. doi: 10.1006/geno.1999.6065. [DOI] [PubMed] [Google Scholar]
- 9.Tao W, et al. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet. 1999;21:177–181. doi: 10.1038/5960. [DOI] [PubMed] [Google Scholar]
- 10.Udan RS, et al. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 11.Pantalacci S, et al. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 12.Jia J, et al. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey KF, et al. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 14.Chan EH, et al. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 15.Zhou D, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho E, et al. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 17.Nishioka N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 20.Sopko R, et al. Phosphorylation of the tumor suppressor fat is regulated by its ligand Dachsous and the kinase discs overgrown. Curr Biol. 2009;19:1112–1117. doi: 10.1016/j.cub.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y, Irvine KD. Processing and phosphorylation of the Fat receptor. Proc Natl Acad Sci U S A. 2009;106:11989–11994. doi: 10.1073/pnas.0811540106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao Y, et al. Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development. 2006;133:2539–2551. doi: 10.1242/dev.02427. [DOI] [PubMed] [Google Scholar]
- 23.Cho E, Irvine KD. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development. 2004;131:4489–4500. doi: 10.1242/dev.01315. [DOI] [PubMed] [Google Scholar]
- 24.Mangeat P, et al. ERM proteins in cell adhesion and membrane dynamics. Trends in cell biology. 1999;9:187–192. doi: 10.1016/s0962-8924(99)01544-5. [DOI] [PubMed] [Google Scholar]
- 25.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 26.McCartney BM, et al. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, et al. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genevet A, et al. Kibra is a regulator of the Salvador/Warts/Hippo signalling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumgartner R, et al. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 30.Ho LL, et al. Mob as tumor suppressor is activated at the cell membrane to control tissue growth and organ size in Drosophila. Dev Biol. 2010;337:274–283. doi: 10.1016/j.ydbio.2009.10.042. [DOI] [PubMed] [Google Scholar]
- 31.Hergovich A, et al. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochemical and biophysical research communications. 2006;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- 32.Hergovich A, et al. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol Cell Biol. 2005;25:8259–8272. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, et al. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren F, et al. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev Biol. 2010;337:303–312. doi: 10.1016/j.ydbio.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao Y, et al. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 39.Zhao B, et al. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- 40.Zhao B, et al. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh H, et al. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badouel C, et al. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, et al. Transcriptional output of the Salvador/warts/hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res. 2009;69:6033–6041. doi: 10.1158/0008-5472.CAN-08-4592. [DOI] [PubMed] [Google Scholar]
- 44.Zhai B, et al. Phosphoproteome analysis of Drosophila melanogaster embryos. Journal of proteome research. 2008;7:1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song H, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez LA, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinhardt AA, et al. Expression of Yes-associated protein in common solid tumors. Human pathology. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Overholtzer M, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 50.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komuro A, et al. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, et al. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68:2789–2794. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- 53.Oka T, et al. Mst2 and Lats Kinases Regulate Apoptotic Function of Yes Kinase-associated Protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 54.Kanai F, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. Embo J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oka T, Sudol M. Nuclear localization and pro-apoptotic signaling of YAP2 require intact PDZ-binding motif. Genes Cells. 2009;14:607–615. doi: 10.1111/j.1365-2443.2009.01292.x. [DOI] [PubMed] [Google Scholar]
- 56.Bertini E, et al. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- 57.Wang K, et al. YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87:77–91. doi: 10.1139/O08-114. [DOI] [PubMed] [Google Scholar]
- 58.Wu S, et al. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 59.Goulev Y, et al. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 60.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao X, et al. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X, et al. Roles for scalloped and vestigial in regulating cell affinity and interactions between the wing blade and the wing hinge. Dev Biol. 2000;228:287–303. doi: 10.1006/dbio.2000.9939. [DOI] [PubMed] [Google Scholar]
- 63.Campbell S, et al. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 1992;6:367–379. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- 64.Peng HW, et al. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 66.Nolo R, et al. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 67.Padgett RW, et al. TGF-beta signaling, Smads, and tumor suppressors. Bioessays. 1998;20:382–390. doi: 10.1002/(SICI)1521-1878(199805)20:5<382::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 68.Ferrigno O, et al. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 2002;21:4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- 69.Alarcon C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varelas X, et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 71.Strano S, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 72.Vassilev A, et al. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hamaratoglu F, et al. The Hippo tumor-suppressor pathway regulates apical-domain size in parallel to tissue growth. J Cell Sci. 2009;122:2351–2359. doi: 10.1242/jcs.046482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Genevet A, et al. The Hippo pathway regulates apical-domain size independently of its growth-control function. J Cell Sci. 2009;122:2360–2370. doi: 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimizu T, et al. The mob as tumor suppressor gene is essential for early development and regulates tissue growth in Drosophila. Genetics. 2008;178:957–965. doi: 10.1534/genetics.107.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silva E, et al. The Tumor-Suppressor Gene fat Controls Tissue Growth Upstream of Expanded in the Hippo Signaling Pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 77.Tapon N, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 78.Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicolay BN, Frolov MV. Context-dependent requirement for dE2F during oncogenic proliferation. PLoS genetics. 2008;4:e1000205. doi: 10.1371/journal.pgen.1000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J, et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baena-Lopez LA, et al. The tumor suppressor genes dachsous and fat modulate different signalling pathways by regulating dally and dally-like. Proc Natl Acad Sci U S A. 2008;105:9645–9650. doi: 10.1073/pnas.0803747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sudol M, et al. Characterization of a novel protein-binding module--the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 83.Macias MJ, et al. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 84.Ilsley JL, et al. The WW domain: linking cell signalling to the membrane cytoskeleton. Cellular signalling. 2002;14:183–189. doi: 10.1016/s0898-6568(01)00236-4. [DOI] [PubMed] [Google Scholar]
- 85.Ohnishi S, et al. Solution structure of an atypical WW domain in a novel beta-clam-like dimeric form. FEBS Lett. 2007;581:462–468. doi: 10.1016/j.febslet.2007.01.008. [DOI] [PubMed] [Google Scholar]