Abstract
Phosphatidylinositol transfer proteins (PITPs) in yeast coordinate lipid metabolism with the activities of specific membrane trafficking pathways. The structurally unrelated metazoan-specific PITPs (mPITPs), on the other hand, are an under-investigated class of proteins. It remains unclear what biological activities mPITPs discharge, and the mechanisms by which these proteins function are also not understood. The soluble class 1 mPITPs include the PITPα and PITPβ isoforms. Of these, the β-isoforms are particularly poorly characterized. Herein, we report the use of zebrafish as a model vertebrate for the study of class 1 mPITP biological function. Zebrafish express PITPα and PITPβ-isoforms (Pitpna and Pitpnb, respectively) and a novel PITPβ-like isoform (Pitpng). Pitpnb expression is particularly robust in double cone cells of the zebrafish retina. Morpholino-mediated protein knockdown experiments demonstrate Pitpnb activity is primarily required for biogenesis/maintenance of the double cone photoreceptor cell outer segments in the developing retina. By contrast, Pitpna activity is essential for successful navigation of early developmental programs. This study reports the initial description of the zebrafish class 1 mPITP family, and the first analysis of PITPβ function in a vertebrate.
Introduction
Phosphoinositides (PIPs), the phosphorylated versions of phosphatidylinositol (PtdIns), are major components of membrane signaling systems (1–4). PIPs take on multiple roles in this regard. These serve as precursors of second messengers (e.g. inositol phosphates and diacylglycerol; 5, 6), and also as binding platforms for recruitment of proteins to appropriate membrane locations (7, 8). The utility of PIPs as binding platforms for the appropriate spatial and temporal recruitment of proteins is, in part, a function of the chemical diversities of PIP headgroups -- diversities encoded by the number and the positional arrangement of phosphates that decorate the inositol headgroup (9). From the perspective of PIPs as binding platforms, further diversification of PIP-centric biological outcomes is regulated by coincidence detection mechanisms that couple the chemical identity of a particular PIP with additional protein or lipid binding activities (10,11).
A newly described coincidence detection mechanism for functional specification of PIP signaling involves Sec14-like PtdIns-transfer proteins (PITPs). Genetic and structural data indicate these proteins possess a lipid sensor role that is coupled to substrate presentation functions required for sufficient PtdIns kinase activity in vivo. The coupling of sensor/presentation activities is realized via heterotypic lipid exchange cycles that allow Sec14-like PITPs to impose an instructive regulation of PtdIns kinases – a level of control that cues activation of lipid kinases to specific metabolic inputs (11). This mechanism is conceptually summarized as Sec14-like PITPs serving as nanoreactors for PIP synthesis (10,11). Predictive bioinformatics suggest Sec14-like proteins link divergent territories of the lipid metabolome to PIP signaling in eukaryotic cells (11,12).
Whereas Sec14 is an ancient eukaryotic structural unit (13), expression of a second unrelated group of PITPs is largely restricted to the Metazoa (mPITPs; 13) – with Dictyostelium presenting an enigmatic exception (14). Whether mPITPs fulfill a nanoreactor function resembling that of Sec14-like PITPs, or whether these proteins function in lipid transfer, remains an open question. Indeed, little is known regarding their biological functions. Our studies focus on the class 1 mPITPs of which there are three in mammals: PITPα, PITPβ and RdgBα (15–17). The α and β isoforms share ca. 77% primary sequence identity, but localize to different intracellular compartments and exhibit distinct phospholipid binding/transfer properties (18). RdgBα is only ca 42% identical to PITPα and PITPβ and remains uncharacterized (19). Whereas Pitpα0/0 mice develop to birth, but suffer from complex postnatal pathologies that result in neonatal death (20), essentially nothing is known regarding PITPβ function. Available data, albeit negative data, suggest PITPβ executes essential housekeeping functions in mice. Given PITPβ localization to the mammalian trans-Golgi complex, such functions are likely executed at the level of the Golgi complex (18).
To address the issue of PITPβ biological function in vertebrates, we used the zebrafish (Danio rerio) model. Zebrafish express a mammalian-like set of mPITPs plus a unique PITPβ-like isoform designated Pitpng. We report Pitpnb dysfunction compromises biogenesis and/or maintenance of the double cone photoreceptor cell outer segments in the developing retina. These defects alter status of a retinal-specific arrestin (Arr3L) in double cone cells. Finally, in contrast to the phenotypes of Pitpnb morphants, knockdown of Pitpna expression results in early developmental failure. Rescue experiments demonstrate: (i) that PtdIns-binding is an essential functional property of PITPβ activity in zebrafish development, and (ii) that a pair of tryptophan residues, previously regarded as critical for class 1 PITP membrane-binding and phospholipids binding/transfer (21), do not play a major role in biological activity. These data provide the initial description of the zebrafish class 1 mPITP family, and document the first functional analysis of a PITPβ in a vertebrate.
Results
The zebrafish cohort of class 1 PITPs
While a presumptive zebrafish PITPβ orthologue has been identified (22), no reports are available for zebrafish PITPβ. However, a protein annotated as homologous to mammalian PITPβ is present in both the ZFIN and NCBI databases (reference numbers ZDB-GENE-040426-895 and NM_200920). The zebrafish PITPβ gene (pitpnb), and its inferred translation product, corresponds in sequence and genomic organization most closely to what we refer to as the mammalian “alternative spliceoform” – as opposed to the “canonical spliceoform” (18, 22). pitpnb resides on chromosome 10 in the genomic region spanning nucleotides 42,255,368 through 42,284,830, and undergoes the same exon-skipping splicing events (involving the last exon of the primary transcript) exhibited by the mammalian PITPβ transcript (18, 23). Thus, two pitpnb mRNA spliceoforms are expressed, and these are designated in a fashion consistent with the nomenclature of the mammalian spliceoforms (i.e. the pitpnb spliceoform is designated pitpnbi2 while the spliceoform that corresponds to the canonical mammalian PITPβ spliceoform is designated pitpnbi1).
Search of the ZFIN database with the rat PITPβ (RnPITPβ) primary sequence as query identified yet another previously unidentified PITPβ-like gene on chromosome 3 (nucleotides 13,898,735 through 13,918,905). The corresponding primary mRNA transcript consists of 11 exons and, when spliced, is predicted to encode a 271 amino acid protein. We designate this gene as pitpng (reference numbers: ZDB-GENE-040426-2791, NM_213443). The coding regions of pitpnbi1 and pitpng share 73% nucleotide sequence identity, and the inferred gene products share 78% identity at the primary sequence level (Fig. 1A).
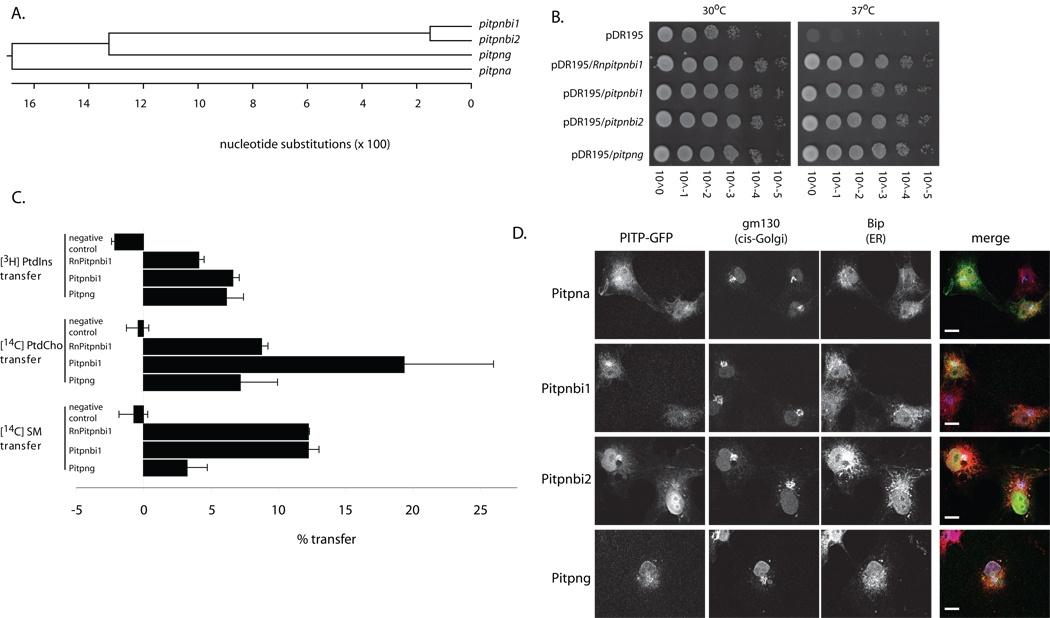
Fig. 1.
Pitpnb and Pitpng isoforms. (A) A ClustalW phylogenetic grouping of presumptive zebrafish Pitpna, Pitpnbi1, Pitpnbi2, and Pitpng is shown. (B) Zebrafish PITP cDNAs were subcloned into the multicopy yeast URA3 expression vector pDR195 where heterologous PITP gene expression is driven by a powerful constitutive promoter. Each individual expression construct was transformed into the yeast strain CTY-1-1A (ura3-52, sec14-1ts). Ten-fold dilution series were prepared from saturated liquid cultures normalized to similar cell densities and dilution spotted onto rich YPD solid media in parallel. One set of YPD plates was incubated for 48 hours at 30° (permissive for sec14-1ts) and the other set at 37°C (restrictive for sec14-1ts). (C) In vitro lipid transfer activities of presumptive zebrafish PITPβ-like proteins. Transfer assays score mobilization of radiolabeled PtdIns, PtdCho, or SM substrate between distinct membrane bilayer systems. Clarified cytosol prepared from yeast strain CTY303 (sec14Δ cki1) expressing each protein of interest served as protein source. Cytosol from CTY303 carrying an empty vector served as a negative control, while corresponding cytosol containing RnPITPβi1 was included as a positive control. The data shown is a single transfer assay, performed in triplicate that is representative of at least three independent experiments. The phospholipid-transfer substrate inputs for these assays were: 21,799 cpm [3H]PtdIns; 13,426 cpm [14C]PtdCho; 12,645 cpm [14C]SM;. Background values (buffer control) for these respective transfer assays were 1324, 1404 and 1038 cpm. (D) Localization of zebrafish PITPβ-like isoforms in mammalian cells. COS7 cells expressing GFP-tagged versions of zebrafish PITPs were fixed, permeabilized, and stained with antibodies that detect the cis-Golgi (gm130; blue) and the ER (BiP; red). Representative profiles for GFP expression or antibody staining are shown in the left panels, and the merged images are shown in the right panel. Scale bars -- 10 µm.
Zebrafish PITPβ-like proteins exhibit PtdIns/PtdCho/SM transfer activities in vitro
A heterologous phenotypic rescue assay was used to determine whether the zebrafish Pitpnb or Pitpng exhibited functional properties consistent with PITPs. This assay is based on previous demonstrations that ectopic expression of rodent PITPα or PITPβ rescues the growth and secretory defects associated with thermosensitive versions of Sec14 -- the major PITP of yeast (17, 24). To this end, a sec14-1ts mutant yeast strain was transformed with yeast episomal plasmids driving constitutive expression of either RnPITPβ (positive control), of Pitpnb spliceoforms 1 or 2, or Pitpng. The ability of these individual plasmids to rescue growth of the sec14-1ts mutant at restrictive temperatures (37°C) was then assessed. The positive control strain expressing the canonical rat PITPβ (RnPITPβ) grew well under these conditions, as expected (Fig. 1B). The sec14-1ts negative control strain carrying vector alone was inviable at 37°C as indicated by its failure to exhibit visible growth. Plasmids driving individual expression of Pitpnb isoforms 1 or 2, or Pitpng, also restored robust growth of the sec14-1ts mutant at 37°C (Fig. 1B).
We tested whether the zebrafish PITPβ-like proteins exhibited intrinsic phospholipid transfer activities with biochemical signatures of PITPβs. Pitpnb isoforms 1 and 2 and Pitpng were individually expressed in the cki1 sec14Δ yeast strain CTY303, a yeast strain that lacks measurable endogenous PtdIns- or PtdCho-transfer activity and permits facile detection of phospholipid transfer activities of heterologous PITPs (24–26). Cytosol was prepared from the appropriate cki1 sec14Δ derivative strains and assayed for PtdIns-, PtdCho- and SM-transfer activities.
Cytosol prepared from the CTY303 vector-only control strain failed to present any measurable PtdIns-, PtdCho- or SM-transfer activity, while lysates containing RnPITPβ (the positive control) exhibited robust transfer activities for all three phospholipid substrates (Fig. 1C). Incorporation of 2 mg of RnPITPβ cytosol into the assay resulted in the in vitro transfer of approximately 16%, 8% and 8% of the total input [3H]-PtdIns, [14C]-PtdCho and [14C]-SM from donor to acceptor membranes, respectively. Similarly, cytosol fractions containing Pitpnbi1 or Pitpnbi2 exhibited comparable transfer activity for all three phospholipid substrates relative to RnPITPβ control cytosol (Fig. 1C). Pitpng cytosol also presented robust PtdIns- and PtdCho-transfer activities, but SM-transfer activity was lower than activities measured for the RnPITPβ or for Pitpnbi1 and Pitpnbi2-containing cytosols.
Mammalian PITPα and PITPβs are further distinguished by their intracellular localization – mammalian PITPβ isoforms target to TGN membranes whereas PITPα localizes to nuclear/cytosolic compartments (18, 27, 28). The three zebrafish PITPβ-like proteins also exhibited PITPβ-like subcellular distributions, rather than PITPα-like distributions. Pitpnbi1, Pitpnbi2 and Pitpng chimeras bearing C-terminal GFP tags distributed to Golgi membranes when expressed in mammalian cells (Fig. 1D) -- as did the control rPITPβ-GFP reporter (data not shown). Pitpng-GFP co-localized with both Golgi and ER markers, while Pitpna exhibited a predominantly a cytosolic/nuclear distribution (Fig. 1D).
Zebrafish PITPβ isoforms stimulate phosphoinositide synthesis in yeast
We exploited the yeast system to assess whether Pitpnbi1, Pitpnbi2 and Pitpng can stimulate activity of a PtdIns 4-OH kinase in its native environment. Using an established strategy (11, 29), we took advantage of yeast ablated for Sac1 PtdIns-4-P phosphatase activity to this end. Yeast defective in Sac1 activity present a ca. 10-fold accumulation of PtdIns-4-P relative to isogenic wild-type strains (30, 31), and this accumulation is attributable to the activity of a particular PtdIns 4-OH kinase in yeast (Stt4; (32, 33). The magnitude of this accumulation is reduced by 50% upon inactivation of the major yeast PITP Sec14 (Fig. 2A,B; (11, 29, 31). Reconstitution of sec14-1ts sac1 yeast with Pitpnbi1, Pitpnbi2 or Pitpng rescued the depressed Stt4 activity of that strain (Fig. 2A,B).
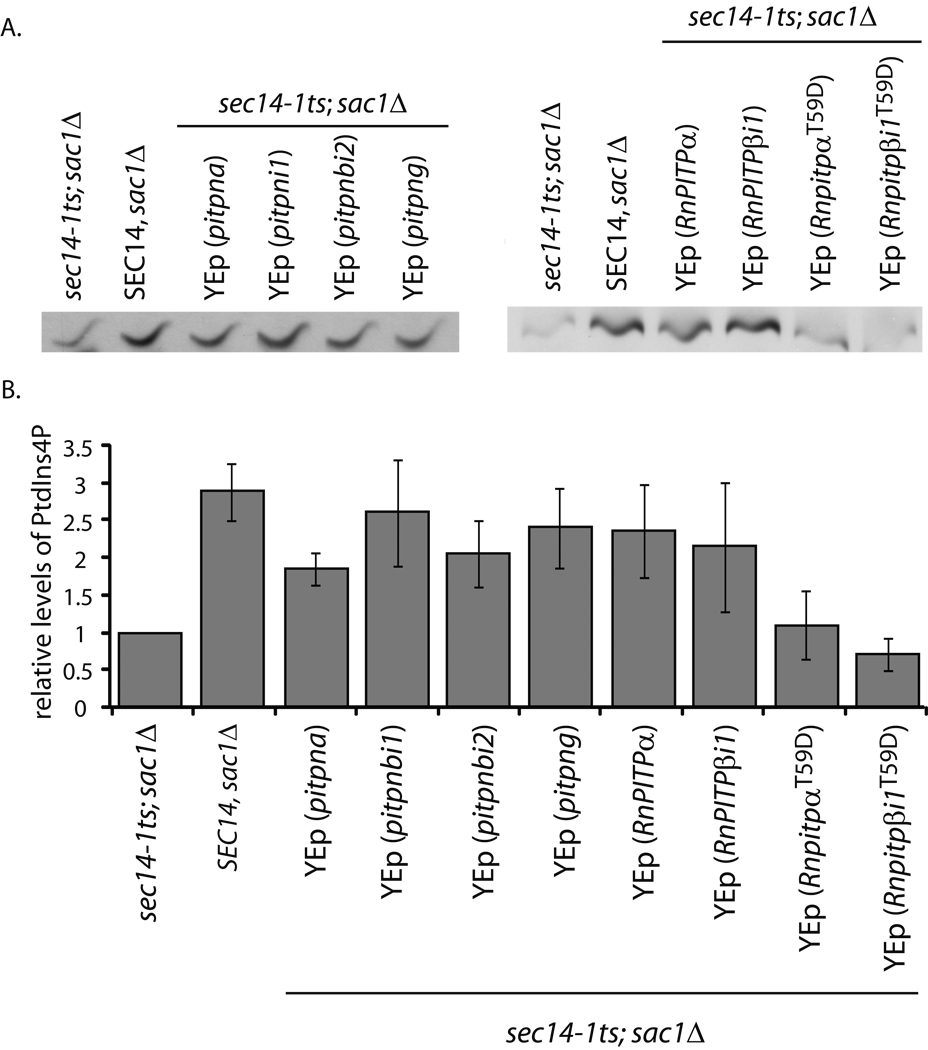
Fig. 2.
Zebrafish and mammalian class 1 PITPs stimulate phosphoinositide synthesis in yeast. (A) Yeast strain CTY100 (sec14-1ts sac1Δ; Cleves et al., 1989) carrying the YEp(URA3) parental expression plasmid pDR195, YEp(pitpna), YEp(pitpnbi1), YEp(pitpnbi2), YEp(pitpng), pDR195(RnPITPα), pDR195(RnPITPβi1), pDR195(RnPITPαT59D), and pDR195(RnPITPβi1T58D) were radiolabeled to steady-state with [3H]-inositol. Included was an isogenic SEC14 strain as positive control (CTY244; SEC14 sac1Δ; Cleves et al., 1989). After a 3 hour shift to 37°C, phospholipids were extracted, resolved by one-dimensional thin layer chromatography (Schaaf et al., 2008). The PtdIns-4-phosphate species are shown. (B) Quantification of PtdIns-4-phosphate. The PtdIns-4-phosphate band intensities as measured by densitometry were expressed as a ratio to PtdIns intensities for purposes of sample normalization, and the ratios are plotted as relative values. The PtdIns-4-phosphate/PtdIns ratio for the sec14-1ts sac1Δ (0.106496 – indicating a ca 10:1 molar ration of PtdIns to PtdIns-4-phosphate in this strain) was set to 1.0 on the relative scale. For comparison, a sec14-1ts strain exhibited a relative value of 0.20075 -- calculated from a PtdIns-4-phosphate/PtdIns ratio of 0.021379.
Similar results were obtained when these assays were performed in yeast sec14-1ts sac1 strains expressing mammalian PITPα and PITPβ. In both cases, reconstitution of a sec14-1ts sac1 strain with mammalian PITP resulted in a ca. 2.5-fold stimulation of Stt4 activity (Fig. 2A,B). These stimulatory effects required PtdIns-binding activity as the T→D missense substitutions for T59 and T58 of PITPα and PITPβ, respectively, completely inactivated these PITPs. PITPαT59D and PITPβT58D are each selectively inactivated for PtdIns binding/transfer without compromise of PtdCho- or, in the case of PITPβ, SM-binding/transfer activity (13, 34). Thus, vertebrate class 1 PITPs are able to stimulate activity of the yeast Stt4 PtdIns 4-OH kinase in its endogenous setting.
pitpnb and pitpng expression profiles
RT-PCR was used as readout for zebrafish PITPβ-like gene expression during development (i.e. at 6 and 24 hour hpf, at 6 dpf, and in adult fish). As shown in Suppl. Fig. S1A, pitpnb and pitpng transcripts were each detected at all four stages. Both pitpnbi1 and pitpng mRNAs were robustly expressed by 24 hpf and, in each case, the levels were maintained into adulthood. Relative to pitpnbi1, expression of pitpnbi2 exhibited a delayed time course -- robust levels were induced at some point between 24hpf and 3dpf. These data indicate that the pitpnb mRNA splicing program, which produces the mRNA for the canonical Pitpnbi1, predominates at the early stages of development. The exon-skipping splicing strategy that generates the mRNA for the alternative Pitpnbi2 spliceoform is engaged later. Whole-mount in situ hybridizations indicate both pitpnb and pitpng are ubiquitously expressed throughout the 28 hpf embryo, and pitpnb expression is particularly robust in the eye (Suppl. Fig. S1B). As to spliceoform specificity, both pitpnbi1 and pitpnbi2 are expressed in the eye, with pitpnbi2 exhibiting higher expression in that organ (Suppl. Fig. S1C). In the experiments described below, all manipulations affect both spliceoforms. As a result, we henceforth refer to these two proteins collectively as Pitpnb.
Characterization of a Pitpnb-specific polyclonal antibody
Polyclonal antibodies were raised against purified recombinant Pitpnbi2, and specificity of the affinity-purified antibodies was tested. First, immunoblotting experiments probing yeast lysates expressing Pitpnbi1, Pitpnbi2 or Pitpng demonstrated the Pitpnb polyclonal antibodies readily detected both Pitpnbi1 and Pitpnbi2, but detected Pitpng only very poorly (Suppl. Fig. S1D). Moreover, Pitpna did not immunoreact with the anti-Pitpnb serum at all. A pan-PITP antibody detected all isoforms in these same lysates (Suppl. Fig. S1D). Second, the immunopurified antibodies detected a protein with an apparent molecular mass of ca. 35 kDa in zebrafish retinal extracts (Suppl. Fig. S1D), an apparent mass consistent with the 32 kDa calculated from the Pitpnbi1 and Pitpnbi2 primary sequences. When zebrafish retinal extracts were probed with pan-PITP antibodies, a single 35 kDa immunoreactive species was detected (Suppl. Fig. S1D). These data demonstrate the affinity-purified Pitpnb antibodies detect Pitpnbi1/Pitpnbi2 without significant cross-reactivity with either Pitpng or Pitpna.
Pitpnb protein expression in adult zebrafish retina is restricted to cone photoreceptor cells
Like other vertebrates, the zebrafish retina is composed of six neuronal cell types whose cell bodies are arranged into three nuclear layers. These nuclear layers are clearly separated from each other by two sharply defined synaptic (plexiform) layers. The photoreceptors, which consist of rod and cone cells, synapse in the outer plexiform layer (OPL) with second order neurons of the inner nuclear layer (INL). Zebrafish possess three different cone cell types: the short single, long single and the double cone cells. Each double cone cell is comprised of a long and short member and, whilst these members are fused along most of their lengths, these maintain individual cell structures such as outer segments and synaptic pedicles. The rod outer segments (ROS) are located adjacent to the pigmented epithelium, while the various cone cell types reside between the elongated ROS and the more distally located rod nuclei of the outer nuclear layer (ONL). Each cone cell type assumes a signature morphology and location within the cone cell layer. Proceeding from the ONL towards the ROS layer, the short single cones, long single cones and double cone cells are each localized in distinct and identifiable rows.
Immunolocalization of Pitpnb in frozen sections of the adult zebrafish retina revealed expression is restricted to the photoreceptor layer with the most intense labeling localized to the OPL (Figs. 3A,B). To determine if the Pitpnb profile corresponded to rod or cone photoreceptors, rhodopsin expression in the ROS was dual-labeled with Pitpnb. The Pitpnb-positive cells (including their relatively short triangular outer segments) were positioned adjacent to the ROS layer (Fig. 3C). These data assign Pitpnb expression to cone photoreceptor cells. To determine whether the most intense Pitpnb staining in the OPL was most highly concentrated in the photoreceptor synaptic terminals, or in the proximal processes of INL cells (e.g. in bipolar or horizontal cells), frozen retinal sections were dual-labeled for Pitpnb and α-tubulin to define the structural organization of the OPL. This labeling clearly identified Pitpnb-positive processes as cone cell synaptic pedicles, rather than the relatively smaller rod spherules. These Pitpnb-labeled processes were intimately associated with the proximal α-tubulin-positive structures, which likely represent bipolar cell dendrites (Fig. 3D). Even at this resolution, invaginating α-tubulin-stained processes were occasionally observed to be partially surrounded by Pitpnb-stained pedicles (data not shown).
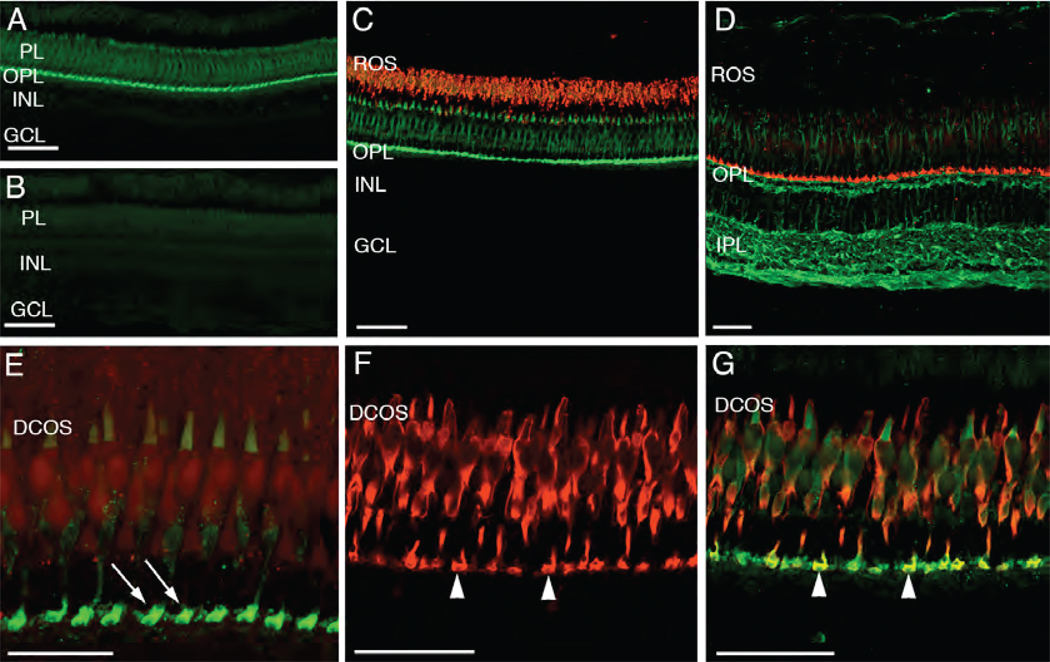
Fig. 3.
Pitpnb immunolocalization in the adult zebrafish retina. (A) Immunolocalization of Pitpnb. An adult retina cryosection labeled with the anti-Pitpnb serum revealed Pitpnb expression is confined to the photoreceptor layer (PL) and the outer plexiform layer (OPL). (B) A negative control section labeled with pre-immune serum shows no staining. (C) A double-label experiment. Retinal sections dual-labeled for Pitpnb (green; anti-DrPITPβ antibodies) and the zpr-3 marker (red; mAb zpr-3; revealed Pitpnb is not expressed in rods. (D) A dual-label image shows staining profiles for Pitpnb (red) and α-tubulin (green). (E) A high magnification image of a Pitpnb staining profile revealing the intense signal in the photoreceptor synaptic pedicles (arrows) in the OPL. (F) Expression of the zpr-1 staining profile (visualized via the mAb zpr-1 antibody) specifically labels double cone cells. (G) A dual-label image. The merged Pitpnb (green) and zpr-1 staining profiles (red) from frozen retinal sections are shown. Arrowheads in Panels F and G point to the double-labeled cone cell synaptic pedicles. Abbreviations: PL, photoreceptor layer; INL, inner nuclear layer; GCL, ganglion cell layer; ROS, rod outer segments; OPL, outer plexiform layer; IPL, inner plexiform layer; DCOS, double cone cell outer segments. Scale bars are 50 µm in all panels except Panel E (25 µm).
The cone photoreceptor localization of Pitpnb was examined at higher resolution (Fig. 3E). Similar to the dual-labeled retinas (Figs. 3C,D), the most intense Pitpnb labeling was associated with expansive photoreceptor terminals (Fig. 3E). Furthermore, the faint outer segment signal was restricted to a single row of cone cells, whose outer segments were located adjacent to the ROS layer. This arrangement is consistent with that of double cone cells. We failed to detect Pitpnb staining in either the long or the short single cone outer segments, although these cell structures were plainly visible within the plane of the sections. The small number of Pitpnb-labeled cone pedicles, relative to the total number of cone cells in this section, corresponded to the number of labeled outer segments.
Pitpnb protein is primarily expressed in double cone photoreceptor cells
To identify the Pitpnb-expressing cone cells, monoclonal antibody (mAb) zpr-1 was used in dual labeling experiments. The mAb zpr-1 specifically decorates an uncharacterized antigen associated with the plasma membrane of double cone cells (termed zpr-1 or Fret43), but not the other photoreceptor cell types (Fig. 3F; 35). Dual-labeling of frozen retinal sections with mAb zpr-1 and affinity-purified anti-Pitpnb serum yielded extensively overlapping signals that were most apparent in the region of the synaptic pedicles (Fig. 3G, arrowheads). We further examined this co-localization between zpr-1 and Pitpnb in retinal whole-mount preparations. At the edge of the retinal tissue, dissociated photoreceptors were observed in vertical profile (Fig. 4A–C). In every case that a double cone cell was identified by mAb zpr-1 staining (Fig. 4B), that cell also expressed Pitpnb (Fig. 4A,C).
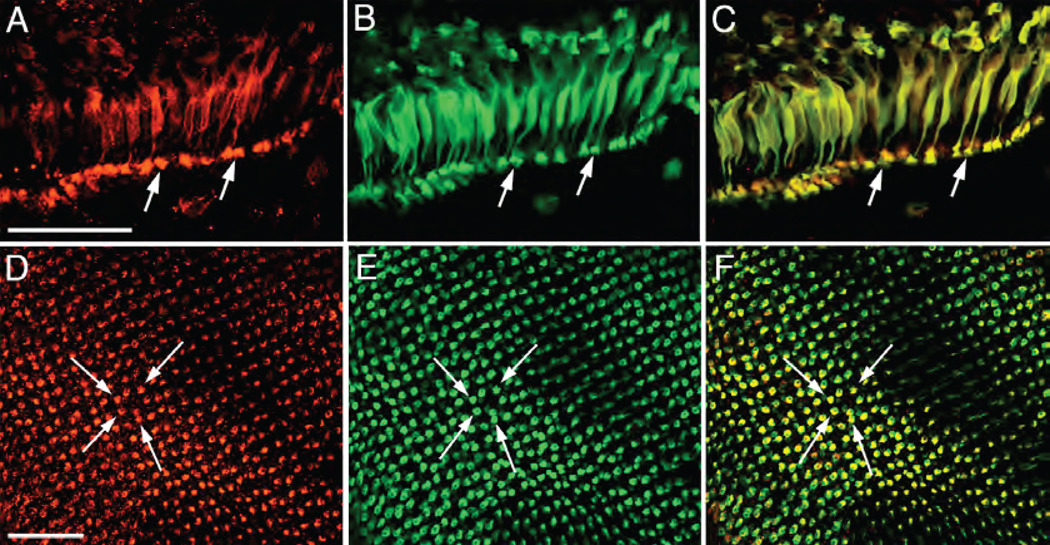
Fig. 4.
Pitpnb localizes to double cone cells. (A–C) Pitpnb and the double cone cell-specific zpr-1 antigen co-localize in the adult retina. Dissociated double cone cells co-labeled with anti- Pitpnb serum and mAb zpr-1. Panel A shows Pitpnb localization, Panel B the zpr-1 profile, and Panel C is a merged image. Arrows identify double cone cell synaptic pedicles. (D–F) Pitpnb antibodies and mAb zpr-1 co-label synaptic pedicles. The staining profiles in retinal wholemounts at the level of cone cell synaptic pedicles; Pitpnb profile (D), mAb zpr-1 antigen (E), and the merged images (F). Arrows highlight the same individual labeled cone pedicles in each panel. There is perfect correspondence between Pitpnb and zpr-1 signals at the level of double cone cell synaptic terminals. Scale bars -- 50 µm.
When viewed from a tangential perspective, the retinal wholemounts revealed that the zebrafish cone cells are uniquely arrayed in a crystalline-like mosaic across the entire retina. In this arrangement, the short and long single cones alternate within single rows. These rows alternate, in turn, with rows composed exclusively of double cone cells. The rod photoreceptors randomly fill in the spaces between the regularly arranged cone cells. Focusing at the level of the photoreceptor cell synaptic termini, zpr-1 and Pitpnb both reside in cells arranged in repeating rows (Fig. 4D–F). Merge of the two images reveals complete correspondence between zpr-1 and Pitpnb-containing cells (Fig. 4F). When viewed from this focal perspective, it was apparent that each labeled pedicle actually consisted of the fused pedicles of the two members of the double cone cell pair. Closer inspection revealed that, in some instances, the zpr-1 and Pitpnb profiles overlapped in the terminal of a single member of a double cone cell pair. These results confirm Pitpnb is primarily expressed in double cone cells, and is not highly expressed in short- or long- single cone cells. The data further suggest that, at least in some cases, Pitpnb expression is primarily restricted to one member of the double cone pair.
Pitpnb insufficiencies result in loss of double cone cell outer segments
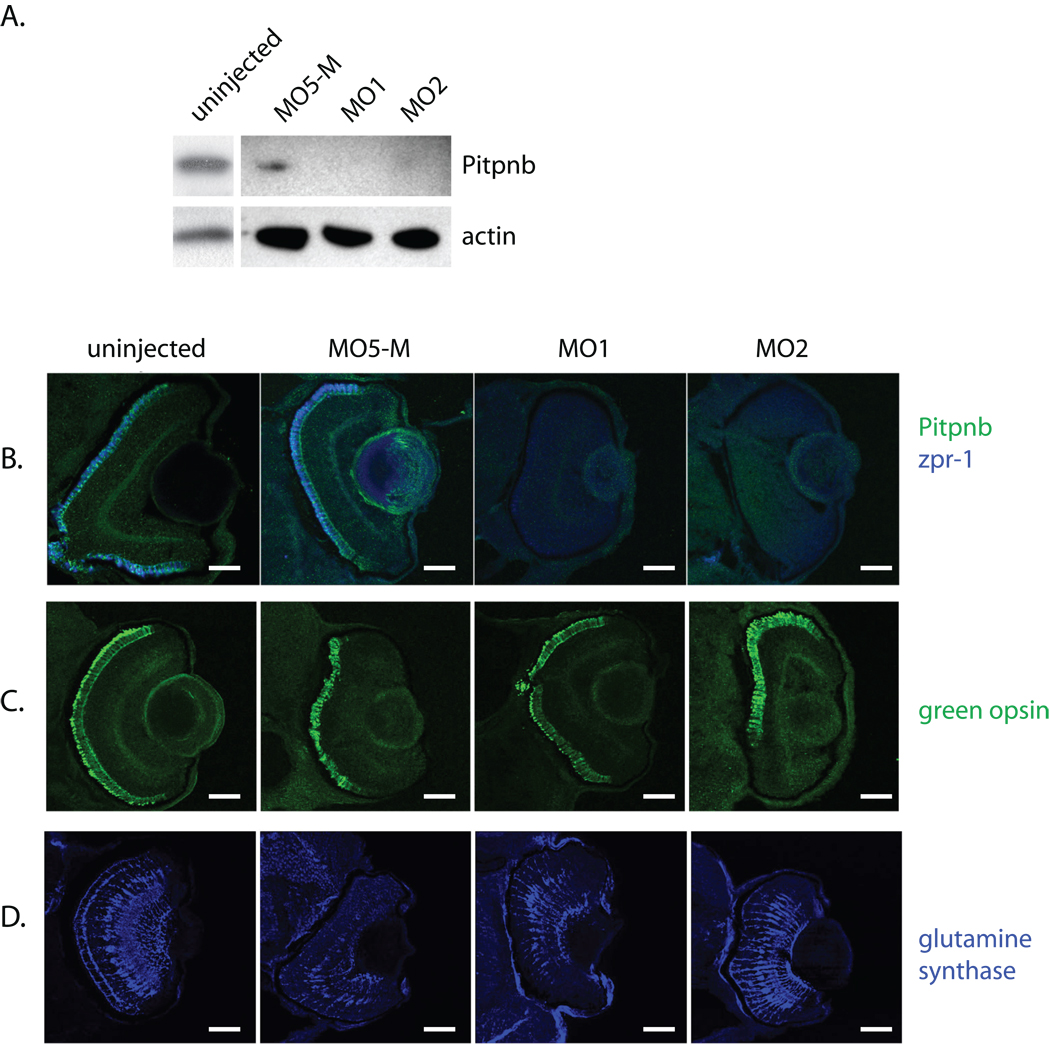
The consequences of reduced Pitpnb protein levels were determined using two different Pitpnb morpholinos (MO1 and MO2; see Materials and Methods). Efficacy of the knockdown was determined by injection of each morpholino at the 1–4 cell stage, preparation of lysates from 3 dpf larvae, and quantification of Pitpnb levels by SDS-PAGE and immunoblotting. Both MO1 and MO2 strongly reduced Pitpnb expression relative to uninjected controls (Fig. 5A). These reductions were not recorded when either of two independent negative control morpholinos were injected – the first a standard control morpholino that is not complementary to any known zebrafish sequence (GeneTools; Philomath, OR), and the second an MO1-like morpholino with 5 mismatches (MO5-M) (Fig. 5A). No differences were recorded in the viabilities of MO1 or MO2-injected morphants at 3 dpf relative to negative controls. There were also no dramatic developmental or morphological defects/delays in Pitpnb morphants relative to controls in that time window (data not shown). That MO1 and MO2 injections effectively compromised Pitpnb expression in retina was indicated by strong reductions in Pitpnb immunostaining in retinal cryosections harvested from 3dpf morphants (Fig. 5B). No derangements in the expression or organization of single cone cell and rod cell opsins (blue opsin and rhodopsin, respectively) were observed in morphant retina (Suppl Fig. S2).
Fig. 5.
Reduced Pitpnb expression results in loss of zpr-1-staining. A) Morpholinos (pitpnb-directed morpholinos 1 and 2, and a 5-base mismatch control version of βMO1, and the standard specific control morpholino) were injected into 1–4 cell stage embryos. Seventy eight hours after fertilization, embryos were disrupted, solubilized in sample buffer, proteins resolved by SDS-PAGE, and transferred to nitrocellulose. Blots probed with anti-Pitpnb immunoglobulin reveal that βMO1 and βMO2, but not the controls, depress Pitpnb protein levels. (B–D) Morphants were fixed at 3 dpf, embedded in OCT, and sectioned. Retinal sections containing retina were stained for (B) Pitpnb and zpr-1 antigen, (C) green opsin, or (D) glutamine synthase. Scale bars -- 100 µm.
Pitpnb insufficiencies were accompanied by strong reductions in staining of double cone cells by mAb zpr-1, however. The majority of Pitpnb morphant retinas presented faint or undetectable zpr-1 staining (Fig. 5B). With a minimum of 14 larvae analyzed for each morpholino injected, 60% of the uninjected and 72% of the MO5-M mismatch control retinas presented strong zpr-1 expression. By contrast, none of the MO1 or MO2 morphant retinas possessed strong zpr-1 staining, and only 21% of the MO1 and MO2 retinas showed moderate zpr-1 expression. Retinal cryosections were also probed for green opsin – another double cone cell marker. With at least 10 morphants examined, 82% and 90% of MO1 and MO2 injected larvae presented robust green opsin staining, respectively, compared to 93% and 81% of the uninjected and the 5-base mismatch control morphant larvae, respectively (Fig. 5C).
A decrement in one double cone cell marker, but not another, might reflect a developmental defect or delay. To address this possibility, the status of Müller glial cells was examined using glutamine synthase as marker. Müller cell development follows that of double cone cells, and these glial cells protect and nourish retinal cells (36). Glutamine synthase expression was unaffected in the either MO1- or MO2-derived Pitpnb morphants (90% and 100% present glutamine synthase staining, respectively) as indicated by comparisons of the morphant staining profiles to those presented by uninjected or MO5-M injected controls (83% and 100%, respectively) (Fig. 5D). These data indicate no general delay in retinal development under conditions of reduced Pitpnb expression.
Pitpnb-deficient double cone cells present outer segment defects
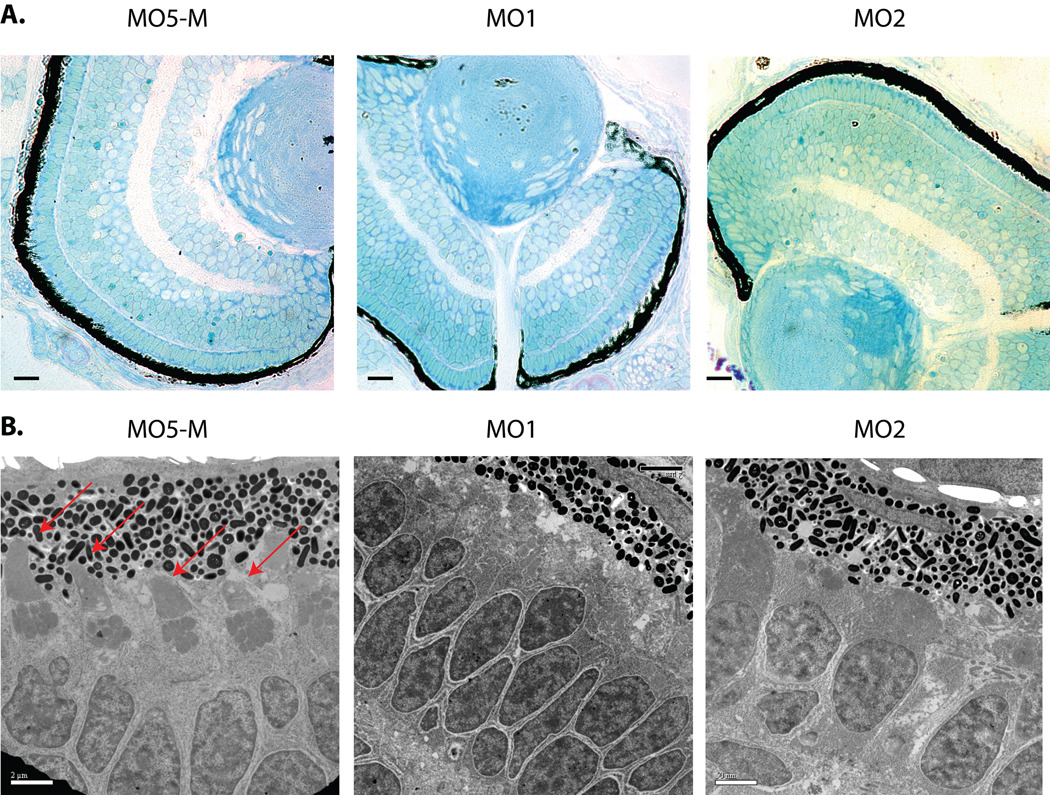
Histological analyses of Pitpnb and control morphant eyes reported no major anatomical abnormalities (6 larvae from each morphant or control class were analyzed). Since the majority of photoreceptors at 3 dpf are cone cells, significant double cone cell loss (by apoptotic or necrotic cell death) was expected to manifest itself in large reductions in numbers of nuclei or obvious signa of degenration. IMF staining of nuclei reported no significant difference in numbers of cone nuclei in Pitpnb morphants relative to controls, nor were structural abnormalities diagnostic of degeneration observed. Histological examinations did reveal a modest disorganization of the ONL in Pitpnb morphants compared to controls -- the distances between the RPE and the ONL exhibited greater variation in Pitpnb morphants compared to controls. The organization of ONL nuclei was also less regular (Fig. 6A) – suggesting subtle double cone cell structural defects.
Fig. 6.
Pitpnb morphants present double cone cell structural defects. (A) Five-base-mismatch control (MO5-M) and Pitpnb morphants (3dpf) were fixed and dehydrated. After embedding in Polybed 812 resin, 3µm sections were cut and stained with 1:1 methylene blue:Azure II. No gross structural defects are observed in morphant eye sections relative to control. Scale bars -- 35 µm. (B) Ultrathin sections were stained with uranyl acetate and lead citrate, and visualized by electron microscopy. Outer cone cells are clearly recognized in the control by their well-formed and distinctive outer-segments (indicated by red arrows). These structures are absent, or otherwise unrecognizable, in the morphant retinas. Scale bars -- 2 µm.
Ultra-thin-section electron microscopy (EM) was used to examine double cone cell morphology in more detail in Pitpnb morphants at 3 dpf. Multiple sections (>25) and at least four morphant and control larvae were examined in these analyses. The EM images consistently revealed a significant decrease in the numbers of properly assembled outer segments in Pitpnb morphants, as opposed to control embryos where these were easily identified (Fig. 6B). The signature triangular structure, a morphological hallmark for double cone cell outer segments, was observed in only 20% and 24% of MO1- and MO2-injected Pitpnb morphant sections, respectively (Fig. 6B). Because the relatively late onset of this phenotype precluded rescue experiments to validate the phenotype as specific consequence of Pitpnb insufficiencies, we also examined double cone cell morphologies as a function of challenge with MO5-M. Indeed, well-formed double cone cell outer segments were obvious in 96% of the thin sections derived from retinas of embryos injected with the MO5-M control morpholino (Fig. 6B). We further note that the double cone cell outer segment deficiencies in morphant larvae disappeared by 5 dpf, at which time morphant retinas were indistinguishable from controls and Pitpnb protein expression was restored to normal levels (data not shown). These data indicate that outer segment defects associated with Pitpnb insufficiencies are corrected upon Pitpnb resupply. Collectively, the EM and histology data suggest Pitpnb morphants produce properly fated double cone cells which present structurally compromised outer segments during the window of time that Pitpnb levels are below functional thresholds.
Impaired outer segment biogenesis/maintenance is expected to manifest itself in compromised phototransduction. To that end, the electrophysiological responses of morphant and control larvae to red light was measured. The technical demands of the experiment require use of 5 dpf larvae, however, and no significant differences in the photo-responses of morphant vs control retinas were recorded (Suppl. Fig. S3). As the outer segment derangements that accompany Pitpnb insufficiencies are no longer apparent at this stage, the data do not address functionality of the structurally impaired outer segments under conditions of Pitpnb deficit. These experiments do demonstrate, however, that the delay in biogenesis of morphologically normal outer segments in retina recovering from Pitpnb deficiencies does not compromise the ultimate production of functional outer segments.
zpr-1 antigen is retinal arrestin 3-like protein
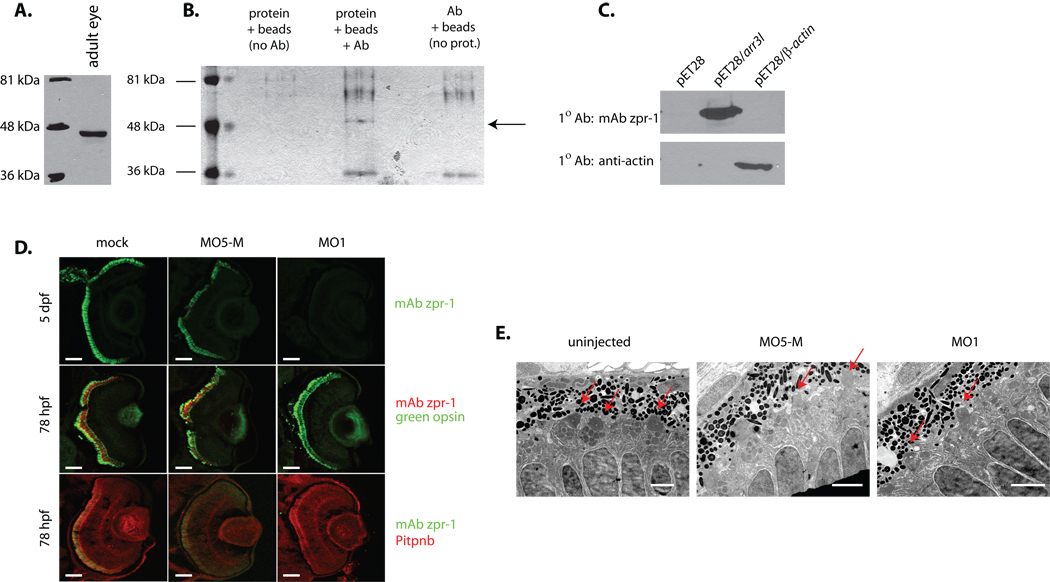
The specific loss of zpr-1 staining in Pitpnb morphant double cone cells is striking given there are no perturbations in expression of other photoreceptor cell markers. These marker analyses suggest some functional linkage between Pitpnb and zpr-1, thereby emphasizing the need to identify the uncharacterized zpr-1 antigen. To this end, mAb zpr-1 was used to immunopurify its cognate ligand from zebrafish retinal homogenate and to subsequently determine its precise identity. Both immunoblotting and immunoprecipitation experiments demonstrate mAb zpr-1 recognizes a 45 kDa protein in lysates of adult zebrafish retina (Fig. 7A,B). MALDI-TOF mass spectrometry analyses of tryptic peptides derived from purified antigen identified two candidate proteins – the 39.5 kDa Danio rerio arrestin 3-like (Arr3L) and the 41.5 kDa Danio rerio β-actin (Suppl. Table S2).
Fig. 7.
Identification of zpr-1 antigen as arrestin-3-like. (A) Lysate of adult zebrafish eye was fractionated by SDS-PAGE, the resolved species transferred to nitrocellulose, and zpr-1 antigen identified by immunoblotting with mAb zpr-1. (B) mAb zpr-1 antigen was immunoisolated from adult zebrafish eye lysates and resolved by SDS-PAGE. Two controls were included: sample without lysate, and sample without mAb zpr-1. The species indicated with an arrow was uniquely recovered from the complete incubation. (C) Recombinant zebrafish β-actin and Arr3L cross-reactivity with mAb zpr-1 or anti-actin immunoglobulin was tested by blotting -- as indicated. Naïve bacterial lystaes served as negative control. (D) Embryos (1–4 cell stage) injected with morpholino against Arr3L (MO1), or the corresponding 5-base Arr3L mismatch control morpholino (MO5-M), were examined at 3 dpf for Arr3L, green opsin, and Pitpnb staining. Uninjected (mock) served as additional control. Scale bars -- 100 µm. (E) Arr3L morpholino-injected (MO1) and uninjected control embryos were fixed and prepared for ultra-thin section electron microscopy. Sections from at least 3 fish were examined and representative images are shown. Outer segments are indicated with a red arrow. Scale bars -- 2 µm.
To distinguish between the two candidates, Arr3L and zebrafish β-actin polypeptides were individually produced in E. coli using a pET28 protein expression vector (see Materials and Methods). Immunoblotting experiments with mAb zpr-1 demonstrated an immunoreactive species of the expected molecular mass in bacterial lysates containing Arr3L (Fig. 7C). No mAb zpr-1 immunoreactive species were detected in lysates prepared from vector-only controls, nor from bacteria expressing zebrafish β-actin. In the latter case, anti-actin serum readily detected the expressed recombinant actin polypeptide. mAb zpr-1 also failed to detect the closely-related arrestin Arr3, the recombinant zebrafish Sag and Zgc:66109 retinal arrestins, or the ubiquitously expressed Arr2a and Arr2b arrestins (Suppl. Fig. S3).
Independent confirmation that zpr-1 antigen is Arr3L was obtained from experiments where Arr3L expression was specifically targeted by morpholino-mediated protein knockdown. Injection of embryos with an Arr3L-directed morpholino exerted no obvious developmental effects (data not shown), but eliminated mAb zpr-1 staining of double cone cells in morphant retina (Fig. 7D). This effect was not recapitulated in either uninjected embryos, or in embryos injected with the cognate 5-base mismatch control morpholino. Moreover, in contrast to the reciprocal case where Pitpnb morphants were strongly impaired for zpr1-immunostaining (see above), Arr3L morphant retinas supported robust expression of both green opsin and Pitpnb (Fig. 7D). Futhermore, Arr3L morphant retinas presented well-formed outer segments (Fig. 7E).
Pitpna morphants arrest early in development
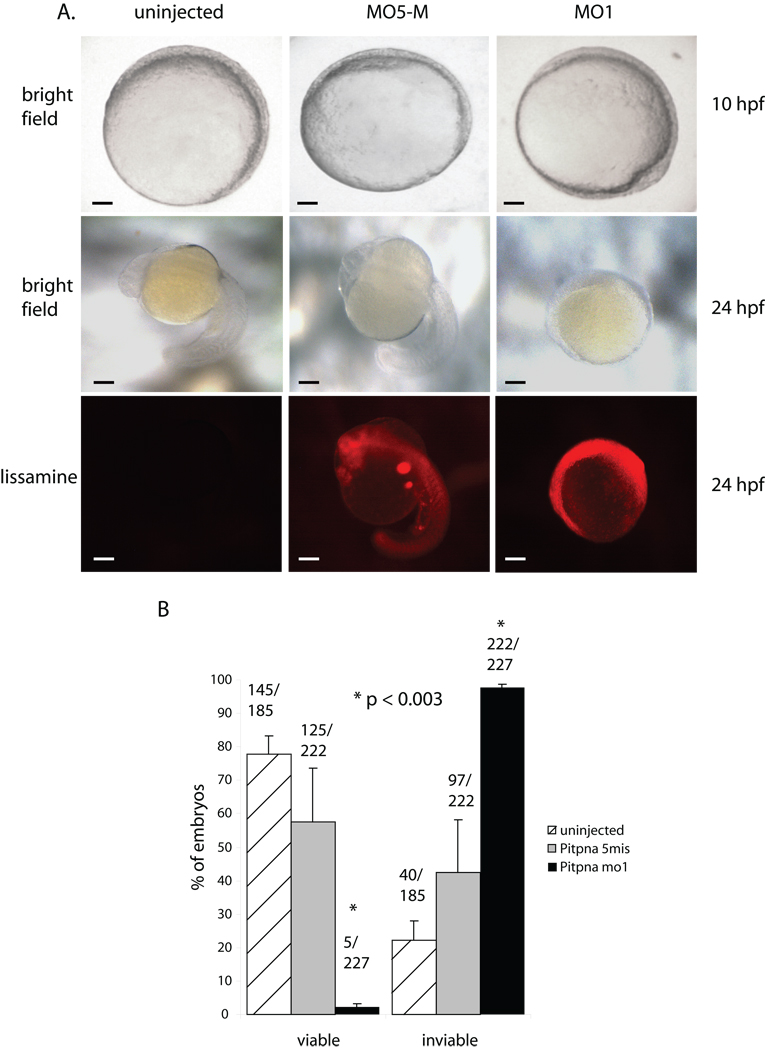
The highly tissue-specific developmental phenotypes associated with Pitpnb morphants raised the question of whether the Pitpng and Pitpna isoforms discharge more significant developmental functions in zebrafish. Embryos injected with morpholinos directed against pitpng mRNA failed to present significant increases in morbidity, or obvious morphological or developmental deficiencies (data not shown). The lack of a suitable Pitpng-specific antibody with which to independently assess Pitpng protein levels in the presumptive morphants precludes confident interpretation of these negative data. By contrast, morpholino-mediated knockdown of Pitpna expression levied an early developmental arrest in the interval between 80% epiboly through the bud stage (8–10 hpf) (Fig. 9B). Until the point of arrest, Pitpna morphants followed developmental trajectories that were indistinguishable from embryos injected with the 5-base mismatch control morpholino. Pitpna morphants remained in this terminal state of developmental arrest through 24 hpf without exhibiting the obvious signs of degeneration or structural decay normally associated with loss of viability. By 48 hpf, however, unmistakable signs of morbidity were on display in all morphants (data not shown).
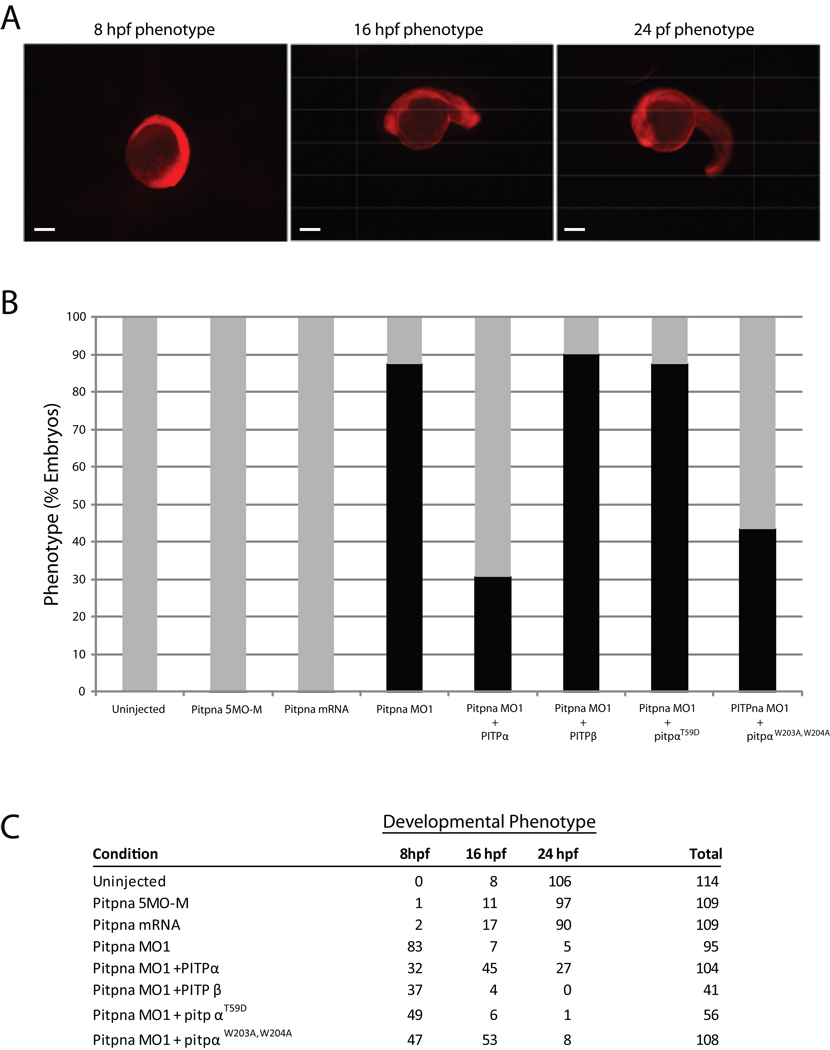
Fig. 9.
Rescue of Pitpna morphant arrest phenotype by rat PITPα. (A) Wild-type 24-,16- and 8 hpf embryos injected at the 1–4 cell stage with a lissamine-tagged Standard Control morpholino, which is not complementary to any known sequence in the zebrafish genome, are shown. The red fluorescence reveals the distribution of the morpholino throughout the embryo, and these developmental phenotypes provide the phenotypic definitions for scoring rescue. Scale bars – 270 µm. (B) Wild-type embryos were either uninjected or injected with an anti-pitpna morpholino containing 5-mismatched bases (Pitpna MO5-M), in vitro transcribed rat PITPα mRNA (PITPa RNA), anti-pitpna morpholino (Pitpna MO1), anti-pitpna morpholino and rat PITPα mRNA (Pitpna MO1 + PITPb RNA), anti-pitpna morpholino and rat PITPα mRNA (Pitpna MO1 + PITPa mRNA), anti-pitpna morpholino and rat pitpW203A, W204A RNA (Pitpna MO1 + PitpaW203A,W204A mRNA), and anti-pitpna morpholino and rat PitpT59D RNA (Pitpna MO1 + PitpaT59D RNA). Preparation of capped mRNAs is described in Suppl. Materials and Methods. Duplicate experiments were performed with each injection mixture and the data were combined from the two independent injection regimes. Gray bars represent the percentage of embryos that developed at least to the 16 hours hpf phenotype by 24 hours after each treatment. The black bars correspond to the percentage of embryos that arrested at 8 hpf developmental stage at 24 hours after each treatment. (C) The total results for each condition in (B), and from which Fig 9B was generated, are tabulated according to the developmental stage achieved by 24 hours after each treatment.
The early developmental arrest observed in the Pitpna morphant was amenable to rescue experiments. In these experiments, either rat PITPα or the canonical rat PITPβ isoform (i.e. the Pitpnbi paralog) were expressed as morpholino-resistant versions of class 1 PITPs in Pitpna morphant embryos. Rescue was scored by the significant increase in the frequency of embryos that progressed beyond the 8 hpf development stage to the 16- and 24 hpf developmental phenotypes (Fig. 9A). Mammalian PITPα expression supported significant rescue of the early developmental arrest characteristic of Pitpna morphants. Whereas 83/95 Pitpna morphants failed to pass the 8 hpf arrest phenotype, co-injection of rat PITPα mRNA permitted 74/104 morphants to develop beyond the 8 hpf arrest (Fig 9B,C). By this same assay, rat PITPβ mRNA co-injection was ineffective in rescue of the Pitpna terminal phenotype -- 37/41 morphants failed to develop past the 8-hpf arrest point (Fig. 9B,C). These functional rescue data not only validate the early developmental arrest as a genuine consequence of Pitpna insufficiencies, but also demonstrate the functional distinction between PITPα and PITPβ isoforms in vertebrate systems.
The rescue assay was also used to dissect the functional properties of PITPα protein required for biological activity in zebrafish. The rat PitpαT59D derivative, which is specifically defective for PtdIns-binding/transfer activity (34), was completely ineffective at rescuing the Pitpna morphant phenotypes (49/56 morphants failed to develop beyond the 8 hpf arrest; Fig 9B,C). Thus, as in the murine system (37), PtdIns-binding is an essential functional property of the zebrafish PITPα paralog. The efficacy of the rat PitpαW203A,W204A was also tested in the morphant rescue assay. This mutant protein is reported to be defective in membrane binding and PtdIns-transfer activity (22), while other studies find this protein competent for PtdIns-transfer activity in vitro, and heterologous rescue of yeast PITP insufficiencies in vivo (18). The former studies predict PitpαW203A,W204A is a nonfunctional protein in vivo (22), while the latter suggest the protein retains substantial biological activity (18). PitpαW203A,W204A expression rescued the early developmental arrest of Pitpna morphants with efficiencies similar to those recorded for ectopic rat PITPα expression; 76/108 morphants developed beyond the 8 hpf arrest stage to at least the 16 hpf developmental stage (Fig 9B,C).
Discussion
Herein, we report zebrafish express a set of mPITPs that recapitulates the cohort found in mammals – i.e. a Pitpna and two directly paralogous spliceoforms of Pitpnb. Zebrafish also express a unique Pitpnb isoform which we designate Pitpng. Morpholino-mediated protein knockdown experiments demonstrate that collective Pitpnb spliceoform function is required for double cone photoreceptor outer segment biogenesis/maintenance during development. The retinal-specific developmental defects levied by reduced Pitpnb function are in stark contrast to the consequences of diminished Pitpna function. Pitpna insufficiencies lead to early developmental failure. The collective data demonstrate Pitpnb and Pitpna isoforms discharge distinct developmental functions in zebrafish. While Pitpna is required for progression through early embryonic development, we propose Pitpnb supports the high capacity membrane trafficking pathway required for outer segment biogenesis and maintenance in double cone photoreceptor cells.
A battery of functional data identifies Pitpnbi1, Pitpnbi2 and Pitpng as PITPs of the β-isoform class. First, each individual protein exhibits unambiguous PtdIns-, PtdCho- and SM-transfer activities in vitro. Of the three, Pitpng harbors the least robust SM-transfer activity, but this activity is significant nonetheless. Second, heterologous expression of each individual protein in yeast with a functionally thermosensitive PITP (the sec14-1ts gene product) phenotypically rescues the conditional growth defects associated with the sec14-1ts lesion. Third, the phenotypic rescue of the sec14-1ts mutation by Pitpnbi1, Pitpnbi2 and Pitpng is accompanied by restoration of stimulated PtdIns-4-phosphate synthesis via the Stt4 PtdIns 4-OH kinase – thereby establishing these particular class 1 mPITPs as functional potentiators of PtdIns kinase activity. That PtdIns-binding is essential for vertebrate class 1 PITP α- and β- isoforms to stimulate PtdIns kinase is amply demonstrated by the inability of mutants specifically impaired for PtdIns-binding/transfer, to effect such a stimulation. While potentiation of PtdIns kinases is a consistent feature of PITPs, the heterologous context of the result has important implications for mechanisms of class 1 mPITP function (see below).
Pitpnb and Pitpng isoforms exhibit similar temporal expression profiles. In situ hybridization experiments, although reporting a ubiquitous spatial expression profile for the pitpnbs and pitpng, score particularly robust pitpnb expression in the developing eye. Restriction of Pitpnb expression to double cone cells, where the protein(s) distribute predominantly to the synaptic pedicles, is striking. Functional interference data are congruent with the Pitpnb expression profile. While Pitpnb morphants progress through larger developmental processes normally, morphologically recognizable double cone cells do not form in these animals. All available data suggest double cone cells are fated correctly in morphant retinas (green opsin, a specific marker for double cone cells, is produced), but these photoreceptor cells present structural derangements associated with impaired biogenesis/maintenance of outer segments. Regardless, the developmental retinal defects associated with Pitpnb insufficiencies are reversible. Pitpnb recovery reconstitutes the retina with morphologically normal and functional double cone cells.
Pitpnb morphants exhibit the unanticipated loss of immunoreactivity of double cone cells with the previously uncharacterized zpr-1 antigen – another specific marker for double cone cells. Herein, zpr-1 is identified as Arr3L, and the linkage between Pitpnb dysfunction, defective outer segment morphology in double cone photoreceptor cells, and altered status of a double cone cell-specific arrestin, is a compelling one. Transcription of the arr3L gene is not compromised in Pitpnb morphants (data not shown), and we believe it most likely that Arr3L protein is destabilized in Pitpnb-insufficient retina. The relationship between Pitpnb and Arr3L expression is not reciprocal. Arr3L morphants do not present loss of Pitpnb expression in the retina, nor do these result in structurally compromised double cone outer segments.
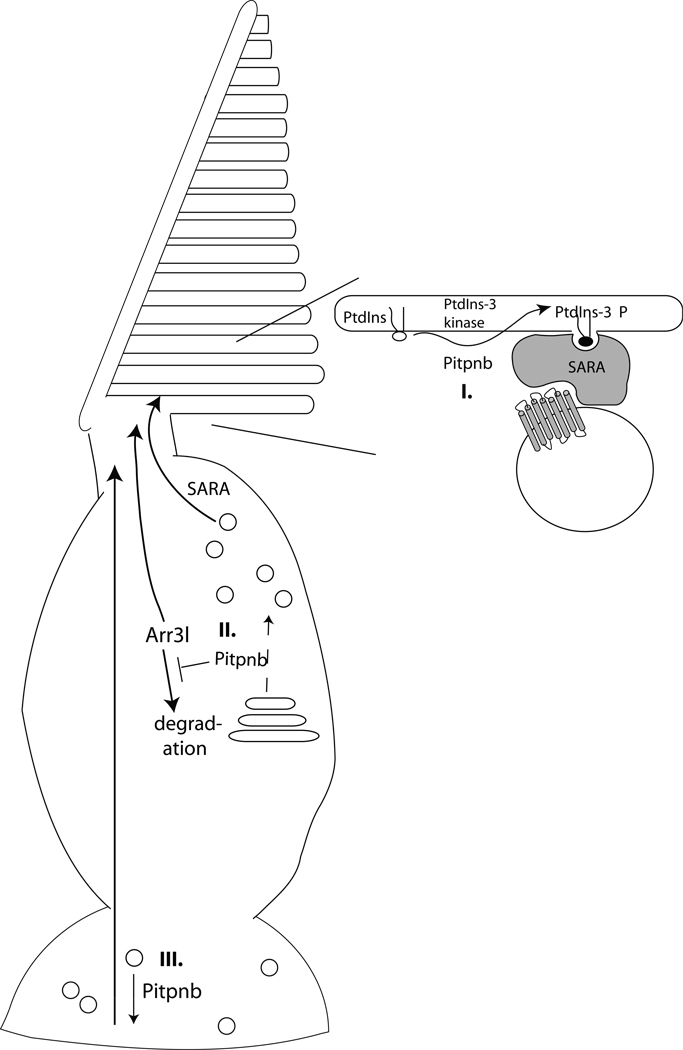
Vertebrate rod and cone cell outer segments are composed of an extensive assembly of intricate membraneous discs/lamellae subject to a vigorous course of continuous self-renewal (38, 39). As such, the outer segment establishment/maintenance program relies on a high capacity membrane trafficking system which renders it exquisitely sensitive to perturbations in membrane trafficking. Localization of mPITPβ isoforms to the trans-Golgi network, and the abilities of Pitpnb isoforms to localize to mammalian Golgi membranes in heterologous expression studies, suggests a potential mechanism for how diminution of Pitpnb activity evokes structural derangement of outer segments. Decreased expression of Pitpnb may subtly compromise Golgi function in double cone cells and evoke significant defects in opsin transport from that organelle. Reduced incorporation of the abundant opsin protein (the major protein expressed in photoreceptor cells) into the outer segment via biosynthetic trafficking will result in outer segment loss if membrane protein turnover in those structures is not sustained at normal rates (Fig. 10). In mammalian rod cells, the SARA protein responds to PtdIns-3-P cues via its FYVE domain, and cooperates with the syntaxin 3 t-SNARE, to drive the vesicle fusion required for outer segment membrane disc formation and maintenance (40). Given Pitpnb potentiates PtdIns kinase activities, one attractive scenario is Pitpnb supports an analogous PtdIns-3-P (or PtdIns-4-P)/SARA-dependent pathway in zebrafish double cone cells. We note an ORF annotated as encoding a zebrafish SARA is present in ZFIN and NCBI databases (ZFYVE9; ref. numbers ZDB-GENE-030131-1426 and CAN88231, respectively).
Fig. 10.
Pitpnb function in double cone cell outer segment biogenesis and maintenance. (I) Pitpnb cooperates with a PtdIns 3-OH kinase to promote PtdIns-3-phosphate production on the outer segment. PtdIns-3-phosphate serves as a landmark on the outer segment membrane that facilitates SARA-regulated docking and fusion of opsin-containing vesicles to the outer segment (acceptor compartment). (II) Pitpnb cooperates with what would likely be a PtdIns 4-OH kinase, at the level of the double cone cell Golgi complex, to promote formation of opsin-containing vesicles that will ultimately fuse to outer segment discs in a SARA-regulated manner. (III) Pitpnb regulates signaling in the synaptic pedicle and the robustness of this signaling is transduced from the synaptic pedicle to outer segments. In this model, Pitpnb may promote formation/fusion of synaptic vesicles and, in the absence of proper synaptic signaling, the outer segment of the cone cell is not maintained. In all three scenarios, Arr3L protein stability is suggested to be dependent on its association with functional outer segments.
While this model has its attractive features, it is not directly congruent with the primary localization of Pitpnb to the synaptic pedicle. The pool of Golgi membranes most proximal to the outer segment lies within the double cone cell inner segment. One possibility is a minor pool of Pitpnb does indeed reside in the inner segment, and is associated with Golgi membranes in that locale, or is transiently associated with the outer segments themselves, but that the steady-state Pitpnb distribution is more concentrated (and more easily detected) in the synaptic pedicle. An alternative possibility is Pitpnb is sequestered from the inner segment and plays no direct role in double cone Golgi function there. Rather, Pitpnb may indirectly coordinate lipid-derived signaling inputs in the synaptic pedicle with inner segment membrane dynamics (Fig. 10).
What level of functional redundancy exists between the various zebrafish PITP isoforms? From the perspective of Pitpng, no clear resolution to this question presents itself at this time. Pitpna and the Pitpnb spliceoforms are functionally distinct proteins, however, as evidenced by the clear phenotypes associated with compromised activities of each. The retinal phenotypes associated with morpholino-mediated Pitpnb protein knockdown are manifest without overt interference from Pitpna function. Reciprocally, interference with Pitpna activity results in an early and terminal developmental arrest in the face of Pitpnb and Pitpng expression. While the late-onset of Pitpnb knockdown phenotypes precluded rescue experiments, the early development failures induced by morpholino-mediated knockdown of Pitpna were amenable to such analyses. We find the Pitpna mutant phenotype is rescued by ectopic expression of mammalian PITPα, but not by mammalian PITPβ expression -- further demonstrating isoform functional specificity. Ectopic expression of the PtdIns-binding-defective mammalian PITPαT59D mutant similarly fails to rescue the early developmental failures associated with Pitpna defects; establishing that PtdIns-binding/exchange activity is an essential functional property of the zebrafish Pitpna – as is the case in the murine model (37). Interestingly, mammalian PITPαW203A,W204A scored as a functional protein as its ectopic expression also rescued early developmental failures in Pitpna morphants. PITPαW203A,W204A is deficient in what is claimed to be a membrane-binding motif essential for class 1 PITP phospholipid exchange activities (22), although other data argue against an essential role for this motif in the transient membrane binding events that accompany phospholipids exchange (18). The rescue experiments reported herein, in a fully vertebrate context, demonstrate Pitpna biological function is not strongly compromised by ablation of the Trp-Trp motif. These data support the view that the motif is not essential for phospholipid exchange by class1 PITPs.
We emphasize that the terminal phenotypes associated with class 1 mPITPα insufficiencies in zebrafish diverge from those documented in mice. In contrast to the early developmental failure observed in Pitpna morphants, the cognate mouse nullizygotes develop to birth, are born alive at Mendelian frequencies as anatomically correct neonates, but expire shortly thereafter (20). Thus, the results reported herein not only demonstrate that Pitpna and Pitpnb discharge different functions during zebrafish development, but also that vertebrates utilize diverse biological strategies for the employ of paralogous class1 PITPs.
Finally, whatever the biological coupling may be for any individual class 1 mPITP, it is a common theme that these proteins potentiate the activities of PtdIns kinases. Do class 1 mPITPs execute this stimulation as bona fide transfer proteins that execute PtdIns-supply (41,42), or do these proteins serve as nanoreactors in the fashion proposed for the Sec14-like PITPs (10)? It is presumed that class 1 mPITPs are carrier proteins that supply membranes engaged in PIP signaling with PtdIns so that the signaling circuit can be recharged (41,42). PtdIns supply models posit mPITP-dependent transport of PtdIns from its site of synthesis in the ER to signaling membranes, and that forward PtdIns flow is counterbalanced by back-transfer of PtdCho to close a vectorial phospholipid transfer loop. There is no evidence to support such a vectorial transfer pathway in vivo, however, and the model poses the additional difficulty that it is effectively impossible to disprove. One distinction between PtdIns-supply and nanoreactor models is that PtdIns surfeit is predicted to abolish requirements for mPITP-mediated PtdIns-supply mechanisms for stimulating PtdIns kinases. By contrast, nanoreactor mechanisms for potentiation of PtdIns-kinases should still be apparent under conditions of PtdIns excess. Our demonstration that both zebrafish and mammalian class 1 mPITPs enhance PtdIns kinase activity in the naturally PtdIns-rich yeast membranes (i.e. bulk yeast membrane PtdIns load is ca. 25% of total glycerophospholipid by mass; 43), in a manner that obligately requires PtdIns-binding, indicates class 1 PITPs can operate in a nanoreactor mode. Whether class 1 mPITPs do so in their native setting is more difficult to discern. Nevertheless, the principle is established. We interpret the fact that vertebrate mPITPs potentiate yeast PtdIns kinase activities, even though these proteins evolved separately from yeast PITPs and PtdIns-kinases, argues strongly against some privileged PITP-PtdIns kinase interaction requirement for productive stimulation of PIP synthesis.
Materials and Methods
Zebrafish
Zebrafish (strains AB and AB/Tu; University of Oregon) were raised in either the Center for Zebrafish Research in the Freimann Life Sciences Center at the University of Notre Dame, or the UNC Zebrafish Aquaculture Core Facility. Fish were fed brine shrimp and flake food 3X daily and maintained at 28.5°C with a light cycle of 14 hours light (250 lux):10 hours dark. All experiments were approved by the animal use committee at the University of Notre Dame and were in compliance with the provisions set forth by the Association for Research in Vision and Ophthalmology (ARVO) for the use of animals in vision research.
Zebrafish PITPβ and rat PITPβ yeast expression vectors
S. cerevisiae strains employed include CTY1-1A (MATa ura3-52 lys2-801 his3Δ200 sec14-1ts), CTY1079 (MATa ura3-52 lys2-801 his3Δ200 sec14-1ts spo14Δ), and CTY303 (MATa ura3-52 his3Δ200 sec14Δcki1::HIS3). Zebrafish cDNA was generated by reverse transcription (kit and protocol from Invitrogen, Carlsbad, CA) from RNA collected from 6 days past fertilization (dpf) zebrafish. pitpna, pitpnbi1, pitpnbi2, and pitpng cDNAs were amplified by PCR from total cDNA using primers clamped with XhoI and SacII restriction sites (Supplementary Table S1). Rat PITPβ was also amplified by PCR using oligonucleotides clamped with XhoI and SacII. The PCR products were verified by nucleotide sequence analysis and subcloned into the XhoI and SacII restriction sites of yeast episomal vector pDR195 to allow expression under control of the powerful and constitutive PMA promoter (41).
Yeast genetic and biochemical methods and phospholipid transfer assays
sec14-1ts complementation experiments, quantification of PtdIns-4-phosphate, and phospholipid transfer assays have been described elsewhere (25, 26, 29, 30, 31, 44). Details are provided in Supplementary Materials and Methods.
Heterologous expression in mammalian cells
Expression and immunofluorescence localization of Pitpnbi1, Pitpnbi2, or Pitpng in mammalian cells was performed as described (18).
Frozen section and whole-mount immunolocalization
Cryosections of adult albino zebrafish eye were prepared and stained as described (45–47). Specific details are provided in Supplementary Materials and Methods. For whole-mount labeling, embryos were fixed at 25°C for 20 min, washed in TBS, post-fixed in methanol (5 min, - 20°C), and washed in TBS. Tissue cryosections (12–16 µm) were blocked with TBS/5% normal goat serum/1% DMSO/0.1% Triton X-100 overnight at 4°C. Retinas and embryos were incubated with primary antibody overnight or for 48 hours, respectively. Antibody staining profiles were developed by washing in several changes of blocking buffer (4 hrs total), and incubation with secondary antibodies overnight at 4°C.
In situ hybridization
In situ hybridizations were performed using standard methods (http://zfin.org/zf_info/zfbook/chapt9/9.82.html) and 28 hpf embryos. Additional details are provided in Supplementary Materials and Methods.
Morpholino injections
Morpholinos designed for specific inhibition of pitpnx translation were from Gene Tools, LLC (Philomath, OR) and had the following sequences: Standard negative control, 5’ –CCTCTTACCTCAGTTAC AATTTATA– 3’; Pitpnb MO1, 5’-ACGATATTCCTTGA TGAGCACCATC– 3’; Pitpnb MO2, 5’-TTGATGAGCA CCATCTTCTTTCCAC –3’; and Pitpnb MO5-M, 5’-AC CATATTCGTTCATGACCACGATC– 3’; Pitpna MO1, 5’-CATGTTATCTCCTTTGCCGCCCCGT-3’; Pitpna 5O-5M, 5’- CATCTTATGTGCTTTCCCCCCC CGT-3’; Arr3L MO1, 5’ – TCTTCTTGTAAACTTTGTCAGCCAT – 3’; Arr3L 5O-5M, 5’- TCTTG TTCTAAAGTTTCTCAC CCAT-3’. All morpholinos were labeled with lissamine at their 3’ ends. Morpholinos were suspended in water to a final concentration of 0.5 mM, and ca. 5 nL was injected into the yolk of 1–4 cell embryos. At 24 hpf, embryos were examined for lissamine fluorescence as indicator of morpholino uptake. Embryos failing to present lissamine staining were discarded. Lissamine-positive embryos were incubated at 28°C until 78 hpf.
To rescue the Pitpnb morphant phenotype, developmental arrest at 8 hpf, embryos (1–4 cell stage) were coinjected with the standard amount of anti-pitpna morpholino and 600 ng of either: i) rat PITPβ RNA, ii) rat PITPα RNA, iii) rat PITPW203A, W204A RNA, or iv) rat PITPT59 RNA. After 24 hours, the embryos were scored for either the Pitpna developmental arrest phenotype or rescue of the morphant phenotype, classified as development through at least 16 hours of development. Control injections of only 600 ng of rat PITPα RNA at the 1–4 cell stage did not affect the normal development of embryos through 24 hours.
Zpr1 immunoblotting and immunoprecipitation
Details regarding immunoblotting and immunopurification procedures for zpr-1 antigen are provided in Supplementary Materials and Methods.
Thin section electron microscopy and histology
Three days after fertilization and morpholino injection, zebrafish larvae were fixed overnight at 4°C in 2% formaldehyde/2.5% glutaraldehyde/100 mM cacodylate, washed 3 times in 100 mM cacodylate and three times in water. Larvae were dehydrated in a 50% to 100% ethanol series, a wash in 1:1 xylene/EtOH, and incubation in 100% xylene. Larvae were embedded in a 1:1 xylene/polybed 812, 1:2 xylene/polybed 812, and 100% polybed 812 series. The polybed 812 embedded samples were baked in molds for two days at 60°C. Thin sections were stained with alcoholic uranyl acetate and Reynold’s lead citrate, and visualized at 80kV on an FEI Tecnai 12 electron microscope. Images were captured using Gatan micrograph 3.9.3 software. For histological analyses, 3 µM sections were cut and stained with 1:1 methylene blue:Azure II. Adult eyes were fixed in 9:1 ethanolic formaldehyde overnight at 4°C. All eyes were cryoprotected in 5% sucrose/1X PBS overnight at 4°C, in 30% sucrose/1X PBS overnight at 4°C, and finally in 30% sucrose/1X PBS:Tissue Freezing Media (Triangle Biomedical Sciences, Durham, NC) (1:1) for 4 hours at room temperature. Tissue was frozen embedded in 100% TFM and sectioned at 18 µm. Slides were dried at 50°C for 2 hours. Tissue sections were rehydrated with 1X PBS, blocked with 1X PBS/5% normal goat serum/0.3% Triton X-100/1% dimethylsulfoxide at room temperature for 1 hour, and incubated overnight at 4°C with one of the following primary sera: 1:5000 dilution of rabbit anti-rod opsin polyclonal antiserum, 1:500 dilution of rabbit anti-green opsin polyclonal antiserum, 1:500 dilution of rabbit anti-blue opsin antibody, 1:1000 dilution of mouse anti-glutamine synthetase monoclonal antibody (Chemicon Corp., Temecula, CA), or 1:500 dilution of monoclonal antibody zpr-1. Sections were subsequently washed three times in 1X PBS/0.1% Tween-20. Goat anti-rabbit or goat anti-mouse Alexa Fluor 488- or 594-conjugated secondary antibodies (Molecular Probes, Carlsbad, CA) were diluted 1:500 in the blocking buffer and incubated on the tissue sections for 1 hour at room temperature. Sections were washed three times in 1X PBS/0.1% Tween-20 and mounted using Vectashield (Vector Laboratories, Burlingham, CA). 10-micron Z-stacked confocal images from the central-dorsal retinal sections were obtained on a Biorad 1024 confocal microscope.
Supplementary Material
Supplementary Fig. S1. Temporal and spatial expression of Pitpnb and Pitpng. (A) Pitpnb and Pitpng are expressed at all ages tested. RNA was collected from zebrafish at 6 and 24 hours past fertilization (hpf), 6 days post-fertilization (dpf), and from adult fish. The RNA was reverse transcribed, and the respective cDNAs (template) amplified using primers directed against Pitpnbi1, Pitpnbi1 and Pitpng sequences. The amplified cDNAs reflect relative mRNA abundance in each sample, and amplification of products was both dependent on template and reverse transcriptase (RT). (B) In situ staining of pitpnb and pitpng expression profiles. In situ hybridization analyses were performed on 28 hpf fish using specific antisense probes for Pitpnb and Pitpng mRNAs, respectively. Sense probes were used as negative controls, as were “mock” controls where probe was omitted. Both pitpnb and pitpng were expressed in a wide range of tissues throughout the larva, and Pitpnb is most highly expressed in the eye. Scale bars -- 93 µm. (C) All PITP isoforms are expressed in the adult zebrafish eye. Total RNA was extracted from an adult zebrafish eye, and reverse transcribed into cDNA. PCR amplification was performed with primers specific for zebrafish pitpna, pitpnbi1, pitpnbi2 or pitpng sequences. (D) Characterization of Pitpnb antibody. Individual DrPITPs were expressed in yeast, lysates were prepared, resolved by SDS-PAGE, transferred to nitrocellulose, and probed with either anti-Pitpnb serum (top panel) or a control pan-PITP antibody (bottom panel) as indicated. To detect total PITPs in the eye, a blot with the same samples was probed with the NT1/3 pan PITP antibody. Protein extracts from zebrafish (Dr) or murine (Mm) eyes were also probed with these respective antibodoes. This blot indicates that at least 2 distinct PITP species are expressed in the zebrafish eye.
Supplementary Fig. S2. Blue opsin and rhodopsin expression are unaffected in Pitpnb morphants. Zebrafish embryos injected with negative control morpholino or pitpnb MO1 were fixed, embedded, and cryosectioned at 3 dpf. Sections through the center of the retina were stained for zpr-1 and the rod cell opsin (rhodopsin) (left panels), or the single cone cell marker blue opsin (right panel). Blue opsin and rhodopsin staining are present in both control and morphant eye sections, while mAb zpr-1-staining is seen only in the control. Scale bars -- 28 µm.
Supplementary Fig. S3. Pitpnb morphants display no defects in electroretinograms (ERGs) measuring their response to light. Zebrafish embryos were injected at the 1–4 cell stage with 5 base-mismatch control (5MIS) or pitpnb morpholinos (morpholino 1 and 2 represent MO1 and MO2, respectively), and an uninjected control was also included. ERGs were recorded at 5 dpf as previously described (Wong KY, Gray J, Hayward CJ, Adolph AR, Dowling JE. Zebrafish. 2004;1(2):121–31.). Following a flash of either white, blue or red light, morphants and controls showed transient corneal changes in potential known as a- wave and b-wave responses. No significant differences in kinetics or amplitude were noted between the averaged responses from different experimental groups. Data were collected from 2 – 5 larvae at each condition using three different sets of injected animals. Representative recordings using red light as the stimulus are presented.
Supplementary Fig. S4. mAb zpr-1 only detects Arr3L, and not other retinal and ubiquitously expressed arrestins. The open reading frame of arr3L and five additional zebrafish arrestins were PCR amplified from an adult zebrafish cDNA library with primers also encoding an HA epitope-tag. The PCR products were subcloned into a yeast expression vector (pDR195) and transformed into yeast. Lysates were prepared from the resulting yeast strains, lysate proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed with mAb zpr-1 and anti-HA. Though all of the arrestins were expressed (as detected by the HA antibody), only Arr3L was detected by mAb zpr-1.
Supplemental Table S1 – Primers used to amplify zebrafish PITP structural genes.
Supplemental Table S2 – MALDI-TOF mass spectrometry of mAb zpr-1 ligand. The gel-purified polypeptide species specifically precipitated from the adult zebrafish eye by mAb zpr-1 (Fig. 7B) was submitted for analysis by mass spectrometry. Two proteins were identified from tryptic digests of the submitted sample, and the summary analytical report is shown.
Fig. 8.
Pitpna morphants fail at an early stage of development. (A) Zebrafish embryos were either injected with pitpna-directed (pitpna MO1) or the corresponding 5-base mismatch control (pitpna MO5-M) morpholinos at the 1–4 cell stage, and brightfield images were taken at the indicated time (hpf). At 24 hpf, images were also taken in the red fluorescence channel to identify embryos containing the lissamine-tagged morpholino. Top row: Scale bars -- 140 µm. Middle and bottom rows: Scale bars -- 210 µm. (B) At 48 hours post-fertilization, control and morphant embryos were classified as viable or inviable based on visual inspection. Data are from three independent experiments, and p-values are indicated.
Acknowledgements
VAB dedicates this paper to the memory of Dennis Shields, a great colleague whose enthusiasm and love for science will be missed by all who had the privilege of knowing him. We thank Beth Morin-Kensicki (UNC) for helpful discussion, and Hal Mekeel for assistance with EM. We thank George Helmkamp Jr. and Lynn Yarbrough (Univ. Kansas Med. Ctr.) for pan-PITP antibody. This work was supported by grants NIH R01NS37723 and NIH R01GM44530 awarded to VAB. SK, TV and DRH were supported by the Univ. of Notre Dame Center for Zebrafish Research. SEB and GWS were supported by NIH R01EY015165. Oligonucleotide synthesis and DNA sequence analyses were performed via the UNC Lineberger Comprehensive Cancer Center Genome Analysis and Nucleic Acids Core Facility. Mass spectrometry was performed at the UNC Proteomics Facility.
References
- 1.Berridge MJ, Irvine RF. Inositol phosphates and cell signalling. Nature. 1989;341:197–205. doi: 10.1038/341197a0. [DOI] [PubMed] [Google Scholar]
- 2.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 3.Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majerus PW. Inositol phosphate biochemistry. Annu Rev. Biochem. 1992;61:225–250. doi: 10.1146/annurev.bi.61.070192.001301. [DOI] [PubMed] [Google Scholar]
- 5.Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;9:484–496. [PubMed] [Google Scholar]
- 6.Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 8.Hurley JH, Meyer T. Subcellular targeting by membrane lipids. Curr Op Cell Biol. 2001;13:146–152. doi: 10.1016/s0955-0674(00)00191-5. [DOI] [PubMed] [Google Scholar]
- 9.Irvine RF, Schell MJ. Back in the water: the return of the inositol phosphates. Nat Rev Cell Mol Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- 10.Ile KE, Schaaf G, Bankaitis VA. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat Chem Biol. 2006;2:576–583. doi: 10.1038/nchembio835. [DOI] [PubMed] [Google Scholar]
- 11.Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, Bankaitis VA. Functional anatomy of phospholipids binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bankaitis VA, Mousley CJ, Schaaf G. Sec14-superfamily proteins and the crosstalk between lipid signaling and membrane trafficking. Trends Bioch. Sci. 2010;35:150–160. doi: 10.1016/j.tibs.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips SE, Vincent P, Rizzieri KE, Schaaf G, Bankaitis VA, Gaucher EA. The diverse biological functions of phosphatidylinositol transfer proteins in eukaryotes. 2006;41:21–49. doi: 10.1080/10409230500519573. [DOI] [PubMed] [Google Scholar]
- 14.Swigart P, Insall R, Wilkins A, Cockcroft S. Purification and cloning of phosphatidylinositol transfer proteins from Dictyostelium discoideum: homologues of both mammalian PITPs and Saccharomyces cerevisiae sec14p are found in the same cell. 2000;347(Pt 3):837–843. [PMC free article] [PubMed] [Google Scholar]
- 15.Dickeson SK, Lim CN, Schuyler GT, Dalton TP, Helmkamp GM, Jr., Yarbrough LR. Isolation and sequence of cDNA clones encoding rat phosphatidylinositol transfer protein. J. Biol. Chem. 1989;264:16557–16564. [PubMed] [Google Scholar]
- 16.Fullwood Y, dos Santos M, Hsuan JJ. Cloning and characterization of a novel human phosphatidylinositol transfer protein, rdgBbeta. J. Biol. Chem. 1999;274:31553–31558. doi: 10.1074/jbc.274.44.31553. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka S, Hosaka K. Cloning of a cDNA encoding a second phosphatidylinositol transfer protein of rat brain by complementation of the yeast sec14 mutation. J Biochem. 1994;115:981–984. doi: 10.1093/oxfordjournals.jbchem.a124448. [DOI] [PubMed] [Google Scholar]
- 18.Phillips SE, Ile KE, Boukhelifa M, Huijbregts RP, Bankaitis VA. Specific and nonspecific membrane-binding determinants cooperate in targeting phosphatidylinositol transfer protein beta-isoform to the mammalian trans-Golgi network. 2006;17:2498–2512. doi: 10.1091/mbc.E06-01-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C, Peng YW, Shang J, Pawlyk BS, Yu F, Li T. The mammalian retinal degeneration B2 gene is not required for photoreceptor function and survival. Neuroscience. 2001;107:35–41. doi: 10.1016/s0306-4522(01)00337-2. [DOI] [PubMed] [Google Scholar]
- 20.Alb JG, Jr., Cortese JD, Phillips SE, Albin RL, Nagy TR, Hamilton BA, Bankaitis VA. Mice lacking phosphatidylinositol transfer protein-alpha exhibit spinocerebellar degeneration, intestinal and hepatic steatosis, and hypoglycemia. J. Biol. Chem. 2003;278:33501–33518. doi: 10.1074/jbc.M303591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Y, Ding YQ, Hong Y, Feng Z, Navarre S, Xi CX, Zhu XJ, Wang CL, Ackerman SL, Kozlowski D, Mei L, Xiong WC. Phosphatidylinositol transfer protein-alpha in netrin-1-induced PLC signalling and neurite outgrowth. Nat Cell Biol. 2005;7:1124–1132. doi: 10.1038/ncb1321. [DOI] [PubMed] [Google Scholar]
- 22.Tilley SJ, Skippen A, Murray-Rust J, Swigart PM, Stewart A, Morgan CP, Cockcroft S, McDonald NQ. Structure-function analysis of phosphatidylinositol transfer protein alpha bound to human phosphatidylinositol. Structure. 2004;12:317–326. doi: 10.1016/j.str.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Morgan CP, Allen-Baume V, Radulovic M, Li M, Skippen A, Cockcroft S. Differential expression of a C-terminal splice variant of phosphatidylinositol transfer protein beta lacking the constitutive-phosphorylated Ser262 that localizes to the Golgi compartment. Biochem J. 2006;398:411–421. doi: 10.1042/BJ20060420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skinner HB, Alb JG, Jr., Whitters EA, Helmkamp GM, Jr., Bankaitis VA. Phospholipid transfer activity is relevant to but not sufficient for the essential function of the yeast SEC14 gene product. EMBO J. 1993;12:4775–4784. doi: 10.1002/j.1460-2075.1993.tb06166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Nature. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Routt SM, Xie Z, Cui X, Fang M, Kearns MA, Bard M, Kirsch DR, Bankaitis VA. Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol. Biol. Cell. 2000;11:1989–2005. doi: 10.1091/mbc.11.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Vries KJ, Heinrichs AA, Cunningham E, Brunink F, Westerman J, Somerharju PJ, Cockcroft S, Wirtz KW, Snoek GT. An isoform of the phosphatidylinositol-transfer protein transfers sphingomyelin and is associated with the Golgi system. Biochem J. 1995;310:643–649. doi: 10.1042/bj3100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Vries KJ, Westerman J, Bastiaens PI, Jovin TM, Wirtz KW, Snoek GT. Fluorescently labeled phosphatidylinositol transfer protein isoforms (alpha and beta), microinjected into fetal bovine heart endothelial cells, are targeted to distinct intracellular sites. Exp Cell Res. 1996;227:33–39. doi: 10.1006/excr.1996.0246. [DOI] [PubMed] [Google Scholar]
- 29.Phillips SE, Sha B, Topalof L, Xie Z, Alb JG, Klenchin VA, Swigart P, Cockcroft S, Martin TF, Luo M, Bankaitis VA. Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol. Cell. 1999;4:187–197. doi: 10.1016/s1097-2765(00)80366-4. [DOI] [PubMed] [Google Scholar]
- 30.Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 31.Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to "bypass Sec14p" and inositol auxotrophy. Mol. Biol. Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemoto Y, Kearns BG, Wenk MR, Chen H, Mori K, Alb JG, Jr., De Camilli P, Bankaitis VA. Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J. Biol. Chem. 2000;275:34293–34305. doi: 10.1074/jbc.M003923200. [DOI] [PubMed] [Google Scholar]
- 33.Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alb JG, Jr., Gedvilaite A, Cartee RT, Skinner HB, Bankaitis VA. Mutant rat phosphatidylinositol/phosphatidylcholine transfer proteins specifically defective in phosphatidylinositol transfer: implications for the regulation of phospholipid transfer activity. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8826–8830. doi: 10.1073/pnas.92.19.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larison KD, Bremiller R. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development. 1990;109:567–576. doi: 10.1242/dev.109.3.567. [DOI] [PubMed] [Google Scholar]
- 36.Bringmann A, Pannicke T, Grosche J, Francke M, Wiedemann P, Skatchkov SN, Osborne NN, Reichenbach A. Muller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006;25:397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Alb JG, Jr., Phillips SE, Wilfley LR, Philpot BD, Bankaitis VA. The pathologies associated with functional titration of phosphatidylinositol transfer protein α activity in mice. J. Lipid Res. 2007;48:1857–1872. doi: 10.1194/jlr.M700145-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Bok D, Young RW. The renewal of diffusely distributed protein in the outer segments of rods and cones. Vision Res. 1972;12:161–168. doi: 10.1016/0042-6989(72)90108-3. [DOI] [PubMed] [Google Scholar]
- 39.Young RW. Biogenesis and renewal of visual cell outer segment membranes. Exp Eye Res. 1974;18:215–223. doi: 10.1016/0014-4835(74)90150-x. [DOI] [PubMed] [Google Scholar]
- 40.Chuang JZ, Zhao Y, Sung CH. SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell. 2007;130:535–547. doi: 10.1016/j.cell.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wirtz KWA. Phospholipid transfer proteins revisited. Biochem. J. 1991;324:353–360. doi: 10.1042/bj3240353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shadan S, Holic R, Carvou N, Ee P, Li M, Murray-Rust J, Cockcroft S. Dynamics of lipid transfer by phosphatidylinositol transfer proteins in cells. Traffic. 2008;10:1743–1756. doi: 10.1111/j.1600-0854.2008.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGee TP, Skinner HB, Whitters EA, Henry SA, Bankaitis VA. A phosphatidylinositol transfer protein controls the phosphatidylcholine content of yeast Golgi membranes. J. Cell Biol. 1994;124:273–287. doi: 10.1083/jcb.124.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB. NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett. 1995;370:264–268. doi: 10.1016/0014-5793(95)00853-2. [DOI] [PubMed] [Google Scholar]
- 45.Vihtelic TS, Doro CJ, Hyde DR. Cloning and characterization of six zebrafish photoreceptor opsin cDNAs and immunolocalization of their corresponding proteins. Vis Neurosci. 1999;16:571–585. doi: 10.1017/s0952523899163168. [DOI] [PubMed] [Google Scholar]
- 46.Barthel LK, Raymond PA. Improved method for obtaining 3-microns cryosections for immunocytochemistry. J Histochem Cytochem. 1990;38:1383–1388. doi: 10.1177/38.9.2201738. [DOI] [PubMed] [Google Scholar]
- 47.Vihtelic TS, Goebl M, Milligan S, O'Tousa JE, Hyde DR. Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J. Cell Biol. 1993;122:1013–1022. doi: 10.1083/jcb.122.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Temporal and spatial expression of Pitpnb and Pitpng. (A) Pitpnb and Pitpng are expressed at all ages tested. RNA was collected from zebrafish at 6 and 24 hours past fertilization (hpf), 6 days post-fertilization (dpf), and from adult fish. The RNA was reverse transcribed, and the respective cDNAs (template) amplified using primers directed against Pitpnbi1, Pitpnbi1 and Pitpng sequences. The amplified cDNAs reflect relative mRNA abundance in each sample, and amplification of products was both dependent on template and reverse transcriptase (RT). (B) In situ staining of pitpnb and pitpng expression profiles. In situ hybridization analyses were performed on 28 hpf fish using specific antisense probes for Pitpnb and Pitpng mRNAs, respectively. Sense probes were used as negative controls, as were “mock” controls where probe was omitted. Both pitpnb and pitpng were expressed in a wide range of tissues throughout the larva, and Pitpnb is most highly expressed in the eye. Scale bars -- 93 µm. (C) All PITP isoforms are expressed in the adult zebrafish eye. Total RNA was extracted from an adult zebrafish eye, and reverse transcribed into cDNA. PCR amplification was performed with primers specific for zebrafish pitpna, pitpnbi1, pitpnbi2 or pitpng sequences. (D) Characterization of Pitpnb antibody. Individual DrPITPs were expressed in yeast, lysates were prepared, resolved by SDS-PAGE, transferred to nitrocellulose, and probed with either anti-Pitpnb serum (top panel) or a control pan-PITP antibody (bottom panel) as indicated. To detect total PITPs in the eye, a blot with the same samples was probed with the NT1/3 pan PITP antibody. Protein extracts from zebrafish (Dr) or murine (Mm) eyes were also probed with these respective antibodoes. This blot indicates that at least 2 distinct PITP species are expressed in the zebrafish eye.
Supplementary Fig. S2. Blue opsin and rhodopsin expression are unaffected in Pitpnb morphants. Zebrafish embryos injected with negative control morpholino or pitpnb MO1 were fixed, embedded, and cryosectioned at 3 dpf. Sections through the center of the retina were stained for zpr-1 and the rod cell opsin (rhodopsin) (left panels), or the single cone cell marker blue opsin (right panel). Blue opsin and rhodopsin staining are present in both control and morphant eye sections, while mAb zpr-1-staining is seen only in the control. Scale bars -- 28 µm.
Supplementary Fig. S3. Pitpnb morphants display no defects in electroretinograms (ERGs) measuring their response to light. Zebrafish embryos were injected at the 1–4 cell stage with 5 base-mismatch control (5MIS) or pitpnb morpholinos (morpholino 1 and 2 represent MO1 and MO2, respectively), and an uninjected control was also included. ERGs were recorded at 5 dpf as previously described (Wong KY, Gray J, Hayward CJ, Adolph AR, Dowling JE. Zebrafish. 2004;1(2):121–31.). Following a flash of either white, blue or red light, morphants and controls showed transient corneal changes in potential known as a- wave and b-wave responses. No significant differences in kinetics or amplitude were noted between the averaged responses from different experimental groups. Data were collected from 2 – 5 larvae at each condition using three different sets of injected animals. Representative recordings using red light as the stimulus are presented.
Supplementary Fig. S4. mAb zpr-1 only detects Arr3L, and not other retinal and ubiquitously expressed arrestins. The open reading frame of arr3L and five additional zebrafish arrestins were PCR amplified from an adult zebrafish cDNA library with primers also encoding an HA epitope-tag. The PCR products were subcloned into a yeast expression vector (pDR195) and transformed into yeast. Lysates were prepared from the resulting yeast strains, lysate proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed with mAb zpr-1 and anti-HA. Though all of the arrestins were expressed (as detected by the HA antibody), only Arr3L was detected by mAb zpr-1.
Supplemental Table S1 – Primers used to amplify zebrafish PITP structural genes.
Supplemental Table S2 – MALDI-TOF mass spectrometry of mAb zpr-1 ligand. The gel-purified polypeptide species specifically precipitated from the adult zebrafish eye by mAb zpr-1 (Fig. 7B) was submitted for analysis by mass spectrometry. Two proteins were identified from tryptic digests of the submitted sample, and the summary analytical report is shown.