Abstract
Regulator of G protein signaling (RGS) complex, Gβ5-RGS7, can inhibit signal transduction via the M3 muscarinic acetylcholine receptor (M3R). RGS7 consists of three distinct structural entities: the DEP domain and its extension DHEX, the G gamma like (GGL) domain, which is permanently bound to the G protein beta subunit Gβ5, and the RGS domain responsible for the interaction with Gα subunits. Inhibition of the M3R by Gβ5-RGS7 is independent of the RGS domain but requires binding of the DEP domain to the 3rd intracellular loop of the receptor. Recent studies identified the dynamic intramolecular interaction between the Gβ5 and DEP domains, which suggested that the Gβ5-RGS7 dimer could alternate between the “open” and “closed” conformations. Here, we identified point mutations that diminish DEP:Gβ5 binding, presumably stabilizing the open state, and tested their effects on the interaction of Gβ5-RGS7 with the M3R. We found that these mutations facilitated binding of Gβ5-RGS7 to recombinant 3rd intracellular loop of M3R, but did not enhance its ability to inhibit M3R-mediated Ca2+ mobilization. This led us to the idea that the M3R can effectively induce the Gβ5-RGS7 dimer to open; such a mechanism would require a region of the receptor distinct from the 3rd loop. Indeed, we found that the C-terminus of M3R interacts with Gβ5-RGS7. Truncation of the C-terminus rendered the M3R insensitive to inhibition by the wild-type Gβ5-RGS7, however, the open mutant of Gβ5-RGS7 was able to inhibit signaling by the truncated M3R. The GST fusion of M3R C-tail could not bind to wild-type Gβ5-RGS7 but could associate with its open mutant as well as to the separated recombinant DEP domain or Gβ5. Taken together, our data are consistent with the following model: interaction of the M3R with Gβ5-RGS7 causes the DEP domain and Gβ5 to dissociate from each other and bind to the C-tail; the DEP domain also binds to the 3rd loop, thereby inhibiting M3R-mediated signaling.
Keywords: G proteins, RGS proteins, DEP domain, Gbeta5, Muscarinic M3 Receptor
G protein coupled receptors (GPCRs) mediate many physiological processes including neuronal transmission and hormonal regulation. According to the classical model of GPCR signaling, an activated receptor interacts with a heterotrimeric G protein causing the exchange of GDP bound to the G protein α subunit (Gα) for GTP (1, 2). Binding of GTP results in the dissociation of Gα from the Gβγ complex (3), and both Gα-GTP and Gβγ influence activities of effector enzymes or ion channels (4–6). The return of the GPCR-mediated pathway to its basal state involves hydrolysis of GTP by the G protein. For most G proteins, rapid GTP hydrolysis requires participation of regulators of G protein signaling (RGS) proteins, which act as GTPase activating proteins (GAPs) for Gα subunits. Over 30 mammalian RGS proteins have been discovered. Outside of the conserved 120 amino acid RGS domain that is responsible for the GAP activity, RGS proteins are very diverse, ranging from ~25 to 120 kDa. According to this diversity, they are classified into distinct subfamilies (reviewed in: (7–11)).
The R7 family of RGS proteins (RGS6, 7, 9 and 11) is defined by the presence of the N-terminal DEP and DHEX domains (Dishevelled, Egl10, Pleckstrin (DEP) and DEP helical extension, respectively), the centrally located GGL domain (G Gamma Like), and the C-terminal RGS domain (12–14). The GGL domain associates permanently with the G protein beta subunit Gβ5 (15–17). The DEP domain binds to R7BP (R7 binding protein), a palmitoylated neuronal protein that can recruit Gβ5-R7 dimers to the plasma membrane (18–20). Biochemical studies and crystallography showed that the DEP domain and the Gβ5 subunit also bind to each other (21, 22).
R7 proteins are implicated in the regulation of neuronal processes such as sensory transduction, locomotor activity and addiction (23). At the molecular level, their function is primarily associated with negative regulation of the Gi family of G proteins. It was shown that R7 proteins do not possess GAP activity for Gq in vitro (24). However, in C. elegans the R7 protein EAT-16 antagonizes the function of Egl-30, the ortholog of Gq (25–27) and in transfected mammalian cells RGS7 was shown to down-regulate signaling via the Gq-coupled muscarinic M3 receptor, M3R (21, 28, 29). In a recent report we showed that this regulation occurs via a novel mechanism that does not require GAP activity, but instead involves direct binding of the RGS7 DEP domain to the 3rd intracellular loop of the M3R (M3Ri3) (30). In the M3R, this loop is unusually long (more than 200 amino acids) and was previously shown to interact with multiple proteins including Gq (31), β-arrestins (32), Gβγ subunits (33, 34), calmodulin (35), and the phosphatase inhibitor SET (36), as well as to contain phosphorylation sites for several kinases (37, 38).
In this study we sought to investigate the significance of the DEP:Gβ5 intramolecular interaction in the association of the Gβ5-RGS7 complex with M3R.
MATERIALS AND METHODS
Reagents and Antibodies
Fura 2AM was obtained from Invitrogen. Unless otherwise noted, all chemicals were obtained from Sigma. Rabbit RGS7 (1:1000), Gβ5 (1:1000), and Gβ1 (1:10,000) antibodies have been described earlier (28). Anti-Flag-M2 HRP conjugate was from Sigma (1:5000) and a mouse anti GFP antibody JL-8 was from Clontech (1:1000). Anti-rabbit (1:5000) and anti-mouse (1:3000) secondary antibodies conjugated to peroxidase were from Jackson laboratories. Anti-rabbit fluorescein-labeled antibodies (1:400) were from Amersham Biosciences and anti-mouse Cy3-labeled antibody (1:400) was from Sigma.
Cell culture, transfection and lysate preparation
CHO-K1 cells were cultured in F-12K Nutrient Mixture (Kaighn’s modification) with 10% FBS and penicillin/streptomycin, and plated to the density of 0.8×106–1.0×106 cells per 100 mm plate 24 hours prior to transfection. Transfection was carried out using Lipofectamine (Invitrogen) in accordance with the manufacturer’s instructions, as described earlier (28). The ratio of RGS7 to Gβ5 DNA was maintained at 5:1, with a total of 8.0 μg of DNA per plate. LacZ DNA was used as a control to ensure that the total DNA per plate remained constant.
After transfection, cells were washed with PBS and lysed in the hypotonic buffer (5 mM Tris-HCL pH 7.6, 0.1 mM MgCl2, 1 mM DTT and protease inhibitors cocktail, Roche). Cells were freeze-thawed twice and centrifuged at 14,000 rpm for 45 minutes. The supernatant (total protein concentration 1.0–1.5 mg/ml) represented the cytosolic fraction and was used for the pull-down assays involving cytosolic proteins.
Cloning of GST fusion proteins
The following GST fusion constructs were generated for bacterial expression and subsequent purification for use in the GST pull-down assay.
R7-DEP
Nucleotides 100–372 (corresponding to amino acids 34–124 of bovine RGS7) were PCR amplified from the full-length RGS7 cDNA and cloned into the pGEX-KG vector, as previously described (21). The mutant forms of the GST-DEP constructs were also generated by PCR-mediated mutagenesis utilizing the primers containing the desired substitutions. The RGS7 amino acids were substituted to the corresponding RGS9 residues in the K52S mutant and triple mutants K52S/E73S/D74G and D29A/R33D/K38D. RGS7 residues were substituted with alanine in the F57A mutant and double mutants F49A/L50A and F107A/F110A. All these R7-DEP mutants were cloned into the pGEX-KG vector linearized with BstBI and HindIII.
The third intracellular loop of the M3 receptor, GST-M3i3
The GST fusion of the third intracellular loop of human muscarinic M3 receptor (amino acids 345–390) was described earlier (30).
The C termini of Muscarinic receptors
The DNA fragment encoding the M3R C terminus (Asn548-Leu590) was amplified from the full length human M3R and cloned into the pGEX-2T vector at the BamH1 and EcoR1 sites. The PCR fragments corresponding to the C-terminal amino acid residues of human M1R (Asn422-Cys460) and M5R (Asn499-Pro532) were cloned in the same manner.
Constructs for expression in mammalian cells
RGS7249-469
This RGS7 construct, described before (21) lacks the DEP and DHEX domains of RGS7 and was generated by PCR amplification of nucleotides 745–1410 (corresponding to amino acids 249–469). The fragment was incorporated into the pcDNA3.0 vector linearized with BamHI and NotI.
YFP-R7
RGS7 cDNA was cloned into the pEYFP-C1 vector and has been previously described (29).
RGS7ED/SG
The double mutation E73S/D74G was introduced into full-length RGS7 using the same primers used for creation of the GST-fusion of the E73S/D74G R7-DEP double mutant (21).
YFP-R7F107A/F110A
RGS7 cDNA was cloned into the pEYFP-C1 vector at the BglII and HindIII sites. The F107A/F110A double mutation was introduced using the same primers used for the generation of the F107A/F110A R7-DEP mutant.
YFP-DEP
Nucleotides 1–372 corresponding to the N terminus and the DEP domain of bovine RGS7, were PCR amplified from full-length RGS7 and cloned into the pEYFP-C1 vector at the BglII and HindIII sites, as previously described (30).
Gβ5 mutants
Gβ5 cDNA was cloned into the pcDNA3.0 vector at the BamHI and NotI sites. Gβ5 mutants K54A, R56A/R57A, K60A, H62A, W107A, D259A I282A/I283A and F284A were generated by substituting the named amino acids for alanines using primers containing the mutations. The PCR generated fragments were then cloned into the pcDNA3.0 vector at the BamHI and NotI sites.
The CFP-Gβ5 construct has been previously described (29, 39).
Truncated M3R (M3R-ΔC)
The human M3R construct that lacks most of the C terminus was produced by introducing a stop codon at position 565 as previously described (40) and was kindly provided by Dr. Andrew Tobin (University of Leicester).
GST pull-down assay
The GST pull-down assays were performed as previously described with minor modifications (21, 30). Glutathione Sepharose 4B beads were pre-washed with PBS + 0.1% CHAPS and incubated at 4°C with purified GST or the GST fusion proteins for 1–2 hours. Purified GST or a GST fusion protein were immobilized on the matrix at the ratio of 0.15–0.3 μg per μl of packed resin; the amount of immobilized protein was determined by a Bradford protein assay (Pierce) and verified by SDS-PAGE stained by Coomassie. After three washes with PBS + 0.1% CHAPS, the slurry was mixed with the cell lysates required by the experiment and incubated overnight at 4°C on a rotary shaker. At the end of the incubation, the agarose beads were settled by gravity and the supernatant was collected as the unbound fraction. The resin was extensively washed with PBS + 0.1% CHAPS and then eluted with SDS-containing sample buffer. In a typical assay, the packed volume of the resin was 30 μl, and the volume of the protein lysate was 300 μl. The beads were washed three times with 600 μl of PBS + 0.1% CHAPS buffer, and eluted with 30 μl of 2x SDS-PAGE sample loading buffer. The input (total cell lysate), the unbound and eluted fractions were resolved on a 10% SDS gel and analyzed by western blotting. In routine experiments where only 5–20% of the protein subject to the pull-down was captured by the beads (i.e., using the GST fusions of the M3R fragments) and there was no appreciable difference between the total and unbound fractions, we only analyzed the unbound and eluted material.
Calcium mobilization assay
The cDNA clone for the human muscarinic M3 receptor was obtained from the Missouri S&T cDNA Resource Center (www.cdna.org). CHO-K1 or CHO-R7BP (30) cells were transiently transfected and plated on 12 mm glass coverslips (Electron Microscopy Sciences). After transfection, cells were washed with Hank’s Buffered Saline Solution (HBSS) supplemented with 2% FBS in and incubated in the same medium containing 1 μM fura-2AM for 45 minutes at ambient temperature in darkness. The cells were then incubated for 30 min in Locke’s buffer (20mM Hepes, 128mM NaCl, 5mM KCl, 1.2mM Na2HPO4, 2.7mM CaCl2 and 10mM Glucose) to allow de-esterification of fura-2AM. The coverslips were then secured in a flow chamber and mounted on the stage of a Nikon TE2000 inverted fluorescence microscope. The chamber was continuously perfused with Locke’s buffer under gravity flow and cells were stimulated with carbachol solution in the same buffer.
The images were collected using a 20x UV objective lens every two seconds using Metafluor software. The excitation wavelengths were 340 and 380 nm and the emission was set at 510 nm. Free Ca2+ concentration was determined from the fluorescence measurements on the basis of calibration performed with the fura-2 Ca2+ imaging kit (Molecular Probes) according to manufacturer’s instructions.
Confocal Microscopy
CHO-K1 or CHO-R7BP cells were plated to achieve a density of 1×105 cells on 22 mm glass coverslips placed into a 6 well plate. For transient transfection, 1μg of total plasmid DNA was used per well, and the RGS7:Gβ5 DNA ratio was maintained at 5:1 for both wild-type and mutant RGS7 and Gβ5 constructs. 24 hours after transfection, cells were fixed in 4% paraformaldehyde in PBS for 10 minutes at room temperature, washed twice with PBS and incubated in blocking solution (1% BSA and 0.1% Triton-X 100 in PBS) for 20 minutes. This was followed by a 1 h incubation with anti-GFP antibody diluted in blocking solution. CHO-R7BP cells were incubated with anti-flag antibody for an additional hour for detection of the tagged R7BP. Fluorescein and Cy3 secondary antibodies were diluted in the blocking solution and used for the detection of the green and red fluorescence, respectively. After staining, the coverslips were then rinsed thoroughly in PBS and water, then mounted on glass slides using Antifade reagent containing Dapi (Invitrogen) and imaged using a Leica TCS SP5 laser scanning confocal microscope. Images shown represent a single optical plane selected from at least 10 stacks that were taken for each cell.
Protein structure modeling
The three-dimensional model of RGS7 was obtained as a homology (comparative) protein structure with SWISS-MODEL and Swiss PdbViewer/Deep View 3.7 (SP5) (41, 42) using Gβ5-RGS9 [Protein Data Bank accession code: 2PBI] (22) as the template. The alignment between bovine RGS7 and residues 1–422 of the mouse RGS9 (22) exhibited 36% identity. The first 42 amino acids in the RGS7 model were automatically omitted by the software during the homology fitting procedure to obtain a “best fit” model. Images, including three dimensional ribbon diagrams were generated using Discovery Studio Visualizer 1.7 (Accelrys, Inc., San Diego, CA, USA).
RESULTS
Our previous studies showed that the DEP domain of RGS7 was responsible for the inhibition of muscarinic M3 receptor signaling, and that the interaction between the DEP domain and the receptor was blocked by the Gβ5 subunit (30). These data led us to hypothesize that the DEP domain can inhibit the M3R once it dynamically dissociates from Gβ5. The main goal of the present study was to test this model by disrupting the DEP:Gβ5 interaction via site-directed mutagenesis and characterization of the interaction of the resulting “open” mutants with the M3R.
Mutational analysis of the interface between the DEP domain of RGS7 and Gβ5
Before the Gβ5-RGS9 crystal structure became available (22), we attempted to locate the residues essential for the Gβ5:DEP interaction through amino acid sequence analysis of different DEP domains and Gβ subunits. We searched for the differences between RGS7, which bound to Gβ5 in our pull-down assay, and RGS9, which did not (21), also taking in consideration the NMR structure of the DEP domain of pleckstrin (43). Since both Gβ5 and Gβ1 bound to the DEP domain of RGS7 we also screened for positively charged conserved residues in Gβ subunits. We identified a double mutation (E73S/D74G, abbreviated as ED/SG) in the DEP domain of RGS7, which diminished its binding to Gβ5 (21). Other tested mutations (K54A, R56A/R57A, K60A and H62A in Gβ5 and D29A/R33D/K38S and K52S in RGS7) had no effect on the Gβ5:DEP interaction (data not shown).
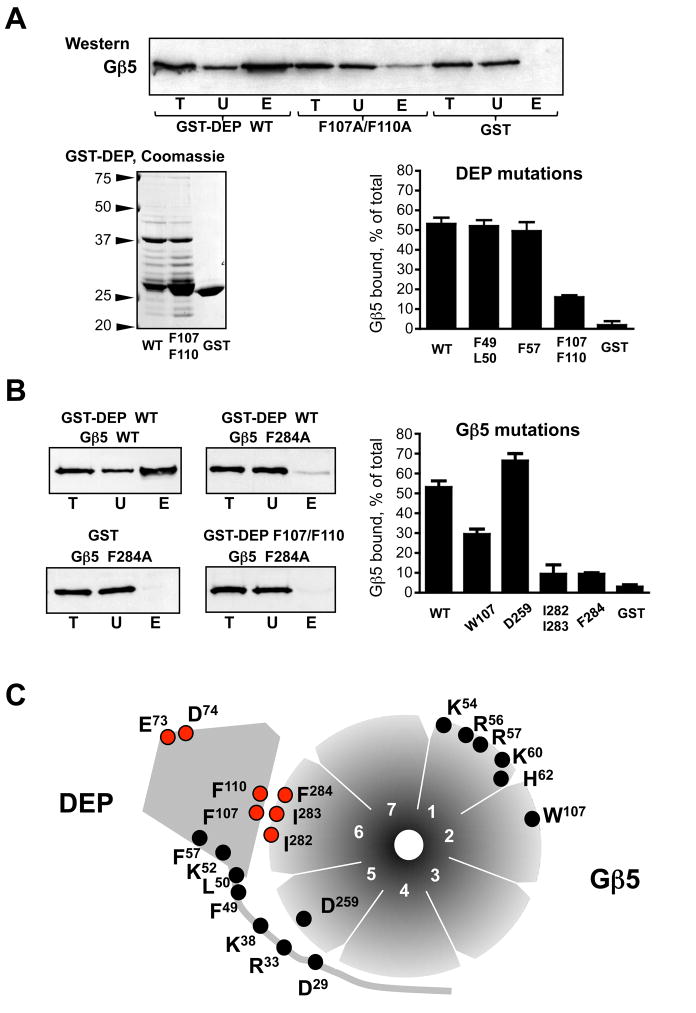
The recently determined crystal structure of the Gβ5-RGS9 dimer (22) allowed us to generate a three-dimensional homology model of the Gβ5-RGS7 complex. As expected, the backbones of RGS7 and RGS9 superimposed very well. Like in the Gβ5-RGS9 complex, in our model, the Gβ5 chain interfaces extensively with the GGL domain of RGS7, and the Gβ5/GGL moiety is sandwiched between the RGS and DEP domains (Fig 1A). We made the assumption that RGS7 and Gβ5 residues located within 1.5 Å from each other could contribute to the interaction between the two proteins. Amino acid residues F49, L50, F57, F107 and F110 of the RGS7 DEP domain and W107, D259, I282, I283 and F284 of Gβ5 fit this criterion (Fig. 1B). We substituted these amino acids for alanines, then expressed and characterized the resulting mutants in vitro (Figures 2–4).
Figure 1. Molecular model of the Gβ5-RGS7 dimer.
(A) Ribbon diagram presentation of the homology model of the Gβ5-RGS7 complex constructed on the basis of PDB coordinates of the crystal structure of the Gβ5-RGS9-1 complex (22). The Gβ5 polypeptide chain is shown in green. The domains of RGS7 are highlighted in the following colors: DEP, blue; DHEX, light brown; GGL, red; RGS, purple. In this projection the doughnut-shaped Gβ5 structure is seen from the side. Gβ5 makes multiple contacts with the GGL domain, and the Gβ5-GGL complex resides between the DEP and RGS domains. The area of contact between the DEP domain and Gβ5 is shown in more detail in (B). (B) The DEP:Gβ5 interface. Amino acids that are found within the 1.5 A radius from the opposite chain are shown in red (RGS7) and purple (Gβ5).
Figure 2. Mutational analysis of the DEP:Gβ5 interface.
Indicated amino acids of the RGS7 DEP domain or Gβ5 were substituted. Residues D29, R33, K38, K52, E73 and D74 were changed to the corresponding RGS9 residues, and all other residues were substituted by alanines, as described in Materials and Methods. (A) Analysis of the RGS7 DEP domain mutants. The DEP domain mutants were expressed as GST fusion proteins and analyzed by SDS-PAGE and Coomassie Blue staining. Along with the full-length GST-DEP fusion protein (~38 kDa) these preparations contained apparent degradation products, which were similar among all the DEP mutants. The wild-type and mutant GST-DEP proteins were immobilized on glutathione beads, so that the amounts of the fusion proteins were equal and corresponded to the amount of GST immobilized on the control beads. The complex of wild-type Gβ5 with the RGS7249-469 construct was expressed in CHO-K1 cells by transient transfection, and the cell lysate was subjected to the GST pull-down, as described in Materials and Methods. The western blot panel shows a representative experiment where the Gβ5-RGS7249-469 complex was tested for binding with the beads containing the GST fusion of wild type DEP domain (WT), the double-mutant F107A/F110A, or GST. The beads were washed and then eluted with SDS-containing sample buffer. The total CHO-K1 cell lysate (T), the unbound (U) and the eluate (E) fractions were probed with the anti-Gβ5 antibody, followed by ECL detection. The films were scanned and analyzed with Scion software. The bar graph summarizes the quantification of the data obtained from the entire series of experiments with all tested RGS7 DEP mutants. Material in the eluate (E) fractions was 5 times more concentrated relative to the unbound or total to ensure that the resulting ECL signal was within the linear range of detection. The values of the amount of Gβ5 present in the analyzed fractions was calculated accordingly and presented as the percentage of Gβ5 eluated from the beads relative to the total amount of Gβ5 in the cell lysate. Shown are the mean values with error bars representing standard deviation from 3–6 independent experiments. (B) Analysis of Gβ5 mutations. The indicated Gβ5 mutants were expressed in CHO cells in a complex with the RGS7249-469 construct and tested for their ability to bind to wild-type GST-DEP using the pull-down described in (A). Representative western blots show the total lysate (T) unbound (U) and eluted (E) fractions analyzed by immunoblot using the antibodies against Gβ5. The lower right panel shows the experiment where the Gβ5 F284A mutant was tested for binding with the F107A/F110A mutant of GST-DEP. The bar graph depicts quantification of the data from 3–4 independent experiments with four Gβ5 mutants (W107A, D259A, F284A and the I282A/I283A double-mutant) binding to wild-type GST-DEP or GST. (C) Diagram summarizing our mutational analysis of Gβ5 and RGS7. The seven blades of the Gβ5 “β-propeller” are numbered 1 through 7. The indicated mutants were tested in the GST pull-down assays described in A and B; the E73S/D74G mutation was described in our earlier study (21). The mutations resulting in a reduction of DEP-Gβ5 interaction are denoted by the red circles, those without a distinct phenotype are marked in black.
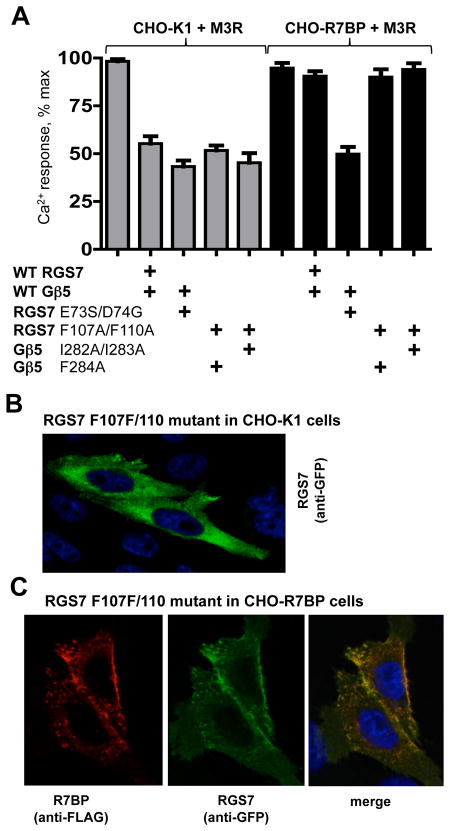
Figure 4. Effect of open mutations and R7BP on Gβ5-RGS7-mediated inhibition of M3R signaling.
(A) The stable cell line expressing flag-tagged R7BP (CHO-R7BP, black bars) was transfected with the M3R together with plasmids encoding the indicated mutants or wild-type Gβ5 and RGS7. CHO-K1 cells (gray bars) were used as the control lacking R7BP. The transfected cells were analyzed to determine the peak Ca2+ responses to application of 100 μM carbachol, as described in the Materials and Methods. The data represent the mean ± the standard deviation of the peak Ca2+ response measured in four independent transfection experiments. (B) CHO-K1 cells were co-transfected with plasmids encoding the F284A mutant of Gβ5 and the F107A/F110A of RGS7, which was fused to the C-terminus of YFP. The cells were then fixed and stained with anti-GFP antibody (1:5000) as described in Materials and Methods. Blue shows staining with Dapi to localize the cell nuclei. (C) The F284A mutant of Gβ5 and the F107A/F110A of RGS7 were co-expressed, via transient transfection, in the stable CHO cell line expressing Flag-tagged R7BP. As in (B), the RGS7 mutant was expressed as the YFP fusion protein. The cells were fixed and stained with anti-GFP antibody (1:5000) to detect the localization of the Gβ5-RGS7 complex (green) and anti-flag antibody (1:5000) to detect R7BP (red).
To study the interaction between RGS7 DEP and Gβ5 (Fig. 2), we used our previously developed pull-down assay (21, 30). The mutants of the DEP domain were expressed as GST fusions in E.coli, and the Gβ5 mutants were expressed in CHO-K1 cells together with the DEP-less C-terminal portion of RGS7 (RGS7249-469). Our data show that the double mutation F107A/F110A in the DEP domain leads to a clear reduction of its interaction with Gβ5. As shown in Figure 2A, more than 50% of Gβ5-RGS7249-469 complex is bound to the beads containing wild-type GST-DEP, whereas the beads with F107A/F110A mutant GST-DEP could retain only about 12% of the Gβ5 complex. Other mutations of the DEP domain, F49A/L50A and F57A, did not influence the interaction with Gβ5. In Gβ5, substitutions of F284 for alanine and the double mutation I282A/I283A also lead to an approximately five-fold reduction in its ability to bind to the RGS7 DEP domain (Figure 2B). The W107A mutation reduced the Gβ5:DEP interaction approximately 2-fold, whereas the D259A mutation in Gβ5 slightly (~10%) increased the fraction of the Gβ5-RGS7249-469 complex absorbed onto the beads with immobilized wild type GST-DEP domain. We also tested association of the Gβ5 F284A mutant with the F107A/F110A mutant of GST-DEP and found that the simultaneous mutation of both partners diminished their interaction to the level of non-specific binding (Figure 2B).
The diagram presented in Figure 2C summarizes all our data on the mutational analysis of the Gβ5:DEP interaction. In our previous study, we identified residues E73 and D74 of RGS7 to be important for association of the DEP domain of RGS7 with the Gβ5 moiety (21). We have now found that residues F107 and F110 of the DEP domain and three adjacent hydrophobic residues of Gβ5, I282, I283 and F284, also contribute to the interaction.
The role of Gβ5:DEP association in the interaction of Gβ5-RGS7 with the M3R
Our previous studies suggested that Gβ5-RGS7 binds to the receptor only when it is in its open conformation, i.e., when Gβ5 does not sequester the DEP domain (30). This idea implied that mutations destabilizing the Gβ5:DEP interaction should facilitate the transition of the Gβ5-RGS7 dimer toward its open conformation, thereby promoting its interaction with the receptor.
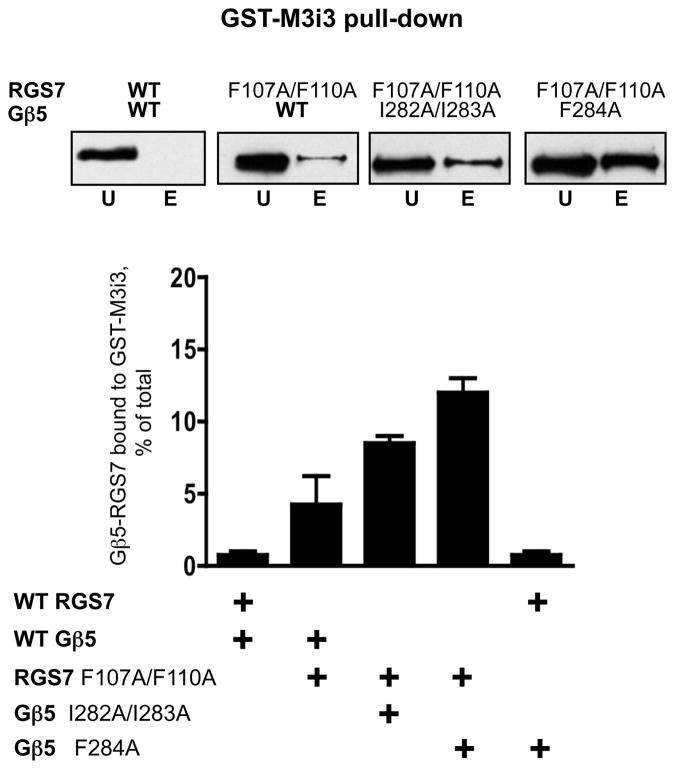
We introduced the F107A/F110A double mutation into the full-length RGS7 cDNA, co-expressed this construct with Gβ5 in CHO-K1 cells and tested it in the pull-down assay with the GST fusion of the 3rd intracellular loop of the M3R (Figure 3). In contrast to the dimer composed of wild-type RGS7 and Gβ5, this mutant could readily bind to the immobilized M3Ri3. Gβ5 mutants F284A and the I282A/I283A, when combined with the wild-type RGS7, could not bind to the M3Ri3 loop. However, the Gβ5 mutations I282A/I283A and particularly F284A augmented the effect of the F107A/F110A mutation of the RGS7. These results corroborate the involvement of residues F107 and F110 of the DEP domain and I282, I283 and F284 of Gβ5 in the DEP:Gβ5 interaction (Figures 1, 2). More importantly, they are consistent with the idea that transition of the Gβ5-RGS7 dimer to its open conformation facilitates its interaction with the M3R.
Figure 3. Interaction of the “open” mutants of Gβ5-RGS7 with the third intracellular loop of the M3R.
The GST fusion of the M3Ri3 was immobilized on the glutathione beads. Wild-type or mutant RGS7 constructs were expressed in CHO-K1 cells as full-length proteins fused to the C-terminus of YFP, together with wild-type or mutant Gβ5. The cell lysates were subjected to the GST pull-down. Following the incubation with the beads, the unbound and eluted material was analyzed by western blotting using the anti-YFP antibody. The different combinations of WT and mutant constructs used are indicated above the four representative immunoblot panels. The amount of RGS7 bound to the M3i3 beads was determined as the fraction of the total amount of RGS7 in the cell lysate. The total was calculated as the sum of the signal in the unbound and eluted fractions, as described in the Materials and Methods. The data show the mean and standard deviation from three or four independent experiments.
In a parallel series of experiments, we tested whether the mutants displaying increased binding to the M3Ri3 loop (i.e., “open” mutants) were also more effective as inhibitors of M3R-signaling. Figure 4A shows the effect of the different open mutants of Gβ5-RGS7 complex on M3R-mediated Ca2+ mobilization elicited by 100 μM carbachol. Contrary to our expectations, the extent of the inhibition conferred by the open mutants was similar to that of the wild-type Gβ5-RGS7 dimer. Recently, we showed that Gβ5-RGS7 could reduce the amplitude of the Ca2+ response to M3R stimulation by up to 90% if carbachol concentration was below its Kd (30). Under such conditions the amplitude of the Ca2+ response is about four times lower compared to the response at 100 μM carbachol, which reduces the overall signal-to-noise ratio, but the inhibitory effect of Gβ5-RGS7 on M3R signaling has a wider dynamic range. We tested the open mutants at 1 μM carbachol, but did not detect a difference between the effect of wild type and mutant Gβ5-RGS7 complexes (data not shown). Our earlier study showed that R7BP precluded wild-type Gβ5-RGS7 from inhibiting M3R signaling, whereas the ED/SG mutant was able to inhibit M3R even in the presence of R7BP (21). Therefore, we expected that the Gβ5:DEP interface mutations identified in this study could also overcome the effect of R7BP and tested the function of the new open mutants in the presence of R7BP. To limit the number of transfected cDNAs to three (M3R, Gβ5 and RGS7), we used the previously constructed CHO cell line that stably expresses Flag-tagged R7BP (21). Like the wild-type Gβ5-RGS7, the open mutants localized to the plasma membranes in the cells (Figure 4B), indicating that these mutations did not diminish the ability of the RGS7 complex to bind to R7BP. We found that in contrast to the ED/SG mutant, the complexes composed of RGS7 F107A/F110A and Gβ5 I282A/I283A or F284A mutants behaved similarly to the wild-type Gβ5-RGS7 both in the absence and in the presence of R7BP.
Thus, whereas the pull-down assay with the M3Ri3 loop revealed a clear difference between the open mutants identified in this study and wild-type Gβ5-RGS7 (Figure 3), the signaling assay with the full-length receptor did not (Figure 4). To explain the apparent discrepancy between the two experiments, we reasoned that the signaling assay involved an additional factor that was absent in the pull-down experiment. The role of this hypothetical factor would be to facilitate the shift of Gβ5-RGS7 to its open conformation, so that the wild-type dimer becomes as effective in interacting with the receptor as its “open” mutants. We proposed that such a factor was a region of M3R, which is present in the full-length receptor but is absent in the 3rd loop.
The C-terminus of the M3R plays a role in the interaction with Gβ5-RGS7
Since the first and second loops of the M3R are short, we expected the C-tail of the receptor to be a more likely candidate region for the interaction with Gβ5-RGS7. To test this hypothesis, we used two complementary approaches. In one series of experiments (Figure 5), we measured Ca2+ responses elicited by the M3R receptor that lacks a portion of its C-terminus (M3R-ΔC). This M3R mutant is truncated by a stop codon at position 565, immediately after the putative helix 8, and was earlier shown to retain the ability to signal via phospholipase C and activate MAP kinases (40). In a complimentary approach, we tested whether Gβ5-RGS7 could directly interact with the GST fusion of the M3R C-terminus (M3R-C) (Figure 6).
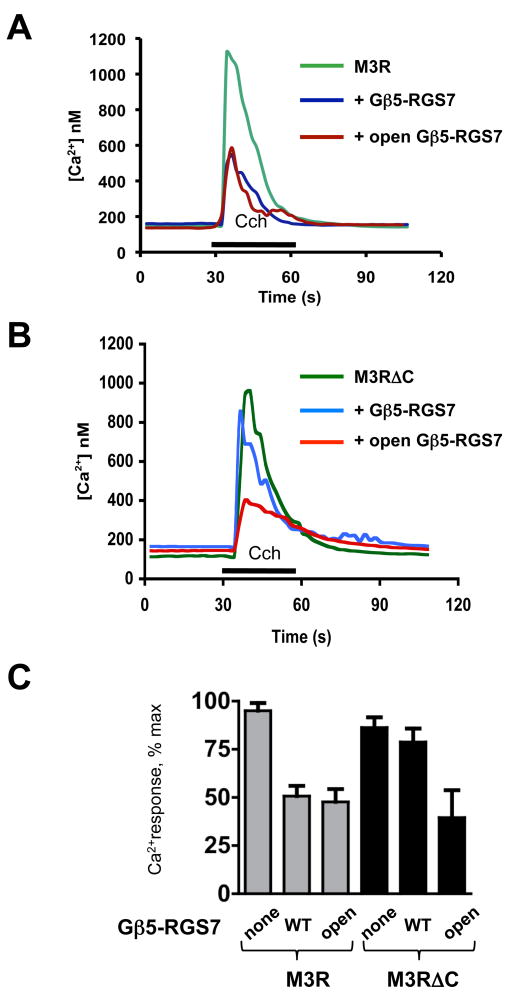
Figure 5. The C terminus of the M3R is essential for the inhibitory action of Gβ5-RGS7 on M3R signaling.
CHO-K1 cells were transiently transfected with M3R, Gβ5 and RGS7 constructs. Cells were grown on glass coverslips, then loaded with fura-2AM and mounted in a flow chamber at the stage of an inverted fluorescence microscope. Changes in free intracellular Ca2+ concentrations in response to stimulation with 100 μM carbachol were recorded in real time from the entire field which contained at least 40 cells, as described in Materials and Methods. The application of carbachol (Cch) is denoted with the black bar. (A) Representative traces from cells transfected with plasmids encoding the M3R and Lac Z cDNAs (green), the M3R together with wild-type Gβ5-RGS7 (blue) or the M3R with “open” Gβ5-RGS7 composed of RGS7 F107A/F110A and Gβ5 F284A mutants (dark red). (B) The M3R construct lacking its C-terminus (M3RΔC) was co-transfected with the wild-type or the open mutant of Gβ5-RGS7. (C). Quantification of the data from four independent experiments showing the mean ± SD of the peak Ca2+ response expressed as the percent of the maximal detected Ca2+ response. Gray bars show inhibition of full-length M3R by wild-type Gβ5-RGS7 or the open mutant, black bars represent the results with the truncated M3R mutant, M3RΔC.
Figure 6. The C terminus of the M3R directly binds to the DEP domain of RGS7, Gβ5 and the open mutant of Gβ5-RGS7.
(A) The C-terminus of the M3R was expressed in E.coli as a GST fusion protein and used to pull down the lysates of CHO cells expressing either the wild-type Gβ5-RGS7 dimer, the Gβ5-RGS7 dimer composed of the RGS7 F107A/F110A and Gβ5 F284A mutants (the open mutant), the DEP domain or Gβ5. In these experiments, RGS7 and Gβ5 were fused to YFP and CFP, respectively, and their presence in the unbound (U) and eluted (E) fractions was detected using an anti-GFP antibody. The pull-down was performed as described in Materials and Methods, and similarly to experiments shown in Figures 2 and 3. (B) The C-termini of muscarinic receptors M1, M3 and M5 were expressed as GST fusion proteins and used to pull down the YFP fusion of the DEP domain of RGS7. Beads with GST were used as a negative control. (C) The GST fusions of the C-termini of M1, M3 and M5 muscarinic receptors were used to pull-down the untagged monomeric Gβ5 or its dimer with the RGS7249-469 construct. Anti-Gβ5 antibody was used to probe the unbound and eluted fractions in the assays. Each western blot is a representative of at least three independent experiments.
Gβ5-RGS7 only marginally reduced the amplitude of the M3R-ΔC-mediated Ca2+ response to carbachol, showing that the C-tail is required for Gβ5-RGS7 to inhibit M3R signaling (Figure 5A, C). However, we found that the open mutant of Gβ5-RGS7 (Gβ5 F284A plus RGS7 F107A/F110A) could inhibit signaling by the truncated receptor (Figure 5B, C). These results indicate that the C-tail is required for inhibition of the M3R by Gβ5-RGS7 and potentially can facilitate the transition of the wild-type Gβ5-RGS7 dimer to its open conformation.
The M3R-C GST fusion, which included the entire C-tail, did not bind to the wild-type Gβ5-RGS7 dimer, but readily bound to the open mutant (Figure 6A). We then asked whether the M3R-C construct could bind to the DEP domain or to Gβ5, and found that, somewhat surprisingly, that it could bind to both these entities. Whereas the DEP domain bound to the C-termini of M1, M3 and M5 muscarinic receptors equally well (Figure 6B), monomeric Gβ5 or its complex with the DEP-less R7249-469 construct bound selectively to the C-terminus of the M3R (Figure 6C).
These results indicate that Gβ5-RGS7 binds to the M3R via both the 3rd intracellular loop and the C-terminal tail.
DISCUSSION
It is now well appreciated that in addition to G proteins, GPCRs interact with a plethora of partners collectively known as GPCR-interacting proteins (GIPs) (44, 45). These diverse proteins modulate G protein signaling and mediate novel pathways that sometimes even bypass the G proteins. Recently, we reported that the muscarinic acetylcholine receptor M3 can directly bind to the regulator of G protein signaling complex, Gβ5-RGS7, and that this interaction occurs between the DEP domain of RGS7 and the 3rd intracellular loop of the M3R (30). In the current paper we focused on further investigating the molecular details of this interaction.
The complexes of the R7 family RGS proteins with Gβ5 were discovered more than a decade ago (15, 17), but the significance of their multi-domain organization and, above all, the reason why they integrate the Gβγ-like entity, remained unclear. It was shown that Gβ5 increases the stability of the associated RGS subunit (28, 46), and that the DEP domains play a role in plasma membrane targeting via R7BP (18, 19). However, stability and membrane anchoring can be achieved via other mechanisms; some RGS proteins function while consisting of little more than the RGS box. A hypothesis explaining the significance of Gβ5-R7 structural organization began to emerge from the finding that the Gβ5 moiety interacts with the DEP domain, and, particularly, from the evidence of the dynamic nature of this interaction (21). In this model, the dynamic interaction of Gβ5 with the DEP domain enables a functional cycle analogous to the Gαβγ ⇔ Gα + Gβγ cycle of heterotrimeric G proteins. As the Gβ5-R7 dimer alternates between the distinct “closed” and “open” conformations, it can interact with different binding partners. In the current paper, we developed this idea further by stabilizing the putative open conformation by introducing mutations that impair the interaction between Gβ5 and the DEP domain.
The first step in our study was to identify the amino acids responsible for the DEP:Gβ5 interaction. On the basis of the homology with the tertiary structure of the Gβ5-RGS9 complex we identified two phenylalanines (F107 and F110) in the DEP domain of RGS7 and the Ile-Ile-Phe triad in Gβ5, as being important for the interaction (Figures 1–2). These amino acids appear to cause the DEP:Gβ5 association through hydrophobic forces. However, other residues may also contribute to this interaction (Figure 2), and, as we previously showed, substitution of E73 and D74 in RGS7 for the corresponding Ser and Gly residues of RGS9 (ED/SG mutation) also diminished DEP:Gβ5 binding (21). The E73 and D74 residues do not localize at the putative DEP:Gβ5 interface (Fig 1B), making it difficult to explain the phenotype. It is possible that our model of RGS7 is not entirely correct, and that the E73 and D74 residues do, in fact, contact the Gβ5 chain. The sequence identity between our template (murine RGS9 crystal structure) and target (bovine RGS7) is only 36%, which might be too low for reliable homology modeling. Therefore, the structure of RGS7, especially in solution, can deviate from RGS9 far more than that afforded by the best match algorithm. An alternative explanation is that the ED/SG mutation allosterically influences the position of F107 and F110 at the interface with Gβ5. The ED/SG mutation and the mutations at the DEP:Gβ5 hydrophobic interface result in a clearly distinct phenotype when tested in the presence of R7BP (Figure 4). Consistent with our previous report, the ED/SG mutation was able to overcome the negative effect of R7BP on the ability of Gβ5-RGS7 to inhibit M3R signaling. In contrast, open mutants identified in this paper were ineffective in inhibiting M3R signaling in the presence of R7BP (Figure 4A). At the same time, all the mutant forms were capable of binding to R7BP, as was indicated by their ability to localize to the plasma membranes in the R7BP-expressing cells (Figure 4B,C). At the moment, the phenotypic difference between the ED/SG mutant and the new mutations identified in this study is difficult to explain, and we can only speculate that the ED/SG mutation is more effective in promoting the open conformation.
The idea brought about by our structure-function analysis is that, regardless of how specific mutations impair the DEP:Gβ5 association, they enhance the interaction of the Gβ5-RGS7 dimer with the M3R. The ED/SG mutation led to a gain of function in terms of inhibition of M3R signaling in the presence of R7BP (21) (Figure 4A). Similarly, mutations of the hydrophobic DEP:Gβ5 interface enabled binding of Gβ5-RGS7 to the 3rd intracellular loop (Figure 3) and the C-tail of the M3R (Figure 6) as well as resulted in the inhibition of the truncated M3R (Figure 5).
In our previous study we identified the 3rd intracellular loop of the M3R as the region responsible for the interaction with Gβ5-RGS7 (30). We found that deletion of this loop rendered the M3R insensitive to Gβ5-RGS7, while recombinant M3Ri3 could directly bind to the isolated DEP domain or the RGS7 monomer. Interestingly, the full-length Gβ5-RGS7 dimer could not bind to the M3Ri3 in vitro, which made it difficult to explain how the dimer could inhibit M3R signaling. We hypothesized that Gβ5-RGS7 can bind to the M3R after assuming the open conformation, the notion supported in this paper (Figures 3, 6A). We also reasoned that mutations that facilitate binding of Gβ5-RGS7 to M3Ri3 should increase the apparent effectiveness of Gβ5-RGS7 as an inhibitor of M3R signaling. However, our assays did not detect such an enhanced activity (Figure 4).
To explain the similarity between the wild-type and mutant Gβ5-RGS7 in their ability to inhibit M3R signaling, we extended our model. We postulated that the receptor has an intrinsic capacity to induce the open conformation in Gβ5-RGS7 and that this capability is associated with an M3R region that is distinct from the third loop. This prediction proved to be correct: we found that the C-tail of the receptor can interact with Gβ5-RGS7 (Figures 5, 6). Truncation of the C-terminus rendered the M3R insensitive to the inhibition by the wild-type Gβ5-RGS7, but interestingly, the open Gβ5-RGS7 mutant could inhibit the truncated receptor (Figure 5). In other words, the Gβ5-RGS7 molecule that has already been opened by a mutation does not require the M3R C-tail to do so.
Our data show that the isolated C-tail cannot bind to the wild-type Gβ5-RGS7 but does bind the open mutant, which implies synergy between the C-tail and the 3rd intracellular loop in the process of Gβ5-RGS7 recruitment. Gβ5 binds to the C-tail of M3R in a selective manner, indicating that it associates with the sequence unique for the M3 subtype. The C-tail of the M3R can now be added to the list of putative binding partners of Gβ5, in addition to Gαi and PLC, which in early studies were shown to interact with Gβ5γ (47–50). On the other hand, the DEP domain of RGS7 appears to bind to both the C-tail and M3Ri3. A more detailed structure-function analysis will be needed to further dissect molecular events involved in the interaction of M3R with Gβ5-RGS7. Since M3R can oligomerize (51), it is possible that Gβ5-RGS7, being a rather large molecule, can simultaneously bind to the C-tail of one M3R molecule and the 3rd loop of another.
Interaction of M3R with Gβ5-RGS7 supports the emerging concept that GPCRs can form complexes with RGS proteins (52–56). For example, yeast RGS protein Sst2 that is unrelated to the R7 family can directly bind to the GPCR Ste2 (57). That study is particularly interesting in relation to the current paper because the Ste2:Sst2 interaction requires the C-tail of the Ste2 receptor and the DEP domain present in Sst2. The results of another group indicated that RGS9 also associated via its DEP domain with the dopamine D2 receptor (58). Our current paper extends our understanding of the role of the DEP and Gβ5/GGL entities in the R7 family by showing that their dissociation in the Gβ5-RGS7 complex can facilitate its interaction with the M3R. The identification of the M3R C-terminus as the site essential for in the interaction with Gβ5-RGS7 provides an additional mechanistic insight into the interaction between this receptor and the Gβ5-RGS7 complex.
Acknowledgments
Supported by NIH grant GM 060019 (V.Z.S.)
We thank Dr. Andrew Tobin (University of Leicester) for DNA constructs.
Abbreviations are
- RGS
regulator of G protein signaling
- R7BP
R7 family RGS protein binding protein
- GPCR
G protein-coupled receptor
- GST
glutathione-S-transferase
- CFP and YFP
cyan and yellow versions of green fluorescent protein (GFP)
- CHO
Chinese hamster ovary
References
- 1.Limbird LE, Gill DM, Lefkowitz RJ. Agonist-promoted coupling of the beta-adrenergic receptor with the guanine nucleotide regulatory protein of the adenylate cyclase system. Proc Natl Acad Sci U S A. 1980;77:775–779. doi: 10.1073/pnas.77.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fung BK, Hurley JB, Stryer L. Flow of information in the light-triggered cyclic nucleotide cascade of vision. Proc Natl Acad Sci U S A. 1981;78:152–156. doi: 10.1073/pnas.78.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Northup JK, Smigel MD, Sternweis PC, Gilman AG. The subunits of the stimulatory regulatory component of adenylate cyclase. Resolution of the activated 45,000-dalton (alpha) subunit. J Biol Chem. 1983;258:11369–11376. [PubMed] [Google Scholar]
- 4.Clapham DE, Neer EJ. New roles for G-protein beta gamma-dimers in transmembrane signalling. Nature. 1993;365:403–406. doi: 10.1038/365403a0. [DOI] [PubMed] [Google Scholar]
- 5.Neer EJ, Clapham DE. Roles of G protein subunits in transmembrane signalling. Nature. 1988;333:129–134. doi: 10.1038/333129a0. [DOI] [PubMed] [Google Scholar]
- 6.Smrcka AV. G protein betagamma subunits: central mediators of G protein-coupled receptor signaling. Cell Mol Life Sci. 2008;65:2191–2214. doi: 10.1007/s00018-008-8006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohlman HG, Thorner J. RGS proteins and signaling by heterotrimeric G proteins. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 8.Berman DM, Gilman AG. Mammalian RGS proteins: barbarians at the gate. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 9.Hepler JR. Emerging roles for RGS proteins in cell signalling. Trends Pharmacol Sci. 1999;20:376–382. doi: 10.1016/s0165-6147(99)01369-3. [DOI] [PubMed] [Google Scholar]
- 10.Chidiac P, Roy AA. Activity, regulation, and intracellular localization of RGS proteins. Receptors Channels. 2003;9:135–147. [PubMed] [Google Scholar]
- 11.Willars GB. Mammalian RGS proteins: multifunctional regulators of cellular signalling. Semin Cell Dev Biol. 2006;17:363–376. doi: 10.1016/j.semcdb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Witherow DS, Slepak VZ. A Novel Kind of G Protein Heterodimer: The Gbeta5-RGS Complex. Receptors Channels. 2003;9:205–212. [PubMed] [Google Scholar]
- 13.Anderson GR, Posokhova E, Martemyanov KA. The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem Biophys. 2009;54:33–46. doi: 10.1007/s12013-009-9052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slepak VZ. Structure, Function, and Localization of Gbeta5-RGS Complexes. In: Fisher Rory A., editor. Progress in Molecular Biology and Translational Science. Vol. 86. Academic Press; 2009. pp. 157–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabrera JL, de Freitas F, Satpaev DK, Slepak VZ. Identification of the Gbeta5-RGS7 protein complex in the retina. Biochem Biophys Res Commun. 1998;249:898–902. doi: 10.1006/bbrc.1998.9218. [DOI] [PubMed] [Google Scholar]
- 16.Levay K, Cabrera JL, Satpaev DK, Slepak VZ. Gbeta5 prevents the RGS7-Galphao interaction through binding to a distinct Ggamma-like domain found in RGS7 and other RGS proteins. Proc Natl Acad Sci U S A. 1999;96:2503–2507. doi: 10.1073/pnas.96.5.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snow BE, Krumins AM, Brothers GM, Lee SF, Wall MA, Chung S, Mangion J, Arya S, Gilman AG, Siderovski DP. A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gbeta5 subunits. Proc Natl Acad Sci U S A. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drenan RM, Doupnik CA, Boyle MP, Muglia LJ, Huettner JE, Linder ME, Blumer KJ. Palmitoylation regulates plasma membrane-nuclear shuttling of R7BP, a novel membrane anchor for the RGS7 family. J Cell Biol. 2005;169:623–633. doi: 10.1083/jcb.200502007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J Biol Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- 20.Jayaraman M, Zhou H, Jia L, Cain MD, Blumer KJ. R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling. Trends Pharmacol Sci. 2008 doi: 10.1016/j.tips.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayanan V, Sandiford SL, Wang Q, Keren-Raifman T, Levay K, Slepak VZ. Intramolecular Interaction between the DEP Domain of RGS7 and the Gbeta(5) Subunit. Biochemistry. 2007;46:6859–6870. doi: 10.1021/bi700524w. [DOI] [PubMed] [Google Scholar]
- 22.Cheever ML, Snyder JT, Gershburg S, Siderovski DP, Harden TK, Sondek J. Crystal structure of the multifunctional Gbeta5-RGS9 complex. Nat Struct Mol Biol. 2008;15:155–162. doi: 10.1038/nsmb.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooks SB, Martemyanov K, Zachariou V. A role of RGS proteins in drug addiction. Biochem Pharmacol. 2008;75:76–84. doi: 10.1016/j.bcp.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 24.Hooks SB, Waldo GL, Corbitt J, Bodor ET, Krumins AM, Harden TK. RGS6, RGS7, RGS9, and RGS11 stimulate GTPase activity of Gi family G-proteins with differential selectivity and maximal activity. J Biol Chem. 2003;278:10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- 25.Hajdu-Cronin YM, Chen WJ, Patikoglou G, Koelle MR, Sternberg PW. Antagonism between G(o)alpha and G(q)alpha in Caenorhabditis elegans: the RGS protein EAT-16 is necessary for G(o)alpha signaling and regulates G(q)alpha activity. Genes Dev. 1999;13:1780–1793. doi: 10.1101/gad.13.14.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chase DL, Patikoglou GA, Koelle MR. Two RGS proteins that inhibit Galpha(o) and Galpha(q) signaling in C. elegans neurons require a Gbeta(5)-like subunit for function. Curr Biol. 2001;11:222–231. doi: 10.1016/s0960-9822(01)00071-9. [DOI] [PubMed] [Google Scholar]
- 27.Patikoglou GA, Koelle MR. An N-terminal region of Caenorhabditis elegans RGS proteins EGL-10 and EAT-16 directs inhibition of G(alpha)o versus G(alpha)q signaling. J Biol Chem. 2002;277:47004–47013. doi: 10.1074/jbc.M208186200. [DOI] [PubMed] [Google Scholar]
- 28.Witherow DS, Wang Q, Levay K, Cabrera JL, Chen J, Willars GB, Slepak VZ. Complexes of the G protein subunit gbeta 5 with the regulators of G protein signaling RGS7 and RGS9. Characterization in native tissues and in transfected cells. J Biol Chem. 2000;275:24872–24880. doi: 10.1074/jbc.M001535200. [DOI] [PubMed] [Google Scholar]
- 29.Witherow DS, Tovey SC, Wang Q, Willars GB, Slepak VZ. G beta 5. RGS7 inhibits G alpha q-mediated signaling via a direct protein-protein interaction. J Biol Chem. 2003;278:21307–21313. doi: 10.1074/jbc.M212884200. [DOI] [PubMed] [Google Scholar]
- 30.Sandiford S, Slepak V. Gβ5-RGS7 selectively inhibits muscarinic M3 receptor signaling via the interaction between the third intracellular loop of the receptor and the DEP domain of RGS7. Biochemistry. 2009;48:2282–2289. doi: 10.1021/bi801989c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wess J, Brann MR, Bonner TI. Identification of a small intracellular region of the muscarinic m3 receptor as a determinant of selective coupling to PI turnover. FEBS Lett. 1989;258:133–136. doi: 10.1016/0014-5793(89)81633-3. [DOI] [PubMed] [Google Scholar]
- 32.Wu G, Krupnick JG, Benovic JL, Lanier SM. Interaction of arrestins with intracellular domains of muscarinic and alpha2-adrenergic receptors. J Biol Chem. 1997;272:17836–17842. doi: 10.1074/jbc.272.28.17836. [DOI] [PubMed] [Google Scholar]
- 33.Wu G, Benovic JL, Hildebrandt JD, Lanier SM. Receptor docking sites for G-protein betagamma subunits. Implications for signal regulation. J Biol Chem. 1998;273:7197–7200. doi: 10.1074/jbc.273.13.7197. [DOI] [PubMed] [Google Scholar]
- 34.Wu G, Bogatkevich GS, Mukhin YV, Benovic JL, Hildebrandt JD, Lanier SM. Identification of Gbetagamma binding sites in the third intracellular loop of the M(3)-muscarinic receptor and their role in receptor regulation. J Biol Chem. 2000;275:9026–9034. doi: 10.1074/jbc.275.12.9026. [DOI] [PubMed] [Google Scholar]
- 35.Lucas JL, Wang D, Sadee W. Calmodulin binding to peptides derived from the i3 loop of muscarinic receptors. Pharm Res. 2006;23:647–653. doi: 10.1007/s11095-006-9784-9. [DOI] [PubMed] [Google Scholar]
- 36.Simon V, Guidry J, Gettys TW, Tobin AB, Lanier SM. The proto-oncogene SET interacts with muscarinic receptors and attenuates receptor signaling. J Biol Chem. 2006;281:40310–40320. doi: 10.1074/jbc.M603858200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budd DC, McDonald JE, Tobin AB. Phosphorylation and regulation of a Gq/11-coupled receptor by casein kinase 1alpha. J Biol Chem. 2000;275:19667–19675. doi: 10.1074/jbc.M000492200. [DOI] [PubMed] [Google Scholar]
- 38.Luo J, Busillo JM, Benovic JL. M3 muscarinic acetylcholine receptor-mediated signaling is regulated by distinct mechanisms. Mol Pharmacol. 2008;74:338–347. doi: 10.1124/mol.107.044750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witherow DS, Slepak VZ. Biochemical purification and functional analysis of complexes between the G-protein subunit Gbeta5 and RGS proteins. Methods Enzymol. 2004;390:149–162. doi: 10.1016/S0076-6879(04)90010-9. [DOI] [PubMed] [Google Scholar]
- 40.Budd DC, McDonald J, Emsley N, Cain K, Tobin AB. The C-terminal tail of the M3-muscarinic receptor possesses anti-apoptotic properties. J Biol Chem. 2003;278:19565–19573. doi: 10.1074/jbc.M211670200. [DOI] [PubMed] [Google Scholar]
- 41.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 42.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 43.Civera C, Simon B, Stier G, Sattler M, Macias MJ. Structure and dynamics of the human pleckstrin DEP domain: distinct molecular features of a novel DEP domain subfamily. Proteins. 2005;58:354–366. doi: 10.1002/prot.20320. [DOI] [PubMed] [Google Scholar]
- 44.Brady AE, Limbird LE. G protein-coupled receptor interacting proteins: emerging roles in localization and signal transduction. Cell Signal. 2002;14:297–309. doi: 10.1016/s0898-6568(01)00239-x. [DOI] [PubMed] [Google Scholar]
- 45.Bockaert J, Fagni L, Dumuis A, Marin P. GPCR interacting proteins (GIP) Pharmacol Ther. 2004;103:203–221. doi: 10.1016/j.pharmthera.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watson AJ, Katz A, Simon MI. A fifth member of the mammalian G-protein beta-subunit family. Expression in brain and activation of the beta 2 isotype of phospholipase C. J Biol Chem. 1994;269:22150–22156. [PubMed] [Google Scholar]
- 48.Watson AJ, Aragay AM, Slepak VZ, Simon MI. A novel form of the G protein beta subunit Gbeta5 is specifically expressed in the vertebrate retina. J Biol Chem. 1996;271:28154–28160. doi: 10.1074/jbc.271.45.28154. [DOI] [PubMed] [Google Scholar]
- 49.Zhang S, Coso OA, Lee C, Gutkind JS, Simonds WF. Selective activation of effector pathways by brain-specific G protein beta5. J Biol Chem. 1996;271:33575–33579. doi: 10.1074/jbc.271.52.33575. [DOI] [PubMed] [Google Scholar]
- 50.Yoshikawa DM, Hatwar M, Smrcka AV. G protein beta 5 subunit interactions with alpha subunits and effectors. Biochemistry. 2000;39:11340–11347. doi: 10.1021/bi0005557. [DOI] [PubMed] [Google Scholar]
- 51.Zeng F, Wess J. Molecular aspects of muscarinic receptor dimerization. Neuropsychopharmacology. 2000;23:S19–31. doi: 10.1016/S0893-133X(00)00146-9. [DOI] [PubMed] [Google Scholar]
- 52.Bernstein LS, Ramineni S, Hague C, Cladman W, Chidiac P, Levey AI, Hepler JR. RGS2 binds directly and selectively to the M1 muscarinic acetylcholine receptor third intracellular loop to modulate Gq/11alpha signaling. J Biol Chem. 2004;279:21248–21256. doi: 10.1074/jbc.M312407200. [DOI] [PubMed] [Google Scholar]
- 53.Georgoussi Z, Leontiadis L, Mazarakou G, Merkouris M, Hyde K, Hamm H. Selective interactions between G protein subunits and RGS4 with the C-terminal domains of the mu- and delta-opioid receptors regulate opioid receptor signaling. Cell Signal. 2006;18:771–782. doi: 10.1016/j.cellsig.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 54.Schwendt M, McGinty JF. Regulator of G-protein signaling 4 interacts with metabotropic glutamate receptor subtype 5 in rat striatum: relevance to amphetamine behavioral sensitization. J Pharmacol Exp Ther. 2007;323:650–657. doi: 10.1124/jpet.107.128561. [DOI] [PubMed] [Google Scholar]
- 55.Langer I, Tikhonova IG, Boulegue C, Esteve JP, Vatinel S, Ferrand A, Moroder L, Robberecht P, Fourmy D. Evidence for a direct and functional interaction between the regulators of G protein signaling-2 and phosphorylated C terminus of cholecystokinin-2 receptor. Mol Pharmacol. 2009;75:502–513. doi: 10.1124/mol.108.051607. [DOI] [PubMed] [Google Scholar]
- 56.Abramow-Newerly M, Roy AA, Nunn C, Chidiac P. RGS proteins have a signalling complex: interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell Signal. 2006;18:579–591. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 57.Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126:1079–1093. doi: 10.1016/j.cell.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 58.Kovoor A, Seyffarth P, Ebert J, Barghshoon S, Chen CK, Schwarz S, Axelrod JD, Cheyette BN, Simon MI, Lester HA, Schwarz J. D2 dopamine receptors colocalize regulator of G-protein signaling 9-2 (RGS9-2) via the RGS9 DEP domain, and RGS9 knock-out mice develop dyskinesias associated with dopamine pathways. J Neurosci. 2005;25:2157–2165. doi: 10.1523/JNEUROSCI.2840-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]