Abstract
Estrogen receptors (ERs) protect pancreatic islet survival in mice through rapid extranuclear actions. ERα also enhances insulin synthesis in cultured islets. Whether ERα stimulates insulin synthesis in vivo and, if so, through which mechanism(s) remain largely unknown. To address these issues, we generated a pancreas-specific ERα knockout mouse (PERαKO−/−) using the Cre-loxP strategy and used a combination of genetic and pharmacologic tools in cultured islets and β cells. Whereas 17β-estradiol (E2) treatment up-regulates pancreatic insulin gene and protein content in control ERαlox/lox mice, these E2 effects are abolished in PERαKO−/− mice. We find that E2-activated ERα increases insulin synthesis by enhancing glucose stimulation of the insulin promoter activity. Using a knock-in mouse with a mutated ERα eliminating binding to the estrogen response elements (EREs), we show that E2 stimulation of insulin synthesis is independent of the ERE. We find that the extranuclear ERα interacts with the tyrosine kinase Src, which activates extracellular signal-regulated kinases1/2, to increase nuclear localization and binding to the insulin promoter of the transcription factor NeuroD1. This study supports the importance of ERα in β cells as a regulator of insulin synthesis in vivo.
Keywords: diabetes, islet
Several lines of evidence suggest that the female hormone 17β-estradiol (E2) improves insulin production. First, a sex dimorphism in diabetic syndromes associated with insulin deficiency has been reported (1). Second, during pregnancy, when circulating E2 concentrations are elevated, an increase in insulin biosynthesis in isolated rat islets of Langerhans has been observed (2, 3). Third, during the estrous cycle in rats, insulin gene expression has been found to positively correlate with serum E2 level (4).
In physiological conditions, short-term glucose stimulation of insulin biosynthesis is regulated predominantly through increased translation of preproinsulin mRNA (5, 6). During prolonged exposure to glucose, increased insulin gene transcription (7–9) and increased preRNA splicing (10) also contribute to maintenance of insulin biosynthesis.
E2 classically exerts its genomic effects by activating nuclear estrogen receptors (ERs) that bind to estrogen response elements (EREs) on the promoter of target genes or through a non-ERE tethering mechanism involving AP1 or SP1 sites (11). E2 also regulates the activity of target genes through activation of extranuclear ERs or the membrane-bound G protein–coupled ER (GPER). ERs are present in the islet β cells (12–14), and we have previously shown that E2 acting through ERα and GPER in β cells favors β cell survival in mice of both sexes (13, 14). Alonso-Magdalena et al. (12) recently reported that E2 also increases pancreatic insulin content through ERα in cultured islets. Whether E2 stimulates insulin synthesis in vivo through a direct effect on ERα in β cells and, if so, through which mechanism(s) remain unknown, however. To address these issues, we generated a pancreas-specific ERα knockout mouse (PERαKO−/−) using the Cre-loxP strategy and used a combination of genetic and pharmacologic tools in cultured islets and MIN6 β cells.
Results
E2 Increases Insulin Synthesis Through ERα.
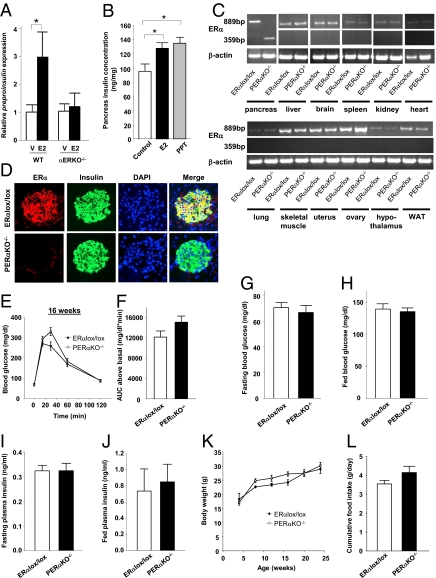
To investigate the roles of ERα, ERβ, and GPER in insulin synthesis, we treated cultured ERα-, ERβ-, and GPER-deficient islets with E2 and studied insulin gene transcription. Whereas E2 stimulated preproinsulin expression by 3-fold in WT islets, this effect was abolished in αERKO−/− mice (Fig. 1A). Conversely, E2 stimulated preproinsulin expression in βERKO−/− and GPERKO−/− islets to a similar extent as that of WT islets (Fig. S1 A and B). To confirm the role of ERα in insulin synthesis in vivo, we treated WT mice with E2 and the selective ERα agonist propyl-pyrazole-triol (PPT) (15). These E2 and PPT treatments produced a 1.3-fold and 1.4-fold increase in pancreas insulin concentration, respectively (Fig. 1B). Taken together, these findings confirm the importance of ERα in mediating E2-induced insulin synthesis.
Fig. 1.
Characterization of the pancreas-specific ERα knockout mice. (A) Relative preproinsulin expression in WT and αERKO−/− islets (n = 4–10) after treatment with E2 (10−8 M) for 48 h. (B) Pancreatic insulin concentration in female WT mice treated with either E2 (4 μg/d) or PPT (100 g/d) for 7 d (n = 6–7/group). (C) ERα mRNA expression in various tissues from PERαKO−/− female mice. (D) Coimmunolocalization of ERα and insulin in islets from female PERαKO−/− and ERαlox/lox mice. (E and F) Glucose tolerance (E) and corresponding (F) area under the curve (AUC) for glucose in 16-wk-old female PERαKO−/− and ERαlox/lox mice (n = 6–12/group). (G–J) Fasting blood glucose (G), fed blood glucose (H), fasting plasma insulin concentration (I), and fed plasma insulin concentration (J) in female PERαKO−/− mice (n = 6–20/group). (K and L) Body weight (K) and cumulative food intake (L) in female PERαKO−/− and ERαlox/lox mice. V, vehicle. Results in A, B, and E–L are expressed as mean ± SEM. *P < 0.05.
Generation of PERαKO−/− Mice.
To investigate whether ERα is involved in insulin synthesis through a direct effect on islets in vivo, we generated PERαKO−/− mice using the Cre-loxP strategy. We confirmed recombination of the ERα gene in the pancreas and the presence of normal ERα transcript in all other organs of the PERαKO−/− mice (Fig. 1C). We observed the absence of ERα protein in PERαKO−/− compared with control ERαlox/lox islets, confirming the efficiency of recombination (Fig. 1D). No significant differences in fasting and fed blood glucose concentrations and i.p. glucose tolerance were observed between PERαKO−/− and ERαlox/lox mice at age 8 wk (Fig. S2 A–D), 16 wk (Fig. 1 E–H), and 24 wk (Fig. S2 E–H). Fasting and fed plasma insulin concentrations were also similar in the PERαKO−/− and ERαlox/lox mice (Fig. 1 I and J). Female PERαKO−/− and ERαlox/lox mice had similar body weight (Fig. 1K) and food intake (Fig. 1L) up to age 24 wk.
ERα in Islets Stimulates Insulin Synthesis in Vivo.
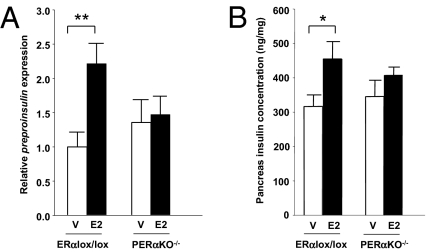
To determine whether E2-induced insulin synthesis was mediated through ERα in the islets in vivo, we treated ERαlox/lox and PERαKO−/− mice with E2. After the E2 treatment, preproinsulin expression (Fig. 2A) and pancreatic insulin concentration (Fig. 2B) were similarly increased in ERαlox/lox mice, but not in PERαKO−/− mice (Fig. 2 A and B). This finding indicates that the islet ERα is essential to E2-induced insulin synthesis in vivo.
Fig. 2.
E2-induced islet insulin synthesis is mediated through ERα in vivo. Preproinsulin expression (A) and pancreas insulin concentration (B) after treatment with E2 (4 μg/d) for 4 d in female ERαlox/lox and PERαKO−/− mice (n = 10–14/group). V, vehicle. Results are expressed as mean ± SEM. *P < 0.05; **P < 0.01.
ERα Amplifies Glucose-Stimulated Insulin Gene Transcription Independently of the Estrogen Response Element.
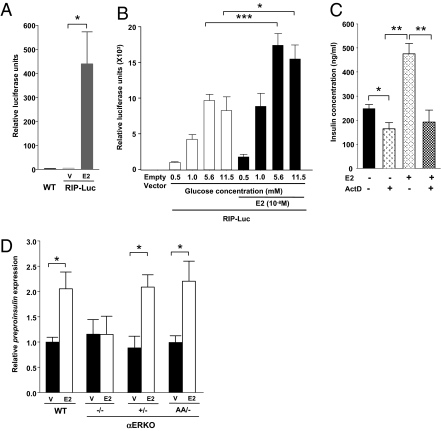
Exposure of pancreatic β cells to elevated glucose concentrations activates the transcriptional control of insulin biosynthesis (7–9). ERα is a ligand-activated transcription factor that regulates the expression of multiple genes in target tissues. To study the effect of E2 on the insulin promoter in vivo, we used a transgenic mouse expressing a rat insulin promoter fused to firefly luciferase selectively in pancreatic islets (16). We observed that in vivo treatment with E2 significantly increased insulin promoter activity in islets (Fig. 3A), demonstrating that the E2-ERα axis activates the insulin promoter in vivo.
Fig. 3.
ERα stimulates insulin promoter activity independently of the ERE. (A) Effect of E2 on insulin promoter activity in vivo. Five-week-old prepubertal female RIP-Luc transgenic mice were injected with E2 (100 μg) 24 h before islet isolation and measurement of luciferase activity from islet homogenates (n = 3–4/group). (B) Effects of glucose and E2 on insulin promoter activity in vitro. INS-1 cells were transfected with RIP-Luc. Relative luciferase activity was measured after 6 h of treatment with indicated glucose concentrations in the absence or presence of E2 (10−8 M). (C) Effects of E2 (10−8 M) and actinomycin D (2 × 10−6 M) treatment for 48 h on insulin concentration in WT islets (n = 3–5/group). (D) Effects of E2 (10−8 M) treatment for 48 h on preproinsulin expression in WT, αERKO−/−, and αERKOAA/- islets (n = 3–6/group). V, vehicle. Results represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
We next examined whether E2-stimulated insulin gene transcription was mediated through a direct effect on the insulin gene promoter in vitro. To do so, we transfected rat INS-1 β cells with a rat insulin promoter (RIP) reporter construct. Consistent with the effect of glucose on insulin gene transcription, glucose increased insulin promoter activity (Fig. 3B). Exposure to E2 at physiological concentrations amplified the effect of glucose at 5.6 mM and 11 mM (Fig. 3B). To confirm that transcription of the insulin gene was the main mechanism of increased insulin content, we incubated E2-treated islets with the transcription inhibitor actinomycin D. At 11 mM glucose, actinomycin D significantly decreased basal islet insulin concentration, consistent with the importance of insulin transcription. We interpreted the failure of actinomycin D to totally suppress basal islet insulin concentration as an indication that insulin mRNA translation is predominant in promoting insulin biosynthesis (Fig. 3C). However, actinomycin D completely abrogated the stimulatory effect of E2 on islet insulin concentration, suggesting that the effect of E2 is transcriptionally mediated (Fig. 3C).
In the classical ER-signaling pathway, E2-activated ERα binds as a homodimer to ERE on the promoter of target genes to initiate gene transcription (11). To investigate whether ERα signals through an ERE to activate insulin transcription, we used a knock-in mouse with a mutation in the first zinc finger of the DNA-binding domain of ERα, which eliminates ERα binding to the ERE (17). We compared preproinsulin expression in cultured αERKOAA/- islets with only one mutant ERα allele and preproinsulin expression in αERKO+/− islets with one functional ERα allele. This allowed us to study the effect of the unique αERKOAA/- mutant allele on preproinsulin expression after E2 treatment. E2 induced a 2-fold increase in preproinsulin expression in WT islets, which was abolished in αERKO−/− islets (Fig. 3D). Conversely, the E2-induced increase in preproinsulin was unaffected in islets from αERKO+/− and αERKO AA/- mice, demonstrating that E2-induced insulin transcription requires only one copy of ERα and is independent of the classical ERE pathway (Fig. 3D).
Extranuclear ERα Stimulates Insulin Synthesis.
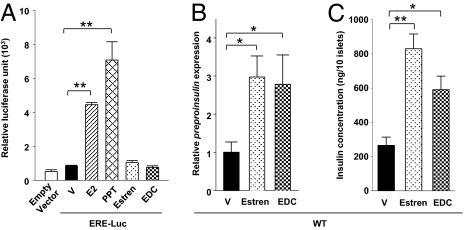
Having established that ERα signaling enhances insulin transcription in β cells through an ERE-independent mechanism increasing insulin promoter activity, we explored the possibility that insulin transcription is regulated by an extranuclear ERα. To address this, we studied islet insulin expression following exposure to two pharmacological probes specific for ER nongenomic actions: (i) estren, a synthetic ER ligand with minor transcriptional activity (18) and (ii) an estrogen dendrimer conjugate (EDC) that remains outside the nucleus and selectively activates extranuclear, nongenomic estrogen actions (19). Compared with E2 and PPT (15), estren and EDC showed no transcriptional activity in MIN6 β cells transfected with the ERE reporter construct (Fig. 4A). When cultured islets from WT female mice were treated with these compounds, estren and EDC similarly increased islet preproinsulin expression (Fig. 4B), which was associated with a corresponding rise in islet insulin concentration (Fig. 4C). Taken together, the findings demonstrate that E2-mediated amplification of glucose-induced insulin transcription is mediated through extranuclear signaling of ERα that results in activation of the insulin promoter.
Fig. 4.
E2-induced insulin synthesis involves extranuclear localization of ERα. (A) MIN6 cells were transfected with an ERE reporter construct (ERE-Luc). Relative luciferase activity was measured after treatment with E2 (10−8 M), PPT (10−8 M), estren (10−8 M), and EDC (10−8 M) for 48 h. (B and C) Effects of estren (10−8 M) and EDC (10−8 M) treatment on preproinsulin expression (B) and insulin concentration (C) WT mouse islets. V, vehicle. Results are reported as mean ± SEM. *P < 0.05; **P < 0.01.
ERα Stimulates Insulin Synthesis through a Src/MAPK Pathway.
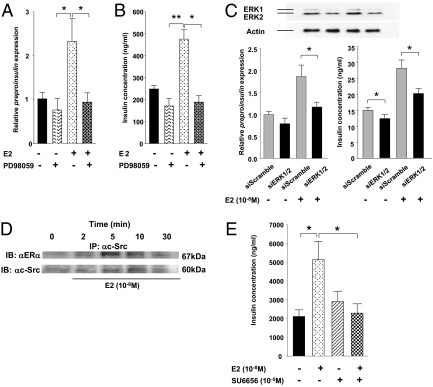
Recently, it was suggested that E2 increases cultured islet insulin content through Src and ERK (12). Having determined that ERα signals outside the nucleus to amplify the stimulatory effect of glucose on preproinsulin transcription, we next investigated whether extranuclear ERα activates ERK1/2 to stimulate insulin synthesis. We found that E2 rapidly increases the phosphorylation of ERK1/2 in MIN6 cells (Fig. S3A). E2-induced ERK1/2 phosphorylation was accompanied by nuclear translocation of a fraction of phosphorylated ERK1/2, which also was observed after treatment with PPT and EDC (Fig. S3B). Accordingly, E2-induced ERK1/2 phosphorylation was blocked by PD98059, the specific inhibitor of phosphorylation of the upstream kinase MEK (Fig. S3C). Thus, inhibition of ERK1/2 phosphorylation by PD98059 abrogated the amplifying effects of E2 on islet preproinsulin expression (Fig. 5A) and islet insulin content (Fig. 5B).
Fig. 5.
ERα amplifies insulin synthesis through the Src/ERK pathway. (A and B) Inhibition of E2-induced preproinsulin expression (A) and rise in insulin concentration by ERK inhibition (B) in WT mice islets. Female mouse islets were treated with E2 (10−8 M) or PD98059 (10−5 M) for 48 h before measurement of preproinsulin expression and insulin concentration. (C) Suppression of insulin synthesis by siRNA of ERK1/2 in MIN6. Cells were transfected with ERK1/2 siRNA for 5 h at 37 °C. ERK1/2 expression (blot), preproinsulin mRNA (Left), and insulin concentration (Right) was determined by Western blot analysis, RT-PCR, and RIA, respectively, after E2 (10−8 M) treatment for 48 h. All blots shown are representative of triplicate experiments. (D) Immunoprecipitation of c-Src in MIN6 cells after E2 stimulation (10−8 M) at 0, 2, 5, 10, and 30 min. (E) Inhibition of E2-induced rise in islet insulin content by SU6656 in WT mice islets (n = 5–6/group). Results represent mean ± SEM. *P < 0.05; **P < 0.01.
To specifically evaluate the involvement of ERK1/2 in ERα-induced insulin expression, we performed ERK1/2 knockdown by siRNA in MIN6 β cells (Fig. 5C). We eliminated 80% of ERK1 proteins and 50% of ERK2 proteins in our siRNA system, because total elimination of ERK2 activates apoptosis (Fig. 5C). In MIN6 cells transfected with control siRNA, E2 induced an 80% increase in both preproinsulin expression and insulin concentration (Fig. 5C). Conversely, in ERK1/2 knockdown cells, E2 demonstrated a significantly impaired ability to augment preproinsulin expression and insulin concentration (Fig. 5C). Furthermore, after E2 stimulation, ERα rapidly complexed with the tyrosine kinase c-Src in MIN6 cells (Fig. 5D). E2-induced ERK1/2 phosphorylation (Fig. S4B) and nuclear translocation (Fig. S4A) were blocked by the Src family tyrosine kinase inhibitor PP1. Furthermore, pharmacologic inhibition of Src by PP1 completely abrogated the stimulatory effects of E2 on islet preproinsulin mRNA expression (Fig. S4C) and islet insulin content (Fig. S4D). The inhibition of E2-mediated increase in islet insulin synthesis was reproduced using the more selective Src family kinase inhibitor SU6656 (20) (Fig. 5E).
ERα Amplifies NeuroD1 Nuclear Localization and Binding to the Insulin Promoter.
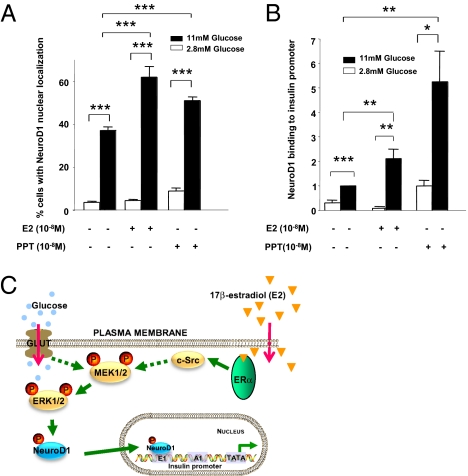
Once activated by glucose, ERK1/2 is instrumental in the activation of insulin gene transcription by phosphorylating the transcription factors NeuroD1 and PDX-1, which then translocate to the nucleus to activate the insulin promoter (21, 22). Taking NeuroD1 as a paradigm of glucose-dependent insulin transcription (21, 22), we sought to determine whether E2 and PPT could amplify glucose-induced NeuroD1 nuclear translocation in MIN6 cells. At low glucose concentrations, NeuroD1 immunoreactivity was mainly cytosolic (Fig. 6A and Fig. S5A). As described previously (23), exposure to 11 mM glucose caused NeuroD1 to translocate into the nucleus, leading to a predominant nuclear localization in ∼40% of cells (Fig. 6A). Treatment with E2 and PPT did not significantly modify NeuroD1 nuclear fraction at low glucose levels; rather, E2 and PPT amplified the effect of 11 mM glucose, leading to a significant increase in MIN6 cells with predominant nuclear localization (Fig. 6A and Fig. S5A).
Fig. 6.
ERα-induced insulin transcription involves NeuroD1 binding to the insulin promoter. (A) Percentage of MIN6 cells with predominant nuclear localization of NeuroD1 in the presence of E2 (10−8 M) or PPT (10−8 M) at 2.8 mM and 11 mM glucose was calculated as described in Materials and Methods. (B) ChIP showing the recruitment of NeuroD1 to the insulin promoter after 24 h of treatment with E2 (10−8 M) or PPT (10−8 M) at 2.8 mM and 11 mM glucose in MIN6 cells. Results were normalized to 11 mM glucose with vehicle (V) and represent mean ± SEM. (C) Proposed mechanism of E2 amplification of insulin gene transcription. *P < 0.05; **P < 0.01; ***P < 0.001.
We next sought to determine whether ERα could amplify glucose-induced binding of NeuroD1 to the insulin promoter by performing ChIP of NeuroD1 in MIN6 cells. We found that NeuroD1 binding to the mouse insulin promoter was dramatically induced after exposure to 11 mM glucose (Fig. 6B), and also that NeuroD1 binding to the insulin promoter at 11 mM glucose was significantly enhanced in the presence of E2 and PPT. To rule out the possibility that ERα binds directly to the insulin promoter, we also performed ChIP of ERα with the insulin promoter (Fig. S6A). At 11 mM glucose, E2 induced the binding of ERα to an IGF1 gene sequence containing an ERE (24) (Fig. S6A). Conversely, under the same conditions, we observed no binding of ERα to the mouse insulin promoter (Fig. S6B). Thus, ERα amplification of preproinsulin transcription involves an increase in NeuroD1 nuclear fraction and binding to the insulin promoter.
Discussion
We report a functional role for ERα in vivo in islets. We have shown that extranuclear ERα amplifies insulin synthesis through direct action on islets by amplifying the effect of glucose on NeuroD1-induced insulin gene transcription. Neither ERβ nor GPER is involved in this process.
The concept of ERα as a nuclear hormone receptor is representative of its functions as a ligand-activated transcription factor. These functions are known to exist in β cells, because E2 and an ERα agonist activate ERE signaling. However, this concept fails to consider other extranuclear roles of ERα, either as a regulator of ion channels or as an initiator of the kinase cascade (25). These nongenomic effects of ERα, which are independent of its direct transcriptional activity, have important biological functions in the proliferation of mammary cancer cells, vasorelaxation in endothelial cells, and prevention of apoptosis in pancreatic β cells (14, 25, 26). A challenge in previous studies of these nongenomic actions of estrogens has been the separation of these actions from those of the genomic pathway, especially in vivo. Using both a mouse lacking ERα in pancreatic islets and a mouse lacking ERE signaling, we have shown that ERα stimulates insulin synthesis in β cells in vivo through an ERE-independent pathway. Using the pharmacologic probe EDC, a molecule that turns on extranuclear ER actions and lacks transcriptional activity in β cells, we have demonstrated that ERα signals outside the nucleus to activate this pathway. Alonso-Magdalena et al. (12) previously reported that ERα increases islet insulin content in culture through the membrane-initiated Src/ERK1/2 pathway. EDC is as potent as E2 in stimulating ERK1/2 phosphorylation through ERα and thus in amplifying insulin expression. We have shown that ERα rapidly associates with the tyrosine kinase Src in β cells. Using more selective pharmacologic and genetic tools to inhibit Src and ERK, we have confirmed the importance of the Src-ERK pathway in mediating ERα amplification of insulin biosynthesis, as was suggested previously (12). The finding that suppression of ERK1 by 80% and ERK2 by only 50% abolished E2-induced insulin synthesis suggests that ERK1 is predominant in the stimulation of insulin synthesis. Both Src and ERK1/2 are clustered in caveolae that are responsible for signal transduction compartmentalization (27). Caveolae are found in β cells (28). Thus, the effect described here might be mediated by a fraction of ERα that is associated with caveolae in the plasma membrane and directly activates Src (25).
Under normal circumstances, glucose regulates insulin biosynthesis through increased translation of preproinsulin mRNA (5, 6) and during prolonged glucose exposure through stimulation of the insulin gene transcription (7–9). Combining an insulin promoter reporter system and experiments of ChIP, we have provided evidence that ERα amplifies glucose-stimulated activation of the insulin promoter in cultured β cells and in mouse islets in vivo without binding to the insulin promoter. Although the effect of E2 appears to be primarily transcriptional, as demonstrated by failure of E2 to increase insulin synthesis in islets after administration of a transcription inhibitor, an effect of E2 on the rate of preproinsulin mRNA translation cannot be ruled out. E2-induced insulin expression occurs predominantly at physiological stimulatory glucose concentrations, suggesting that E2 amplifies the glucose signal. Glucose stimulates insulin transcription by increasing the binding to the insulin promoter of at least three β cell–specific transcription factors: PDX-1, NeuroD1, and MafA (29–31). ERK1/2 is instrumental in glucose-stimulated insulin gene transcription by phosphorylating the transcription factors Neuro-D1 and PDX-1, which directly activate the insulin promoter (21, 22). The mechanistic advance of the present study is the demonstration that activation of ERα potentiates the effect of glucose on NeuroD1 nuclear localization and binding to the insulin promoter, thereby amplifying insulin gene transcription (Fig. 6B).
This study has implications for human physiology. During late pregnancy, when both levels of both E2 and progestin are high, insulin resistance and compensatory β cell hyperplasia develop, amplified by prolactin and placental lactogens (32). E2 amplification of insulin biosynthesis may synergize with prolactin and placental lactogens to boost β cells insulin production to meet the increased metabolic demand of pregnancy. Thus, ERα appears to integrate nutrient-sensing information in the nucleus of β cells to maintain insulin homeostasis. E2, the primary female reproductive hormone, has evolved to favor nutrient storage for pregnancy. In this regard, it is not surprising that E2 amplifies the biosynthesis of the main storage hormone, insulin.
In conclusion, ligand-activated extranuclear ERα amplifies glucose-stimulated insulin gene transcription through a direct islet effect in vivo through the Src/ERK signaling pathway and NeuroD1 activation of the insulin promoter. This study expands our understanding of the regulation of insulin gene transcription and identifies ERα as a target in β cells to enhance insulin synthesis.
Materials and Methods
Generation of Mutant Mice.
The generation, characterization, and genotyping strategy of αERKO−/−, αERKOAA/- (provided by Larry Jameson, Northwestern University), GPERKO (provided by Debbie Clegg, University of Texas Southwestern Medical Center, Dallas), and RIP-Luc mice lines were done as described previously (13, 16, 17). The pancreas-specific ERα knockout mice (PERαKO−/−) were generated by crossing ERαlox+/− with the ERαlox+/− Pdx1-Cre+/− mice (provided by Doug Melton). The primers used to confirm recombination of ERα are listed in Table S1. For GTT analysis, mice were fasted overnight before glucose injection (2 g/kg i.p.). Blood was drawn from tail veins for glucose determination before and at 15, 30, 60, and 120 min after injection using a One-Touch Ultra glucometer (Lifescan). All animal experiments were approved by Northwestern University's Animal Care and Use Committee.
In Vivo Drug Administration and Measurement of Pancreas Insulin.
E2 (4 μg/d; Tocris Biosciences), PPT (100 μg/d) (15), and vehicle (10% ethanol and 90% sesame oil) were administered s.c. twice daily for 4 d. At the completion of drug treatment, pancreases were rapidly excised and mouse islets isolated as described previously (14). Total pancreas and/or islet protein was extracted in acid ethanol overnight at 4 °C before insulin measurement by RIA (Linco).
Measurement of Islet Insulin Concentration.
Mouse islets were cultured in phenol red-free RPMI (11 mM glucose, 10% charcoal-stripped FBS) for 24 h before the experiments. Islets (10 islets/condition) were incubated with E2 (10−8 M), PPT (10−8 M), EDC (10−8 M), estren (10−8 M; Steraloids), PD 98059 (10−5 M; Cayman Chemicals), SU6656 (10−6 M), actinomycin D (8 × 10−6 M; Biomol International), or vehicle (ethanol) for 48 h. Insulin concentration was measured by RIA (Linco).
Immunohistochemistry.
Deparaffinized pancreatic sections were heat-treated for antigenic retrieval in citrate buffer (pH 6) for 1 h before blocking with 5% normal donkey serum and 2% MOM reagent (Vectashield). Sections were incubated overnight at 4 °C with antibodies against insulin (1:1,000; Linco) and ERα (1:100, MC-20; Santa Cruz Biotechnology). The next day, after several washes, sections were incubated with FITC- or Cy3-conjugated antibodies (1:200) at room temperature for 1 h. After several washes, sections were counterstained with DAPI (1:50,000) at room temperature for 5 min and visualized under a fluorescent microscope (Zeiss).
Morphometry.
MIN6 cells were treated with E2 (10−8 M) or PPT (10−8 M) in 2.8 mM or 11 mM glucose DMEM for 16 h. Cells were fixed in acetone for 10 min at 4 °C), blocked with 5% normal donkey serum at room temperature for 1 h, incubated overnight at 4 °C) with goat α-NeuroD1 antibodies (1:100; Santa Cruz Biotechnology) and subsequently with Cy3-conjugated donkey α-goat antibodies (1:200) at room temperature for 1 h, and then counterstained with DAPI (1:50,000) for 10 min before morphometric analysis. Cells with predominant nuclear localization of NeuroD1, as defined previously (23), were counted by two independent observers in four independent experiments using a confocal microscope (Zeiss LSM 510 Meta).
Western Blot Analysis.
MIN6 cells were starved overnight in serum-free phenol red-free RPMI before treatment with E2 (10−8 M). After treatment, cells were homogenized in lysis buffer as described previously (14). ERK1/2 phosphorylation (1:100; Cell Signaling), c-Src (1:100, N-16; Santa Cruz Biotechnology), and ERα (1:100, MC-20; Santa Cruz Biotechnology) were determined by Western blot analysis.
RIP-Luc and ERE-Luc Transfection and Luciferase Assay.
INS-1 cells were cultured in RPMI and plated in 24-well plates (10 × 104cells/well). Cells were transfected with 2 μL of Lipofectamine 2000 (Invitrogen) and 0.8 μg of RIP1-Luc containing a 410-bp DNA sequence upsteam of the transcription start site of the rat insulin promoter 1. Before drug treatment, cells were cultured in RPMI phenol red-free medium supplemented as above for 6 h. For RIP-Luc transfection experiments, cells were treated with glucose (0.5 mM, 1 mM, 5.6 mM, or 11.5 mM) with or without E2 (10−8 M) for 6 h. For ERE-Luc transfection experiments, cells were treated for 24 h with E2 (10−8 M), PPT (10−8 M), estren (10−8 M), or EDC (10−8 M). For measurement of luciferase activity, INS-1 cells or isolated islets from RIP-Luc mice were washed and lysed with Promega Cell Culture Lysis Reagent. After centrifugation, 2–6 μg of protein was used in the Promega Luciferase Assay System. Values are reported as relative luciferase units corrected for protein concentration.
Real-Time RT-PCR and Regular PCR.
Preproinsulin gene expression were quantified in mouse islets by real-time RT-PCR (iCycler; BioRad) and normalized to β-actin expression. In brief, total RNA was extracted in TRIzol (Invitrogen), then 1μg of RNA was reversed-transcribed using the Bio-Rad iScript cDNA Synthesis Kit. RT-PCR primer sequences are available on request. For the characterization of PERαKO−/− mice, after total tissue RNA extraction by TRIzol, 3 μg of cDNA was reverse-transcribed using the Invitrogen SuperScript II kit. After regular PCR, products were separated on 1.2% agarose gel.
ChIP.
MIN6 cells were cultured in phenol red-free DMEM overnight before treatment with V, E2 (10−8 M), or PPT (10−8 M) for 24 h at 2.8 mM or 11 mM glucose. ChIP was performed using a commercially available kit (Upstate Laboratories). After IP of NeuroD1 (N3663; Sigma-Aldrich), real-time qPCR amplification of mouse insulin was performed using Sybr Green (BioRad). The primer sequences are listed in Table S1.
ERK1/2 Silencing in MIN6 Cells.
ERK1/2 expression was silenced in MIN6 cells using mouse 20- to 25-nucleotide prevalidated small interfering RNA (siRNA) duplexes (Santa Cruz Biotechnology). Cells were plated in 12-well plates and grown to 50% confluence in media containing 15% FCS without antibiotics overnight. The next day, transfectant siRNA was prepared according to the manufacturer's instructions. Transfectant siRNA complexes were added to each well to a final concentration of 25 nM/well (0.239 μg). MIN6 cells were transfected with ERK1/2 siRNA at 37 °C for 6 h before being transferred to fresh media containing antibiotics. Cells were treated with E2 (10−8 M) for 48 h and lysed at the completion of the experiment. The amounts of siRNA, siRNA transfection reagent, and siRNA transfection medium were proportionally scaled down relative to the surface area of 12-well plates for insulin quantitation by RIA and preproinsulin mRNA by RT-PCR.
Statistical Analysis.
Results are presented as mean ± SEM unless stated otherwise. Data were analyzed using either the unpaired Student t test or one-way ANOVA, as appropriate. A P value < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Surabhi Bhatt for technical assistance. This reseach was supported by grants from the National Institutes of Health (NIH; RO1 DK074970, P50 HD044405), the Juvenile Diabetes Research Foundation (1-2006-837), and the March of Dimes Birth Defects Foundation (6-FY07-312) (to F.M.J.). The synthesis and characterization of PPT and EDC were funded by NIH Grants R37 DK15556 (to J.A.K.) and CA18119 (to B.S.K.). The generation of the ERαlox/lox mice was funded by the National Institute of Environmental Health Sciences, Division of Intramural Research Grant Z01ES70065 (to K.S.K.). C.L.M. was the recipient of a Juvenile Diabetes Research Foundation postdoctoral fellowship, and J.T. was a trainee funded by Endocrinology Training Grant T32 DK007169.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914501107/-/DCSupplemental.
References
- 1.Louet JF, LeMay C, Mauvais-Jarvis F. Antidiabetic actions of estrogen: Insight from human and genetic mouse models. Curr Atheroscler Rep. 2004;6:180–185. doi: 10.1007/s11883-004-0030-9. [DOI] [PubMed] [Google Scholar]
- 2.Costrini NV, Kalkhoff RK. Relative effects of pregnancy, estradiol, and progesterone on plasma insulin and pancreatic islet insulin secretion. J Clin Invest. 1971;50:992–999. doi: 10.1172/JCI106593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bone AJ, Taylor KW. Mitabolic adaptation to pregnancy shown by increased biosynthesis of insulin in islets of Langerhans isolated from pregnant rat. Nature. 1976;262:501–502. doi: 10.1038/262501a0. [DOI] [PubMed] [Google Scholar]
- 4.Morimoto S, Cerbón MA, Alvarez-Alvarez A, Romero-Navarro G, Díaz-Sánchez V. Insulin gene expression pattern in rat pancreas during the estrous cycle. Life Sci. 2001;68:2979–2985. doi: 10.1016/s0024-3205(01)01100-6. [DOI] [PubMed] [Google Scholar]
- 5.Itoh N, Okamoto H. Translational control of proinsulin synthesis by glucose. Nature. 1980;283:100–102. doi: 10.1038/283100a0. [DOI] [PubMed] [Google Scholar]
- 6.Permutt MA. Effect of glucose on initiation and elongation rates in isolated rat pancreatic islets. J Biol Chem. 1974;249:2738–2742. [PubMed] [Google Scholar]
- 7.Alarcón C, Leahy JL, Schuppin GT, Rhodes CJ. Increased secretory demand rather than a defect in the proinsulin conversion mechanism causes hyperproinsulinemia in a glucose-infusion rat model of non–insulin-dependent diabetes mellitus. J Clin Invest. 1995;95:1032–1039. doi: 10.1172/JCI117748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giddings SJ, Chirgwin J, Permutt MA. Effects of glucose on proinsulin messenger RNA in rats in vivo. Diabetes. 1982;31:624–629. doi: 10.2337/diab.31.7.624. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen DA, Welsh M, Casadaban MJ, Steiner DF. Control of insulin gene expression in pancreatic beta cells and in an insulin-producing cell line, RIN-5F cells, I: Effects of glucose and cyclic AMP on the transcription of insulin mRNA. J Biol Chem. 1985;260:13585–13589. [PubMed] [Google Scholar]
- 10.Wang J, et al. Regulation of insulin preRNA splicing by glucose. Proc Natl Acad Sci USA. 1997;94:4360–4365. doi: 10.1073/pnas.94.9.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 12.Alonso-Magdalena P, et al. Pancreatic insulin content regulation by the estrogen receptor ERa. PLoS ONE. 2008;3:1–11. doi: 10.1371/journal.pone.0002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le May C, et al. Estrogens protect pancreatic beta cells from apoptosis and prevent insulin-deficient diabetes mellitus in mice. Proc Natl Acad Sci USA. 2006;103:9232–9237. doi: 10.1073/pnas.0602956103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, et al. Importance of extranuclear estrogen receptor-alpha and membrane G protein–coupled estrogen receptor in pancreatic islet survival. Diabetes. 2009;58:2292–2302. doi: 10.2337/db09-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stauffer SR, et al. Pyrazole ligands: Structure–affinity/activity relationships and estrogen receptor-alpha–selective agonists. J Med Chem. 2000;43:4934–4947. doi: 10.1021/jm000170m. [DOI] [PubMed] [Google Scholar]
- 16.Smith SJ, et al. In vivo monitoring of pancreatic beta cells in a transgenic mouse model. Mol Imaging. 2006;5:65–75. [PubMed] [Google Scholar]
- 17.Jakacka M, et al. An estrogen receptor (ER)alpha deoxyribonucleic acid–binding domain knock-in mutation provides evidence for nonclassical ER pathway signaling in vivo. Mol Endocrinol. 2002;16:2188–2201. doi: 10.1210/me.2001-0174. [DOI] [PubMed] [Google Scholar]
- 18.Kousteni S, et al. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: Dissociation from transcriptional activity. Cell. 2001;104:719–730. [PubMed] [Google Scholar]
- 19.Harrington WR, et al. Estrogen dendrimer conjugates that preferentially activate extranuclear, nongenomic versus genomic pathways of estrogen action. Mol Endocrinol. 2006;20:491–502. doi: 10.1210/me.2005-0186. [DOI] [PubMed] [Google Scholar]
- 20.Blake RA, et al. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol Cell Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoo S, et al. Regulation of insulin gene transcription by ERK1 and ERK2 in pancreatic beta cells. J Biol Chem. 2003;278:32969–32977. doi: 10.1074/jbc.M301198200. [DOI] [PubMed] [Google Scholar]
- 22.Petersen HV, Jensen JN, Stein R, Serup P. Glucose-induced MAPK signaling influences NeuroD1-mediated activation and nuclear localization. FEBS Lett. 2002;528:241–245. doi: 10.1016/s0014-5793(02)03318-5. [DOI] [PubMed] [Google Scholar]
- 23.Andrali SS, Qian Q, Ozcan S. Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J Biol Chem. 2007;282:15589–15596. doi: 10.1074/jbc.M701762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hewitt SC, Li Y, Li L, Korach KS. Estrogen-mediated regulation of Igf1 transcription and uterine growth involves direct binding of estrogen receptor alpha to estrogen-responsive elements. J Biol Chem. 2010;285:2676–2685. doi: 10.1074/jbc.M109.043471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin ER. Plasma membrane estrogen receptors. Trends Endocrinol Metab. 2009;20:477–482. doi: 10.1016/j.tem.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Mauvais-Jarvis F. Rapid, nongenomic estrogen actions protect pancreatic islet survival. Islets. 2009;1:273–275. doi: 10.4161/isl.1.3.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 28.Nevins AK, Thurmond DC. Caveolin-1 functions as a novel Cdc42 guanine nucleotide dissociation inhibitor in pancreatic beta cells. J Biol Chem. 2006;281:18961–18972. doi: 10.1074/jbc.M603604200. [DOI] [PubMed] [Google Scholar]
- 29.Melloul D, Ben-Neriah Y, Cerasi E. Glucose modulates the binding of an islet-specific factor to a conserved sequence within the rat I and the human insulin promoters. Proc Natl Acad Sci USA. 1993;90:3865–3869. doi: 10.1073/pnas.90.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 31.Sharma A, Fusco-DeMane D, Henderson E, Efrat S, Stein R. The role of the insulin control element and RIPE3b1 activators in glucose-stimulated transcription of the insulin gene. Mol Endocrinol. 1995;9:1468–1476. doi: 10.1210/mend.9.11.8584024. [DOI] [PubMed] [Google Scholar]
- 32.Brelje TC, et al. Effect of homologous placental lactogens, prolactins, and growth hormones on islet B cell division and insulin secretion in rat, mouse, and human islets: Implication for placental lactogen regulation of islet function during pregnancy. Endocrinology. 1993;132:879–887. doi: 10.1210/endo.132.2.8425500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.