Abstract
Very little is currently known about how nucleotide excision repair (NER) functions at the ends of chromosomes. To examine this, we introduced the URA3 gene into either transcriptionally active or repressed subtelomeric regions of the yeast genome. This enabled us to examine the repair of ultraviolet (UV)-induced cyclobutane pyrimidine dimers (CPDs) in identical sequences under both circumstances. We found that NER is significantly more efficient in the non-repressed subtelomere than the repressed one. At the non-repressed subtelomere, UV radiation stimulates both histones H3 and H4 acetylation in a similar fashion to that seen at other regions of the yeast genome. These modifications occur regardless of the presence of the Sir2 histone deacetylase. On the other hand, at the repressed subtelomere, where repair is much less efficient, UV radiation is unable to stimulate histone H4 or H3 acetylation in the presence of Sir2. In the absence of Sir2 both of these UV-induced modifications are detected, resulting in a significant increase in NER efficiency in the region. Our experiments reveal that there are instances in the yeast genome where the maintenance of the existing chromatin structures dominates over the action of chromatin modifications associated with efficient NER.
INTRODUCTION
Efficient repair of DNA damage induced by extracellular and intracellular agents is vital for the maintenance of genome integrity. Nucleotide excision repair (NER) is a highly conserved repair pathway among various organisms that removes bulky DNA lesions, including ultraviolet (UV)-induced cyclobutane pyrimidine dimers (CPDs), 6-4 photoproducts (6-4PPs) and other chemical adducts (1–4). There are two NER sub-pathways: transcription-coupled repair (TC-NER) that operates on the transcribed strand (TS) of active genes, and global genome repair (GG-NER) that operates on the overall genome. A great deal is known about the molecular mechanism of the ‘core reaction’ of NER (5,6) and much of the recent attention on NER has been focused on how DNA lesions are detected and repaired in the chromatin environment in living cells (7).
The majority of the eukaryotic genome is organized into a structural hierarchy of chromatin. The basic structural unit of chromatin is the nucleosome where about 146 bp of DNA is wrapped around an octamer of histones H2A, H2B, H3 and H4. Nucleosomes are connected in a ‘beads on a string’ manner by linker DNA, and are subsequently compacted further into higher-order chromatin structures (8). The packaging of DNA into nucleosomes and chromatin provides a template considerably different from naked DNA and this influences all DNA based processes, including DNA repair. In vitro studies using reconstituted nucleosomes as templates showed that nucleosomes exert an inhibitory effect on the NER of DNA damage since the overall repair of DNA damage by NER is less efficient in nucleosomes than in naked DNA (9–11). In vivo, both the static and dynamic aspects of nucleosome structure/behaviour have been shown to have an influence on NER. First, the default state of chromatin structure within which DNA damage is located significantly affects the efficiency of lesion removal by NER. High-resolution analysis of CPD removal by NER revealed a faster repair of lesions in linker DNA and towards the 5′-end of positioned nucleosomes and a slower repair in the centre of the nucleosomes. This modulation was found in the non transcribed strand of active genes and both strands of inactive genes in several loci in Saccaromyces cerevisiae, including URA3 (12,13), the GAL1-10 promoter (14), MET16 (15) and MET17 (16). Secondly, histone modifications, especially acetylation and chromatin remodelling, have been shown to occur during NER. Indications of this stem from observations made decades ago. First, following NER newly synthesized DNA in human fibroblasts showed enhanced nuclease sensitivity (17) and, second, treatment of non-replicating human cells with sodium butyrate, an inhibitor of histone deacetylases to enhance the overall histone acetylation, promotes repair synthesis following UV irradiation (18). More recently, histone H3 was found to be hyperacetylated in the MFA2 promoter and this hyperacetylation of histone H3 is necessary for efficient repair of CPDs in this region (19,20). UV treatment also stimulates both histones H3 and H4 hyperacetylation globally, but at MFA2 H3 hyperacetylation dominates, with little change occurring in acetylation at H4 (19). This, together with the study showing that Sir2 selectively influences NER at a specific locus but not at others (21), further re-enforces our proposals that domains or regions exist where different histone modifications can influence NER (22). Subunits of the yeast chromatin remodelling complex SWI/SNF were also found to co-purify with Rad4 and Rad23, factors that are involved in the early stage of UV damage recognition in NER (23), whereas the SWI/SNF complex stimulates NER both in vivo and in reconstituted nucleosomes in vitro (23,24).
Epigenetic silencing represents a unique mechanism of transcriptional regulation in S. cerevisiae and it occurs at the mating-type loci HML, HMR, telomeres and the rDNA repeats (25). It distinguishes itself from promoter specific gene repression in that the proteins involved in silencing appear to target distal regulatory sites (rather than gene specific promoters) to generate a large domain of repressive chromatin, i.e. heterochromatin (26). At telomeres, the formation of silencing and heterochromatin initiates by the binding of Rap1 to its binding site. Rap1 then recruits the Sir proteins, including Sir2, Sir3 and Sir4 (26). Sir2 is a NAD+-dependent histone deacetylase, with a preference for removing the acetyl group from K9 and K14 of histone H3 and K16 of histone H4 (27,28). The hypoacetylated form of K16 in histone H4 particularly intensifies the binding of Sir3 to histone tails, which further recruits Sir2 and Sir4. This process repeats itself to permit the spreading of the Sir proteins into the chromosome and further away from telomere ends (29–31). The spreading of the Sir proteins propagates the silencing and genes near the telomeres are transcriptionally repressed. This phenomenon is referred to as ‘telomere position effect’ (TPE) (32). Both the Sir proteins and histone modifications, including acetylation and methylation, are important for proper silencing (33–35). The transcriptional repression by TPE diminishes gradually with increasing distance from the terminus and that varies at individual telomere ends. This is illustrated in studies where reporter genes are inserted into various subtelomeric locations and in different chromosome ends (36,37). In accordance with this, both the distribution of the Sir proteins (38) and histone H4 acetylation (39) appear to spread in a gradient fashion from the telomere end, ranging from the highest binding of the Sir proteins and a hypoacetylated state of histone H4 K16 in regions near telomeres to the lowest binding of the Sir proteins and a hyperacetylated state of H4 K16 in regions further away from the telomere. This reflects the nature of local structural elements in individual chromosome ends and their association with the Sir and other proteins which govern the spread (37).
The system whereby a URA3 reporter gene is placed in subtelomeric regions has proven to be very informative both in studies of gene repression by silencing (34) and its associated chromatin features (40). Here, we take advantage of this system and focus on NER in the same URA3 sequence, either in repressive or in non-repressive subtelomeric regions. This enables us to examine the repair of UV-induced CPDs from identical sequences under both circumstances and to correlate NER with the status of the chromatin. In this study, we show that chromatin structure, gene expression and repair efficiency are inter-related. Intriguingly, we report that in the repressed subtelomere Sir2p suppresses the UV-induced histones H3 and H4 acetylations that are linked to efficient NER in other regions of the genome.
MATERIALS AND METHODS
Yeast strains, growth conditions and UV treatment
The strains in this study include FEP178 (MATa, ura3-52::KanMX can1-1 ade2Δ leu2Δ URA3 at CHRIII-R), FEP100-10 (MATa, ura3-52::KanMX can1-1 ade2Δ URA3 at CHRXI-L), FEP178sir2Δ (same as FEP178, except sir2::LEU2), FEP100-10sir2Δ (same as FEP100-10, except sir2::LEU2). W303 (MATa, ada2-1 leu2-3, 112 his3-1 ura3-52 trp1-100 can1-100), rad9Δ (same as W303, except rad9::HIS3). In FEP178 and FEP100-10, URA3 is inserted into the subtelomeric region of chromosome IIIR and chromosome XIL where the promoter is ∼1.75 kb proximal to TG1-3 sequence and 1 kb proximal to the Core X-ACS (37). The genomic ura3-52 in all above strains was replaced with KanMX. To achieve this and to disrupt SIR2, the required polymerase chain reaction (PCR) product with the marker sequence in the middle flanked by sequence homologous to the flanking sequence of the target gene was used to transform the relative strains via the lithium acetate method (41). Positive colonies from the selective plates were further confirmed by PCR. Yeast strains were grown in yeast complete medium (YPD) at 30°C. Cells at exponential phase (2–4 × 107 cells/ml) from overnight culture in YPD were collected and resuspended in phosphate-buffered saline (PBS) to a final concentration of 2 × 107 cells/ml. UV treatment was carried out as previously described (19) at a dose of 150 J/m2. After the UV treatment, cells were resuspended in YPD and incubated at 30°C in the dark for 1–4 h for the repair experiment and 0.5–1 h for the chromatin immunoprecipitation (ChIP) experiment.
DNA isolation and determination of the rate of NER
Genomic DNA was extracted as previously described (19). About 30 µg of DNA was digested with 120 U of MseI restriction enzyme overnight at 37°C. The purified DNA was then treated with a crude extract of Micrococcus luteus (ML) which contains CPD specific endonuclease. The strands were separated using biotin-labelled probes for the MseI fragment of the URA3 gene and labeled with radioactive dATP as previously described (30). The DNA fragments for specific strands were loaded onto a 6% polyacrylamide gel and resolved by electrophoresis at 70 W for 2.5 h. The probes used are: for the NTS of MseI fragment, 5′-biotin-GATAGCTTTTTTAATTGAAGCTCTAATTTGTGAGTTTAGTATAC-3′; for the TS of MseI fragment, 5′-biotin-GATAGCTTTTTTAGCCGCTAAAGGCATTATCCGCCAAGTACA-3′. The sequences in italics are the overhangs in the probes.
Autoradiographs were scanned with a Typhoon PhosphorImager (GE Healthcare) and the signal for each band was quantified using ImageQuant 5.0 software. The damage at particular sites in the sequence was calculated as the percentage of radioactivity in a band in relation to the total damage signal in each lane and the signals seen immediately after UV (0 samples) were taken as 100% damage. The repair rate was presented as the time needed to repair 50% of the initial lesions (T50%). The bands which are close to each other in the gel and have same repair rate were treated as a single band. The data plotted in the graph represent the average from three to five independent experiments. The statistical analysis to compare the repair rate between different strains was carried out using the Mann–Whitney test.
Nucleosome mapping
Chromatin mapping was carried out as previously described (42). Exponential phase cells (2–4 × 107cells/ml) from an overnight culture in YPD were harvested and permeabilized with zymolyase. Extracted chromatin was digested with micrococcal nuclease (MNase) for 10 min at 37°C. Naked DNA was also extracted and similarly treated with MNase. After MNase treatment, samples were digested with MseI restriction enzyme. Both strands were separated, labeled and analysed as for DNA repair experiments.
Chromatin immunoprecipitation
ChIP experiments were carried out as previously described (19). Cross-linking was carried out with 1% of formaldehyde in final concentration for 10 min at room temperature. The chromatin was sonicated to obtain DNA fragments with lengths between 400 and 1000 bp. One-hundred microlitres of sheared chromatin solution was precipitated overnight at 4°C in a total volume of 500 μl with 5 µl of anti-histone H3 (Upstate Biotechnology), 3 µl of anti-acetyl histone H3 (against H3 acetylated at K9, K14, Upstate Biotechnology) and 3 µl of anti-acetyl histone H4 (against H4 acetylated at K16, Upstate Biotechnology) antibodies, respectively.
Quantitative PCR
Quantitative PCR was carried out using iQ SYBR Green Supermix (Bio-Rad Laboratories) in the Bio-Rad iCycler. Both the immunoprecipitated and input samples were used so the amplification efficiency for each set of primers was adjusted. The samples were diluted appropriately and triplicates of each sample were used to perform PCR. Melting curves were performed to ensure that there was only one product. Data were analysed using Bio-Rad iQ5 software version 3.0a (Bio-Rad Laboratories). The primers used are: 5′TAAGCCGCTAAAGGCATTAT3′ (forward) and 5′ACCGTGTGCATTCGTAA3′ (reverse) for the region from +222 to +369 bp of URA3; 5′GGCTTTATTGCTCAAAGAGAC3′ (forward) and 5′CTTGTCATCTAAACCCACA3′ (reverse) for the region from +540 to +669 bp.
Western analysis of nucleosomal histone acetylation
Chromatin from individual samples was prepared as above. After limited MNase digestion the resultant supernatant containing polynucleosomes was denatured and separated on 12% polyacrylamide–sodium dodecyl sulphate (SDS) gel. Western blotting was carried out according to the standard procedure. Anti-histone H3 (Upstate Biotechnology), anti-acetyl histone H3 (against H3 acetylated at K9, K14, Upstate Biotechnology) and anti-acetyl histone H4 (against H4 acetylated at K16, Upstate Biotechnology) antibodies were used following the manufacturer’s instruction. The signal was detected using an ECL advance (GE Healthcare) and the final image was taken and processed using a LabWorks Bioimaging System.
Measurement of URA3 expression
Appropriate amount of cells from overnight culture at exponential phase were spread onto YPD plates for each strain, with and without 5-fluoro-orotic acid (5-FOA). Colonies were scored after incubation at 30°C for 3–4 days. The percentage of 5-FOA-resistant colonies was calculated.
RESULTS
URA3 is alternatively transcriptionally active or repressed at different chromosome ends
In this study, we employed a set of established yeast strains in which URA3 is inserted into the subtelomeric region of chromosome III-R and chromosome XI-L, both about 1 kb away from their CoreX-ACS sequence positioned towards the centromere (37). In addition, we eliminated the endogenous ura3-52 sequence at its natural location. This provides strains with a unique URA3 sequence in the specific subtelomeric regions, a requisite for our technology to examine the incidence and fate of UV-induced CPDs at nucleotide resolution (see ‘Materials and methods’ section).
In these strains, we measured the expression of URA3 at these chromosome ends to determine the extent of TPE. The expression of URA3 was determined by measuring the fraction of cells that are able to grow on YPD plates containing 5-FOA versus those on plates without 5-FOA. 5-FOA is only toxic when the URA3 product is present in the cell. Therefore, cells expressing URA3 are unable to grow on 5-FOA containing plates and cells that do not express URA3 become 5-FOA resistant and form colonies.
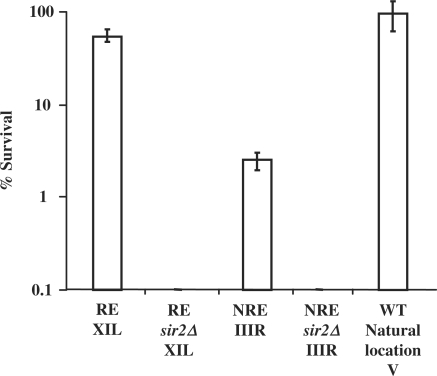
The results are presented in Figure 1 and they confirm that different chromosome ends have different extents of silencing as was previously reported (37). When URA3 is inserted at chromosome XI-L about 47% of the cells survived on 5-FOA plates, whereas only about 2% of the cells survived on 5-FOA plates when URA3 is inserted at chromosome III-R. When the SIR2 gene is deleted, no colonies were seen on 5-FOA plates for both strains. This indicates that the URA3 gene is repressed when at chromosome XI-L, but not so when at chromosome III-R. Hence, thereafter we term the chromosome III-R end as the non-repressive end (NRE) and the chromosome XI-L end as the repressive end (RE). When SIR2 is absent, the repression of URA3 is relieved at the RE. Cells with URA3 at its natural location grow normally on YPD plates with 5-FOA as URA3 is not normally expressed when these cells grow in complete medium containing uracil (43,44).
Figure 1.
Survival of the NRE, NREsir2Δ, RE, REsir2Δ on 5-FOA plates. Survival is presented as the fraction of cells that form colonies on 5-FOA plates versus non-5-FOA plates. The strains used are NRE (URA3 inserted at the non-repressive end), NREsir2Δ (URA3 inserted at the non-repressive end with SIR2 deletion), RE (URA3 inserted at the repressive end) and REsir2Δ (URA3 inserted at the repressive end with SIR2 deletion).
URA3 chromatin is more sensitive to MNase at the NRE
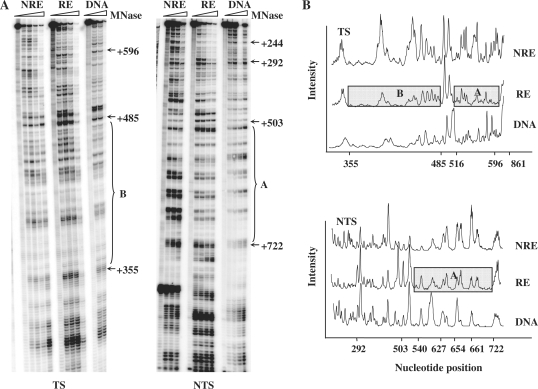
Nucleosomes and other proteins that bind to the DNA can inhibit the DNA repair machinery (7,45,46). In order to obtain information about the chromatin structure of the inserted URA3 at these chromosome ends and its potential impact on NER, we investigated the MNase sensitivity at URA3 by high-resolution analysis (42). When URA3 was at the NRE and RE, extracted chromatin was treated with increasing amount of MNase and naked DNA was used as a control. The fragment selected for examination was the MseI restriction fragment. This locates in the coding region of the subtelomeric URA3 and contains 640 bp (from +221 to +861 bp). Figure 2A shows gels for the mapping of MNase sensitive sites in the TS and NTS of the MseI fragments of URA3 at the RE and NRE. The relative intensity of each band is represented graphically in Figure 2B. In the graph, the higher the band peaks, the greater is the accessibility of the DNA to MNase. The results indicate that overall, at the RE, the chromatin in URA3 is less amenable to MNase cleavage than when it is at the NRE. This becomes especially apparent in two regions, designated as A and B in Figure 2. Region A spans from +355 to +485 (+1 is designated as the first A of URA3 start codon,) and region B spans from +540 to +722. Thus, increased MNase sensitivity correlates well with increased URA3 expression at these ends.
Figure 2.
MNase sensitivity of the chromatin containing the MseI restriction fragment of URA3 at the NRE and RE. (A) Typical footprinting autoradiographs showing MNase-sensitive sites in the TS and NTS of the MseI restriction fragment of URA3. DNA and chromatin samples from each strain (NRE and RE) were treated with increasing amount of MNase. Protected regions in chromatin at the repressive end are indicated by A and B. Nucleotide positions were indicated alongside the gels as numbers relative to the ATG start codon of URA3. (B) Relative MNase sensitivity after scanning the TS and NTS gels as shown in (A). The boxes indicate the regions that are less sensitive to MNase.
The repair of CPDs is faster in URA3 when at the NRE
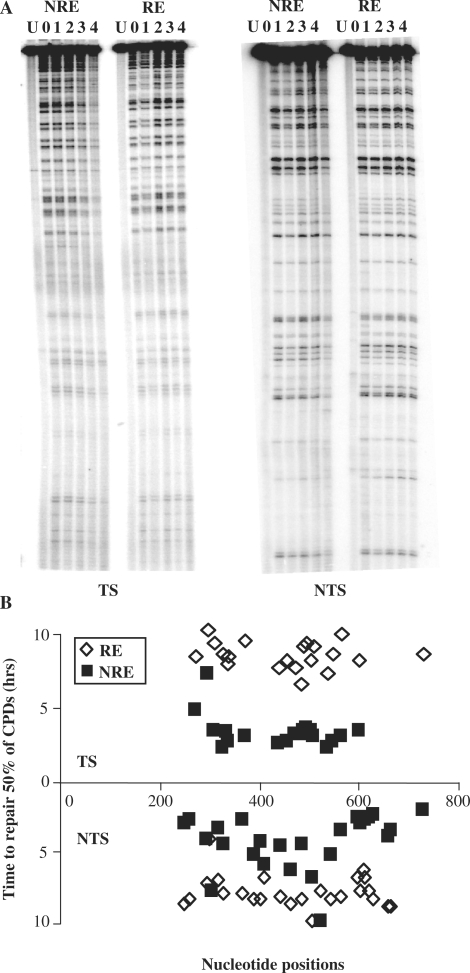
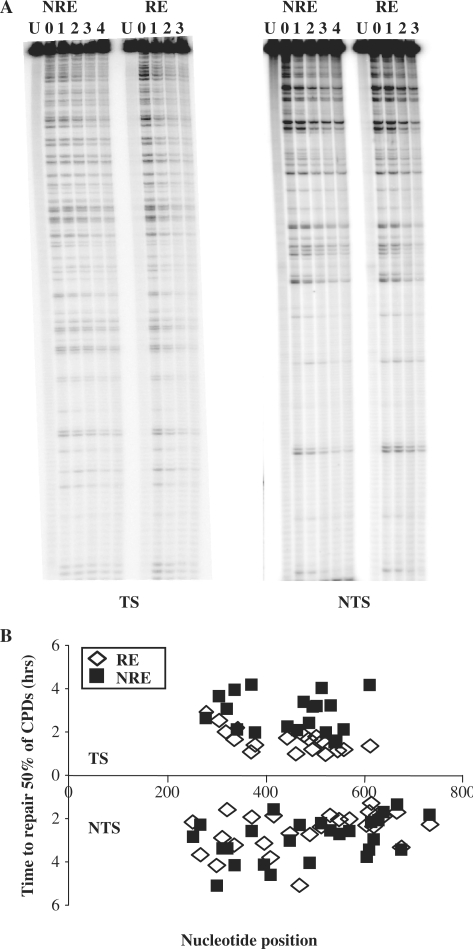
The NER of CPDs in the URA3 gene when at the NRE and the RE was investigated at nucleotide resolution. Figure 3A shows DNA sequencing gels detecting CPDs in the TS and NTS of the MseI restriction fragment. As expected the unirradiated sample (U) provided a unique top band which is indicative of the full-length MseI fragment. UV-irradiated samples were taken at various times after 150 J/m2 of UV to determine the incidence of CPDs, as described previously (19). The multiple bands below the top bands in each lane reflect the CPD incidence at specific dipyrimidine sites within the sequence and are of varying intensities. They were quantified and CPD repair rates at the MseI fragment were summarized in Figure 3B, which shows the time in hours needed to repair 50% of the initial lesion (T50%) at each CPD site.
Figure 3.
The repair of CPDs in the TS and NTS of URA3 at the NRE and RE. (A). Gels depicting the incidence of CPDs in the MseI fragment of URA3 in the NRE and RE. U, UV-untreated samples; 0, samples that were UV treated but without repair time; 1–4, UV-treated samples that were allowed to repair the damage for 1–4 h. (B) Graph representing the repair rate for individual CPDs at each nucleotide position for the NRE and RE. The repair rate is represented as the time required to remove 50% of the initial damage (T50%).
When we compare the difference in repair of CPDs from URA3 at the RE and NRE, it is clear that repair is much faster when URA3 is at the NRE for both the NTS (P < 0.0001) and TS (P < 0.0001). In terms of strand bias, there is little difference between the TS and NTS of URA3 for the overall repair of CPDs when URA3 is at the RE. The average T50% for the entire fragment is 8.27 h for the TS and 8.07 h for the NTS. However, the repair is markedly faster for the TS when URA3 is at the NRE, with an average T50% of 2.97 h for the TS and 4.46 h for the NTS. Here, the faster repair of CPDs in the TS as opposed to the NTS of URA3 is suggestive of TC-NER. A key point to be noted is that the NTS of the URA3 fragment in the NRE is repaired more rapidly when compared to the same sequence at the RE.
When URA3 is at the RE, the chromatin showed different sensitivity to MNase along the sequence, being particularly resistance to MNase in two regions (Figure 2A and B). The repair of CPDs in these regions showed no clear disadvantage compared to other regions. However, for the same site at different chromosome ends the repair correlates well with the MNase sensitivity described above. In general, the chromatin of URA3 is less accessible to MNase at the RE and the repair of CPDs is slower when compared to those events at the NRE. For instance, one of the maximal differences for individual repair of CPDs was obtained around +561 bp, +554/TTTCTCT/+560 (3.7-fold difference) and one of the minimal differences were found around +478 bp, +477/TC/+478 (2.30-fold difference). In the NTS, one of the maximal differences for repair of CPDs was obtained around +616 bp, +615/TTT/+618 (2.8-fold difference) and the minimal difference was around +397 bp, +395/TTT/+398 (1.14-fold difference). The extent of NER at the RE does not correlate with the MNase sensitivity since it is equally slow in the MNase sensitive regions.
Deletion of SIR2 renders the repressed URA3 chromatin more sensitive to MNase
Sir2p is the enzymatic component of the SIR complex which is involved in silencing. We deleted the SIR2 gene to study how the disruption of silencing affected the expression of the URA3 gene, its chromatin structure and NER. As mentioned above, deletion of SIR2 results in the derepression of URA3 when it is at the RE as indicated by the data shown in Figure 1.
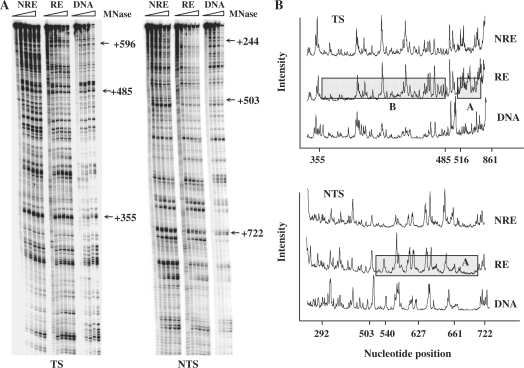
The same MseI restriction fragment of URA3 was digested with various amounts of MNase as described earlier. The gels and graphs, showing the relative intensity of individual bands and hence the extents of MNase cutting, are provided in Figure 4A and B. In general, we observed the URA3 chromatin at the RE in the sir2 mutant became more sensitive to MNase when compared with that in the wild type, especially in the regions A and B (Figures 2B and 4B). At the NRE, the URA3 chromatin was as sensitive to MNase as in the wild-type strain. This indicates SIR2 deletion relieves the repression of URA3 at the RE (Figure 1), and this corresponds to a change of the chromatin structure rendering it more sensitive to MNase.
Figure 4.
MNase sensitivity of URA3 chromatin at the NRE and RE in the sir2Δ mutant. (A) Gels showing MNase sensitive sites in the TS and NTS of URA3 chromatin in the RE and NRE in the sir2Δ mutant. DNA and chromatin samples from each strain were treated with increasing amounts of MNase. (B) Graph representing the relative MNase sensitivity after scanning the gels in (A) Regions A and B are the same regions as in Figure 2.
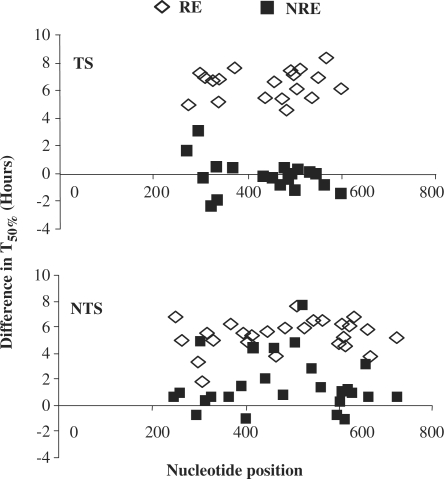
Deletion of SIR2 has differential effects on NER in URA3 at the RE and the NRE of chromosomes
Next, we examined the influence of deletion of SIR2 on the repair of UV-induced CPDs in the URA3 sequence at the two chromosomal ends. The same MseI restriction fragment of the URA3 gene as above was examined. The resultant gels and T50% measurements are presented in Figure 5A and B. SIR2 deletion enhances the overall NER in the URA3 gene both at the RE and the NRE (Figure 6). In Figure 6, we plotted the difference in T50% (the numerical difference between time required to repair 50% of the initial damage) between the wild type and the sir2Δ mutant. The effect of Sir2 varies at these locations. There are significant differences in the repair rates in the URA3 at the RE (P < 0.0001) for both TS and NTS: repair is faster when SIR2 is deleted (Figure 6). There are also differences in repair rates for the TS (P < 0.05) and the NTS (P < 0.0001) in URA3 at the NRE. However, the differences are smaller than those at the RE, especially for the TS (Figure 6). The average repair rates in the TS and NTS at the RE are 3-fold and 4.8-fold slower in the wild type than that in the sir2Δ mutant, respectively. In contrast, at the NRE these numbers are 1.03-fold for the TS and 1.5-fold for the NTS.
Figure 5.
Repair of CPDs in the TS and NTS of URA3 in the NRE and RE in the sir2Δ mutant. (A) Typical gels showing the incidence of CPDs in the TS and NTS of URA3 in the NRE and RE in the sir2Δ mutant. U, UV-untreated samples; 0, samples that were UV treated but without repair time; 1–4, samples that were UV-treated and repair time (1–4 h) was given. (B) Time required to repair 50% of the initial CPDs (T50%) at individual sites.
Figure 6.
The difference of T50% for the TS and NTS of URA3 in wild type and the sir2Δ mutant. Data plotted are the difference in T50% of each CPD (the T50% of each CPD in URA3 at the RE and NRE in the wild-type cells subtracting the corresponding data in the sir2Δ mutants).
When URA3 is at different chromosome ends in the sir2Δ mutant, there was no difference statistically in the overall repair of CPDs in the NTS (P > 0.05). However, the repair in the TS at the RE is 1.8-fold faster than that at the NRE (P < 0.0001) (Figure 5A and B). The average T50% at the RE was 1.71 h compared with 3.07 h at the NRE.
Again for the same CPD site, the repair correlated well with the MNase sensitivity of the chromatin around the damage site. With respect to the NTS, the maximum difference in repair of individual CPDs occurs around +605bp, +603/CCC/+606. This region is more sensitive to MNase at the RE than at the NRE in the sir2Δ mutant, and the repair is 2.7-fold faster. At positions +637bp, +636/TT/+638, where in both strains the chromatin is similarly sensitive to MNase, the repair rate is also the same. For the TS the maximum difference in repair of individual CPDs is at positions +358 bp, +357/CTT/+360, being 3.3-fold faster in the absence of Sir2, yet at around +331 bp, +331/TCTT/335, there is no difference in the repair rate irrespective of Sir2. In this case, at both ends the chromatin is sensitive to MNase. The results imply that SIR2 has a strong effect on NER at the RE but this effect is reduced for the NRE. This correlates well with the role of SIR2 in silencing, and is supported by MNase mapping analysis and expression of the gene as previously described.
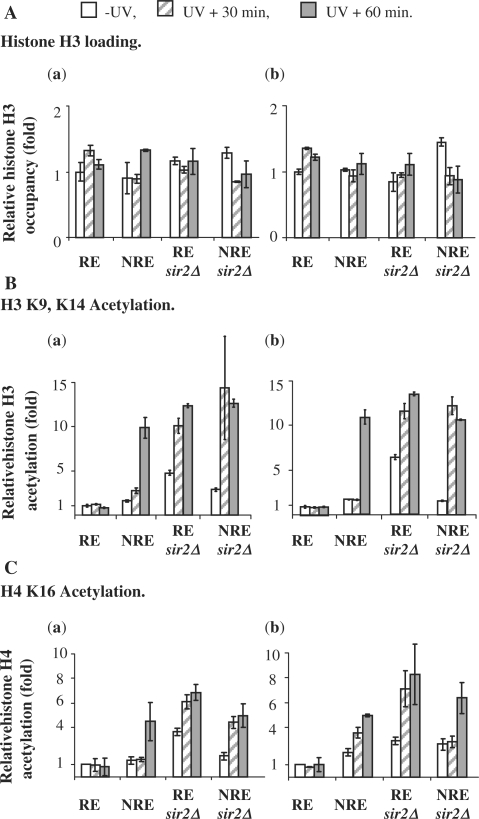
SIR2 regulates UV-induced histone H3 and H4 hyperacetylation, but only at the repressive end
Our previous studies showed that UV irradiation stimulates histone H3 and H4 acetylation and this response in histone acetylation varies for different regions in the genome (19). Sir2p contains histone deacetylase activity (27,28) and it removes the acetyl group from histone H4 K16 and H3 K9, K14 (47,48). To examine how histone acetylation in the subtelomeric region responds to UV and whether Sir2p plays a role in it, we examined the acetylation levels of histone H3 at K9, K14 (Figure 7B) and histone H4 at K16 (Figure 7C) in the relocated URA3 gene before and after UV (30 and 60 min). We also monitored the histone H3 occupancy (Figure 7A) and used this to normalize the acetylation levels of H3 and H4 across the samples. All the data are presented as fold increases relative to that in the RE before UV treatment. The regions selected for the analysis were from +222 to +369 bp and from +540 to 669 bp in the coding region of the subtelomerically located URA3 gene.
Figure 7.
Histone acetylation at the coding region of the subtelomeric URA3 in response to UV irradiation. The histone H3 loading (A), H3 K9, K14 (B) and H4 K16 (C) acetylation are represented as a relative level to that in the RE before UV treatment. The acetylation level of H3 K9, K14 and H4 K16 was normalized against histone H3 data. a. in the region from +222 to +369; b. in the region from +540 to +669.
As indicated in Figure 7A, the histone H3 occupancy at the two regions in all the samples remains almost the same. However, the levels of H3 (K9, K14) and H4 (K16) acetylation are different (Figure 7B and C). First, the NRE has naturally occurring slightly higher levels of both H3 (K9, K14) and H4 (K16) acetylation than the RE (between 1.36 ± 0.20- and 1.97 ± 0.35-fold increases in the two regions compared to in the RE). Deletion of SIR2 results in significant increases in the levels of H3 (K9, K14) and H4 (K16) acetylation at the RE (4.78 ± 0.28-fold increases in the region from +222 to +369bp and 6.42 ± 0.50-fold increases in the region from +540 to +669 bp in the sir2Δ mutant for H3 acetylation; 3.67 ± 0.11-fold increases in the region from +222 to +369 bp and 2.93 ± 0.42-fold increases in the region from +540 to +669 bp for H4 acetylation). At the NRE, deletion of SIR2 results in only slight increases in the levels of H3 (K9, K14) and H4 (K16) acetylation (between 1.57 ± 0.06- and 2.91 ± 0.18-fold increases in the two regions). Second, UV treatment triggers hyperacetylation of H3 (K9, K14) and H4 (K16) at the NRE. This stimulation is not Sir2p dependent because the sir2Δ mutant responds equally well when compared to the wild type. UV radiation increases H3 (K9, K14) acetylation about 10- fold in the two regions, both in the wild type and in the mutant 60 min after UV. For histone H4 (K16) acetylation, the increases are between 4.50 ± 1.55- and 6.30 ± 1.24-fold. In contrast, when URA3 is at the RE there is no detectable UV-induced hyperacetylation of histones H3 (K9, K14) and H4 (K16) in the wild type. Only when SIR2 is deleted, does the UV-induced histone hyperacetylation occur (Figure 7 B and C). With increased acetylation levels of both H3 (K9, K14) and H4 (K16) in the sir2Δ mutant even prior to UV exposure, there are further increases in these levels after UV, reaching levels similar to those seen in the NRE. This indicates that at the RE, Sir2p inhibits the acetylation of histones H3 (K9, K14) and H4 (K16) to maintain the silencing of this region, thus overriding the hyperacetylation of histones H3 and H4 that can enable efficient NER.
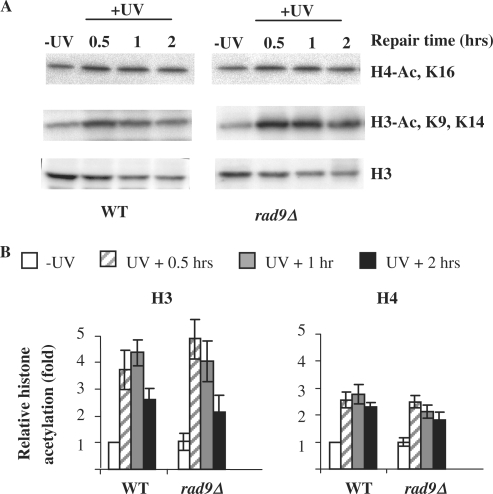
UV-induced histones H3 and H4 acetylation does not relate to cell cycle arrest after UV
One of the major events occurs in cells after DNA damage is cell cycle arrest and Rad9 plays a critical role in this event. In rad9Δ mutant cells, there is a failure to arrest in the cell cycle after UV irradiation (49–51). To determine whether UV-induced histone acetylation relates to cell cycle arrest and its associated events, we monitored the acetylation of total histones H3 and H4 in chromatin by western blotting in the wild type and rad9Δ mutant cells. As indicated in Figure 8, UV stimulates both H3 and H4 acetylation in the rad9Δ mutant similarly as in the wild type. This finding precludes the possibility that UV-induced histone hyperacetylation observed is primarily due to cell cycle arrest.
Figure 8.
Total histone acetylation in response to UV irradiation. (A) Western images of nucleosomal histones with anti-H3, anti-acetyl H3 (K9, K14) and anti-acetyl H4 (K16) in the wild-type and rad9Δ mutant. (B) relative abundances of acetylated nucleosomal H3 at K9, K14 and H4 at K16. The acetylation levels are presented as fold increases relative to that of the wild-type cells before UV irradiation.
DISCUSSION
Here we present a study which examined the repression, the chromatin structure and the NER of UV-induced CPDs in the same URA3 sequence at two different subtelomeric regions. This study has enabled us to determine how these events are inter-related. The results show that when URA3 is at the NRE, the repair efficiency, the MNase accessibility and the expression of the gene are elevated compared with when it is inserted at the RE. When SIR2 is deleted, the repression of URA3 at the RE is relieved. The chromatin accessibility to MNase, the expression of URA3 and the NER efficiency are more similar to that of URA3 at the NRE. Most importantly, we found that at the RE, Sir2p suppresses the UV-induced histone H3 and H4 hyperacetylation which was found in the NRE and in other regions of the genome (19,20) and which was linked to efficient repair of UV induced CPDs in those regions.
In our strains, the different levels of URA3 expression at the two chromosome ends was retained and was consistent with that previously reported (37). Although it is not known why different chromosomes display different levels of silencing at telomeres, several studies suggest that telomere length and certain structural elements with the telomere affect the amount of Rap1p protein recruited, the longer the telomere the higher is the amount of recruited Rap1p (52–56). There are also other proteins such as Rif1p and Rif2p involved in regulating the length of a telomere, and these proteins are recruited to the telomere by an interaction with the C-terminal domain of Rap1p. This is the same domain involved in the recruitment of the Sir proteins. Therefore, these proteins compete with the Sir proteins to interact with Rap1p, making the amount of Sir proteins critical for the appropriate silencing (57). The concentration of these proteins can differ at distinct telomeres, so creating a variation in gene repression (37,58).
The expression of URA3 at the RE is subjected to TPE (32,37,59). TPE-associated repression is believed to result from the generation of a large domain of repressive chromatin. Here, the accessibility of the URA3 chromatin at the two chromosome ends was measured using MNase digestion. Overall, the chromatin is more sensitive to MNase at the NRE than at the RE. The main changes in MNase accessibility between RE and NRE are found at two regions (Figure 2A and B), Region A covers 182 bp of DNA, and is likely occupied by a nucleosome. Region B consists of 130 bp and is slightly shorter for the usual length of nucleosomal DNA, so it is possible that this region may be occupied by an unusual nucleosome or by other proteins involved in silencing. When silencing is disrupted by deletion of SIR2, the MNase resistant feature in these two regions disappeared and chromatin at the RE becomes as sensitive to MNase as that at the NRE. This MNase accessibility correlates with the expression of URA3 and it is comparable with a previous report where transcriptional status of a gene and the accessibility to the DNA in chromatin are associated (60).
NER is more efficient at the NRE that at the RE. However, the NER extent at the RE does not mirror MNase sensitivity; it is equally slow in the MNase sensitive regions. It is possible that the chromatin structure at this tightly repressed chromatin has other factors that can influence the accessibility of NER over larger domains. Genes such as URA3 at its endogenous location are repressed but are poised for transcription and their repair profiles exhibit faster repair in linker as opposed to nucleosome cores (12,13). The disruption of silencing in the sir2Δ mutant leads to the repair of the NTS of URA3 at the RE becoming similar to that in the NRE. Surprisingly, the repair at the TS is slightly faster in the RE than in the NRE, suggesting that the loss of Sir2p has a greater effect at the RE than at NRE. Moreover, when silencing is disrupted in the sir2Δ mutant, the only change in repair at the NRE occurs at the NTS, this being slightly faster than in the wild type.
Previously Thoma’s group examined NER in strains containing the URA3 gene inserted 2 kb from a telomere and URA3 was transcriptionally active in sir3Δ mutants, partially silenced in SIR3 cells, or completely silenced by overexpression of SIR3 or deletion of RPD3 (61). The active URA3 gene showed efficient repair. Partial silencing reduced NER, whereas complete silencing inhibited NER in the promoter, the coding region and the 3′-end. Conventional low-resolution nuclease footprinting revealed subtle changes in the promoter proximal nucleosome when partially silenced but a pronounced reorganization of chromatin over the whole gene occurred in silenced chromatin. NER was deemed sensitive to chromatin changes associated with silencing, but no correlations were examined with respect to NER and changes in UV-induced histone acetylations.
Acetylation and other covalent modifications can influence the chromatin packaging and therefore its accessibility to various metabolic processes. Transcription and fast repair have been associated with histone hyperacetylation. We previously reported that histone H3 is hyperacetylated at K9 and/or K14 at the MFA2 promoter independently of transcription following UV. At MFA2, this H3 acetylation is primarily undertaken by the HAT Gcn5p (19,20). UV-induced histone acetylation also occurs more widely in the genome and at histones H3 and H4 (19,20). Importantly, UV-induced H3 acetylation improves the efficiency of NER and pre-UV hyperacetylated regions can undergo fast NER independently of Rad16, the GG-NER specific protein which is somehow needed for post-UV increases in H3 acetylation (62). This provides a direct link between GG-NER and histone hyperacetylation. Whether this UV-induced histone hyperacetylation is completely specific to NER still needs further investigation. Histone hyperacetylation could regulate NER either directly through generating a suitable binding surface for proteins involved in NER or indirectly through changing the compaction of nucleosomes. The enzymes that acetylate and deacetylate histone tails are likely in equilibrium. For instance, Sas2p counteracts the deacetylation function of Sir2p and blocks the spread of silencing (35,39,63). This equilibrium could regulate the deposition of histone variants and the acetylation at a particular position. Hence, if this is disturbed by mutating one of the enzymes involved in the control of this equilibrium, such as a HAT or HDAC, then an effect on repair capacity is likely.
At telomeres our results show that UV-induced histone hyperacetylation at H3 (K9, 14) and H4 (K16) occurs only at the NRE, but not at the RE in NER competent cells. Sir2p does not regulate the UV-induced histone hyperacetylation at the NRE, since the sir2 mutant and the wild type have a similar UV response for histone acetylation. On the other hand, histones in the RE are hypoacetylated at both H3 K9, K14 and H4 K16 and these acetylation levels do not change in response to UV; they are likely linked to the slow repair. There is a huge increase in the histone H3 and H4 acetylation after UV at the RE when SIR2 is deleted. This indicates a prominent role for Sir2p in regulating histone acetylation in the RE in response to UV. Perhaps the primary role of Sir2p at the RE is to maintain a repressive chromatin, and this takes priority over modifications to increase NER. Therefore, only when SIR2 is deleted can the activity of a histone acetyltransferase(s), prevail after UV to acetylate H4 K16 and H3 K9, K14. An interesting question currently being pursued is does this Sir suppression reduce the occupancy of the Rad16/Rad7/Abf1 GGR complex at the RE and thus reduce the ability of HATs to operate? The damage at the RE has to be repaired, either by NER or by other repair pathways, e.g. translesion synthesis. One possible scenario for NER is that ‘windows of opportunity’ occur for the action of Rad16 and GG-NER. For example, the reversal of histone hypoacetylation could occur at a certain stage of the cell cycle, such as prior to DNA replication and GG-NER may be more efficient at this stage
Our data show that Sir2p, which is involved in silencing, suppresses the efficient NER of UV-induced CPDs. Thus, NER as well as gene expression can be altered by chromatin modifications that occur at different chromosome ends due to silencing. As the increase in histone acetylation after UV leads to chromatin remodelling which enables a more efficient removal of CPDs at other regions of the genome (19,62) it is likely that the inability to increase the low acetylation levels of histones H3 and H4 at the RE post-UV in the wild type is directly linked to the reduced NER. Hence, there appear to be instances in the yeast genome where the maintenance of existing chromatin structures dominates over the action of chromatin modifications associated with efficient NER.
FUNDING
The UK Medical Research Council (Programme Award to R.W., Career Establishment Grant to S.H.R. and Studentship to A.I.). Funding for open access charge: The UK Medical Research Council.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank our colleagues for their suggestions and technical assistance.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, DC: American Society of Microbiology Press; 1995. [Google Scholar]
- 2.Prakash S, Prakash L. Nucleotide excision repair in yeast, Mutat. Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 3.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 4.Reed SH, Waters R. DNA repair. In: Cooper DN, editor. Nature Encyclopaedia of the Human Genome. London: Nature Publishing Group; 2003. pp. 148–154. [Google Scholar]
- 5.Guzder SN, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 6.Aboussekhra A, Biggerstaff M, Shivji MKK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hübscher U, Egly J-M, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 7.Waters R, Smerdon MJ. Preface. DNA Repair. 2005;8:853–854. doi: 10.1016/j.dnarep.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 9.Wang ZG, Wu XH, Friedberg EC. Nucleotide excision repair of DNA by human cell extracts is suppressed in reconstituted nucleosomes. J. Biol. Chem. 1991;266:22472–22478. [PubMed] [Google Scholar]
- 10.Sugasawa K, Masutani C, Hanaoka F. Cell-free repair of UV-damaged simian virus 40 chromosomes in human cell extracts. I. Development of a cell-free system detecting excision repair of UV-irradiated SV40 chromosomes. J. Biol. Chem. 1993;268:9098–9104. [PubMed] [Google Scholar]
- 11.Ura K, Araki M, Saeki H, Masutani C, Ito T, Iwai S, Mizukoshi T, Kaneda Y, Hanaoka F. ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J. 2001;20:2004–2014. doi: 10.1093/emboj/20.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wellinger RE, Thoma F. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 1997;16:5046–5056. doi: 10.1093/emboj/16.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tijsterman M, de Pril R, Tasseron-de Jong JG, Brouwer J. RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6-4) photoproduct and cyclobutane pyrimidine dimer repair in yeast. Mol. Cell. Biol. 1999;19:934–940. doi: 10.1128/mcb.19.1.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Smerdon MJ. Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 15.Ferreiro JA, Powell NG, Karabetsou N, Kent NA, Mellor J, Waters R. Cbf1p modulates chromatin structure, transcription and repair at the Saccharomyces cerevisiae MET16 locus. Nucleic Acids Res. 2004;32:1617–1626. doi: 10.1093/nar/gkh324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell NG, Ferreiro J, Karabetsou N, Mellor J, Waters R. Transcription, nucleosome positioning and protein binding modulate nucleotide excision repair of the Saccharomyces cerevisiae MET17 promoter. DNA Repair. 2003;2:375–386. doi: 10.1016/s1568-7864(02)00239-2. [DOI] [PubMed] [Google Scholar]
- 17.Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA- repair synthesis. Proc. Natl Acad. Sci. USA. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smerdon MJ, Lan SY, Calza RE, Reeves R. Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. J. Biol. Chem. 1982;257:13441–13447. [PubMed] [Google Scholar]
- 19.Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc. Natl Acad. Sci. USA. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y, Waters R. Histone acetylation, chromatin remodelling and nucleotide excision repair: hint from the study on MFA2 in Saccharomyces cerevisiae. Cell Cycle. 2005;4:1043–1045. doi: 10.4161/cc.4.8.1928. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhuri S, Wyrick JJ, Smerdon MJ. Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:1690–1700. doi: 10.1093/nar/gkp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waters R, Teng Y, Yu Y, Yu S, Reed SH. Tilting at windmills? The nucleotide excision repair of chromosomal DNA. DNA Repair. 2009;8:146–152. doi: 10.1016/j.dnarep.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat. Struct. Mol. Biol. 2006;13:902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- 24.Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol. 2002;22:6779–6787. doi: 10.1128/MCB.22.19.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moazed D. Common themes in mechanisms of gene silencing. Mol. Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 26.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 27.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 28.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rusche LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae, Mol. Biol. Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoppe G, Tanny J, Rudner A, Gerber S, Danaie S, Gygi S, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudner AD, Hall BE, Ellenberger T, Moazed D. A nonhistone protein–protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 2005;25:4514–4528. doi: 10.1128/MCB.25.11.4514-4528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandell LL, Zakian VA. Telomeric position effect in yeast. Trends Cell Biol. 1992;2:10–14. doi: 10.1016/0962-8924(92)90138-d. [DOI] [PubMed] [Google Scholar]
- 33.Gross DS. Sir proteins as transcriptional silencers. Trends Biochem. Sci. 2001;26:685–686. doi: 10.1016/s0968-0004(01)01985-5. [DOI] [PubMed] [Google Scholar]
- 34.Katan-Khaykovich Y, Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 2005;24:2138–2149. doi: 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 36.Aparicio OM, Gottschling DE. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 37.Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sperling AS, Grunstein M. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc. Natl Acad. Sci. USA. 2009;106:13153–13159. doi: 10.1073/pnas.0906866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 40.Loney ER, Inglis PW, Sharp S, Pryde FE, Kent NA, Mellor J, Louis EJ. Repressive and non-repressive chromatin at native telomeres in Saccharomyces cerevisiae. Epigenetics Chromatin. 2009;2:18. doi: 10.1186/1756-8935-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 42.Teng Y, Yu S, Waters R. The mapping of nucleosomes and regulatory protein binding sites at the Saccharomyces cerevisiae MFA2 gene: a high resolution approach. Nucleic Acids Res. 2001;29:E64–4. doi: 10.1093/nar/29.13.e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy A, Exinger F, Losson R. cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol. Cell. Biol. 1990;10:5257–5270. doi: 10.1128/mcb.10.10.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flynn PJ, Reece RJ. Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol. Cell. Biol. 1999;19:882–888. doi: 10.1128/mcb.19.1.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 2004;271:2335–2349. doi: 10.1111/j.1432-1033.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 46.Ataian Y, Krebs JE. Five repair pathways in one context: chromatin modification during DNA repair. Biochem. Cell. Biol. 2006;84:490–504. doi: 10.1139/o06-075. [DOI] [PubMed] [Google Scholar]
- 47.Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 49.Toh GW, Lowndes NF. Role of the Saccharomyces cerevisiae Rad9 protein in sensing and responding to DNA damage. Biochem. Soc. Trans. 2003;31:242–246. doi: 10.1042/bst0310242. [DOI] [PubMed] [Google Scholar]
- 50.Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in. Saccharomyces cerevisiae Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 51.Siede W, Friedberg AS, Friedberg EC. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 53.Kyrion G, Liu K, Liu C, Lustig AJ. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 54.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 55.Levy DL, Blackburn EH. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol. Cell. Biol. 2004;24:10857–10867. doi: 10.1128/MCB.24.24.10857-10867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji H, Adkins CJ, Cartwright BR, Friedman KL. Yeast Est2p affects telomere length by influencing association of Rap1p with telomeric chromatin. Mol. Cell. Biol. 2008;28:2380–2390. doi: 10.1128/MCB.01648-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parsons XH, Garcia SN, Pillus L, Kadonaga JT. Histone deacetylation by Sir2 generates a transcriptionally repressed nucleoprotein complex. Proc. Natl Acad. Sci. USA. 2003;100:1609–1614. doi: 10.1073/pnas.0434064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandell LL, Gottschling DE, Zakian VA. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl Acad. Sci. USA. 1994;91:12061–12065. doi: 10.1073/pnas.91.25.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 60.de Bruin D, Kantrow SM, Liberatore RA, Zakian VA. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol. Cell. Biol. 2000;20:7991–8000. doi: 10.1128/mcb.20.21.7991-8000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livingstone-Zatchej M, Marcionelli R, Moller K, De Pril R, Thoma F. Repair of UV lesions in silenced chromatin provides in vivo evidence for a compact chromatin structure. J. Biol. Chem. 2003;278:37471–37479. doi: 10.1074/jbc.M306335200. [DOI] [PubMed] [Google Scholar]
- 62.Teng Y, Liu H, Gill HW, Yu Y, Waters R, Reed SH. Saccharomyces cerevisiae Rad16 mediates ultraviolet-dependent histone H3 acetylation required for efficient global genome nucleotide-excision repair. EMBO Rep. 2008;9:97–102. doi: 10.1038/sj.embor.7401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shia WJ, Li B, Workman JL. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 2006;20:250. doi: 10.1101/gad.1439206. [DOI] [PMC free article] [PubMed] [Google Scholar]