SUMMARY
DNA synthesis by classical polymerases can be blocked by many lesions. These blocks are overcome by translesion synthesis, whereby the stalled classical, replicative polymerase is replaced by a non-classical polymerase. In eukaryotes, this polymerase exchange requires PCNA monoubiquitination. To better understand the polymerase exchange, we have developed a novel means of producing monoubiquitinated PCNA, by splitting the protein into two self-assembling polypeptides. We determined the X-ray crystal structure of monoubiquitinated PCNA and found that the ubiquitin moieties are located on the back face of PCNA and interact with it via their canonical hydrophobic surface. Moreover, the attachment of ubiquitin does not change PCNA’s conformation. We propose that PCNA ubiquitination facilitates non-classical polymerase recruitment to the back of PCNA by forming a new binding surface for non-classical polymerases, consistent with a “tool belt” model of the polymerase exchange.
DNA damage, caused by radiation and a variety of chemical agents, can lead to mutations, genomic instability, cancer, and cell death. Genetic studies in the yeast Saccharomyces cerevisiae have revealed three general pathways for coping with radiation-induced DNA damage in eukaryotes 1. Proteins in the Rad3 pathway catalyze nucleotide excision repair, which removes bulky, helix-distorting lesions. Proteins in the Rad52 pathway catalyze double-strand break repair through homologous recombination. Proteins in the Rad6 pathway catalyze post-replication repair, a multi-faceted process that includes translesion synthesis.
Post-replication repair is regulated by the monoubiquitination and polyubiquitination of proliferating cell nuclear antigen (PCNA), the eukaryotic sliding clamp processivity factor. Rad6 is an ubiquitin conjugating enzyme that associates with Rad18, a ubiquitin ligase 2–4. The Rad6-Rad18 complex catalyzes the monoubiquitination of PCNA on Lys-164, which promotes translesion synthesis 5,6. The Mms2-Ubc13 dimer is an ubiquitin conjugating enzyme that associates with Rad5, another ubiquitin ligase, and this complex catalyzes the formation of polyubiquitin chains via Lys-63 linkages 7–9. These proteins convert monoubiquitinated PCNA (UbiPCNA) to polyubiquitinated PCNA, which promotes a currently uncharacterized error-free pathway of post-replication repair 5,6.
Translesion synthesis is a process that occurs when a classical DNA polymerase (i.e., one that synthesizes DNA during normal replication and repair) is blocked at a DNA lesion in the template strand. In translesion synthesis, the stalled classical polymerase is replaced by a non-classical DNA polymerase which then carries out replication through the damage. Eukaryotes possess several non-classical DNA polymerases, which differ from their classical counterparts in the ability to accommodate damaged DNA templates 10,11. DNA polymerase zeta (pol ζ), for example, functions in the mutagenic bypass of a wide range of lesions 12–15. By contrast, DNA polymerase eta (pol η) functions in the error-free translesion synthesis of thymine dimers 16,17. In humans, lack of pol η causes the variant form of xeroderma pigmentosum, a cancer-prone genetic disorder 18,19.
Several lines of evidence demonstrate that the monoubiquitination of PCNA plays a critical role in recruiting non-classical polymerases to sites of DNA damage and in orchestrating the polymerase exchange step between the classical and non-classical polymerases during translesion synthesis. First, most non-classical polymerases, including pol η, possess ubiquitin binding motifs, and mutations in these motifs lead to loss of protein function in vivo 20,21. Second, in human cells, pol η and UbiPCNA co-localize to replication foci following DNA damage 22. Moreover, pol η specifically interacts with UbiPCNA, but not with unmodified PCNA in these cells following DNA damage 22. Third, purified yeast pol η can replace the classical DNA polymerase delta (pol δ) on the DNA when it stalls in vitro in the presence of UbiPCNA, but not in the presence of unmodified PCNA 23.
Despite the obvious importance of UbiPCNA in facilitating the polymerase exchange step of translesion synthesis, the structural and biochemical basis by which it does this remains unknown. Efforts to better understand the polymerase exchange have been hampered by the inability to produce sufficient quantities of UbiPCNA for structural and biochemical studies. Here we report a novel strategy to produce large quantities of yeast UbiPCNA by splitting the protein into two polypeptides that self-assemble in vivo. We show that UbiPCNA produced in this manner stimulates pol η activity in vitro and fully supports translesion synthesis in vivo. We have determined the X-ray crystal structure of UbiPCNA and found that the ubiquitin moieties are located on the back face of the PCNA ring. Moreover, the attachment of ubiquitin to PCNA does not change the conformation of PCNA. This strongly suggests that ubiquitination of PCNA facilitates non-classical polymerase recruitment to the back face of the PCNA ring by forming a new interacting surface for the non-classical polymerases. This is consistent with a “tool belt” model of the polymerase exchange in which classical and non-classical polymerases simultaneously bind to UbiPCNA.
RESULTS
Production of Split PCNA and UbiPCNA
The production of sufficient quantities of monoubiquitinated proteins for structural and biochemical studies is challenging. Here we report a novel strategy for easily producing large quantities of monoubiquitinated proteins. This strategy, which could be applied to a variety of systems, is (1) to split the target protein at the site of monoubiquitination into two polypeptides, (2) to fuse ubiquitin in frame at the N-terminus of the C-terminal fragment of the target protein, and (3) to co-express the two polypeptides and allow them to self-assemble in vivo. We have successfully used this approach to produce monoubiquitinated PCNA, and we produced a split, non-ubiquitinated form of PCNA as well.
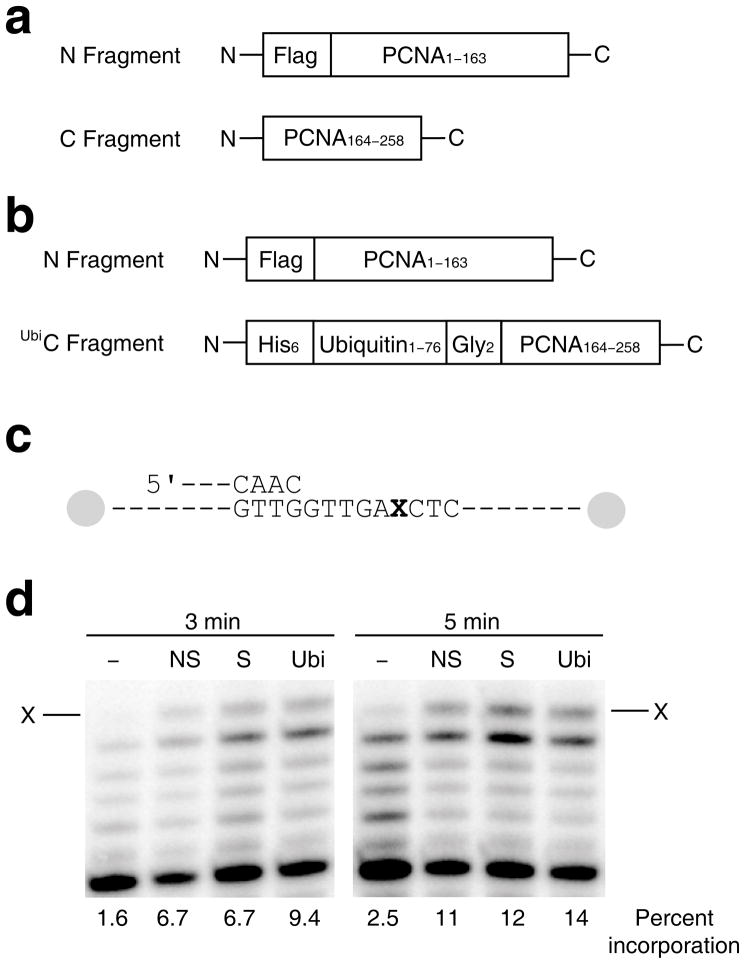
The polypeptides used to over-express split PCNA and split UbiPCNA are shown in Fig. 1a and Fig. 1b. For production of split PCNA, the first polypeptide (the N fragment) contained amino acid residues 1 to 163 of PCNA and was N-terminally FLAG™-tagged. The second polypeptide (the C fragment) contained residues 164 to 258 of PCNA. For production of split UbiPCNA, the first polypeptide (the N fragment) was identical to the one used to produce split PCNA. The second polypeptide (the UbiC fragment) contained the entire ubiquitin sequence (residues 1–76) fused via a short linker to residues 165 to 258 of PCNA and was N-terminally His6-tagged. The short linker consisted of two glycine residues because this is nearly isosteric with the side chain of Lys-164 and the isopeptide bond to the C-terminus of ubiquitin.
Figure 1.
Stimulation of pol η activity by split PCNA and UbiPCNA. (a) Diagram of the two polypeptides used to generate split PCNA is shown. (b) Diagram of the two polypeptides used to generate UbiPCNA is shown. (c) Diagram of the running start DNA substrate used. The ‘X’ represents an abasic site. Both ends of the template strand are capped with biotin-streptavidin blocks. (d) Autoradiograph of the products of the running start reaction of pol η and the indicated DNA substrate after 3 min. and 5 min. The arrow represents incorporation opposite the abasic site. Lanes labeled ‘–’ contain no PCNA, lanes labeled ‘NS’ contain non-split PCNA, lanes labeled ‘S’ contain split PCNA, and lanes labeled ‘Ubi’ contain UbiPCNA. The percent incorporation is shown below each lane.
We were able to purify milligram quantities of both split PCNA and split UbiPCNA. In both cases, the two polypeptides fragments were present in a one-to-one ratio. Size exclusion chromatography showed that split PCNA had a Stokes radius of 45 Å, which was identical to the Stokes radius of non-split PCNA and closely agreed with the actual radius of the PCNA trimer (46 Å). The Stokes radius of split UbiPCNA was 50 Å, which was slightly larger than the Stokes radius of unmodified PCNA. This demonstrated that, like non-split PCNA, split PCNA and split UbiPCNA formed stable, ring-shaped trimers. Furthermore, both mass spectrometry and Western blotting confirmed the presence of the ubiquitin moiety in the split UbiPCNA preparations.
Effect of split PCNA and UbiPCNA on DNA polymerase η activity
Before carrying out structural determinations, we first examined whether split PCNA and split UbiPCNA could function in vitro to stimulate the catalytic activity of DNA polymerase η (pol η), a prototypical non-classical DNA polymerase. It has been shown previously that both non-split PCNA and non-split UbiPCNA stimulate the ability of pol η to incorporate nucleotides opposite template abasic sites 24–26. Thus we examined the ability of split PCNA and split UbiPCNA to stimulate pol η in a running start assay (Fig. 1c and Fig. 1d). The different PCNA proteins (non-split PCNA, split PCNA, and split UbiPCNA) were loaded onto the DNA substrate by replication factor C (the ATP-dependent clamp loading complex), and both ends of the DNA were blocked with biotin/streptavidin to prevent the PCNA proteins from sliding off the substrate. Figure 1d shows the incorporation of nucleotides by pol η opposite an abasic site under running start conditions. Pol η alone had very low activity in this context and incorporated opposite the lesion on 2.5% of the substrates in 5 min. In the presence of non-split PCNA, split PCNA, and split UbiPCNA, pol η had greater activity and incorporated opposite the lesion on 11%, 12%, and 14% of the substrates in 5 min., respectively. We observed no full length, run-off products under these conditions. Although full-length products were observed previously for pol η in experiments with both unmodified PCNA and UbiPCNA 25, the enzyme was in excess over the DNA in that study compared to the conditions used here in which the DNA was in a 10-fold excess over the enzyme.
To quantify the effects of non-split PCNA, split PCNA, and split UbiPCNA on pol η activity, we carried out steady-state kinetic studies of nucleotide incorporation opposite a template abasic site (Supplementary Table 1). Non-split PCNA stimulated the catalytic efficiency (Vmax/Km) of nucleotide incorporation by pol η by a factor of 2.5 relative to the efficiency of incorporation in the absence of PCNA. Similarly, split PCNA stimulated the catalytic efficiency of pol η by a factor of 2.7. Split UbiPCNA stimulated nucleotide incorporation opposite the abasic site to a slightly greater extent (by a factor of 3.8) than did non-split PCNA and split PCNA. These results show that both split PCNA and UbiPCNA retained the ability to stimulate the catalytic activity of pol η, and that UbiPCNA stimulated the activity of pol η to a slightly greater extent than did unmodified PCNA.
Effects of split PCNA and UbiPCNA on cell growth and UV sensitivity
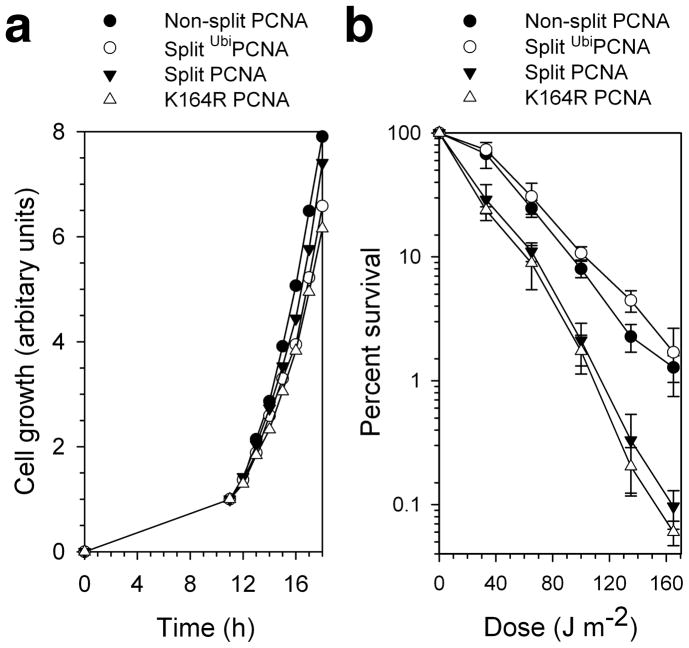
We next determined whether the split PCNA and split UbiPCNA proteins would support cell viability and normal cell growth. We generated four pol30Δ yeast strains (POL30 encodes PCNA) harboring plasmids encoding different versions of PCNA. One strain produced the wild-type PCNA protein; another produced the mutant K164R PCNA protein, which served as a negative control because it cannot be monoubiquitinated by the Rad6-Rad18 complex. The other two strains produced the split PCNA and split UbiPCNA proteins. The POL30 gene is essential, and all four PCNA variants supported cell viability. Moreover, all four strains grew at the same rate (Fig. 2a). Consequently, no serious defects in normal DNA replication occurred in the presence of split PCNA or UbiPCNA.
Figure 2.
Viability and UV sensitivity of yeast cells expressing split PCNA and UbiPCNA. (a) The growth of cells producing only non-split PCNA, the K164R mutant PCNA protein, split PCNA, or UbiPCNA is graphed as a function of time. (b) UV sensitivity of cells producing only non-split PCNA, the K164R mutant PCNA protein, split PCNA, or UbiPCNA is shown by graphing the percent of surviving cells as a function of the UV dose. Error bars represent standard deviation.
To determine whether the split PCNA and split UbiPCNA proteins functioned in translesion synthesis in vivo, we examined the UV sensitivity of these four yeast strains (Fig. 2b). The strain producing the wild-type PCNA protein was substantially more resistant to UV radiation than was the strain producing the K164R mutant PCNA protein. This was because the K164R mutant PCNA protein cannot be monoubiquitinated, and this eliminates translesion synthesis. The strain producing the split PCNA protein was as sensitive to UV radiation as the strain producing the K164R mutant protein. This suggests that splitting PCNA between residues 163 and 164 prevented the monoubiquitination of PCNA by the Rad6-Rad18 complex. Interestingly, the strain producing the split UbiPCNA protein was at least as resistant to UV radiation as the strain producing the non-split PCNA protein. These results clearly demonstrate that split UbiPCNA fully supported translesion synthesis in vivo.
Structure of split PCNA
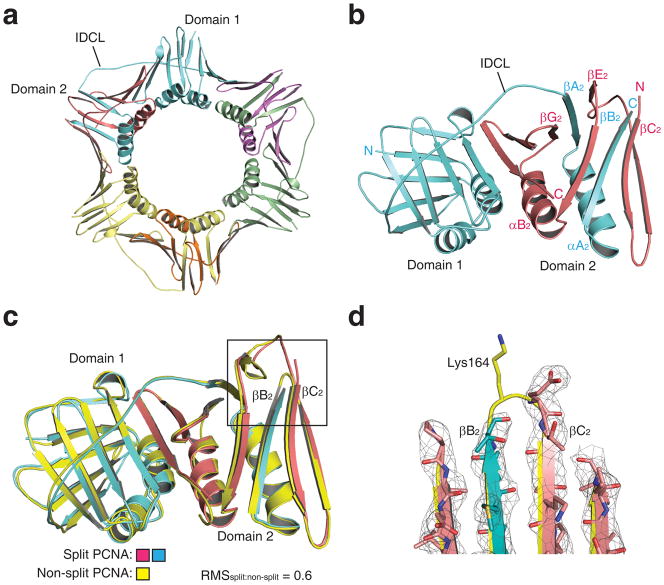
Confident that split PCNA both stimulated the activity of pol η in vitro and supported cell viability in vivo, we proceeded to determine the X-ray crystal structure of split PCNA to a resolution of 3.0 Å (Table 1). There was a single PCNA subunit in the asymmetric unit, so the structure of the biologically relevant trimer (Fig. 3a) was obtained by generating the symmetry related neighboring subunits as was done previously for non-split PCNA 27. The structure of a single monomer of split PCNA with the N fragment colored blue and the C fragment colored red is shown in Figure 3b. Each PCNA monomer had two domains, domain 1 (residues 1–118) and domain 2 (residues 135–258), joined by long, flexible linker called the inter-domain connector loop (IDCL, residues 119–134). This structure shows that these two polypeptides self-assembled with the N fragment and the C fragment interdigitating in domain 2. The N fragment contained all of domain 1 and portions of domain 2, specifically β strands βA2 (residues 135–140) and βB2 (residues 157–163) and α helix αA2 (residues 141–153); the C fragment contained the remainder of domain 2. Three of the four α helices from each monomer that line the inside of the central cavity of the ring-shaped trimer were from the N fragment; only helix αB2 (residues 157–163) was from the C fragment. (A diagram of the protein topology is provided as Supplementary Fig. 1.)
Table 1.
Data collection and refinement statistics (molecular replacement)
| Split PCNA | UbiPCNA | |
|---|---|---|
| Data collection | ||
| Space group | P213 | P213 |
| Cell dimensions | ||
| a, b, c (Å) | a=b=c=123.00 | a=b=c=122.52 |
| α, β, γ(°) | α=β=γ=90 | α=β=γ=90 |
| Resolution (Å) | 43.5–3.0 (3.1–3.0) | 43.3–2.8 (2.9–2.9) |
| Rsym or Rmerge | 10.2 (42.8) | 10.9 (60.4) |
| I/σI | 12.9 (3.8) | 10.5 (2.4) |
| Completeness (%) | 100 (3.1–3.0: 99.8) | 100 (2.9–2.8: 99.4) |
| Redundancy | 9.50 (6.99) | 8.74 (5.38) |
| Refinement | ||
| Resolution (Å) | 43–3.0 | 43–2.8 |
| No. reflections | 12666 | 15333 |
| Rwork/Rfree | 24.3/26.7 | 27.7/31.4 |
| No. atoms | ||
| Protein | 1994 | 3816 |
| Ligand/ion | NA | NA |
| Water | 0 | 0 |
| B-factors | ||
| Protein | 80.7 | 75.8 |
| Ligand/ion | NA | NA |
| Water | NA | NA |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.017 |
| Bond angles (°) | 1.164 | 1.587 |
Values in parentheses are for highest-resolution shell.
Figure 3.
Structure of split PCNA. (a) Structure of the split PCNA trimer is shown with the three N fragments colored blue, green, and yellow and the three C fragments colored red, purple, and orange. Domain 1, domain 2, and the interdomain connector loop (IDCL) are indicated. (b) Structure of a single split PCNA monomer is shown (viewed from the opposite side of the ring relative to panel a) with the N fragment colored blue and the C fragment colored red. The interdigitating β strands of the two fragments in domain 2 are labeled. (c) The backbone of split PCNA, which is colored blue (N fragment) and red (C fragment), is superimposed on the backbone of non-split PCNA, which is colored yellow. (d) Close up of the loop between β strands βB2 and βC2 showing the position of Lys-164 in non-split PCNA and the break in the backbone between the N fragment and C fragment of split PCNA. The electron density (level=2.0) is shown for split PCNA.
To ensure that splitting PCNA did not result in appreciable changes to its structure, we overlaid the backbone of split PCNA and non-split PCNA (Fig. 3c). The root-mean-square deviation between these two structures was 0.6 Å over the 254 Cα atoms showing that the break in the protein backbone between residues 163 and 164 did not substantially affect the structure of the PCNA monomer. In fact, the break in the protein backbone did not alter the structures of the β strands immediately adjacent to the break (βB2 and βC2), except at the position of Lys-164. This residue was disordered in split PCNA, but was not disordered in non-split PCNA (Fig. 3d). This likely explains why split PCNA did not support translesion synthesis in vivo.
Structure of UbiPCNA
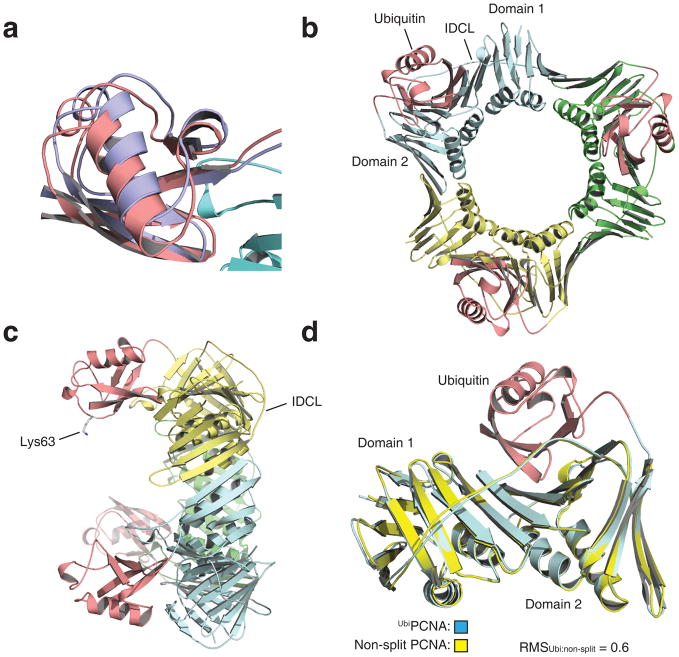
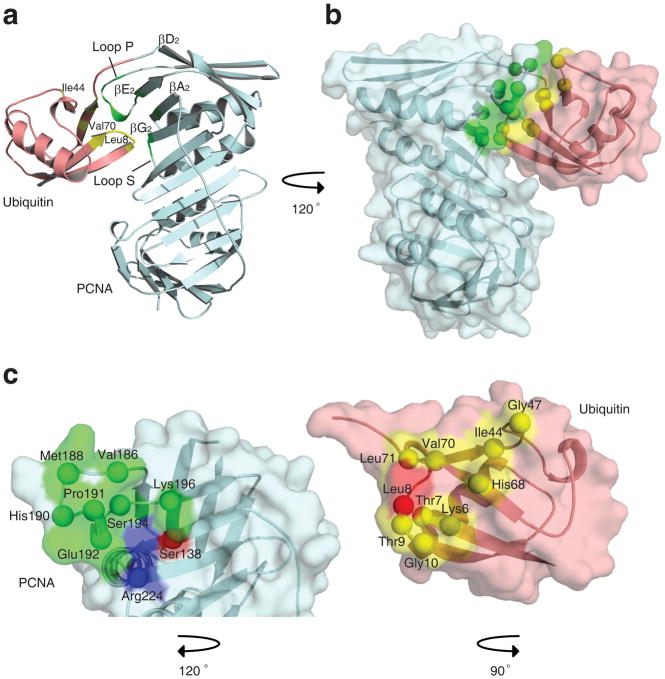
Confident that split UbiPCNA both stimulated the activity of pol η in vitro and supported cell viability and translesion synthesis in vivo, we then determined the X-ray crystal structure of UbiPCNA to a resolution of 2.8 Å (Table 1). While there was a single PCNA subunit and a single ubiquitin moiety in each asymmetric unit, the ubiquitin moiety occupied two distinct, yet very similar positions within the asymmetric unit. This means that the ubiquitin was capable of moving around somewhat in the protein crystal, but preferred to be in one of these two positions. Ubiquitin moieties in these preferred positions were both oriented the same way and were separated by only 2.5 Å (Fig. 4a). Thus we can safely conclude that the ubiquitin was located on the back face of the PCNA ring on the opposite side from the IDCL (Fig. 4b and Fig. 4c). (Stereo images of the electron density of the ubiquitin are provided as Supplementary Fig. 2.)
Figure 4.
Structure of UbiPCNA. (a) Overlay of the two preferred positions of the ubiquitin moiety. Position 1 is shown in red and position 2 is shown in blue. The corresponding atoms in these positions are separated by 2.5 Å. (b) Structure of the UbiPCNA trimer is shown with the three PCNA subunits shown in blue, green, and yellow and the three ubiquitin moieties (in position 1) shown in red. (c) Side view of the UbiPCNA trimer with the side chain of Lys-63 of the ubiquitin (the site of polyubiquitination) indicated. (d) The backbone of UbiPCNA, which is colored blue (PCNA portion) and red (ubiquitin moiety), is superimposed on the backbone of non-split PCNA, which is colored yellow.
To determine whether the monoubiquitination of PCNA altered the conformation of the PCNA portion of the molecule, we overlaid the backbones of UbiPCNA and non-split PCNA (Fig. 4d). The root-mean-square deviation between these two structures was 0.6 Å over the 254 Cα atoms of the PCNA. In addition, we did not detect any local differences between the structures of UbiPCNA and non-split PCNA. This shows that the attachment of ubiquitin did not alter the conformation of PCNA in any notable way.
The surface of the ubiquitin moiety that interacts with PCNA was the canonical hydrophobic surface centered on Leu-8, Ile-44, and Val-70 that interacts with a variety of other proteins 28,29. The regions of PCNA that interact with ubiquitin were all in domain 2 and included residues on β strand βA2 (residues 135–140), loop P (residues 184–196), β strand βE2 (residues 196–199), loop S (residues 222–223), and β strands βG2 (residues 224–229) (Fig. 5). In addition to hydrophobic contacts, there were several electrostatic and hydrogen bonding interactions. For example, the backbone carbonyl oxygen of Leu-8 of ubiquitin interacted with a nitrogen atom on the side chain of Arg-224 of PCNA. (Diagrams of the hydrophobic, electrostatic, and hydrogen bonding interactions are provided as Supplementary Fig. 3.)
Figure 5.
Interactions between ubiquitin and PCNA within UbiPCNA. (a) Ribbon representation showing the ubiquitin-PCNA interface. The ubiquitin moiety is shown in red, and the PCNA is shown in blue. Regions of the ubiquitin moiety contacting the PCNA are shown in yellow, and regions of the PCNA contacting the ubiquitin moiety are shown in green. (b) Space filled representation of the ubiquitin-PCNA interface shown from a different angle. (c) Close up of the interfaces on the ubiquitin moiety and PCNA. The ubiquitin moiety and the PCNA have been separated and rotated relative to the orientation in panel B to show the binding surfaces on each. Residues forming hydrophobic contacts are shown in green and yellow for the PCNA and ubiquitin moiety, respectively. Residues forming electrostatic contacts are shown in blue and red.
DISCUSSION
Arguably the least understood step of translesion synthesis is the polymerase exchange step between the classical and the non-classical polymerase. Insight into the structural and mechanistic basis of the polymerase exchange has come from studies of prokaryotic systems. An X-ray crystal structure of the polymerase-associated domain (PAD) of non-classical DNA polymerase IV (pol IV) from E. coli bound to the β sliding clamp has been determined 30. This structure shows that the C-terminal tail of pol IV binds to the front of the clamp in a hydrophobic pocket while the remainder of the PAD interacts with the side of the clamp at the subunit interface. Further biochemical studies showed that pol IV and the clamp form a tool belt on the DNA with classical DNA polymerase III (pol III) 31. In this tool belt mechanism, pol IV binds to the side of the clamp and rides piggy back while pol III synthesizes DNA in front of the clamp. When replication by pol III is blocked at a lesion in the template, these two polymerases switch places and pol IV begins synthesizing DNA.
One crucial difference between the prokaryotic and eukaryotic systems is that the polymerase exchange in eukaryotes requires the monoubiquitination of PCNA. This was shown with an in vitro reconstituted system comprised of classical pol δ, non-classical pol η, and PCNA or UbiPCNA 23. In this system, pol η could not exchange with pol δ at the replication fork unless synthesis by pol δ was stalled. Moreover, the exchange between pol η and pol δ occurred in the presence of UbiPCNA, but not in the presence of unmodified PCNA. Precisely how UbiPCNA facilitated the polymerase exchange reaction in this system, however, was not clear.
To better understand the polymerase exchange in eukaryotes, we determined the X-ray crystal structure of UbiPCNA. We found two very similar preferred positions for the ubiquitin moiety on the back side of PCNA. These preferred positions were not the result of crystal contacts, but rather formed by specific interactions between ubiquitin and PCNA. Support for this comes from the fact that there is substantially more buried surface area (1900 Å2) between ubiquitin and the PCNA subunit to which it is attached than there is between ubiquitin and other symmetry related molecules (650 Å2). Moreover, the high solvent content of the protein crystal (70%) – combined with the high degree of flexibility of the C terminus of ubiquitin – suggests that the ubiquitin would be free to orient many different ways if there was not a specific interaction between ubiquitin and PCNA. The fact that ubiquitin has a preferred orientation shows that the interaction between ubiquitin and PCNA is specific.
Despite the fact that the interaction between ubiquitin and PCNA is specific, it appears that this interaction is rather weak. Support for this comes from the fact that there are two principal positions for the ubiquitin. We suggest that the weakness of this interaction affords ubiquitin the flexibility to re-orient itself so that it can bind other interacting partners via the same canonical hydrophobic surface with which it binds PCNA. For example, NMR titrations have shown that the UBZ motif of pol η interacts with this same surface on ubiquitin 32. Thus, for UbiPCNA to bind to the UBZ of pol η, the ubiquitin moiety must undergo a rotation of approximately 60° in order to expose its binding site for the UBZ motif.
There are four general models by which the monoubiquitination of PCNA alone could facilitate the polymerase exchange reaction. These four general models are not intended to be mutually exclusive, and any combination of them is possible in principle. Model 1: ubiquitination directly reduces the binding affinity for the classical polymerase to PCNA and promotes its dissociation via interactions between the ubiquitin and the classical polymerase. Model 2: ubiquitination indirectly reduces the binding affinity for the classical polymerase via allosteric effects on PCNA. Model 3: ubiquitination directly enhances the affinity for the non-classical polymerase and promotes its recruitment via interactions between the ubiquitin and the non-classical polymerase. Model 4: ubiquitination indirectly enhances the affinity for the non-classical polymerase via allosteric effects.
The structure of UbiPCNA provides compelling reasons to reject three of these four models of the polymerase exchange reaction. First, the attachment of ubiquitin to PCNA does not alter the conformation of the PCNA. There are no detectible changes to the structure of the hydrophobic pocket on the front face of PCNA near the IDCL to which the conserved PCNA-interacting protein (PIP) motifs of various proteins, including classical and non-classical polymerases, bind. This implies that monoubiquitination of PCNA does not induce allosteric effects resulting in either a reduction of the affinity of the classical polymerase or an enhancement of the affinity of the non-classical polymerase for PCNA as had been suggested previously 33. This argues against model 2 and model 4. Second, the ubiquitin is bound on the back face of the PCNA ring, presumably far away from where the classical polymerase sits in front of the PCNA ring. This strongly suggests that the ubiquitin does not promote classical polymerase dissociation by directly interacting with the classical polymerase. This argues against model 1. Consequently, the structure of UbiPCNA supports only model 3 – namely that UbiPCNA directly facilitates non-classical polymerase recruitment to the back face of the PCNA ring by forming a new interacting surface for the non-classical polymerase.
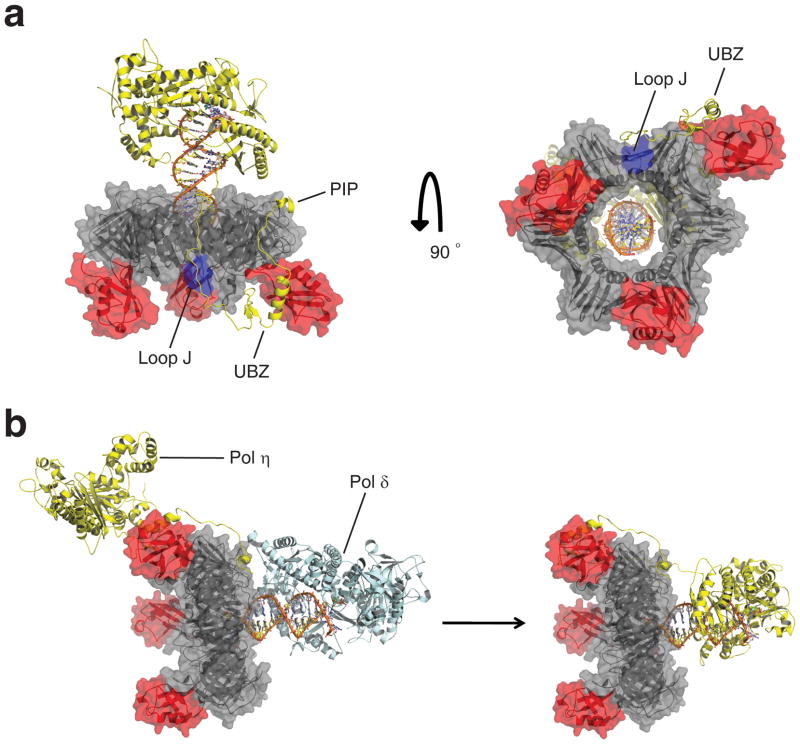
The structure of UbiPCNA reported here represents the form of the protein to which pol η is recruited. Although we do not know exactly what the complex of UbiPCNA bound to pol η and DNA looks like, we are now in an excellent position to model this complex (Fig. 6a). This model is based on the X-ray crystal structure of the catalytic core of pol η 34,35, the X-ray crystal structure of the PIP motif of pol η bound to PCNA 36 and the NMR structure of the UBZ motif of pol η 32. In this structural model, the PIP motif of pol η at its extreme C terminus (residues 617–632) binds in the hydrophobic pocket on the front face of the UbiPCNA ring near the IDCL. The pol η protein chain then makes its way to the back face of the UbiPCNA ring where the UBZ motif (residues 566–577) interacts with the ubiquitin, which has been rotated 60° in order to expose its binding site for the UBZ motif. From there, the protein chain makes its way back to the front side of the PCNA ring where the catalytic core of pol η (residues 1–513) binds to the DNA primer terminus. It should be noted that the pol η protein chain, somewhere between the catalytic core and the UBZ motif, likely passes nearby and interacts with loop J of PCNA (residues 105–110), which has previously been shown by structural and biochemical studies to be important for pol η function 37.
Figure 6.
Model of the complex between UbiPCNA and pol η. (a) Two views of the model of the UbiPCNA-pol η complex. The PCNA portion is colored grey, loop J of PCNA is colored blue, the ubiquitin moieties are colored red, and the pol η is colored yellow. The PCNA-interacting peptide (PIP) and ubiquitin-binding, zinc-binding (UBZ) motifs of pol η and loop J of PCNA are indicated. (b) A possible tool belt model showing the recruitment of pol η to the side and back face of UbiPCNA while pol δ (colored blue) sits in front of UbiPCNA. Eventually, pol δ is displaced from the front of UbiPCNA by the catalytic core of pol η. For simplicity sake, pol δ is shown as dissociating from the complex, although this need not be the case. The structure of pol δ is from 38.
According to this structural model, with the exception of the PIP motif, the entire C-terminal region of pol η interacts exclusively with the side and back face of UbiPCNA. This is consistent with, although does not by itself imply, a tool belt model of translesion synthesis. In essence, PCNA ubiquitination could set up the tool belt by recruiting pol η to the side and back of UbiPCNA via the C-terminal region of pol η while pol δ synthesizes DNA in front of the UbiPCNA ring (Fig. 6b). The catalytic core of pol η could ride piggy back on the UbiPCNA ring because the catalytic core is connected to the C-terminal region by a long, flexible linker. When pol δ encounters a template lesion and stalls, it could be displaced by the catalytic core of pol η, which would then begin synthesizing DNA in front of the UbiPCNA ring. Whether pol δ would dissociate at this point or remain bound to UbiPCNA and ride piggy back while pol η synthesizes DNA – the latter option being analogous to the prokaryotic system – is unclear. While there is compelling evidence that prokaryotes utilize a tool belt mechanism to carry out translesion synthesis 31, it remains to be seen whether eukaryotes utilize such a mechanism.
METHODS
Protein Expression and Purification
Non-split yeast PCNA with an N-terminal FLAG™ tag was over-expressed in E. coli Rosetta-2 (DE3) cells from a pET11a plasmid as described previously 37. To produce split PCNA and UbiPCNA, the gene encoding the N-terminally Flag™ tagged N fragment was cloned into multi-cloning site 1 of the pET-Duet1 plasmid. The gene encoding either the C fragment or the N-terminally His6-tagged UbiC fragment was cloned into multi-cloning site 2 of the same plasmid. The two fragments of split PCNA or of UbiPCNA were simultaneously over-expressed in E. coli Rosetta-2 (DE3) cells. Cells were lyzed and the cell lysate was subjected to ultracentrifugation as described previously 37. Non-split PCNA and split PCNA were purified using an Anti-Flag™ M2 affinity chromatography column (Sigma) and a Superose 6 size exclusion chromatography column (Pharmacia GE Healthcare). UbiPCNA was purified the same way, except that an NTA-agarose affinity chromatography column (Qiagen) was used before the anti-Flag™ affinity column.
Polymerase Activity Assays
All polymerase activity assays were carried out as described previously 37. In the steady state kinetic assays, the reaction mixtures contained various concentrations of dATP (0 to 600 μM). Reactions were initiated by adding 1 nM pol η and were quenched after 10 min. In the running start bypass assays, the reaction mixtures contained 20 μM of each dNTP, and reactions were quenched after 3 and 5 min.
Genetic Complementation Assays
Because the POL30 gene (which encodes PCNA) is essential, we carried out a plasmid shuffle. The wild-type PCNA gene under control of its native promoter was subcloned into pTB366 (URA3) and transformed into wild-type EMY74.7 yeast cells. The genomic POL30 gene was then replaced by the TRP1 gene through homologous recombination. Verification was carried out with PCR. Genes for wild-type, non-split PCNA, the K164R mutant PCNA protein, and the N fragment of split PCNA and split UbiPCNA were sub-cloned into the p425 GPD vector (LEU2). The C fragments of split PCNA or the UbiC fragment of split UbiPCNA were sub-cloned into the p423 GPD vector (HIS3). Combinations of these plasmids or empty p423 GPD vector were transformed into the pol30Δ cells to generate the strains producing only wild-type PCNA, only the mutant K164R PCNA, only split PCNA, and only split UbiPCNA. Following counter-selection with FOA, the absence of full length PCNA genes in strains producing split PCNA and split UbiPCNA were confirmed by both PCR and DNA sequencing. These strains were assayed for UV resistance as previously described 39. Growth rates for these strains were examined by inoculating 100 ml liquid media with 1 × 105 cells from overnight cultures. The growth rate at 30 °C was monitored by measuring absorbance at 600 nm.
Crystallization of Split PCNA and UbiPCNA
Crystallization of the split PCNA protein was performed manually using the hanging drop method with 2 μl drops. The best diffracting crystals were obtained within 3 days at 18 °C by combining an equal volume of protein (20 mg ml−1) with reservoir solution containing 1.9 M ammonium sulfate and 0.1 M sodium citrate (pH 6.0). Crystallization of the UbiPCNA protein was set up using a TTP LabTech Mosquito by the hanging drop method with 400 μl drops. The best diffracting crystals were obtained within 60 days at 18 °C by combining an equal volume of protein (18 mg ml−1) with a reservoir solution containing 2.04 M ammonium sulfate, 0.1 M sodium citrate (pH 6.2), and 3% (v/v) ethanol.
Data Collection and Structural Determination
Both split PCNA and UbiPCNA protein crystals were pre-soaked in a mother liquor containing 10% (v/v) glycerol prior to being flash cooled in liquid nitrogen. Mounted crystals were subsequently used for data collection at 100 K at the 4.2.2 synchrotron beamline at the Advanced Light Source in the Lawrence Berkeley National Laboratory. The data were collected with a crystal to detector distance of 150 mm. The data were processed and scaled using d*trek 40 to a resolution of 2.9 Å, and the space groups were determined to be P213 for both proteins. Molecular replacement was performed using the structure of non-split PCNA (PDB ID: 1PLQ) 27 and PHASER 41 to produce the initial model. For split PCNA, simulated annealing was performed to remove any structural bias using PHENIX 42 prior to refinement with REFMAC5 from the CCP4 package 43. Model building was carried out using Coot 44.
For UbiPCNA, we initially obtained clear electron density for only the PCNA portion of UbiPCNA. A difference map between the split PCNA and UbiPCNA revealed clear extra density suggesting the position of the ubiquitin moiety, but the density was not good enough to orient the ubiquitin. To improve the maps, we first refined only the PCNA portion of the complex using REFMAC5 43 followed by maximum entropy refinement as implemented in Buster 45. ESSENS 46 and SOLEX were used to determine the orientation and position of the ubiquitin in this improved electron density map in a non-biased manner following the approach used previously to determine the structure of the acetylcholinesterase-fasciculin complex 46. The top two orientations of the ubiquitin (with scores of 3.7 and 2.8, which represent the number of standard deviation above the mean) were similarly oriented and structurally possible. These two orientations were assigned equal occupancy and subjected to a final round of refinement using REFMACS5. It should be noted that there is precedent for alternative domain conformations, as a 120-amino acid domain of the ISP protein in the structure of the eleven-subunit mitochondrial cytochrome bc1 complex showed a mixture of three different conformations 47.
Ramachandran analyses showed that in the split PCNA structure, 90.9% of the residues were in their most favored conformations and 8.1% of the residues were in allowed conformations. In the split UbiPCNA structure, 87.5% of the residues were in their most favored conformations and 12.5 % of the residues were in allowed conformations. In neither structure were any residues in disallowed conformations.
Supplementary Material
Acknowledgments
The project described was supported by Award Number R01GM081433 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
We thank Edward Cho and Stephanie Perkins for technical assistance. We thank Christine Kondratick, Lynne Dieckman, John Pryor, and Marc Wold for valuable discussions.
Footnotes
AUTHOR CONTRIBUTIONS
B.D.F. performed the biochemical and genetics experiments. B.D.F. and L.G. collected the X-ray diffraction data. B.D.F., L.G., and S.R. analyzed the X-ray diffraction data. M.T.W. supervised the study. B.D.F. and M.T.W. wrote the manuscript. All authors discussed the results and approved the manuscript.
ACCESSION CODES
Protein Data Bank coordinates for split PCNA, split UbiPCNA with ubiquitin in position 1, and split UbiPCNA with ubiquitin in position 2 have been deposited with accession codes 3L0X, 3L10, and 3L0W, respectively.
References
- 1.Prakash S, Sung P, Prakash L. DNA-Repair Genes and Proteins of Saccharomyces-Cerevisiae. Annual Review of Genetics. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 2.Jentsch S, McGrath JP, Varshavsky A. The Yeast DNA-Repair Gene Rad6 Encodes a Ubiquitin-Conjugating Enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 3.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific Complex-Formation between Yeast Rad6 and Rad18 Proteins - a Potential Mechanism for Targeting Rad6 Ubiquitin-Conjugating Activity to DNA-Damage Sites. Genes & Development. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 4.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. Journal of Biological Chemistry. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 5.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked tomodification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 6.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 7.Broomfield S, Chow BL, Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 9.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. Embo Journal. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes & Development. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 11.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annual Review of Biochemistry. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 12.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 13.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 14.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair. 2002;1 doi: 10.1016/s1568-7864(02)00038-1. PII S1568–7864(02)00038–1. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. DNA Repair and Replication. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 16.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol eta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 17.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase eta. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 19.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 20.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 21.Guo CX, et al. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Molecular and Cellular Biology. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Molecular Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhuang ZH, et al. Regulation of polymerase exchange between Pol eta and Pol delta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Molecular Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 25.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishna TSR, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 28.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochemical Journal. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nature Reviews Molecular Cell Biology. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 30.Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. Embo Journal. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Indiani C, McInerney P, Georgescu R, Goodman MF, O’Donnell M. A sliding-clamp toolbelt binds high-and low-fidelity DNA polymerases simultaneously. Molecular Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Bomar MG, Pai MT, Tzeng SR, Li SSC, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase eta. Embo Reports. 2007;8:247–251. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acharya N, et al. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trincao J, et al. Structure of the catalytic core of S-cerevisiae DNA polymerase eta: Implications for translesion DNA synthesis. Molecular Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 35.Alt A, et al. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 36.Hishiki A, et al. Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen. Journal of Biological Chemistry. 2009;284:10552–10560. doi: 10.1074/jbc.M809745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freudenthal BD, Ramaswamy S, Hingorani MM, Washington MT. Structure of a Mutant Form of Proliferating Cell Nuclear Antigen That Blocks Translesion DNA Synthesis. Biochemistry. 2008;47:13354–13361. doi: 10.1021/bi8017762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nature Structural & Molecular Biology. 2009;16:979–U107. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howell CA, Kondratick CM, Washington MT. Substitution of a residue contacting the triphosphate moiety of the incoming nucleotide increases the fidelity of yeast DNA polymerase zeta. Nucleic Acids Research. 2008;36:1731–1740. doi: 10.1093/nar/gkn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallographica Section D-Biological Crystallography. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 41.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallographica Section D-Biological Crystallography. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 43.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 46.Harel M, Kleywegt GJ, Ravelli RBG, Silman I, Sussman JL. Crystal structure of an acetylcholinesterase-fasciculin complex: Interaction of a three-fingered toxin from snake venom with its target. Structure. 1995;3:1355–1366. doi: 10.1016/s0969-2126(01)00273-8. [DOI] [PubMed] [Google Scholar]
- 47.Iwata S, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc(1) complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.