Abstract
The molecular clock maintains energy constancy by producing circadian oscillations of rate-limiting enzymes involved in tissue metabolism across the day and night1–3. During periods of feeding, pancreatic islets secrete insulin to maintain glucose homeostasis, and while rhythmic control of insulin release is recognized to be dysregulated in humans with diabetes4, it is not known how the circadian clock may affect this process. Here we show that pancreatic islets possess self-sustained circadian gene and protein oscillations of the transcription factors CLOCK and BMAL1. The phase of oscillation of islet genes involved in growth, glucose metabolism, and insulin signaling is delayed in circadian mutant mice, and both Clock5,6 and Bmal17 mutants exhibit impaired glucose tolerance, reduced insulin secretion, and defects in size and proliferation of pancreatic islets that worsen with age. Clock disruption leads to transcriptome-wide alterations in the expression of islet genes involved in growth, survival, and synaptic vesicle assembly. Remarkably, conditional ablation of the pancreatic clock causes diabetes mellitus due to defective β-cell function at the very latest stage of stimulus-secretion coupling. These results demonstrate a role for the β-cell clock in coordinating insulin secretion with the sleep-wake cycle, and reveal that ablation of the pancreatic clock can trigger onset of diabetes mellitus.
The circadian clock drives cycles of energy storage and utilization in plants, flies, and mammals in anticipation of changes in the external environment imposed by the rising and setting of the sun1. In mammals, the transcription factors CLOCK and BMAL1 drive the central oscillator within the hypothalamus and even in peripheral tissues, yet a major question remains regarding the link between cellular rhythms and organismal homeostasis, including constancy of energy and fuel utilization cycles8,9. In humans, one of the most pronounced rhythmic aspects of physiology involves the daily variation of glucose tolerance and insulin sensitivity across the 24 hr day, and importantly, disruption of circadian oscillation of glucose metabolism is a hallmark of type 2 diabetes4. Moreover, there is growing evidence that metabolic and circadian systems are interconnected at the transcriptional level, as genomic analyses have revealed that both neural and peripheral clocks regulate the 24 hr periodicity of RNAs that mediate rate-limiting steps in glycolysis, fatty acid oxidation, and oxidative phosphorylation, indicating that these processes are primed to occur at the optimal time during glucose and fatty acid utilization cycles2,10–13.
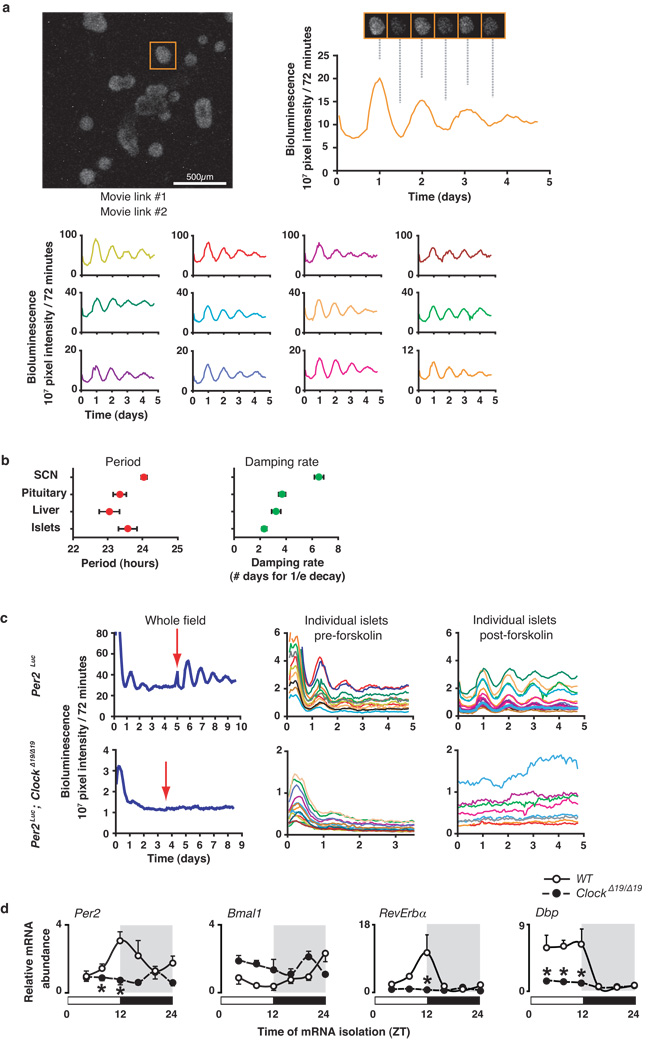
Prompted by the hypothesis that the circadian clock exerts effects on metabolism through cellular actions, we sought to dissect the impact of clock function within the pancreatic islet, a principle regulator of glucose homeostasis. We used real-time bioluminescence imaging in isolated pancreatic islets from Period2Luciferase (Per2Luc) knock-in mice to determine whether the clock is expressed autonomously within pancreas14. Continuous monitoring of light emission from individual islets revealed a self-sustained high-amplitude rhythm of PER2::LUC expression with a period length of 23.58 ± 0.3 hrs (Fig 1a–b and Suppl Movies S1–2), which closely matched that of other peripheral tissues and the SCN (Fig 1b)14,15. The oscillation gradually dampened after three days, similar to pituitary and liver (Fig 1b), and addition of 10µM forskolin to islets led to immediate reinitiation of robust rhythms (Fig 1c). Bioluminescence from individual islets from ClockΔ19/Δ19 mutant mice lacked a circadian rhythm, even after forskolin stimulation (Fig 1c). Quantitative real-time PCR showed that Per2 RNA expression was reduced and rhythmicity abolished in islets from ClockΔ19/Δ19 mutant mice (Fig 1d). Together, the PER2 protein and mRNA oscillation in WT islets, as well as the loss of rhythmicity of Per2 in ClockΔ19/Δ19 islets, provide evidence for a self-sustained clock in endocrine pancreas.
Figure 1. Cell autonomous oscillator in pancreas.
(a) Islets from Per2Luc mice were imaged, and orange trace at right represents bioluminescence rhythm collected from the islet in the orange square (left). Traces from other islets are shown below. See also Supplemental Movies 1–2. (b) Periods of luminescence and damping rate in multiple tissues (mean ± S.E.M., n=6 mice/genotype). (c) Whole field and individual traces from WT and ClockΔ19/Δ19 islets. Red arrow indicates exposure to 10 µM forskolin for 1 hour. (d) Oscillation of clock genes in WT and ClockΔ19/Δ19 mutant islets across 24 hrs (mean ± S.E.M., n=4 mice/genotype/time point, 2-way ANOVA, *p<0.05).
Since a major mechanism of circadian regulation involves the cycling of genes involved in cell metabolism and proliferation, we examined 24 hr RNA rhythms of essential transcripts involved in insulin secretion and β-cell growth in isolated islets (Suppl Fig 1). ClockΔ19/Δ19 mutant islets showed decreased expression levels of genes downstream of CLOCK:BMAL1 that comprise the core circadian loop, as well as the D-box and ROR feedback loops (Fig 1d). ClockΔ19/Δ19 mutant animals also showed decreased levels of expression and/or phase shifts of RNA oscillation of genes involved in insulin signaling (InsR, Irs2, Pi3K p85, Akt2), glucose sensing (Glut2, Gck), and islet growth and development (CyclinD1, Gsk3β , Hnf4α , Hnf1α , Pdx1, NeuroD1) (Fig Suppl 1). The alterations in temporal patterns of gene expression in ClockΔ19/Δ19 mutant islets were distinct from those in ClockΔ19/Δ19 mutant liver, reflecting partitioning of metabolic functions within these two tissues at different times of day (Fig Suppl 1–2 and Supporting Description 1).
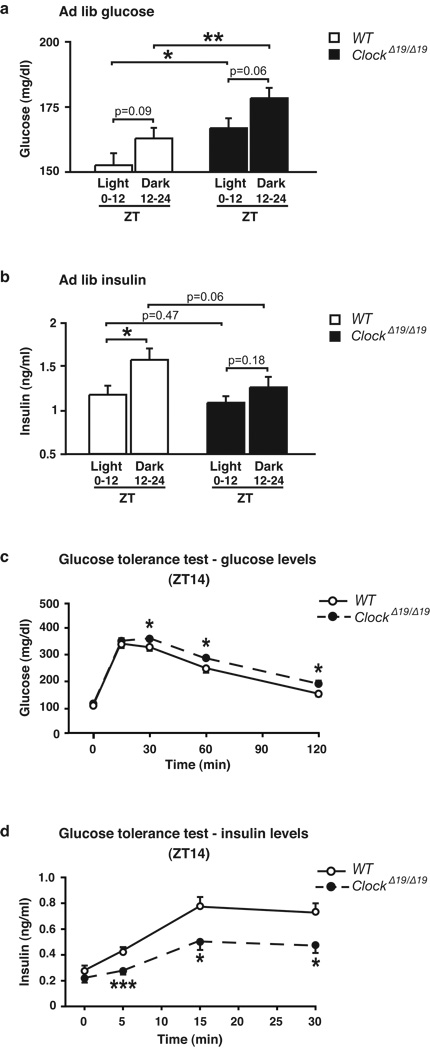
To determine whether molecular disruption of the clock in pancreas corresponds with abnormalities in the temporal control of glucose metabolism, we analyzed 24 hr glucose and insulin profiles in 8 month old ClockΔ19/Δ19 mutant mice and their WT littermates during ad lib feeding. In ClockΔ19/Δ19 mutants, glucose levels were elevated across the entire light-dark (LD) cycle without a rise in insulin levels, whereas insulin rises in WT animals during the beginning of the feeding period (Fig 2a–b). ClockΔ19/Δ19 mutant mice also displayed significantly elevated fasting glucose levels at both ZT2 and ZT14 (Fig Suppl 3e–f). Glucose tolerance tests further revealed a 50% reduction in insulin release, corresponding with increased glucose excursion in ClockΔ19/Δ19 mutant mice particularly at the beginning of the dark period (Fig 2c–d and Supporting Description 2). The likelihood that impaired glucose tolerance in ClockΔ19/Δ19 mutant mice involves a primary defect in pancreatic function was further supported by the finding that these animals have normal insulin tolerance (Fig Suppl 3c–d).
Figure 2. Diabetic phenotypes in 8 month old circadian mutant mice.
Ad lib fed (a) glucose and (b) insulin in ClockΔ19/Δ19 animals, shown as the average for time points during the light and dark periods (n=17). (c) Glucose tolerance (n=15–18) and (d) insulin secretion (n=8–10) in ClockΔ19/Δ19 mice at ZT14 following intraperitoneal glucose administration of 2 or 3g/kg body weight, respectively n=15–18). Data was analyzed by Student’s t-test (a–b) and 1-way ANOVA (c–d). p<0.05; **p<0.01; ***p<0.001. All values represent mean ± S.E.M.
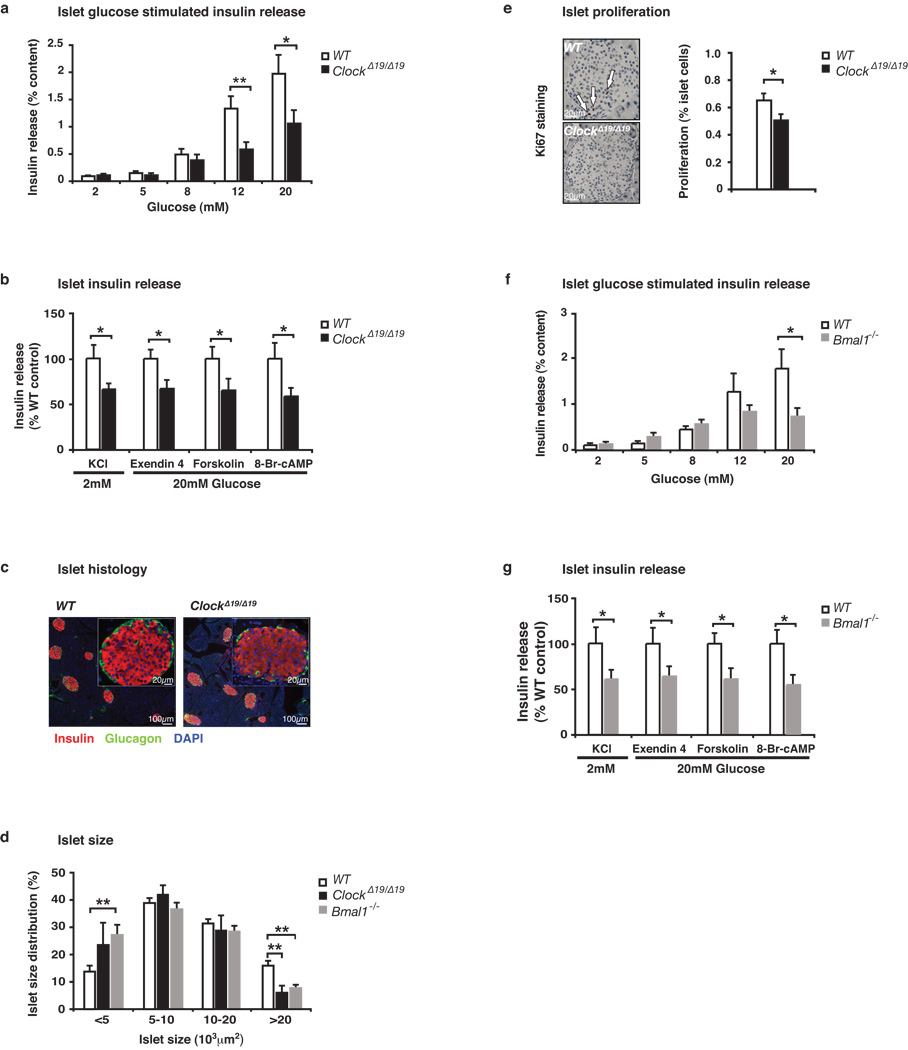
To better understand the impact of the circadian gene mutation on pancreatic function, we examined glucose-stimulated insulin secretion (GSIS) in isolated size-matched pancreatic islets from 8 month old mice. Islets from ClockΔ19/Δ19 mice displayed a ~50% reduction in GSIS (Fig 3a) and failed to respond to KCl (Fig 3b), indicating a defect in insulin exocytosis. Consistent with a predominant defect in insulin release rather than glucose metabolism, we observed normal calcium flux in response to 12 mM glucose in ClockΔ19/Δ19 mutant compared to WT islets (Fig Suppl 5d–e). Further consistent with defects in exocytosis, islets from ClockΔ19/Δ19 mutant mice displayed diminished insulin secretory responses to the cyclase activators forskolin and exendin-4, as well as 8-Bromo-cyclic AMP, localizing the impaired function of ClockΔ19/Δ19 mutant islets to a late stage in stimulus-secretion coupling16 (Fig 3b). Lastly, in agreement with an exocytic defect as the cause of decreased insulin release in circadian mutant animals, we did not observe a significant difference in either absolute insulin mRNA levels (Fig Suppl 1b) or in islet insulin content (WT 38.2 ng insulin/islet vs ClockΔ19/Δ19 32.9 ng insulin/islet, p=0.08).
Figure 3. Reduced islet size, proliferation, and insulin release in 8–10 month old circadian mutant mice.
(a) Glucose-stimulated insulin release in isolated ClockΔ19/Δ19 islets compared to similar-sized WT islets (n=9–10 mice/genotype), normalized to % insulin content. (b) Insulin secretion from ClockΔ19/Δ19 islets in response to secretagogues (n=6–14). Insulin release was calculated as in (a), and ClockΔ19/Δ19 values are expressed as a percentage of WT. (c) Representative islet morphology in ClockΔ19/Δ19 and WT pancreata (body weight and pancreata weight were not different). (d) Size of islets isolated from ClockΔ19/Δ19 and Bmal1−/− mice compared to WT (n=6–9). (e) Ki67 staining of islet proliferation (ClockΔ19/Δ19 and WT, n=5–6). (f) Glucose-stimulated insulin secretion in Bmal1−/− islets compared to WT (n=5). (g) Insulin secretion from Bmal1−/− islets in response to secretagogues (n=6–10). Five islets per mouse were analyzed in triplicate for each test condition; data was analyzed by Student’s t-test. *p<0.05; **p<0.01, and values represent mean ± S.E.M.
In contrast to 8 month old ClockΔ19/Δ19 mutant mice, we found that fasting and fed glucose levels in 3 month old ClockΔ19/Δ19 mutants were normal and that they had enhanced insulin sensitivity, possibly due to clock disruption at the level of either liver, skeletal muscle or adipose tissue (Fig Suppl 4a–f). Nonetheless, isolated islets even from young ClockΔ19/Δ19 mutant mice displayed impaired GSIS (Fig Suppl 4g). These observations are consistent with early onset of a primary islet cell defect that only manifests as overt diabetes in >8 month old ClockΔ19/Δ19 mutants, likely due to the gradual onset of insulin resistance during aging (compare Fig Suppl 3c,d and 4e,f), unmasking the β-cell defect and thereby resulting in age-associated hyperglycemia and hypoinsulinemia.
Functional defects in insulin secretion have previously been shown to arise due to impairment of genetic pathways that alter islet proliferation and survival17–21. In this regard, although indirect immunofluorescence revealed normal overall architecture of islets of ClockΔ19/Δ19 mice (Fig 3c), light microscopy of isolated islets revealed that ClockΔ19/Δ19 mutant islets were smaller than those of WT (Fig 3d). Total islet area was also reduced by ~20% in ClockΔ19/Δ19 mice based upon morphometric analysis of intact pancreata, corresponding with a trend towards decreased total pancreatic insulin content (Fig Suppl 5a–b). To determine whether the observed decrease in islet size corresponded with decreased islet proliferation, we stained pancreatic sections with the proliferation marker Ki67. Surprisingly, we found a 23% decrease in proliferation in islets from ClockΔ19/Δ19 mutant animals (Fig 3e), together with a trend towards increased islet apoptosis (Fig Suppl 5c). Microarray analysis revealed decreased levels of genes encoding components of E-box, D-box, and ROR transcription modules (Per1, Per3, Rev-erbα Tef, Dbp) (Supporting Description 3, Fig Suppl 6b and Suppl Tables 1–2). Gene Ontogeny enrichment revealed significant alterations in vesicular docking and trafficking factors (including Vamp3 and Syntaxin6) and increases in the cell death factor S100a6 (Fig Suppl 7 and Supporting Description 4), consistent with reduced function and size of islets from ClockΔ19/Δ19 mutant mice.
To determine whether the defects of islet function and size are unique to the ClockΔ19/Δ19 animal, or instead reflect a generalized role for the core circadian network in islet function, we analyzed both GSIS and islet size in Bmal1−/− mutant mice7. Bmal1−/− islets exhibited up to a 60% reduction in insulin secretion compared to littermate controls in response to glucose, KCl, exendin4, forskolin, and 8-Br-cAMP (Fig 3f–g). Furthermore, there was a 2-fold reduction in the percentage of large islets in Bmal1−/− mice compared with WT littermates (Fig 3d). The similarity of defects in islets from ClockΔ19/Δ19 mutant and Bmal1−/− mice suggests that multiple core circadian genes impact β-cell function and development.
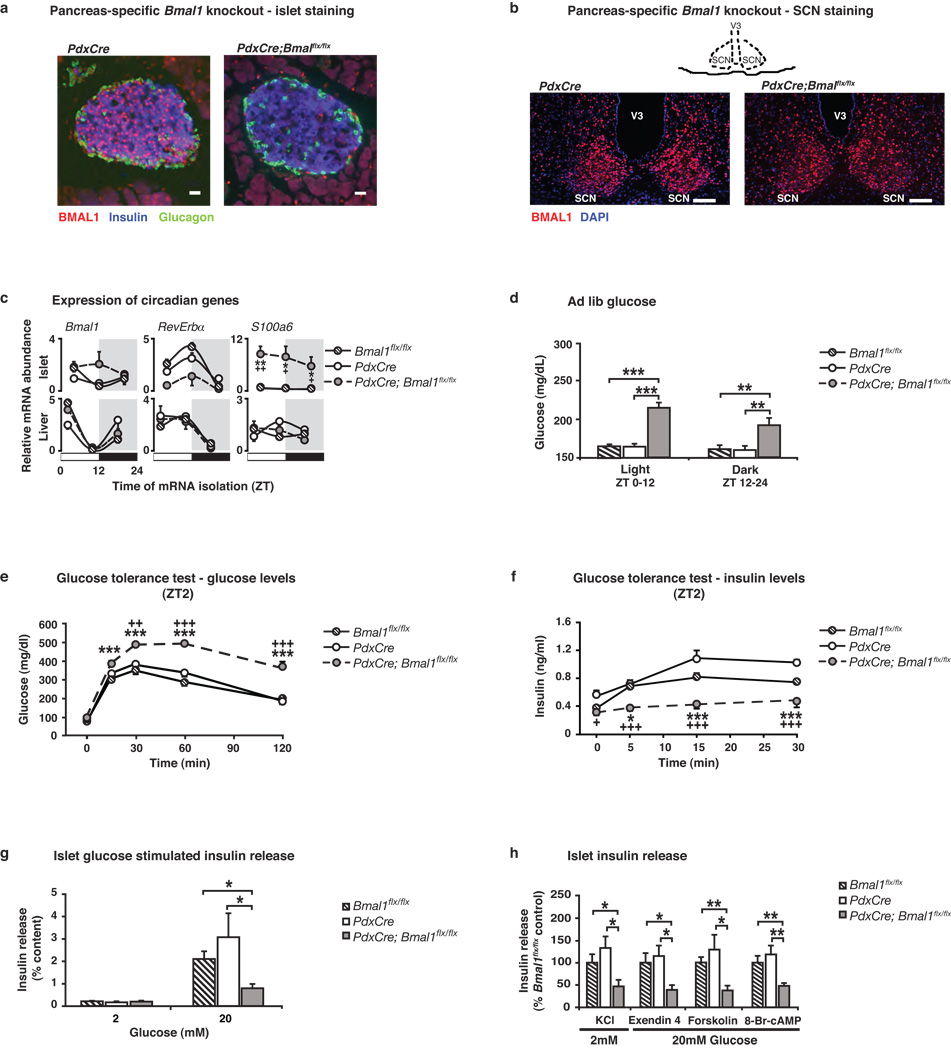
To further evaluate the contribution of the pancreatic clock to both whole body glucose metabolism and to islet function, we generated pancreas-specific Bmal1 mutant mice using the promoter of the homeodomain transcription factor PDX1 to drive expression of CRE-recombinase22,23 (Fig 4). We performed immunofluorescent staining to confirm loss of BMAL1 expression specifically within pancreatic islets (Fig 4a) and not within brain regions such as the suprachiasmatic nucleus (SCN), arcuate nucleus (ARC), dorsomedial hypothalamus (DMH), and paraventricular nucleus (PVN) (Fig 4b and Fig Suppl 8). Quantitative real-time PCR analysis of key circadian genes in islet and liver from PdxCre;Bmal1flx/flx and control mice revealed islet-specific alterations in gene expression profiles (Fig 4c and Supporting Description 5), further confirming the specificity of the Bmal1 mutation to the islet. Importantly, the pancreas-specific Bmal1 knockout mice showed normal circadian activity, feeding rhythms, and body weight and composition (Fig Suppl 8b–h). Remarkably, however, we found that 2–4 month old PdxCre; Bmal1flx/flx mice displayed significantly elevated ad lib glucose levels throughout the day (Fig 4d), as well as dramatically impaired glucose tolerance and decreased insulin secretion (Fig 4e–f, Fig Suppl 9c–d and Suppl Table 3). It is important to note that these phenotypes develop in PdxCre;Bmal1flx/flx mice at a young age (2–4 mo) compared to the global ClockΔ19/Δ19 mutant mice. Furthermore, the impaired glucose tolerance in the pancreas-specific Bmal1 mutant mice was much more pronounced than in either of the global ClockΔ19/Δ19 or Bmal1 nullizygous mice, consistent with compensation occurring in the global mutant mice and emphasizing the primary role of the islet clock in maintaining euglycemia. Finally, because the pancreas-specific Bmal1 mutant mice have normal activity and feeding rhythms, as well as normal body weight (Fig Suppl 8), the metabolic phenotypes that develop must be due to disruption of the clock network within the islet, rather than due to secondary changes in activity or behavior. Interestingly, our isolated islet studies in the pancreas-specific Bmal1 knockout mice revealed diminished insulin responsiveness to glucose, KCl, exendin 4, forskolin, and 8-Br-cAMP, indicating a block in exocytosis (Fig 4g–h), similar to that of ClockΔ19/Δ19 and Bmal1 mutant mice. Thus, ablation of the pancreatic clock is sufficient to render the islet refractory to glucose and insulin secretagogues, resulting in hyperglycemia.
Figure 4. Impaired glucose tolerance and islet insulin secretion in 2–4 mo old pancreas-specific circadian mutant mice.
(a) Immunofluorescent staining of BMAL1 (red), insulin (blue), and glucagon (green) in PdxCre; Bmal1flx/flx and control islets (Scale bar, 25 m). (b) Immunofluorescent staining of BMAL1 (red) and DAPI (blue) in SCN of PdxCre; Bmal1flx/flx and control mice (Scale bar, 50 µm). (c) Oscillation of Bmal1, Rev-erbα, and s100a6 in islets and liver of PdxCre; Bmal1flx/flx mice at three sequential 8 hr time points (n=4 mice/genotype/time). (d) Blood glucose levels in ad lib fed PdxCre; Bmal1flx/flx mice, shown as the average for values in light and dark (n=9–10). (e) Glucose tolerance (n=6–8) and (f) insulin secretion (n=9–11) in PdxCre; Bmal1flx/flx mice at ZT2 following intraperitoneal glucose administration of 2 or 3g/kg body weight, respectively. (g) Insulin release in response to glucose in PdxCre; Bmal1flx/flx islets compared to size-matched control islets (n=5–7). (h) Insulin secretion from PdxCre; Bmal1flx/flx islets in response to secretagogues (n=5–7). For all studies, five islets per mouse were analyzed in triplicate for each concentration of glucose and secretagogue. Data was analyzed by 1-way (e,f) and 2-way (c) ANOVA, and Student’s t-test (d,g,h). *p<0.05; **p<0.01; ***p<0.001. For c,e,f, * denotes significance between Bmal1flx/flx and PdxCre;Bmal1flx/flx, and + denotes significance between PdxCre and PdxCre;Bmal1flx/flx. All values represent mean ± S.E.M.
The recent availability of both global and tissue-specific genetic models of circadian disruption offers unique opportunities to define the role of circadian oscillators in physiology. For instance, our previous studies in ClockΔ19/Δ19 mutant mice showed that these mice are susceptible to obesity and certain features of metabolic syndrome including steatosis, adipose hypertrophy, and hyperlipidemia, due to primary defects within both hypothalamus (decreased anorexigenic peptidergic signaling), and also within liver. Yet ClockΔ19/Δ19 animals have sufficient β-cell reserve early in life to drive adiposity, although eventually their weight gain plateaus coincident with progressive β-cell failure. In contrast, ablation of the clock within liver results in hypoglycemia24,25, while clock function within liver is also tied to reduced cholesterol extrusion as bile, steatosis, and hypertriglyceridemia, all features of metabolic syndrome26–28. In summary, obesity in circadian mutants is separable from insulin resistance, and the same defects that cause changes in β-cell function can ameliorate insulin resistance in the liver, through mechanisms that require further investigation. Our findings in β-cell conditional knockouts underscore the intimate integration of circadian and metabolic systems within multiple tissues and the complex effect of clock function in separate tissues on energy and metabolic homeostasis in the whole animal. In addition to orchestrating the daily cycles of glucose metabolism, the islet circadian transcription network also participates in the determination of islet cell mass, similar to previously reported effects of the clock on growth and regeneration of bone29 and liver30. Clinical evidence further suggests that temporal defects in insulin secretion are early indicators of β-cell dysfunction. Therefore, consideration of the dynamics of clock function across temporally distinct phases of the day-night cycle, and within different tissues as the cycle progresses, may also explain obesity and metabolic pathologies that emerge in states of circadian disruption including shift work and night eating.
METHODS SUMMARY
Islet Isolation and in Vitro Insulin Secretion Analysis
Pancreatic islets were isolated and secretion assays performed as described in Supplementary Material. Briefly, for insulin release, 5 islets were statically incubated in Krebs-Ringer Buffer and stimulated for 1hr at 37°C with various glucose concentrations, 30mM KCl, 100nM exendin 4, 2.5µM forskolin, or 1mM 8-Br-cAMP.
Circadian Expression of PER2::LUC
Bioluminescence from Per2Luc and Per2Luc;ClockΔ19/Δ19 islets was continuously imaged using an XR/MEGA-10Z cooled CCD camera, as islets were cultured at 37°C in a Lucite environmentally controlled chamber (full details in Supplementary Material). Period and damping rate were calculated using Lumicycle Analysis software (Actimetrics).
Full methods accompany this paper.
Supplementary Material
Acknowledgements
We thank Drs. Fred Turek, Ravi Allada, and Graeme Bell for helpful discussions and comments on the manuscript. We thank Akira Kohsaka, Emily Chen, Joseph Doering, and Carl Radosevich for their technical support, as well as the Biological Imaging Facility at Northwestern University and the Islet Biology Core of the University of Chicago DRTC. We thank Dr. Douglas Melton for the PdxCre mice. Work was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases to K.M.R. and L.H.P.; the National Institute of Health, Chicago Biomedical Consortium Searle Funds, and Juvenile Diabetes Research Foundation to J.B.; Grant R37-ES-005703 from the National Institute of Health to C.A.B.; and the National Institute of Mental Health to J.S.T.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: B.M. performed and analyzed most of the experiments in this study, with technical assistance from Y.K., G.I., S.M., and C.O. E.D.B. and C.K. conducted and analyzed real-time bioluminescence imaging experiments in isolated pancreatic islets. H.S. conducted immunostaining experiments. M.V. performed statistical analysis. J.L. conducted and analyzed Ca2+ influx experiments. S.D.C. and L.J.B. performed statistical and gene ontogeny analysis of microarray data. C.A.B. provided Bmal1flx/flx mice. J.S.T., L.P., X.W., K.M.R., B.M., and J.B. provided critical intellectual input in the preparation of the manuscript. K.M.R., B.M., J.S.T., and J.B. wrote the paper.
Author Information: Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. J.S.T. is an Investigator in the Howard Hughes Medical Institute and a cofounder of ReSet Therapeutics Inc., and J.S.T. and J.B. are members of its scientific advisory board. J.B. is also an advisor and receives support from Amylin Pharmaceuticals. Correspondence and requests for materials should be addressed to J.B. (j-bass@northwestern.edu).
REFERENCES
- 1.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 3.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 4.Polonsky KS, et al. Abnormal patterns of insulin secretion in non-insulin-dependent diabetes mellitus. N Engl J Med. 1988;318:1231–1239. doi: 10.1056/NEJM198805123181903. [DOI] [PubMed] [Google Scholar]
- 5.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornmann B, et al. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(e34):0179–0189. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarthy JJ, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:89–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 13.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 14.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 16.Drucker DJ. The biology of incretin hormones. Cell Metab. 2006;3:153–165. doi: 10.1016/j.cmet.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Gao N, et al. Foxa2 controls vesicle docking and insulin secretion in mature Beta cells. Cell Metab. 2007;6:267–279. doi: 10.1016/j.cmet.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Rulifson IC, et al. Wnt signaling regulates pancreatic beta cell proliferation. Proc Natl Acad Sci U S A. 2007;104:6247–6252. doi: 10.1073/pnas.0701509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih DQ, et al. Loss of HNF-1alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 2001;50:2472–2480. doi: 10.2337/diabetes.50.11.2472. [DOI] [PubMed] [Google Scholar]
- 20.Stoffers DA, et al. Insulinotropic glucagon-like peptide 1 agonists stimulate expression of homeodomain protein IDX-1 and increase islet size in mouse pancreas. Diabetes. 2000;49:741–748. doi: 10.2337/diabetes.49.5.741. [DOI] [PubMed] [Google Scholar]
- 21.Withers DJ, et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 22.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 23.Westgate EJ, et al. Genetic components of the circadian clock regulate thrombogenesis in vivo. Circulation. 2008;117:2087–2095. doi: 10.1161/CIRCULATIONAHA.107.739227. [DOI] [PubMed] [Google Scholar]
- 24.Lamia KA, Storch KF, Weitz C. J. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(e377):1893–1899. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duez H, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 27.Noshiro M, et al. Multiple mechanisms regulate circadian expression of the gene for cholesterol 7alpha-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J Biol Rhythms. 2007;22:299–311. doi: 10.1177/0748730407302461. [DOI] [PubMed] [Google Scholar]
- 28.Kudo T, Kawashima M, Tamagawa T, Shibata S. Clock mutation facilitates accumulation of cholesterol in the liver of mice fed a cholesterol and/or cholic acid diet. Am J Physiol Endocrinol Metab. 2008;294:E120–E130. doi: 10.1152/ajpendo.00061.2007. [DOI] [PubMed] [Google Scholar]
- 29.Fu L, et al. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 30.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.