Abstract
Defective RNA metabolism is an emerging mechanism involved in ALS pathogenesis and possibly in other neurodegenerative disorders. Here, we show that microRNA (miRNA) activity is essential for long-term survival of postmitotic spinal motor neurons (SMNs) in vivo. Thus, mice that do not process miRNA in SMNs exhibit hallmarks of spinal muscular atrophy (SMA), including sclerosis of the spinal cord ventral horns, aberrant end plate architecture, and myofiber atrophy with signs of denervation. Furthermore, a neurofilament heavy subunit previously implicated in motor neuron degeneration is specifically up-regulated in miRNA-deficient SMNs. We demonstrate that the heavy neurofilament subunit is a target of miR-9, a miRNA that is specifically down-regulated in a genetic model of SMA. These data provide evidence for miRNA function in SMN diseases and emphasize the potential role of miR-9–based regulatory mechanisms in adult neurons and neurodegenerative states.
Keywords: ALS, Dicer, microRNA, motor neuron, neurodegeneration
Regulation by micro-RNA (miRNA) appears to be the most abundant mode of posttranscriptional regulation (1). This is because hundreds of miRNA genes, each regulating a diverse set of downstream targets, take part in practically all cellular processes, whether in health or disease.
Genome-encoded miRNAs are transcribed as long RNA transcripts that fold back on themselves to form distinctive hairpin structures. The long miRNA precursor is first digested by the Drosha microprocessor complex (2–4) and then by Dicer1 (5). The mature miRNA is loaded onto the Argonaute silencing complex (6) that directs posttranscriptional repression through miRNA:mRNA pairing. The two main mechanisms for repression of gene expression by miRNA are miRNA-directed translational repression and mRNA destabilization [reviewed in (7, 8)].
Work over the past years has documented a crucial role for miRNA-dependent posttranscriptional gene regulation in the development and function of neurons [e.g., (9–14); recently reviewed in (15–17)]. For example, miR-9 is an ancient neuronal gene involved in flies in selection of neuronal precursors from the neuroepithelium (10). The miR-9 gene is conserved to vertebrates, wherein it specifies the midbrain-hindbrain boundary (18) and, with miR-124, plays a role in neuronal differentiation (19, 20).
Furthermore, alterations in the function of miRNA contribute to susceptibility to neuronal disease. Although this may be associated with loss of neurons (21–27), behavioral and neuroanatomical phenotypes in the absence of neurodegeneration were also reported (28). The expression of specific miRNA was also linked to neurodegeneration; for example, a significant decrease in miR-9 and miR-9* expression was noted in patients with Huntington's disease (20), miR-9 and miR-132 are downregulated in Alzheimer's disease brains (29) and loss of miR-133 expression were suggested to play a role in Parkinson's disease (23).
ALS is a neurodegenerative disease that specifically affects upper and lower motor neurons (MNs), leading to progressive paralysis and death. Recently discovered mutations in the genes encoding the RNA-binding proteins FUS/TLS [ALS6 locus (30, 31)] and TARDBP/TDP43 [ALS10 locus (32, 33)] suggest important roles for regulatory RNA in the pathogenesis of ALS (34). Intriguingly, these disease-related RNA-binding proteins were identified in neuronal RNA granules (35, 36) and with miRNA-associated complexes (3, 37). Similarly, juvenile forms of motor neuron diseases (MND) are related to posttranscriptional regulators of gene expression, namely, SETX [ALS4 locus; (38)], IGHMBP2 (39) and SMN1 (40, 41), the latter functionally engaged in miRNA-protein complexes (42, 43). Plausibly, a considerable portion of the MND spectrum may be directly related to RNA metabolism and posttranscriptional regulation of gene expression.
This emerging appreciation of RNA regulatory function in neurons encouraged us to hypothesize that miRNA may be involved in the pathogenesis of MNDs. In this work, we show that miRNA dysfunction causes spinal muscular atrophy (SMA). Furthermore, we show that the neurofilament heavy subunit (NEFH) previously implicated in MND is specifically up-regulated in Dicer1-deficient MNs.
We additionally relate the down-regulation of the miR-9 gene to changes in neurofilament stoichiometry in both the Dicer1 model and in a murine SMN1 model of SMA. These data provide direct evidence for miRNA malfunction in MNDs and promote further evaluation of miR-9 in neurodegeneration.
Results
Loss of miRNA Activity in the MNDicermut Causes Progressive Locomotor Dysfunction.
Because miRNA makes up the largest group of regulatory RNA (1) and has previously been associated with neurodegenerative states (22, 23, 25, 28), we sought to evaluate its involvement in MN pathologies. To this end, we specifically ablated Dicer1 in postmitotic postnatal MNs, crossing a Dicer1 conditional allele (44) with a Cre-recombinase transgene driven by a cholinergic-specific promoter [vesicular acetyl-choline transporter (VAChT)-Cre.fast (45–47)]. This transgene is expressed in postmitotic somatic MNs as early as postnatal day 7 but is not expressed during development. Because Dicer1 activity is required for miRNA processing in vivo (1, 25, 28), VAChT-Cre.fast;Dicerflx/flx animals (referred to below as “MNDicermut”) lose the ability to make functional miRNA in a subset of postmitotic SMNs, and therefore provide a compelling model for miRNA loss of function in SMNs (details of the mouse genetic system are shown in Fig. S1).
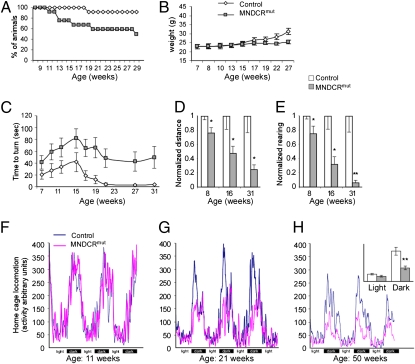
Whereas VAChT-Cre.fast;Dicerflx/+ heterozygous animals (“controls”) are apparently normal and their survival is comparable with that of WT mice, the median life of MNDicermut mice is 29 wk, with the last moribund animal euthanized at 50 wk of age. In addition, MNDicermut mice gained weight slower than control littermates (Fig. 1 A and B). To understand the pathology of these mice better, we conducted a broad series of functional tests to evaluate their locomotor activity. From the age of 7 wk, the MNDicermut mice did not perform as well as controls on a “vertical pole test” (Fig. 1C). Furthermore, a video-monitored “open-field” assay revealed that MNDicermut mice progressively travel shorter distances and rear less than controls (Fig. 1 D and E). In addition, a home-cage study across the circadian cycle indicated that the locomotor activity of MNDicermut mice gradually deteriorates compared with that of controls (Fig. 1 F and H). We hypothesized that this apparent deterioration in activity and locomotor parameters is likely the consequence of muscular atrophy.
Fig. 1.
Inferior survival and motor activity of the MNDicermut mice. (A) Kaplan–Meier survival curve for controls and conditional Dicer1 KO mice (control, n = 12; MNDCRmut, n = 12). The median survival of MNDicermut mice is 29 wk, and it is >60 wk for controls. (B) Weight gain of controls and MNDicermut mice. (C) Time to complete a turn in the pole task for controls and MNDicermut mice. (D and E) Open-field measures at 8, 16, and 31 wk of age. (D) Ratio of distance traveled in the open-field arena compared with the mean of controls (control, n = 12; MNDCRmut, n = 12). (E) Ratio of rearing events performed in the open-field arena compared with the mean of controls (control, n = 12; MNDCRmut, n = 12). Home-cage locomotion of MNDicermut and controls at 11 (F), 21 (G), and 50 (H) wk of age (control, n = 12; MNDCRmut, n = 12). (Inset) Average of activity throughout the measured period. *P < 0.05; **P < 0.01.
MNDicermut Mouse Exhibits Denervation Muscular Atrophy.
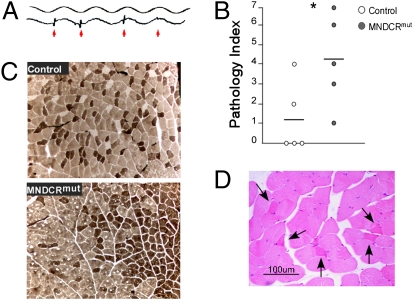
To characterize the muscle phenotype directly, we performed a needle electromyography (EMG) study on the hind-limb interosseous and gastrocnemius muscles. EMG performed on anaesthetized animals revealed frequent fibrillation potentials in the MNDicermut mouse relative to controls (Fig. 2 A and B). These data are consistent with an ongoing denervation process, which probably underlies the progressive locomotive deterioration of the MNDicermut animals and also their observable tremor.
Fig. 2.
MNDCRmut mice exhibit muscular atrophy with signs of denervation. Hind-limb interosseous and gastrocnemius muscle bipolar EMG recording. (A) Representative EMG traces of control (Upper) and MNDicermut (Lower) mice under anesthesia. Frequent fibrillation potentials are annotated by red arrows. (B) EMG Pathology Index was evaluated for individual controls (○; n = 5) and MNDicermut mice (●; n = 5). This scale (range: 0–7) reflects the intensity and frequency of fibrillation potentials in coded mice, noting that the electromyographer was blinded as to the genotype of the mouse tested. (C) Basic ATPase staining of transverse section through control and MNDicermut tibialis anterior muscles. Fiber grouping events were observed only in mutant muscles. (D) H&E staining of transverse section through the tibialis anterior MNDicermut muscle. Angular fibers are marked by arrows. (Scale bar: 100 μm.) *P < 0.05; **P < 0.01.
Histological examination of the MNDicermut tibialis anterior muscle by myosin ATPase reaction further supports neuropathy, because MNDicermut mice frequently exhibited fiber type grouping. This finding is characteristic of a denervation/reinnervation pathology (48) and was not seen in the controls (Fig. 2C). Further, muscle fibers with a large cross-sectional area are specifically lost in the MNDicermut mouse, and the total fiber diameter of the MNDicermut mouse is reduced relative to controls, although this failed to reach statistical significance (Fig. S2). Finally, we depicted angular myofibers on muscle histology, a pathognomonic sign of denervation-related muscular atrophy (Fig. 2D). Taken together, we conclude that MNDicermut animals suffer from denervation muscular atrophy, which suggests loss of spinal motor neurons (SMNs).
MNs Are Lost in the MNDicermut Mouse.
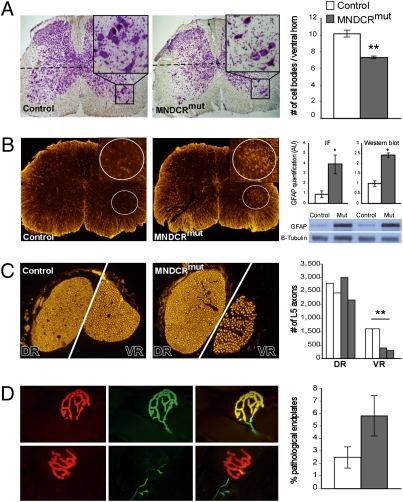
On spinal cord histology, we studied by Nissl staining the numbers of large perikaryon numbers (>20 μm in diameter) in the ventral horn of the lumbar (L4-L5) level of the spinal cord. We noted a significant decrease in large MN numbers in multiple sections throughout the lumbar regions of the MNDicermut mouse relative to controls (average of 7 and 10.1 MNs per section, respectively; depicted in 15 lumbar sections per animal; n = 5 and n = 5, respectively; Fig. 3A).
Fig. 3.
MNDicermut mice exhibit spinal cord ventral horn sclerosis and axonopathy. (A) (Left) Representative Nissl staining of lumbar (L4-L5) sections from a MNDicermut mouse and a control littermate. (Insets) Enlargements of a ventral horn area in each section. The dashed line represents the border under which large-diameter cells (>20 μm) were counted. (Right) Average number of MNs counted per ventral horn section in lumbar spinal cord of 4-mo-old MNDicermut mice and controls (average of 7 and 10.1 MNs per section, respectively; 15 lumbar sections per animal; n = 5 and n = 5, respectively). (B) (Left) Representative lumbar section from 4-mo-old MNDicermut mice and controls immunostained for GFAP. (Right) Quantification of the GFAP immunofluorescence (IF) signal (arbitrary units, 3 lumbar sections per animal; n = 5 and n = 5, respectively) and quantification of GFAP by Western blot analysis of lower spinal cord extracts from MNDicermut mice (“mut”) and controls, normalized to the expression of β-tubulin (arbitrary units; n = 3 and n = 4, respectively). A representative capture from the Western blot analysis is provided for two mutants and two controls. (C) (Left) Representative dorsal (sensory, Left) and ventral (motor, Right) roots used for axon number measurements, stained with anti-NEFM antibody. (Right) Average axon number in dorsal and ventral roots of MNDicermut mice and controls (n = 2 and n = 2, respectively). (D) (Left) Representative hind-limb tibialis anterior NMJ, demonstrating complete overlap (Upper) or partial overlap (Lower) between the postsynapse (red, rhodamine-labeled bungarotoxin) and presynapse (green, mixture of anti-neurofilament and synaptophysin antibodies; yellow, merged channels) components. (Right) Percentage of pathological end plates in MNDicermut mice and controls. These represent 17 aberrant NMJs of 651 NMJs that were individually screened in control mice and 43 aberrant NMJs of 760 NMJs in MNDicermut mice (n = 2 and n = 2, respectively). *P < 0.05; **P < 0.01.
Negative immunoreactivity for both TUNEL and activated caspase-3 was consistent with a typical slow death profile of SMNs encountered in many MNDs.
Reactive astrocytosis is often taken as an indication of neuronal toxicity or neuronal death (49); therefore, we immunoquantified GFAP expression levels by immunofluorescence and Western blotting. We detected enhanced GFAP immunoreactivity in sections of the lateroventral aspect of the lumbar spinal cord of the MNDicermut mice and substantiated this by Western blot analysis that revealed higher levels of GFAP expression in spinal cord extracts of the MNDicermut animals relative to controls (Fig. 3B). These data support reactive astrocytosis and MN loss.
We further evaluated the discrete population of proximal motor axons at the ventral root before they are joined by sensory axons. MNDicermut mice exhibit a significant decrease in MN axon numbers when compared with controls, whereas dorsal root sensory axons remain intact as expected (Fig. 3C).
Signs of Axonopathy in the MNDicermut Mouse.
Dysfunction and/or degeneration of the neuromuscular junction (NMJ) accompanies or even precedes the loss of MN bodies in a few models of ALS (50–52). We went on to evaluate potential distal axonal defects in the MNDicermut mouse. Detailed evaluation of the pre- and postsynaptic compartments of 350 individual NMJs in the hind-limb tibialis anterior revealed that aberrant architecture was twice as frequent in the NMJs of MNDicermut mice relative to controls (Fig. 3D). This is intriguing, because miRNA is known to have distal perisynaptic functions (11, 53–55), suggesting that miRNA-related neuropathy may exist while the axons are still occupying the end plate.
MNDicermut Mouse Fails to Coordinate Neurofilament Subunit Stoichiometry.
Dysregulation of the coordinated expression of the light neurofilament (NEFL), medium neurofilament (NEFM), and NEFH subunits causes axonal cytoskeletal defects (56, 57). For example, NEFL mutations cause type 2E Charcot–Marie–Tooth motor neuropathy (58). Furthermore, experimental perturbation of the fine neurofilament balance in mouse models results in phenotypes closely resembling human MN pathologies (59, 60) and has previously been suggested as a component of human ALS (61–63).
More specifically, posttranscriptional regulation of neurofilament gene expression plays a key role in neuronal well-being (64), and deletion of the NEFH tail was suggested as a component of ALS (61).
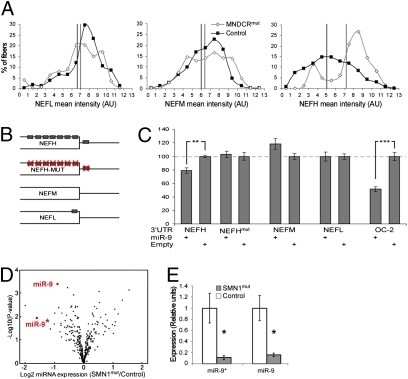
Previous work revealed that neurofilament expression is regulated by the 3′UTR of the mRNA. Further, the 3′UTR appears to interact with an uncharacterized trans-acting factor (65–67) that is attenuated in ALS (68). We reasoned that this ill-characterized trans-acting factor may, in fact, be a miRNA. Thus, we analyzed the relative expression levels of the neurofilament subunit proteins in MNDicermut mice and sibling controls. Quantification of the neurofilament immunofluorescent signal in approximately 2,000 lumbar axons revealed that the expression levels of NEFL and NEFM were comparable with the WT. However, the expression of the heavy subunit (NEFH) is specifically up-regulated in the MNDicermut mouse (Fig. 4A).
Fig. 4.
miR-9 is specifically down-regulated in a model of SMA and is located upstream of coordinated expression of the neurofilament subunits. (A) Binned distribution of neurofilament subunit expression intensity. The percentage of axons at any intensity bin is mentioned on the y axis. NEFL (Left), NEFM (Center), and NEFH (Right). Black and gray lines represent the global mean intensity of control and MNDicermut axons, respectively. (B) Illustration of sequences cloned into luciferase reporter constructs used for functional evaluation of miR-9 interactions with neurofilament subunit mRNAs, wherein NEFHmut stands for seed-mutated NEFH. Gray boxes represent miR-9–binding sites (C) Heterologous luciferase reporter assay reveals that miR-9 may function upstream of the NF subunits. Levels of luciferase activity in HEK293 cells transfected with either an empty vector or a vector overexpressing miR-9. Data are normalized to the activity of a cotransfected β-galactosidase reporter and presented as the percentage of luciferase activity in the absence of miR-9. OC-2 (a fragment of the Onecut2 3′UTR) is used as a positive control. (D and E) WT control mouse ES cells (mESCs) and SMN1mut mESCs harboring a homozygous mSMN1 mutation and two copies of an hSMN2 transgene were differentiated in vitro into MNs. The cells were FACS-purified according to the expression of GFP transgene, driven by the Hlxb9 promoter. (D) Volcano plot exemplifying the log2 ratio of SMN1mut/WT miRNA expression on the x axis and the log10 P value obtained by a two-tailed Student's t test on the y axis. (E) Quantitative PCR analysis of miR-9 and miR-9* expression in MNs derived from SMN1mut mESCs (gray bars) and WT mESCs (empty bars). *P < 0.05; **P < 0.01.
Coordinated Expression of the Neurofilament Subunits Is Achieved by miR-9.
The up-regulation of NEFH in the MNDicermut mouse, and consequent loss of the coordinated expression of the three neurofilament subunits could, in principle, have been attributable to a direct or indirect requirement for miRNA to mediate the levels of these genes. To assess the possibility of direct miRNA regulation, we searched the neurofilament sequences for potential miRNA-binding sites. We found a single miR-9–binding site on the NEFL mRNA. In contrast, the NEFH mRNA harbors nine miR-9–binding sites, dispersed over the 3′UTR of NEFH mRNA and the 3′-portion of the coding region (Fig. 4B). Next, we obtained a heterologous reporter assay to demonstrate that miR-9 is able to modulate the expression of NEFH and that this is dependent on the presence of miR-9–binding sites at the NEFH mRNA (Fig. 4C). These data strongly suggest a model in which the loss of miR-9 expression or activity may result in derepression of NEFH and, subsequently, dysregulation of neurofilament stoichiometry.
miR-9 Is Specifically Down-Regulated in a Model of SMA.
To relate these results to the pathogenesis observed in other models of MND, we profiled miRNA expression levels in MNs carrying an SMN1mut allele, which is characteristic of the pediatric form of SMA (69). Notably, dysregulation of neurofilament expression in the Dicer1 model is reminiscent of the SMN1 mutant phenotype (70), and SMN1 is functionally engaged in miRNA–protein complexes in human cells (42, 43). Thus, we have carried out in vitro differentiation of ES cells harboring an SMN1 mutation into MNs (71, 72). Next, we screened a miRNA microarray (LNA oligo platform; Exiqon) with labeled RNA extracted from FACS-purified SMN1mut MNs. Direct comparison of RNA from WT and SMN1mut MNs revealed that the expression of only a few miRNAs is significantly decreased in SMN1mut MNs. Intriguingly, the most significantly down-regulated miRNAs were miR-9 and miR-9* (Fig. 4D). These two miRNA species are processed from the same hairpin, and quantitative PCR assay revealed up to a 15-fold decrease in the expression of both miR-9 and miR-9* in SMN1mut MNs relative to control (Fig. 4E).
Taken together, we present here a model for SMN disease based on Dicer1 loss of function. In this model, SMN-specific loss of miRNA activity results in denervation muscular atrophy. Additionally, changes in the expression levels of the neurofilament subunits likely contribute to the disease. This phenotype is attributed to dysregulation of miR-9, an upstream regulator of the neurofilament mRNAs. The relevance of miR-9 to MNDs originates from its neuron-specific expression and its dramatic down-regulation in SMN1-deficient MNs. It will be important to explore how miR-9 acts as an effector gene downstream of SMN1 and what are the specific subsets of SMA phenotypes governed by miR-9.
Discussion
The role of miRNA in neurons and the ways by which miRNA is involved in neurological diseases are gradually being uncovered. Thus, the loss of miRNA activity, through recombination of a Dicer1 conditional allele, was shown to cause progressive neurodegeneration in several neuronal systems (21–26, 28). Additionally, changes in the expression of specific miRNAs were reported in neurodegenerative states such as Huntington's chorea and Parkinson's disease.
Consistently, the data presented in this work suggest that miRNA dysfunction results in neurodegeneration of SMNs. As a result of miRNA malfunction in SMNs, mice develop denervation-dependent muscle atrophy. Although MNs die in all forms of ALS and SMA, Dicer1 inactivation in neurons does not always lead to cell death. For example, targeted deletion of Dicer1 in striatal neurons did not result in overt neuronal loss (28).
Early changes in animal activity were noted as early as 2 mo, and signs of the disease can be documented by EMG or histology at 4 mo of age. This suggests relatively slow progression of the disease, which is consistent with the slow onset of cell death in other Dicer1 KO models (25).
The loss of MNs and their dysfunction are often related to aggregation pathologies and to defects in their intermediate filament system (60, 64, 73). In order for the neuron to function properly, neurofilament gene expression must be tightly coordinated. We demonstrated that the coordinated expression of the neurofilament subunits is perturbed by Dicer1 loss of function, because there is specific up-regulation of just the NEFH. We were further able to link this observation to dysregulation of miR-9. The unequal affinity of miR-9 to the three neurofilament subunit mRNAs is apparently attributable to the differing number of miRNA-binding sites (seed matches) on these mRNAs. Thus, the NEFH mRNA harbors nine miR-9–binding sites, whereas only a single seed match is positioned within the NEFL mRNA. Consequentially, loss of miRNA activity affects these target genes differentially, as is revealed by direct axon immunostaining in the Dicer1 model and by a reporter assay, wherein miR-9 overexpression affects NEFH expression only in the presence of miR-9 seed-match sequences on NEFH 3′UTR. Together, these data imply a unique role for miRNA in parallel fine-tuning of related genes, whose coordinated expression should be tightly controlled. Our data on miR-9 may explain previous observations describing the probable presence of a trans-acting factor, acting at a posttranscriptional level in the regulation of proper neurofilament stoichiometry (61, 65–68).
miR-9 is a highly conserved neuronal-specific miRNA that has been shown to be involved in many facets of neurobiology. In flies, miR-9 was shown to be important for proper selection of neuronal precursors from the neuroepithelium (10), and in zebrafish, it was shown to be involved in the setting of the midbrain-hindbrain boundary (18). In mammals, the miR-9 gene is involved in a feedback loop with the repressor element-1 silencing transcription factor and its co-factor complex (REST/Co-REST) complex and acts alongside miR-124 in switching of BAF chromatin-remodeling complexes in neural development (19, 20).
The potential role of miR-9 in coordinated regulation of the neurofilament subunits corresponds to previous observations. Specifically, the neurofilament 3′UTR is essential for regulation of their expression (64). Further, we have shown that miR-9 is located downstream of SMN1, and may therefore mediate intermediate filament defects reported in SMA (70).
In the future, gain- and loss-of-function studies may help to clarify whether the neurofilament defects that are observed in many MN diseases may be modified by manipulations of miR-9 expression and what additional facets of SMN1-dependent SMA are attributed to miR-9 function.
A functional role for miRNA in specific neurological processes emerges when our observations of miR-9 action upstream of neurofilament expression are considered together with reports suggesting roles for other miRNAs, such as miR-132, miR-134, miR-124 and miR-138 (9, 11, 13, 14), in mature neurons. Importantly, Williams et al. (74) recently showed that deleting the gene encoding miR-206 in G93A-Sod1 mice accelerated the progression of ALS symptoms and shortened survival, suggesting that miR-206 has a neuroprotective role in the postsynaptic compartment after nerve damage. This is likely, because miR-206 normally represses histone deacetylase 4, an established inhibitor of muscle reinnervation. Thus, miRNA dysfunction has direct relevance for our understanding of neurodegenerative disorders perturbing the regulation of specific target genes at the neuron or in the innervated myofiber.
The RNA-binding capability of proteins involved in MN pathologies (i.e., TDP-43 and FUS/TLS as well as SETX, SMN1, and IGHMBP2) implies that a considerable number of diseases within the MN spectrum may be directly related to RNA metabolism and posttranscriptional regulation of gene expression. Indeed, in our study, we were able to show that loss of SMN1 activity in cultured ES cell-derived MNs affects the specific expression of a subset of miRNAs, including expression of the neuronal miR-9 gene. Taken together with SMN1 physical engagement in miRNA–protein complexes (7, 71), it is plausible that SMN1 functions in miRNA bioprocessing in neurons.
The proteins TDP43 and FUS/TLS have recently revolutionized the way in which ALS is viewed, implying a pivotal role for defects in RNA regulation (30–34). Strikingly, these two proteins appear to interact physically with Drosha (3). Therefore, one possibility is that either FUS/TLS, TDP-43, or both are involved in microprocessing. This is further supported by a recent report suggesting direct role for TDP-43 in the processing of at a few miRNA (37). However, these proteins were reported to be associated with RNA transport in neurons (35, 36), suggesting that they may have RNA-related regulatory roles in the cytoplasm. Finally, TDP-43 binds and regulates expression of the NEFL subunit through its 3′UTR (75), providing an intriguing hypothesis that TDP-43 may work as a cofactor of the Argonaute silencing complex.
In summary, the data presented in this work provide direct evidence for the role of miRNA in MND and substantiate our understanding of miRNA-related neurodegenerative states in general. Initial support for a functional relationship of miRNA with proteins, such as TDP-43, FUS/TLS, and SMN1, should encourage revision of MN pathologies and further exploration of miRNA-based mechanisms in ALS pathogenesis and related diseases.
Materials and Methods
Animals.
We crossed a Dicer1 conditional allele (43) with a Cre-recombinase transgene driven by a cholinergic-specific promoter (VAChT-Cre) (45) (Fig. S1). Protocols for the behavioral examinations are described in SI Text. Needle EMG was performed with a bipolar electrode inserted into the hind-limb interosseous and gastrocnemius muscles. A scale (range: 1–7) designated the “EMG Pathology Index,” which reflects the intensity and frequency of fibrillation potentials, is described in SI Text. Spinal cord, ventral root, and muscle tissue preparation; staining protocols; and the antibodies used are described in SI Text.
Differentiation of MNs in Culture.
Mouse ES cells from a Tg(Hlxb9-GFP)1Tmj Tg(SMN2)89Ahmb Smn1tm1Msd/J mouse (stock no. 006570; Jackson Laboratory) (69) were differentiated into MNs as previously described (71, 72). Labeled RNA was hybridized onto a miRCURY LNA microarray (Exiqon). quantitative PCR assays for miR-9 and miR-9* were performed with Taqman (Applied Biosystems). Constructs for the neurofilament luciferase assays are described in SI Text.
Supplementary Material
Acknowledgments
We thank Menachem Segal, Eithan Galun, Avraham Yaron, and Yoram Groner for feedback on the work; Mike Fainzilber and Benny Shilo for remarks on the manuscript; Tali Zimmerman and Judith Chermesh for mouse husbandry; Dena Leshkowitz and Ester Feldmesser for statistics; and Cherill Banks for editorial assistance. E.H. is the incumbent of the Helen and Milton A. Kimmelman Career Development Chair. A.C. is the incumbent of the Philip Harris and Gerald Ronson Career Development Chair. This work was supported by research grants (to E.H.) from the Israel Science Foundation Legacy program, Britain-Israel Research and Academic Partnership, Nella and Leon Benoziyo Center for Neurological Disease, Estate of Flourence Blau, and Wolfson Family Charitable Trust for miRNA and by research grants (to A.C.) from the Israel Science Foundation, Israel Ministry of Health, Nella and Leon Benoziyo Center for Neurosciences, Roberto and Renata Ruhman, Mr. and Mrs. Mike Kahn, Mr. Jorge David Ashkenazi, and Mr. and Mrs. Barry Wolfe. Work at the lab of L.L.R. is supported by the Spinal Muscular Atrophy Foundation and by the Harvard Stem Cell Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006151107/-/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 3.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 6.Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 7.Kim VN, Jinju Han J, Siom MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 8.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 9.Vo N, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 12.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314:2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel G, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 17.Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Leucht C, et al. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 19.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 22.Hebert SS, De Strooper B. Molecular biology: miRNAs in neurodegeneration. Science. 2007;317:1120–1224. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1179–1180. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson PT, Keller JN. RNA in brain disease: No longer just “the messenger in the middle.”. J Neuropathol Exp Neurol. 2007;66:461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- 25.Schaefer A, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuellar TL, et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci USA. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cogswell , et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimer's Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 30.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 31.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 34.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: The FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiebler MA, Bassell GJ. Neuronal RNA granules: Movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 36.Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 37.Buratti E, et al. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010;277:2268–2281. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen YZ, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grohmann K, et al. Mutations in the gene encoding immunoglobulin mubinding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat Genet. 2001;29:75–77. doi: 10.1038/ng703. [DOI] [PubMed] [Google Scholar]
- 40.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 42.Dostie J, et al. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mourelatos Z, et al. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misawa H, et al. VAChT-Cre.Fast and VAChT-Cre.Slow: Postnatal expression of Cre recombinase in somatomotor neurons with different onset. Genesis. 2003;37:44–50. doi: 10.1002/gene.10224. [DOI] [PubMed] [Google Scholar]
- 46.Yamanaka K, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Misawa H, et al. Conditional knockout of Mn superoxide dismutase in postnatal motor neurons reveals resistance to mitochondrial generated superoxide radicals. Neurobiol Dis. 2006;23:169–177. doi: 10.1016/j.nbd.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 48.Morris CJ, Woolf AL. Mechanism of type 1 muscle fibre grouping. Nature. 1970;226:1061–1062. doi: 10.1038/2261061a0. [DOI] [PubMed] [Google Scholar]
- 49.Hall ED, Oostveen JA, Gurney ME. Relationship of microglial and astrocytic activation to disease onset and progression in a transgenic model of familial ALS. Glia. 1998;23:249–256. doi: 10.1002/(sici)1098-1136(199807)23:3<249::aid-glia7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 50.Kennel PF, Finiels F, Revah F, Mallet J. Neuromuscular function impairment is not caused by motor neurone loss in FALS mice: An electromyographic study. NeuroReport. 1996;7:1427–1431. doi: 10.1097/00001756-199605310-00021. [DOI] [PubMed] [Google Scholar]
- 51.Frey D, et al. Early and selective loss of neuromuscular synapse subtypes with low sprouting competence in motoneuron diseases. J Neurosci. 2000;20:2534–2542. doi: 10.1523/JNEUROSCI.20-07-02534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer LR, et al. Amyotrophic lateral sclerosis is a distal axonopathy: Evidence in mice and man. Exp Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Corbin R, Olsson-Carter K, Slack F. The role of microRNAs in synaptic development and function. BMB Rep. 2009;42:131–135. doi: 10.5483/bmbrep.2009.42.3.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiore R, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Julien JP. Neurofilament functions in health and disease. Curr Opin Neurobiol. 1999;9:554–560. doi: 10.1016/S0959-4388(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 57.Liem RK, Messing A. Dysfunctions of neuronal and glial intermediate filaments in disease. J Clin Invest. 2009;119:1814–1824. doi: 10.1172/JCI38003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mersiyanova IV, et al. A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am J Hum Genet. 2000;67:37–46. doi: 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Côté F, Collard JF, Julien JP. Progressive neuronopathy in transgenic mice expressing the human neurofilament heavy gene: A mouse model of amyotrophic lateral sclerosis. Cell. 1993;73:35–46. doi: 10.1016/0092-8674(93)90158-m. [DOI] [PubMed] [Google Scholar]
- 60.Lariviere RC, Julien JP. Functions of intermediate filaments in neuronal development and disease. J Neurobiol. 2004;58:131–148. doi: 10.1002/neu.10270. [DOI] [PubMed] [Google Scholar]
- 61.Al-Chalabi A, et al. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum Mol Genet. 1999;8:157–164. doi: 10.1093/hmg/8.2.157. [DOI] [PubMed] [Google Scholar]
- 62.Figlewicz DA, et al. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum Mol Genet. 1994;3:1757–1761. doi: 10.1093/hmg/3.10.1757. [DOI] [PubMed] [Google Scholar]
- 63.Tomkins J, et al. Novel insertion in the KSP region of the neurofilament heavy gene in amyotrophic lateral sclerosis (ALS) NeuroReport. 1998;9:3967–3970. doi: 10.1097/00001756-199812010-00036. [DOI] [PubMed] [Google Scholar]
- 64.Thyagarajan A, Strong MJ, Szaro BG. Post-transcriptional control of neurofilaments in development and disease. Exp Cell Res. 2007;313:2088–2097. doi: 10.1016/j.yexcr.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 65.Lin H, Zhai J, Schlaepfer WW. RNA-binding protein is involved in aggregation of light neurofilament protein and is implicated in the pathogenesis of motor neuron degeneration. Hum Mol Genet. 2005;14:3643–3659. doi: 10.1093/hmg/ddi392. [DOI] [PubMed] [Google Scholar]
- 66.Nie Z, et al. Untranslated element in neurofilament mRNA has neuropathic effect on motor neurons of transgenic mice. J Neurosci. 2002;22:7662–7670. doi: 10.1523/JNEUROSCI.22-17-07662.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cañete-Soler R, Silberg DG, Gershon MD, Schlaepfer WW. Mutation in neurofilament transgene implicates RNA processing in the pathogenesis of neurodegenerative disease. J Neurosci. 1999;19:1273–1283. doi: 10.1523/JNEUROSCI.19-04-01273.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ge WW, Leystra-Lantz C, Wen W, Strong MJ. Selective loss of trans-acting instability determinants of neurofilament mRNA in amyotrophic lateral sclerosis spinal cord. J Biol Chem. 2003;278:26558–26563. doi: 10.1074/jbc.M302886200. [DOI] [PubMed] [Google Scholar]
- 69.Monani UR, et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 70.Cifuentes-Diaz C, et al. Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model. Hum Mol Genet. 2002;11:1439–1447. doi: 10.1093/hmg/11.12.1439. [DOI] [PubMed] [Google Scholar]
- 71.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 72.Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiao S, McLean J, Robertson J. Neuronal intermediate filaments and ALS: A new look at an old question. Biochim Biophys Acta. 2006;1762:1001–1012. doi: 10.1016/j.bbadis.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Williams AH, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strong MJ, et al. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.