Abstract
Background:
C35 is a 12 kDa membrane-anchored protein endogenously over-expressed in many invasive breast cancers. C35 (C17orf37) is located on the HER2 amplicon, between HER2 and GRB7. The function of over-expressed C35 in invasive breast cancer is unknown.
Methods:
Tissue microarrays containing 122 primary human breast cancer specimens were used to examine the association of C35 with HER2 expression. Cell lines over-expressing C35 were generated and tested for evidence of cell transformation in vitro.
Results:
In primary breast cancers high levels of C35 mRNA expression were associated with HER2 gene amplification. High levels of C35 protein expression were associated with hallmarks of transformation, such as, colony growth in soft agar, invasion into collagen matrix and formation of large acinar structures in three-dimensional (3D) cell cultures. The transformed phenotype was also associated with characteristics of epithelial to mesenchymal transition, such as adoption of spindle cell morphology and down-regulation of epithelial markers, such as E-cadherin and keratin-8. Furthermore, C35-induced transformation in 3D cell cultures was dependent on Syk kinase, a downstream mediator of signalling from the immunoreceptor tyrosine-based activation motif, which is present in C35.
Conclusion:
C35 functions as an oncogene in breast cancer cell lines. Drug targeting of C35 or Syk kinase might be helpful in treating a subset of patients with HER2-amplified breast cancers.
Keywords: breast cancer, C35, HER2, epithelial to mesenchymal transition, ITAM, Syk kinase
The gene C35 (C17orf37) is located within the smallest region of amplification of the HER2 amplicon, between HER2 and GRB7. It is a 12 kDa membrane-anchored protein over-expressed in 40–50% of invasive breast cancers (Evans et al, 2006). C35 has recently been implicated in conferring invasive potential in prostate cancer cell lines (Dasgupta et al, 2009). It contains a canonical immunoreceptor tyrosine-based activation motif (ITAM; Evans et al, 2006), a motif common in receptors of the immune system (Underhill and Goodridge, 2007), which has been associated with cell transformation through the activation of downstream Syk signalling. This raises the possibility that C35 can function as a transforming oncogene. The ability of ITAM-containing proteins to transform non-haematopoietic cells was previously shown using viral glycoproteins, such as the murine mammary tumour virus envelope protein (MMTV Env; Katz et al, 2005). Other examples of non-haematopoietic transformation by ITAM-containing proteins include latent membrane protein 2A of Epstein–Barr virus in skin keratinocytes (Lu et al, 2006) and K1 protein of Kaposi's sarcoma-associated herpes virus in endothelial cells (Wang et al, 2006). Particularly relevant were the observations that ITAM-containing proteins contribute to mammary epithelial cell (MEC) transformation and development of mammary carcinomas (Katz et al, 2005; Grande et al, 2006; Ross et al, 2006).
Using the ITAM-containing envelope protein of MMTV Env and a chimeric B-cell receptor protein, many researchers have made several key observations (Katz et al, 2005; Grande et al, 2006; Ross et al, 2006): (1) ITAM-containing protein expression can transform immortalised normal MECs in three-dimensional (3D) culture; (2) ITAM-induced transformation is dependent on its tyrosine phosphorylation and is associated with downstream Src and Syk kinase activation and (3) mutation of the ITAM tyrosines reduces tumour induction markedly by MMTV in vivo and influences its genomic integration. Therefore, ITAM-containing protein expression can switch on an intrinsic transformation programme in MECs. This programme is closely associated with epithelial to mesenchymal transition (EMT). Whereas epithelial markers such as E-cadherin and keratin-18 are down-regulated, mesenchymal markers such as N-cadherin and vimentin are up-regulated (Katz et al, 2005; Grande et al, 2006).
In this study, we determined the co-expression of C35 and HER2 proteins in human breast cancers. High levels of C35 expression were shown to induce invasion mediated by EMT in vitro 3D cultures using cell lines. Mutation of ITAM of C35 (or downstream Syk inhibition) was sufficient for the reversal of C35-induced transformation. Syk inhibition in combination with anti-HER2 therapy was shown to be effective in BT474 cell line model, offering a possible therapeutic approach to treat HER2+ tumours.
Materials and methods
Tissue microarray construction and AQUA analysis
The population characteristics of the trastuzumab-treated cohort are summarised in Supplementary Table S1. HER2 gene amplification status was confirmed by fluorescence in situ hybridisation (FISH) according to the manufacturer's recommendations (HER2 FISH PharmDx; Dako, Ely, Cambridge, UK). The use of this cohort was approved by the Lothian Research Ethics Committee (08/S1101/41). After H&E sectioning of representative tumour blocks, tumour areas were marked for TMA construction and 0.6 mm2 cores were placed into three separate TMA replicates for each sample, as previously described (Kononen et al, 1998).
Immunofluorescence was carried out using methods previously described (Camp et al, 2002). Pan-cytokeratin antibody was used to identify infiltrating tumour cells and normal epithelial cells, DAPI counterstain to identify nuclei and Cy-5-tyramide detection for target (C35, 1 : 500 dilution; Vaccinex, Rochester, NY, USA) for compartmentalised (tissue and subcellular) analysis of tissue sections. Monochromatic images of each TMA core were captured at × 20 objective using an Olympus AX-51 epifluorescence microscope (Olympus, Southend-on-Sea, UK), and high-resolution digital images analysed by the AQUAnalysis software (HistoRx Ltd., Branford, CT, USA). Briefly, a binary epithelial mask was created from the cytokeratin image of each TMA core. Similar binary masks were created for cytoplasmic and nuclear compartments on the basis of DAPI staining of nuclei. C35 expression was quantified by calculating the Cy5 fluorescent signal intensity on a scale of 0–255 within each image pixel, and the AQUA score was computed by dividing the sum of Cy5 signal within the epithelial mask by the area of the cytoplasmic compartment.
AQUA scores were averaged from replicate cores. If the tumour epithelium comprised <5% of total core area, the core was excluded from analysis. To determine the cut-point value for C35 expression in Kaplan–Meier analysis (Altman et al, 1994), we used X-Tile (Yale University New Haven, CT, USA), which allows determination of an optimal cut point while correcting for the use of minimum P statistic (Camp et al, 2004). Overall survival was subsequently assessed by Kaplan–Meier analysis with log-rank for determining statistical significance. Comparison of differences in means of C35 according to HER2 status was carried out using the Student's t-test. All calculations and analyses were two tailed where appropriate using SPSS 14.0 for Windows (SPSS Inc., Chicago, IL, USA).
Immunohistochemistry
The following antibodies were used: C35, an affinity-purified rabbit polyclonal antibody 78.2 (Vaccinex) at 0.42 μg ml−1; cytokeratins 5/6 (CK5/6), rabbit polyclonal antibody (Dako) at 1 : 50 dilution; E-cadherin, mouse monoclonal (BD Biosciences, Oxford, UK) at 1 : 450 dilution; Twist, mouse monoclonal (Abcam, Cambridge, UK) at 1 : 100 dilution and claudin-7, rabbit polyclonal (Abcam) at 1 : 100 dilution.
Antigen retrieval for C35, E-cadherin and claudin-7 was carried out using sodium citrate buffer (18 μM citric acid, 82 μM sodium citrate, pH 6.0). Antigen retrieval for Twist was carried out using Tris/EDTA buffer (1 mM EDTA, 10 mM Tris-HCl base, pH 8.0). Standard immunohistochemistry protocol was carried out using the REAL EnVision mouse/rabbit kit (Dako), according to the manufacturer's instructions. For C35, comparative staining showed that automated AQUA immunofluorescence and manual immunohistochemistry scores correlated as follows: <100 : 0; 100–200 : 1+ 201–300 : 2+ and >300 : 3+.
HER2 immunohistochemistry was carried out using HercepTest (Dako), according to the manufacturer's instructions; with antigen retrieval at 96°C for 40 min. Staining was carried out on Autostainer (Dako). HER2 assessment was carried out according to the ASCO/CAP guidelines (Wolff et al, 2007). HER2 tumours were defined as positive only when the immunohistological score was 3+ and HER2 amplification was subsequently verified by FISH.
Cell lines, transfection and foci formation
The BT474, T47D, MBA-MD-231 and SKBr3 cell lines were obtained from the American Type Culture Collection. BT474, MBA-MD-231 and SKBr3 cells were cultured in RPMI 1640 (Invitrogen, Paisley, UK) supplemented with 10% donor bovine serum, 50 U ml−1 penicillin and 50 mg ml−1 streptomycin. T47D cells were cultured in DMEM (Invitrogen) supplemented with 10% donor bovine serum, 50 U ml−1 penicillin and 50 mg ml−1 streptomycin.
H16N-2 is an immortalised cell line derived from normal breast epithelium that does not over-express C35 (a kind gift from Dr V Band; Band and Sager, 1991). H16N-2 cells were cultured in DFCI media (Evans et al, 2006) or commercial MEGM (Lonza, Slough, UK) supplemented with 5% serum. The culture media were supplemented with 0.5 mg ml−1 G418 for vector selection. For detection of foci formation, we stained confluent monolayers with crystal violet (0.1% crystal violet, 20% ethanol) for 5 min, followed by de-stain rinse with water.
C35 and ITAM mutants through transfection
The coding region for human C35 protein was cloned into plasmid vector pIRESneo3 (Clontech, Mountain View, CA) at BsiWI and BamHI restriction sites. Plasmid DNA encoding wild-type (wt), Y39F/Y50F ITAM mutant or empty vector was transfected into host cells using Lipofectamine 2000 (Invitrogen) in OptiMem transfection medium following the manufacturer's protocol. Transfection medium was replaced with growth medium after 6 h. Transfectants were selected on G418, 48 h after transfection. Bulk transfected lines were cloned using cloning discs.
C35 recombinant cells by retroviral transduction
The coding region for human C35 protein (Evans et al, 2006) was cloned into retroviral vector pLXSN. To make a stable retrovirus producing line, we transfected pLXSN encoding wt C35 or empty vector into PA317 cells. Viral supernatants were collected, filter sterilised (0.45 μM) and titrated in the range of approximately 105 PFU per ml. H16N-2 were seeded at 3- to 5 × 106 cells in a T75 flask and incubated with 3 ml of viral supernatant and 2 μg ml−1 polybrene at 37°C for 6 h. Infection media were replaced with DFCI growth media and 0.5 μg ml−1 G418 was added at 48 h after infection. Bulk transduced lines were cloned by limiting dilution. Cell lines were assessed for C35 expression by western blot and/or immunofluorescence staining with C35 mouse monoclonal antibody (clone 1F2.4.1; Vaccinex) on fixed and permeabilised cells.
Soft agar colony formation assays
Triplicate wells of a six-well plate were seeded with uniform H16N-2 or MDA-MB-231 cell suspension diluted in DFCI, 0.33% agar (4 × 103 cells per well), which was layered over a bottom layer containing 0.625% agar. Plates were incubated up to 5 weeks at 37°C, fresh media were added to each well every week to replenish nutrients and moisture. Presence of colonies was detected under light microscope and visual inspection, at which point colonies were stained with P-iodonitrotetrazolium violet dye (Sigma-Aldrich, Gillingham, UK). Iodonitrotetrazolium violet stock (dissolved in 95% ethanol at 20 mg ml−1) was diluted to 1 mg ml−1 in PBS and 0.25 ml was added to each well. After overnight incubation at 37°C, visible colonies were counted in each well; counts from three wells were averaged. The number of colonies was normalised by multiplying the average number of soft agar colonies by the ratio of attached growth colonies normal to attached growth colonies transfectant. The attached growth assay was carried out at the same time as the soft agar assay, where 1/100 of each soft agar dilution was seeded into 100 mm dish. At 10–12 days after seeding, the dishes were stained with crystal violet and colonies were counted (Foos et al, 1998). Similar results were obtained in three independent experiments.
Collagen invasion assays
To characterise the mode of invasion of C35-expressing cells, we carried out collagen invasion assays essentially as previously described (Amjad et al, 2007). These assays are different from traditional Boyden chambers in several aspects: (1) the material used is a mixture of collagen and fibroblasts, generating a lattice of stroma-like substance; (2) the presence of live fibroblasts allows for continuing interaction with the epithelial cells; (3) importantly, the cells are examined as they invade the collagen lattice, not only measuring the number that have invaded right through the material.
Briefly, rat collagen I solution was mixed with 105 human breast fibroblasts (obtained from reduction mammoplasty, referenced in Amjad et al, 2007) per lattice and left to contract in fibroblast media (DMEM (Invitrogen) supplemented with 10% serum, 50 U ml−1 penicillin and 50 mg ml−1 streptomycin) for 4–7 days. When the lattices were of the required size (approximately four-fold contraction), 3 × 105 H16N-2 cells from the desired lines were seeded on top of the lattices and incubated as submerged cultures for 3–4 days in H16N-2 media. To induce invasion, we raised the lattices to the air/liquid interface and incubated for further 7 days before the they were fixed in 10% phosphate-buffered formalin and embedded in wax.
RNA extraction and RT–PCR
RNA was extracted by RNeasy Mini kit (Qiagen, Crawley, UK), evaluated on Agilent (South Queensferry, UK) Bioanalyzer (RIN>9.5) and labelled using Illumina TotalPrep RNA amplification kit (Applied Biosystems/Ambion, Austin, TX, USA) according to the manufacturers' instructions. Triplicate samples from whole invasion assays (1500 ng cRNA each) were hybridised to Illumina BeadChips, according to the manufacturer's instructions. Whole-genome gene expression analysis was performed using Illumina HumanRef-8 v3 Expression BeadChip and BeadArray Reader. Microarray data were analysed using packages within Bioconductor (Gentleman et al, 2004; http://www.bioconductor.org) implemented in the R statistical programming language (http://www.r-project.org/). The gene expression data were normalised using quantile normalisation within the bead array package (Dunning et al, 2007) and differential gene expression was assessed using significance analysis of microarrays (SAM; Tusher et al, 2001) using the siggenes package. The data set of Herschkowitz et al (2007) was downloaded from the UNC Microarray Database (https://www.genome.unc.edu/).
Confirmation of gene expression patterns from biological triplicates of invasion assays was carried out using the QuantiTect SYBR Green kit (Corbett/Qiagen, Crawley, UK) on a Corbett Rotor-Gene 3000. Primers for CDH1 were: forward 5′-CGGAGAAGAGGACCAGGACT-3′, reverse 5′-GGTCAGTATCAGCCGCTTTC-3′ for CLDN7: forward 5′-AAAATGTACGACTCGGTGCTC-3′, reverse 5′-AGACCTGCCACGATGAAAAT; for TBP: forward 5′-GGGGAGCTGTGATGTGAAGT-3′, reverse 5′-CCAGGAAATAACTCTGGCTCA-3′ for ACTB: forward 5′-CCTTCCTGGGCATGGAGTCCT-3′, reverse 5′-GGAGCAATGATCTTGATCTT-3′. QuantiTect Primer Assays (Qiagen) were used for KRT8, MAL2, TACSTD1 and SPINT2. PCR programme was identical for all genes: 95°C, 15 min (94°C, 15 s; 56°C, 30 s; 72°C, 30 s) × 50 cycles; 72°C, 5 min. Standard reference human cDNA was from Clontech (catalogue no. 639654), random primed, ∼50 ng RNA equivalent per μl was used for quantification of mRNA expression. Final normalisation as shown in Figure 4 was performed against the geometrical mean of ACTB and TBP levels.
Flow cytometry
shRNA constructs were cloned into Open Biosystems/ThermoFisher, Huntsville, AL) lentiviral inducible system; cell lines generated using non-silencing and shRNA-598 (agagagacactctccatgaaca) were evaluated for both C35 and Her2 expression. FACS analysis: cells were cultured in complete medium in the presence or absence of 0.5 μg ml−1 doxcycline for at least 7 days, collected with trypsin and re-suspended in FACS buffer (PBS (pH 7.2), 1% BSA). For HER2 staining, cells were incubated with 2 μg ml−1 biotinylated Herceptin or Remicade as human IgG1 isotype control, for 20 min on ice, followed by washing and incubation with 2 μg ml−1 streptavidin–APC. For C35 staining, cells were fixed and permeabilised according to the manufacturer's instruction using Invitrogen Fixation and Permeabilization kit GAS-004, and stained with 0.5 μg ml−1 C35 monoclonal antibody 1F2 or mouse IgG (BD Biosciences, catalogue no. 557732) conjugated to Alexa 647 for 45 min at room temperature. Cells were washed in FACS buffer and analysed on FACSCalibur. Samples were run in triplicate and averaged; ratio of median fluorescence intensity was plotted.
Three-dimensional cultures
Three-dimensional cultures have been used to study the behaviour of MECs in the presence of reconstituted basement membrane (Debnath and Brugge, 2005). This assay is particularly useful in observing oncogenic potential, by measuring morphological changes of the acinar structures formed in the culture. Such changes include enlarged acinar structures, local invasion and lack of lumen formation (Debnath and Brugge, 2005). We previously studied the effects of ITAM-containing proteins using 3D cultures (Katz et al, 2005; Grande et al, 2006), accurately predicting their contribution to tumour formation in vivo (Ross et al, 2006).
Cells (5 × 103 cells per chamber) were cultured on Matrigel (BD Biosciences) cushions following the precise protocol published previously (Debnath et al, 2003) using the usual cell culture media with the addition of 2% Matrigel. The structures were analysed, at a magnification of × 20, on a Leitz (Microscope Co., Glasgow, UK) Dialux 20 equipped with an Insight 4 video camera and SPOT software (Diagnostic Instruments, Sterling Heights, MI, USA). Quantification of structure size was carried out using a 10 × 50 μm grid reticule (Fisher Scientific, ThermoFisher, Huntsville, AL, USA), with 20–50 structures counted from each chamber. The inhibitors BAY61-3606 and piceatannol (Merck, Nottingham, UK) and trastuzumab/Herceptin (Roche Diagnostics, Penzberg, Germany) were added as follows (Figures 5 and 6):
(5a) T47D cells treated for 14 days with the Syk inhibitors BAY61-3606 (100 nM) or piceatannol (1 μg ml−1) (added twice: at days 8 and 11).
(5b) BT474 cells treated for 13 days with trastuzumab (20 μg ml−1) and/or BAY61-3606 (50 nM) (twice: at days 7 and 10).
(6b) Y39F/Y50F ITAM mutant or wt C35-expressing cell lines were treated for 14 days with the Syk inhibitor BAY61-3606 (50 nM, twice: at days 8 and 11).
For siRNA experiments, 105 cells were plated 48 h before the 3D culture. After 24 h, the cells were transfected with 100 nM non-targeted or Syk siRNA SmartPOOLs (Dharmacon, Cramlington, UK), using Lipofectamine (Invitrogen). At 24 h after the transfection cells were collected, counted and seeded on Matrigel as described above. The effectiveness of the SmartPOOLs vs single siRNA was measured by qPCR (all reagents from Dharmacon) after 48 h on plastic (Supplementary Figure S3).
Statistical analysis
For comparisons of means of structure diameters, two-tailed unpaired t-test was used. P-values were as follows:
(3c) C35, null vs C35pool: <0.0001; C35pool vs C35hi: 0.0041. E-cadherin, C35: null vs C35pool: <0.0001; C35pool vs C35hi: <0.0001.
(5a) None vs piceatannol: 0.0343; none vs BAY61-3606: 0.0119.
(5b) None vs trastuzumab/Herceptin: not significant; none vs BAY61-3606: 0.0356; none vs trastuzumab +BAY61-3606: <0.0001; BAY61-3606 vs trastuzumab +BAY61-3606: 0.0005; trastuzumab vs trastuzumab +BAY61-3606: <0.0001.
(5d) Non-targeted siRNA vs C35 siRNA: <0.0001; non-targeted siRNA vs HER2 siRNA: 0.0329; non-targeted siRNA vs Syk siRNA: 0.0104.
(6a) Neo vs Y39F/Y50F C35: not significant; neo vs wt C35: 0.0053; Y39F/Y50F C35 vs wt C35: 0.0026.
(6b) Y39F/Y50F C35 vs wt C35: 0.0111; Y39F/Y50F C35, none vs BAY61-3606: not significant; wt C35, none vs BAY61-3606: 0.0111.
(6c) Neo non-targeted vs Syk siRNA: not significant; neo non-targeted vs C35 non-targeted: 0.0011; C35 non-targeted vs Syk: 0.0018.
Results
C35 protein is co-expressed with HER2 in human breast cancer cells
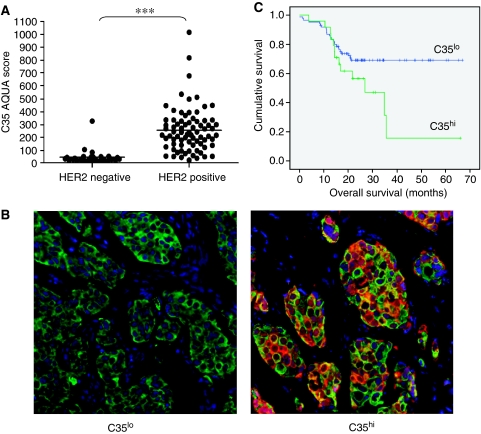
C35 protein expression was analysed by quantitative immunofluorescence using the HistoRx AQUA image analysis system (Camp et al, 2002) (1) to determine whether it is co-expressed with HER2 in the same cancer cells and (2) to investigate whether level of expression of protein was associated with therapeutic response to trastuzumab (Herceptin) in a retrospective clinical cohort of 122 treated patients, 32 of which were found later to be HER2 negative (Faratian et al, 2009). Pre-treatment C35 protein levels measured by immunofluorescence were significantly associated with HER2 copy number amplification assessed by FISH (Figure 1A and B; mean AQUA score HER2 not amplified=47.8 (s.d. 55.2; range 17.4–327.7), mean AQUA score HER2 amplified=255.2 (s.d. 170.9; range 40.1–1014.9); Mann–Whitney U-test, P<0.0001). In cancers with no HER2 amplification expression of C35 was uniformly low in all but two cases, with AQUA scores of less than 100.
Figure 1.
Clinical profile of C35 expression in human breast cancer. (A) Distribution of C35 immunofluorescence according to HER2 amplification status, as determined by fluorescence in situ hybridisation (***P<0.001). (B) representative examples of C35hi and C35lo immunofluorescence. Green: epithelial cell mask (pan-keratin); red: C35. Immunohistochemistry of C35 in primary breast cancers is shown in Supplementary Figure S1. (C) Kaplan–Meier survival curves according to optimal C35 cutpoint determined by minimum P-value method (log-rank test, P=0.028).
We next sought to establish whether quantitative C35 expression was associated with response to trastuzumab as measured by the overall survival time. In univariate analysis high C35 expression (cut point AQUA score 304; minimum P-value method) was associated with worse overall survival (Figure 1C; log-rank test P=0.0285), along with stage, ER status and chemotherapy regimen (Supplementary Table S1). However, only stage was associated with overall survival in a Cox regression multivariate analysis. Analysis of C35 expression in HER2-amplified tumours similarly did not yield a significant association with survival (Figure 1C).
Over-expression of C35 leads to EMT-mediated cell invasion
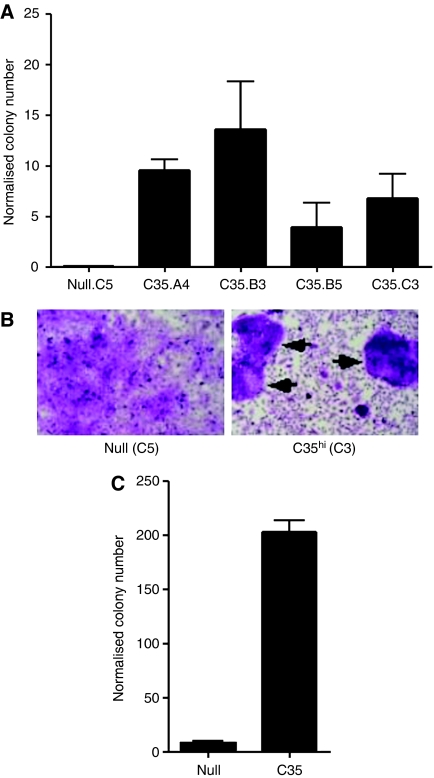
We carried out colony formation assays in soft agar to test whether C35 can induce MEC transformation. For this purpose, the normal MEC line H16N-2, which has been used previously for cell transformation assays (Burwell et al, 2007; Rhodes et al, 2009), was retrovirally transduced with wt C35. Colonies expressing high levels of C35 consistently formed enlarged structures in soft agar, whereas empty vector-expressing controls did not (Figure 2A). In contrast to the C35 transfectant pool, two of H16N-2 clones expressing high levels of wt C35 protein also showed foci formation when grown on plastic (Figure 2B; data not shown). In the breast cancer cell line MDA-MB-231, which normally expresses very low levels of C35, similar to those in the H16N-2 parental line (Evans et al, 2006), C35 expression was able to transform the MDA-MB-231 cell line at levels exceeding the transforming potential observed in the H16N-2 cell line (Figure 2C).
Figure 2.
C35 expression in mammary epithelial cells leads to colony formation in soft agar. Quantification of soft agar assays in (A) H16N-2 cells expressing empty retroviral construct (null clone) or C35 (clones A4, B3, B5 and C3) and in (C) MDA-MB-231 cells (null or C35 transfectant pools). (B) Foci formation (marked by arrows) assay results are shown for null.C5 and the C35.C3 (high expressing) clones.
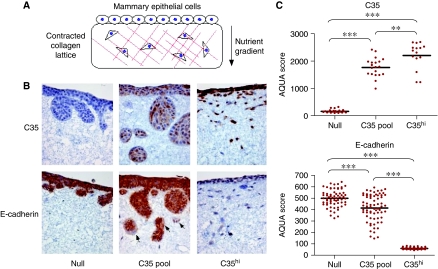
We previously reported that ITAM-containing proteins such as MMTV Env can induce an invasive phenotype in human MECs (Katz et al, 2005), likely to be caused by an EMT (Katz et al, 2005; Grande et al, 2006). It has also been shown that C35 promotes migration and invasion in prostate cancer cell lines (Dasgupta et al, 2009). To determine whether C35 expression also results in a similar behaviour in MEC, we used an invasion assay that used collagen lattices closely resembling breast stroma in vivo (Amjad et al, 2007; data not shown). The stroma-like lattices were generated by rat collagen I, contracted by seeding breast fibroblasts into the collagen gel. After the lattices contracted, MECs were seeded on top and invasion was induced by a nutrient gradient (Figure 3A). Although vector only (null) cells did not significantly invade the lattice, expression of C35 induced invasion. The C35 transfectant pool, which had variable levels of C35 expression, invaded mostly in large clusters of cells (Figure 3B). Three high-expressing clones showed complete transition to spindle cell phenotype, with single cells invading deep into the lattice (Figure 3B; Supplementary Figure S2). We chose one high-expressing clone, C35.C3, for further molecular characterisation (Figure 3C). Gradual loss of E-cadherin was apparent, occasionally in the C35-expressing pool and entirely within the C35hi clone (Figure 3B and C). Finally, all three major transcription factors known to be involved in EMT were examined. Slug expression was not detected and the level of Snail expression did not change in any C35-expressing cells. In contrast, Twist protein expression correlated positively with C35 expression (data not shown).
Figure 3.
C35-expression leads to invasive phenotype, associated with epithelial to mesenchymal transition. (A) Schematic illustration of the invasion assay set-up in a collagen gel containing fibroblasts overlaid with epithelial cells. (B) H16N-2 cells expressing empty vector (null, left panels), C35-expressing pool (middle panels) and C35hi-expressing cells were stained by immunohistochemistry for C35 and E-cadherin. Note specific areas of E-cadherin loss in C35 pool (arrows). (C) Quantification of C35 and E-cadherin by AQUA is shown. Bar indicates mean of measurements (P-value indicators: *<0.05; **<0.01; ***<0.001).
We carried out whole-genome expression array analysis to examine which transcripts correlate with C35 expression in the collagen invasion assays (raw gene expression files are publicly available from the caBIG-supported Edinburgh Clinical Research Facility Data Repository: https://www.catissuesuite.ecmc.ed.ac.uk/caarray/). Of the top 100 ranked differentially expressed genes by SAM (Tusher et al, 2001), the majority of the genes were down-regulated (62 of 98 probes, 63%, excluding a duplicate and a discontinued probe). First, we examined using KEGG analysis pathways activated or deactivated by C35 expression. The KEGG pathway that was most significantly over-represented by the most consistently differentially expressed genes by SAM analysis was cell communication (P=4.33E−08, FDR=5.43E−05). The genes responsible were KRT15, GJB2, COL17A1, DSG3, KRT13, KRT6A, KRT6B, KRT14, KRT16, KRT8 and LAMA3. Using the DAVID Bioinformatics database (Huang et al, 2009), we found that the processes highlighted by this pathway are cell–cell contact (adherens junctions, tight junctions, desmosomes) and ECM–receptor interactions, including focal adhesions. Interestingly, gene expression of PLAU (uPA), MMP9, VEGFA and VEGFB did not correlate with C35 levels. This suggests involvement of a different set of activated signalling pathways in MECs compared with prostate cancer cells (Dasgupta et al, 2009).
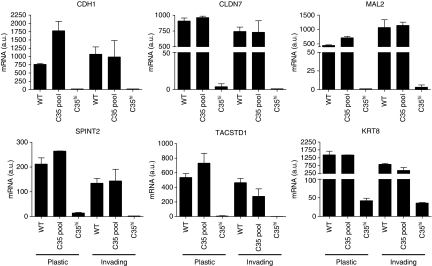
When the most consistently differentially expressed genes were compared with those identified in two molecular subtypes of breast cancers linked recently to EMT, claudin low (Herschkowitz et al, 2007) and metaplastic breast cancers (Hennessy et al, 2009), a number of interesting results were discovered. Of the 23 commonly changed genes, 5 (22%) were among most changed by C35 expression: E-cadherin (CDH1), claudin-7 (CLDN7), MAL2, EpCAM (TACSTD1) and HAI-2 (SPINT2). Validation by quantitative PCR confirmed that all five genes were down-regulated by high expression of C35 in the invasion assays (Figure 4). Cells expressing high levels of C35 also down-regulated eight cytokeratin genes (out of 98 top ranked, 8%), also consistent with loss of epithelial phenotype. These included keratin-8 (KRT8, Figure 4), which is often down-regulated in EMT-like breast tumours (Herschkowitz et al, 2007; Hennessy et al, 2009). Both loss of cell–cell contact and down-regulation of cytokeratins have been linked with EMT and are thought to enable cancer cell invasion (Levayer and Lecuit, 2008).
Figure 4.
Genes down-regulated in C35-induced transformed phenotype. C35-induced down-regulation of CDH1, CLDN7, KRT8, MAL2, TACSTD1 and SPINT2 was observed in cells grown on plastic and in the invasion assays. Biological triplicate mRNA expression data are shown for empty vector, C35 expressing pool and C35hi-expressing clone (C35.C3).
C35-induced cell transformation is dependent on the function of its ITAM
Previous studies have used 3D cell culture, in which MECs are grown on reconstituted basement membrane (Matrigel) and form spherical structures resembling the terminal ductal lobular units in the breast. These cell cultures show many in vivo properties of MECs. This model has been extensively used to study oncogenic phenotypes (Debnath and Brugge, 2005). C35 contains an ITAM, a motif found in glycoproteins of oncogenic retroviruses, that is linked to epithelial cell transformation through the protein tyrosine kinase Syk (Katz et al, 2005; Lu et al, 2006; Wang et al, 2006). Syk binds to the ITAM through its tandem SH2 domains and activates multiple growth signalling pathways, including PI3K, PLCγ, Ras/MAPK and NFκB, among others (Underhill and Goodridge, 2007).
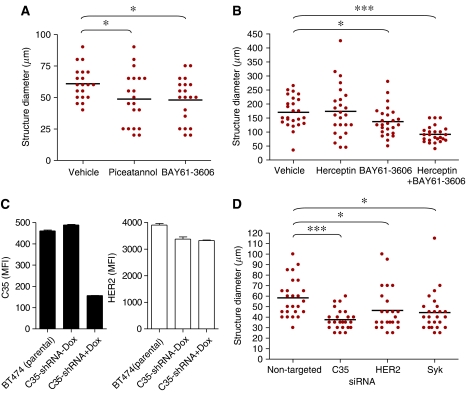
We determined the C35 and HER2 status of three breast cancer cell lines, as well as Syk expression. BT474 and SKBr3 lines harbour HER2 and C35 gene amplification and show high levels of mRNA expression of these genes (Supplementary Figure S4). T47D cells have no HER2 gene amplification and they express moderate levels of C35 (21-fold less than SKBr3 cells, 4-fold more than MCF10A cells). T47D cells are sensitive to Syk inhibition, by piceatannol or BAY61-3606 treatment (Yamamoto et al, 2003; Figure 5A), similar to that of the H16N-2 C35-expressing line in 3D culture (Figure 6B). Therefore, the response of HER2-amplified cells to Syk inhibition was determined. BT474 cells were chosen as they form non-polarised but well-defined ‘mass’ 3D structures (Kenny et al, 2007), similar to those generated by T47D cells. Treatment with Syk inhibitors, or Syk siRNA, reduced the size of BT474 3D structures (Figure 5D). This effect was unlikely due to changes in HER2 expression (Figure 5C). Syk inhibition combined with Herceptin (trastuzumab) resulted in even smaller structures, similar in size to those seen with immortalised, but non-transformed, cell lines (Figure 5B).
Figure 5.
Inhibition of C35 and Syk reduces mammary epithelial cells acinar structure size. (A) Quantification of 3D structure size in T47D cells at day 14 after treatment with the Syk inhibitors BAY61-3606 or piceatannol. (B) Quantification of structure size in BT474 cells at day 13 after treatment with Herceptin (trastuzumab) and/or BAY61-3606. (C) Knockdown of C35 by siRNA in BT474 cells (left panel) has no effect on HER2 surface expression (right panel) as determined by flow cytometry. (D) Quantification of structure size of BT474 cells treated with non-targeted, C35, HER2 or Syk siRNA, at day 6 of 3D culture.
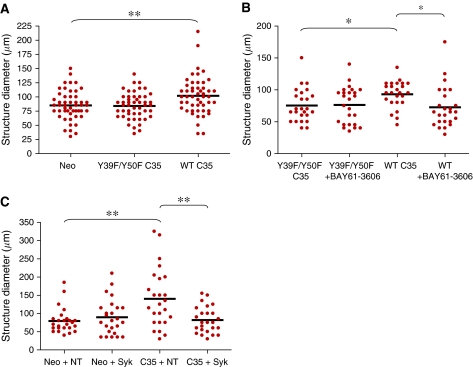
Figure 6.
C35 expression in normal mammary epithelial cells leads to cell transformation in 3D cultures. (A) H16N-2 acinar structures (at day 11) expressing empty vector (Neo), Y39F/Y50F ITAM mutant or wild-type (wt) C35 protein. (B) Quantification of structure diameter in Y39F/Y50F ITAM mutant or wt C35-expressing cell lines at day 14 after treatment with the Syk inhibitor BAY61-3606 (50 nM, twice: at days 8 and 11). (C) Quantification of structure size of Neo and wt C35-expressing H16N-2 cells treated with non-targeted, Syk siRNA, at day 6 of 3D culture.
We generated H16N-2 cell transfectant pools expressing the wt C35 protein or its Y39F/Y50F ITAM mutant. When grown in reconstituted basement membrane (3D culture), MECs expressing ITAM-containing proteins showed a transformed phenotype. This phenotype included enlargement of the acinar structures and was dependent on functional ITAM in these proteins (Katz et al, 2005; Grande et al, 2006). Consistent with these previous observations, when cultured in 3D, C35-expressing cells formed enlarged structures in comparison to empty vector-expressing cells (t-test, P=0.0053). Immunoreceptor tyrosine-based activation motif mutant C35-expressing cells formed similar structures to those of vector-expressing cells (Figure 6A).
Growth of C35-expressing H16N-2 cells was sensitive to Syk inhibition in 3D culture (Figure 6B) similar to other cell lines expressing ITAM-containing proteins (Katz et al, 2005; Grande et al, 2006). This was confirmed by siRNA knockdown for both Syk and C35 (Figure 6C). We also observed down-regulation of Syk mRNA in H16N-2 expressing the Y39F/Y50F ITAM mutant, compared with those expressing wt C35 (data not shown). This observation supports the view that Syk interacts with functional ITAM-containing C35.
Discussion
HER2/ErbB2 amplification is a frequent and well-studied event in breast and other cancers. The genetic fragment being amplified is commonly known as the HER2 amplicon. The smallest region of amplification of the HER2/ERBB2 amplicon on human chromosome 17q12 contains 14 core genes, of which STARD3, TCAP, PNMT, PERLD1, ERBB2, GRB7, GSDML and C17orf37/C35 are over-expressed when amplified (Kauraniemi and Kallioniemi, 2006; Marchio et al, 2008). The function of HER2, in breast cancer in particular, has been subject to intense research efforts, culminating in the design of both small molecule inhibitors and monoclonal antibodies in treatment of HER2+ patients (Bublil and Yarden, 2007). Recent efforts have concentrated on understanding how co-amplification of HER2 with the non-core amplicon gene Topoisomerase II (TOP2A) may affect response to chemotherapy (Pritchard et al, 2008). Much less is known about the functional importance of the core genes co-amplified with HER2. One of the best studied of these core genes is GRB7. Co-expression of Grb7 and HER2 facilitates HER2 signal transduction and functions synergistically for tumour formation (Stein et al, 1994; Bai and Luoh, 2008). Tumours co-expressing high levels of Grb7 and HER2 have a worse outcome than those with only higher levels of HER2 (Nadler et al, 2010), in line with the clinical data presented here. Both GRB7 and another core gene, STARD3, contribute to the growth of HER2-amplified cell lines in vitro (Kao and Pollack, 2006).
Here, we show that primary breast cancers have high levels of C35 protein expression when harbouring HER2 gene amplification, and that over-expression of C35 and HER2 protein is correlated in both breast cancer cell lines and primary tumours, in agreement with previous findings (Evans et al, 2006). It is estimated that tumours can express 70–100 times the normal breast tissue C35 transcript level (Evans et al, 2006). Cell lines expressing high levels of C35 showed high invasive behaviour in vitro. The overall phenotype is consistent with EMT, including down-regulation of E-cadherin and up-regulation of Twist. Interestingly, more gene transcripts were down-regulated than up-regulated among the 100 most changed transcripts. This raises the possibility of common suppression mechanism of transcription, downstream of C35 expression. A study in a pancreatic cancer cell line has suggested that the protein inhibitor specific for HGF activator-1 (HAI-1), an HAI-2 homologue, may activate an EMT programme in these cells by up-regulating the transcription factor SIP-1/ZEB-2 and consequently repressing E-cadherin (Cheng et al, 2009).
We found that tyrosine mutation in the ITAM of C35, or Syk kinase inhibition, is sufficient to abolish the potential of C35 protein to cause enlargement of acinar structures in 3D cell culture. Studies in DLBCL lines have shown that some, but not all tumours, expressing ITAM-containing proteins may respond to Syk inhibition (Chen et al, 2008). Evidence in this study using C35-expressing MEC lines has supported this strategy in vitro. Syk expression and activation are also modulated by extracellular matrix, through integrin signalling (Zhang et al, 2009). Syk promotes cell–cell contact on plastic (Zhang et al, 2009) and its genetic knock-down promotes cell mobility and invasion (Sung et al, 2009; Zhang et al, 2009). Syk may also have a tumour suppressor function in breast cancer through its kinase activity in the nucleus (Coopman et al, 2000; Sung et al, 2009). A plausible mechanism is that the interaction of C35 with Syk mimics global knock-down of Syk by changing its localisation away from integrins (Zhang et al, 2009) or the nucleus (Coopman et al, 2000). When activated in the cytoplasm, Syk functions as a promoter of cell growth (Zhou and Geahlen, 2009), consistent with the function postulated in this study. Our study results indicate that recently described Syk inhibitors (Braselmann et al, 2006; Chen et al, 2008) may be effective in C35 over-expressing breast cancer cells and thus have therapeutic value.
Other therapeutic approaches may be developed to take advantage of these findings in the treatment of human breast cancer, including the development of inhibitors of C35 interaction with proteins other than Syk, such as the novel ITAM-interacting protein Shb (Matskova et al, 2007). The Src kinase Lyn is unlikely to be involved in C35-induced EMT, because Lyn mRNA levels are reduced by approximately five-fold in C35hi cells, in comparison with both null and C35 transfectant pool cells.
In conclusion, we show here that the HER2 amplicon contains a second oncogene, C35, in the context of breast cancer. Our observations suggest that targeting C35 as well as HER2 may be beneficial for patients with HER2-amplified breast cancers. C35/C17orf37 has recently been included in an expression signature predicting metastatic risk in node-negative breast cancer after chemotherapy (Jezequel et al, 2009). This signature does not include HER2, therefore suggesting a possible autonomous role for C35, and warrants further investigation.
Acknowledgments
We thank Alexey Larionov (Edinburgh Breakthrough Research Unit), Amy Sutton and Alan Howell (Vaccinex) for their technical assistance. We thank Jorge Reis-Filho for critically reading the manuscript. Supported by Scottish Funding Council and Breakthrough Breast Cancer. Clinical materials were obtained through the auspices of the Edinburgh Experimental Cancer Medicine Centre. For YMK.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
ESS and EEE are employed by Vaccinex Inc., which identified C35 as a biomarker in human breast cancer. All other authors declare that they have no competing interests.
Supplementary Material
References
- Altman DG, Lausen B, Sauerbrei W, Schumacher M (1994) Dangers of using ‘optimal’ cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst 86: 829–835 [DOI] [PubMed] [Google Scholar]
- Amjad SB, Carachi R, Edward M (2007) Keratinocyte regulation of TGF-beta and connective tissue growth factor expression: a role in suppression of scar tissue formation. Wound Repair Regen 15: 748–755 [DOI] [PubMed] [Google Scholar]
- Bai T, Luoh SW (2008) GRB-7 facilitates HER-2/Neu-mediated signal transduction and tumor formation. Carcinogenesis 29: 473–479 [DOI] [PubMed] [Google Scholar]
- Band V, Sager R (1991) Tumor progression in breast cancer. In Neoplastic Transformation in Human Cell Culture. Rhim JS, Dritschilo A (eds), pp 169–178. The Human Press: Totowa, New Jersey [Google Scholar]
- Braselmann S, Taylor V, Zhao H, Wang S, Sylvain C, Balloum M, Qu K, Herlaar E, Lau A, Young C, Wong BR, Lovell S, Sun T, Park G, Argade A, Jurcevic S, Pine P, Singh R, Grossbard EB, Payan DG, Masuda ES (2006) R406, an orally available Syk kinase inhibitor blocks Fc receptor signaling and reduces immune complex-mediated inflammation. J Pharmacol Exp Ther 319(3): 998–1008 [DOI] [PubMed] [Google Scholar]
- Bublil EM, Yarden Y (2007) The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr Opin Cell Biol 19: 124–134 [DOI] [PubMed] [Google Scholar]
- Burwell EA, McCarty GP, Simpson LA, Thompson KA, Loeb DM (2007) Isoforms of Wilms' tumor suppressor gene (WT1) have distinct effects on mammary epithelial cells. Oncogene 26: 3423–3430 [DOI] [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL (2002) Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 8: 1323–1327 [DOI] [PubMed] [Google Scholar]
- Camp RL, Dolled-Filhart M, Rimm DL (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 10: 7252–7259 [DOI] [PubMed] [Google Scholar]
- Chen L, Monti S, Juszczynski P, Daley J, Chen W, Witzig TE, Habermann TM, Kutok JL, Shipp MA (2008) SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood 111: 2230–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Fukushima T, Takahashi N, Tanaka H, Kataoka H (2009) Hepatocyte growth factor activator inhibitor type 1 regulates epithelial to mesenchymal transition through membrane-bound serine proteinases. Cancer Res 69: 1828–1835 [DOI] [PubMed] [Google Scholar]
- Coopman PJ, Do MT, Barth M, Bowden ET, Hayes AJ, Basyuk E, Blancato JK, Vezza PR, McLeskey SW, Mangeat PH, Mueller SC (2000) The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature 406: 742–747 [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Wasson LM, Rauniyar N, Prokai L, Borejdo J, Vishwanatha JK (2009) Novel gene C17orf37 in 17q12 amplicon promotes migration and invasion of prostate cancer cells. Oncogene 28: 2860–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Brugge JS (2005) Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer 5: 675–688 [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS (2003) Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30: 256–268 [DOI] [PubMed] [Google Scholar]
- Dunning MJ, Smith ML, Ritchie ME, Tavare S (2007) Beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23: 2183–2184 [DOI] [PubMed] [Google Scholar]
- Evans EE, Henn AD, Jonason A, Paris MJ, Schiffhauer LM, Borrello MA, Smith ES, Sahasrabudhe DM, Zauderer M (2006) C35 (C17orf37) is a novel tumor biomarker abundantly expressed in breast cancer. Mol Cancer Ther 5: 2919–2930 [DOI] [PubMed] [Google Scholar]
- Faratian D, Goltsov A, Lebedeva G, Sorokin A, Moodie S, Mullen P, Kay C, Um IH, Langdon S, Goryanin I, Harrison DJ (2009) Systems biology reveals new strategies for personalizing cancer medicine and confirms the role of PTEN in resistance to trastuzumab. Cancer Res 69: 6713–6720 [DOI] [PubMed] [Google Scholar]
- Foos G, Garcia-Ramirez JJ, Galang CK, Hauser CA (1998) Elevated expression of Ets2 or distinct portions of Ets2 can reverse Ras-mediated cellular transformation. J Biol Chem 273: 18871–18880 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande SM, Katz E, Crowley JE, Bernardini MS, Ross SR, Monroe JG (2006) Cellular ITAM-containing proteins are oncoproteins in nonhematopoietic cells. Oncogene 25: 2748–2757 [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, Liu W, Stivers D, Baggerly K, Carey M, Lluch A, Monteagudo C, He X, Weigman V, Fan C, Palazzo J, Hortobagyi GN, Nolden LK, Wang NJ, Valero V, Gray JW, Perou CM, Mills GB (2009) Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69: 4116–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, Backlund MG, Yin Y, Khramtsov AI, Bastein R, Quackenbush J, Glazer RI, Brown PH, Green JE, Kopelovich L, Furth PA, Palazzo JP, Olopade OI, Bernard PS, Churchill GA, Van Dyke T, Perou CM (2007) Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jezequel P, Campone M, Roche H, Gouraud W, Charbonnel C, Ricolleau G, Magrangeas F, Minvielle S, Geneve J, Martin AL, Bataille R, Campion L (2009) A 38-gene expression signature to predict metastasis risk in node-positive breast cancer after systemic adjuvant chemotherapy: a genomic substudy of PACS01 clinical trial. Breast Cancer Res Treat 116: 509–520 [DOI] [PubMed] [Google Scholar]
- Kao J, Pollack JR (2006) RNA interference-based functional dissection of the 17q12 amplicon in breast cancer reveals contribution of coamplified genes. Genes Chromosomes Cancer 45: 761–769 [DOI] [PubMed] [Google Scholar]
- Katz E, Lareef MH, Rassa JC, Grande SM, King LB, Russo J, Ross SR, Monroe JG (2005) MMTV Env encodes an ITAM responsible for transformation of mammary epithelial cells in three-dimensional culture. J Exp Med 201: 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauraniemi P, Kallioniemi A (2006) Activation of multiple cancer-associated genes at the ERBB2 amplicon in breast cancer. Endocr Relat Cancer 13: 39–49 [DOI] [PubMed] [Google Scholar]
- Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ (2007) The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol 1: 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4: 844–847 [DOI] [PubMed] [Google Scholar]
- Levayer R, Lecuit T (2008) Breaking down EMT. Nat Cell Biol 10: 757–759 [DOI] [PubMed] [Google Scholar]
- Lu J, Lin WH, Chen SY, Longnecker R, Tsai SC, Chen CL, Tsai CH (2006) Syk tyrosine kinase mediates Epstein–Barr virus latent membrane protein 2A-induced cell migration in epithelial cells. J Biol Chem 281: 8806–8814 [DOI] [PubMed] [Google Scholar]
- Marchio C, Natrajan R, Shiu KK, Lambros MB, Rodriguez-Pinilla SM, Tan DS, Lord CJ, Hungermann D, Fenwick K, Tamber N, Mackay A, Palacios J, Sapino A, Buerger H, Ashworth A, Reis-Filho JS (2008) The genomic profile of HER2-amplified breast cancers: the influence of ER status. J Pathol 216: 399–407 [DOI] [PubMed] [Google Scholar]
- Matskova LV, Helmstetter C, Ingham RJ, Gish G, Lindholm CK, Ernberg I, Pawson T, Winberg G (2007) The Shb signalling scaffold binds to and regulates constitutive signals from the Epstein–Barr virus LMP2A membrane protein. Oncogene 26: 4908–4917 [DOI] [PubMed] [Google Scholar]
- Nadler Y, Gonzalez AM, Camp RL, Rimm DL, Kluger HM, Kluger Y (2010) Growth factor receptor-bound protein-7 (Grb7) as a prognostic marker and therapeutic target in breast cancer. Ann Oncol 21: 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard KI, Messersmith H, Elavathil L, Trudeau M, O'Malley F, Dhesy-Thind B (2008) HER-2 and topoisomerase II as predictors of response to chemotherapy. J Clin Oncol 26: 736–744 [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Ateeq B, Cao Q, Tomlins SA, Mehra R, Laxman B, Kalyana-Sundaram S, Lonigro RJ, Helgeson BE, Bhojani MS, Rehemtulla A, Kleer CG, Hayes DF, Lucas PC, Varambally S, Chinnaiyan AM (2009) AGTR1 overexpression defines a subset of breast cancer and confers sensitivity to losartan, an AGTR1 antagonist. Proc Natl Acad Sci USA 106: 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SR, Schmidt JW, Katz E, Cappelli L, Hultine S, Gimotty P, Monroe JG (2006) An immunoreceptor tyrosine activation motif in the mouse mammary tumor virus envelope protein plays a role in virus-induced mammary tumors. J Virol 80: 9000–9008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein D, Wu J, Fuqua SA, Roonprapunt C, Yajnik V, D'Eustachio P, Moskow JJ, Buchberg AM, Osborne CK, Margolis B (1994) The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. EMBO J 13: 1331–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YM, Xu X, Sun J, Mueller D, Sentissi K, Johnson P, Urbach E, Seillier-Moiseiwitsch F, Johnson MD, Mueller SC (2009) Tumor suppressor function of Syk in human MCF10A in vitro and normal mouse mammary epithelium in vivo. PLoS One 4(10): e7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DM, Goodridge HS (2007) The many faces of ITAMs. Trends Immunol 28: 66–73 [DOI] [PubMed] [Google Scholar]
- Wang L, Dittmer DP, Tomlinson CC, Fakhari FD, Damania B (2006) Immortalization of primary endothelial cells by the K1 protein of Kaposi's sarcoma-associated herpesvirus. Cancer Res 66: 3658–3666 [DOI] [PubMed] [Google Scholar]
- Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF (2007) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 25: 118–145 [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Takeshita K, Shichijo M, Kokubo T, Sato M, Nakashima K, Ishimori M, Nagai H, Li YF, Yura T, Bacon KB (2003) The orally available spleen tyrosine kinase inhibitor 2-[7-(3,4-dimethoxyphenyl)-imidazo[1,2-c]pyrimidin-5-ylamino]nicotinamide dihydrochloride (BAY61-3606) blocks antigen-induced airway inflammation in rodents. J Pharmacol Exp Ther 306: 1174–1181 [DOI] [PubMed] [Google Scholar]
- Zhang X, Shrikhande U, Alicie BM, Zhou Q, Geahlen RL (2009) Role of the protein tyrosine kinase Syk in regulating cell-cell adhesion and motility in breast cancer cells. Mol Cancer Res 7(5): 634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Geahlen RL (2009) The protein-tyrosine kinase Syk interacts with TRAF-interacting protein TRIP in breast epithelial cells. Oncogene 28: 1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.