Abstract
Cocaine is considered to be the most addictive of all substances of abuse and mediates its effects by inhibiting monoamine transporters, primarily the dopamine transporters. There are currently no small molecules that can be used to combat its toxic and addictive properties, in part because of the difficulty of developing compounds that inhibit cocaine binding without having intrinsic effects on dopamine transport. Most of the effective cocaine inhibitors also display addictive properties. We have recently reported the use of cocaine esterase (CocE) to accelerate the removal of systemic cocaine and to prevent cocaine-induced lethality. However, wild-type CocE is relatively unstable at physiological temperatures (τ1/2 ∼13 min at 37°C), presenting challenges for its development as a viable therapeutic agent. We applied computational approaches to predict mutations to stabilize CocE and showed that several of these have increased stability both in vitro and in vivo, with the most efficacious mutant (T172R/G173Q) extending half-life up to 370 min. Here we present novel X-ray crystallographic data on these mutants that provide a plausible model for the observed enhanced stability. We also more extensively characterize the previously reported variants and report on a new stabilizing mutant, L169K. The improved stability of these engineered CocE enzymes will have a profound influence on the use of this protein to combat cocaine-induced toxicity and addiction in humans.
Keywords: cocaine esterase, computational, drug abuse, thermostable
Introduction
Cocaine is the most addictive of all abused substances. Estimates for 2006 suggest that over half of all illicit drug-based emergency department visits involved cocaine, with 548 608 occurrences, and accounts for 181 visits per 100 000 people in the USA (SAMHSA, 2006). The devastating medical and social cost of cocaine addiction and overdose make discovery of pharmacological agents to block the addictive effects of cocaine an important goal. The mechanism by which cocaine exerts its effect is through binding monoamine transporters and blocking the reuptake of dopamine, norepinephrine and serotonin in the synaptic junctions and potentiating the effects of neurotransmitters in the synapse. Chronic and prolonged blockade of dopamine transporters can lead to reinforcement of self-administration, and thereby to various forms of addiction (Benowitz, 1993). At higher concentrations, cocaine also blocks norepinephrine and serotonin reuptake transporters, which contributes to its toxic effects, including seizures, tachyarrhythmias and sudden death (Johanson and Fischman, 1989; Crumb and Clarkson, 1990; Benowitz, 1993; Uhl et al., 2002).
Unfortunately, developing a classical antagonist to block the actions of a pleiotropic inhibitor such as cocaine has proven difficult (Gorelick, 1997), as classical small molecules that compete with cocaine binding, have neurological effects themselves and could potentially be abused (Gorelick et al., 2004). To circumvent this problem, agents that intercept cocaine in the bloodstream have been developed to alter its distribution or accelerate its clearance (Gorelick, 1997; Baird et al., 2000; Carrera et al., 2001, 2004; Deng et al., 2002; Kantak, 2003; Meijler et al., 2005; Rogers et al., 2005). In humans, cocaine is primarily metabolized to inactive products benzoic acid and ecgonine methyl ester by serum butyryl cholinesterase (BchE), leading to an in vivo half-life (τ1/2) of ∼116 min. The τ1/2 can be shortened to 10 min following intravenous administration of exogenous BchE, suggesting that cocaine-degrading agents may represent a potential approach to treat cocaine abuse (Lynch et al., 1997; Browne et al., 1998). The administration of BchE or engineered mutants with enhanced catalytic activity reduces cardiovascular and psychomotor stimulant effects of cocaine in rats and mice (Carmona et al., 1996; Lynch et al., 1997; Mattes et al., 1997; Gao et al., 2005) and can partially protect against the lethal doses of cocaine administration (Lynch et al., 1997).
Catalytic antibodies against cocaine (mAb antibody 15A10) at high doses (15–50 mg/kg) (Mets et al., 1998; Deng et al., 2002), and more recently, cocaine esterase from the Rhodococcus sp. strain MB1 (CocE) (Bresler et al., 2000) at lower doses (1–10 mg/kg), have proven profoundly effective against cocaine-induced lethality in vivo (Cooper et al., 2006; Ko et al., 2007). CocE hydrolyzes the benzoyl ester of cocaine (Turner et al., 2002) with a catalytic efficiency ∼800-fold greater than that of BChE. Although intravenous injection of purified CocE prior to cocaine administration protects against cocaine-induced lethality, its duration of effectiveness is very short. CocE administration in rats or mice as little as 10 min prior to lethal doses of cocaine leads to only 67% or 50% survival, respectively (Cooper et al., 2006; Ko et al., 2007). Similarly, in vitro incubation of CocE in rat plasma or in buffer at 37°C reveals a dramatic time-dependent inactivation with τ1/2 ∼ 13 min (Cooper et al., 2006) Clearly, a more stable form of CocE is necessary to exploit the superior catalytic efficiency of the enzyme for the treatment of cocaine toxicity and addiction.
In our previous study, we described structure-based and computational approaches to predict mutants of CocE that have increased stability (Gao et al., 2009). We employed a combination of molecular modeling, energy minimization and molecular dynamics (MD) using the Rosetta Design (Kuhlman and Baker, 2000; Korkegian et al., 2005) and AMBER programs (Vagin et al., 2004) and provided preliminary studies of two mutants, T172R and G173Q, that display improved thermostability both in vitro and in vivo. Here we report the elucidation of the crystal structures of these mutants and analyze the mechanism by which they are stabilized. We also report more detailed analysis of these two mutants, and report our findings on a third stabilizing mutant, L169K, including its structure. Importantly, we reveal the likely molecular basis for how these three substitutions, out of a total of 34 substitutions originally predicted, dramatically improve the τ1/2 of the enzyme both in vitro and in vivo.
Materials and methods
Materials
Cocaine was purchased from Mallinckroft Inc. All other reagents are of analytical grade and were obtained from Fisher Scientific or Sigma-Aldridge Corp.
Site-directed mutagenesis
pET22b(+)CocE plasmid was kindly provided by Dr Nick Larsen (The Scripps Institute, CA, USA) (Turner et al., 2002). Wild-type (wt-CocE) and CocE mutants were expressed as C-terminal hexahistidine-tagged proteins containing the exogenous sequence KLAAALEHHHHHH at the C-terminus. Point mutations were generated using a modified QuikChange™ (Stratagene) mutagenesis protocol. To generate double mutants, cDNAs with single-point mutations were used as templates for a second round of mutagenesis. Oligonucleotide primer sequences are available on request. All mutants were confirmed by sequencing of both strands over the entire coding region. CocE preparations were expressed in E.coli BL-21 Gold (DE3) cells grown at 37°C to an OD600 of 0.8. Protein expression was induced with 1 mM isopropyl-β-thiogalactopyranoside and cells were harvested after incubation for 12 h at 18°C.
Purification of cocaine esterase and mutants
Cells were pelleted, resuspended in 50 mM Tris–HCl, pH 8.0, 150 mM NaCl supplemented with protease inhibitors (3 µg/ml each of leupeptin and lima bean or soybean trypsin inhibitor) and lysed using a French press. Wild-type or mutant CocE was enriched using Talon™ metal chelate affinity chromatography (Clontech Laboratories, Inc.), followed by anion-exchange chromatography on a Q-Sepharose fast performance liquid chromatography column (GE Healthcare). CocE was eluted from the Q-Sepharose column with a 150–450 mM NaCl linear gradient in buffers containing 20 mM HEPES pH 8.0, 2 mM MgCl2, 1 mM EDTA and 1 mM DTT (unless mentioned otherwise). The peak fractions were pooled and concentrated typically to 5 mg/ml using Centricon-30 concentrators (Millipore), and then snap frozen in liquid nitrogen and stored at −80°C.
Michaelis–Menten kinetics of cocaine hydrolysis
A spectrophotometric real-time assay was used to monitor cocaine hydrolysis (Xie et al., 1999). The initial rates of decay were determined by following the change in intrinsic absorbance of cocaine at 240 nm (6700 M−1cm−1) on a SpectraMax Plus 384 UV plate reader (Molecular Devices) using SOFTmax Pro software (Version 3.1.2). The reaction was initiated by the addition of 100 μl of wt-CocE or mutant CocE (50, 25, 10 or 5 ng/ml) made in phosphate-buffered saline (PBS) pH 7.4 to 100 μl of a 2X cocaine solution made in PBS. The final enzyme concentration was 25, 12.5, 5 or 2.5 ng/ml and the final cocaine concentrations were: 125, 62.5, 31.25, 15.63, 7.81, 3.91, 1.95, and 0.98 μM. Vmax and Km values were calculated using Prism (GraphPad Software). For stability measurements, wt-CocE or mutant CocE was diluted to 50 ng/ml (2×) concentration and incubated at 37°C. Aliquots were removed at varying time points and assayed for activity against cocaine as described above (n of 5). Temperature-dependent decay in esterase activity was measured by pre-incubating wt-CocE and mutant forms of CocE (50 ng/ml) at various temperatures (0, 25, 35, 37, 42, 45, 48, 50 and 55°C) for 30 min and then the activity remaining was measured as described above. These experiments were done in duplicates at two separate times (n = 2).
Melting temperatures (tm) measurements using thermofluor
Purified wt-CocE and mutants T172R, G173Q, T172R-G173Q and L169K were buffer exchanged into 50 mM Tris, pH 8.0. Melting temperatures were determined through monitoring 1-anilinonaphthalene-8-sulfonic acid (1, 8-ANS) (Sigma) binding to CocE during protein unfolding (Mezzasalma et al., 2007). The fluorescence of 1, 8-ANS is greatly enhanced if bound to hydrophobic environments such as hydrophobic surfaces that are exposed during the unfolding process. Wild-type CocE or mutants (0.2 mg/ml) were incubated with 1, 8-ANS (100 μM) in a total volume of 10 μl in triplicate using ABgene 384-well PCR (Thermo-Fisher) microtiter plates. Fluorescence at different temperatures (25–85°C) was measured using a ThermoFluor 384 reader (Johnson and Johnson). The fluorescence emission data were analyzed using ThermoFluor Acquire 3.0 software as per manufacturer's guidelines.
In vivo protection against cocaine-induced lethality
Male NIH-Swiss mice (25–32 g) were obtained from Harlan Inc. (Indianapolis, IN, USA) and housed in groups of six mice per cage. All mice were allowed ad libitum access to food and water, and were maintained on a 12 h light–dark cycle with lights on at 06:30 AM in a room kept at a temperature of 21–22°C. Experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the National Institutes of Health. The experimental protocols were approved by the University Committee on the Use and Care of Animals at the University of Michigan.
Cocaine-induced toxicity was characterized by the occurrence of lethality, as defined by the cessation of observed movement and respiration. Mice were placed individually in Plexiglas observation chambers (16 × 28 × 20 cm high) to be habituated for 15 min before drug administration. Following intraperitoneal (i.p.) cocaine administration, the mouse was immediately placed back in the same chamber for observation. The presence or absence of lethality was recorded for 60 min following cocaine administration.
For injections, mice were individually placed in a small restraint chamber (outer tube diameter: 30 mm; inner tube diameter: 24 mm) that left the tail exposed. The tail was cleansed with an alcohol wipe and a 30G1/2 precision glide needle (Fisher Scientific) was inserted into one of the side veins for infusion. The intravenous (i.v.) injection volume of wt-CocE or mutant CocE was 0.2 ml per mouse. Sterile gauze and pressure were applied to the injection site to staunch bleeding.
The potency of CocE mutants to protect against cocaine-induced toxicity was assessed following i.v. enzyme administration (0.3 or 1 mg) 1 min prior to administration of several doses of i.p. cocaine (180, 320, 560 and 1000 mg/kg, n = 6/dose). Dose-response curves of cocaine-induced lethality in the absence or presence of a single dose of the enzyme were determined to demonstrate the in vivo protective effects of CocE mutants.
The duration of protection against cocaine toxicity was assessed by monitoring lethality following i.v. enzyme administration (0.1, 0.3 and 1 mg) prior to i.p. cocaine (LD100, 180 mg/kg). Cocaine hydrochloride was dissolved in sterile water and administered i.p. at a volume of 0.01 ml/g. Wild-type CocE or CocE mutants were diluted to different concentrations in phosphate-buffered saline and administered i.v. at a volume of 0.2 ml per mouse. Lethality was monitored following injection at 1, 5, 10, 30, 60, 120, 180, 240 or 300 min after enzyme administration. Each treatment used six mice to assess the percent of lethality in mice pretreated with a single dose of wt-CocE or mutant CocE at a single time point.
To express the potency value of each enzyme preparation, a group LD50 value was calculated by least-squares regression using the portion of the dose-response curve spanning 50% occurrence of lethality. These values were used to compare the degree of rightward shifts of the dose-response curve in the absence or presence of the enzyme pretreatment. The duration of protection of wt-CocE and CocE mutants against cocaine-induced lethality was expressed as the pretreatment time point where 50% of the animals were protected.
Crystallization and structure determination
Crystals were grown by the hanging drop vapor diffusion method as previously described (Larsen et al., 2002). To harvest crystals, 2 μl of cryoprotectant (5 mM Tris–HCl, pH 7.0, 1.5 M ammonium sulfate, 10 mM HEPES, pH 7.5, 2 mM MgCl2, 1 mM EDTA, 825 mM NaCl and 25% glycerol) were added to each 2 μl hanging drop, and then crystals were transferred to 100% cryoprotectant and flash-frozen on nylon cryo-loops in liquid nitrogen. Crystals were harvested within 3 days of tray set-up.
Diffraction intensities were collected at the Advanced Photon Source at beamlines supported by GM/CA- and LS-CAT, and then reduced and scaled using HKL2000 (Otwinowski and Minor, 1997). Models were refined using REFMAC5 (Vagin et al., 2004) using a previously determined structure of CocE (PDB entry 1JU3) (Larsen et al., 2002), with side chains of mutated residues omitted, as the starting coordinates. Model building and water addition were performed with O (Jones et al., 1991) and COOT (Emsley and Cowtan, 2004) followed by maximum likelihood refinement in REFMAC5. Unambiguous electron density was observed for all mutated side chains. In total, data sets from 23 crystals were collected. At least three data sets from each mutant were analyzed to examine the variability of the H2-H3 loop. Figures were generated with PyMol [http://www.pymol.org]. Coordinates and structure factors are deposited in the PDB under accession codes 3i2i (T172R), 3i2g (G173Q), 3i2f (T172R/G173Q), 3i2h (L169K), 3i2j (wt-CocE without DTT) and 3i2k (wt-CocE with DTT adduct).
Results
Prediction and enzymatic characterization of thermostable mutants
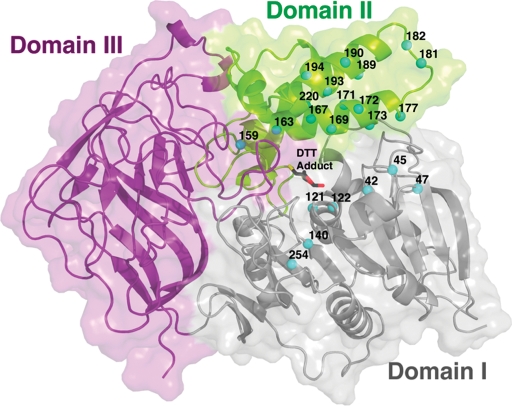
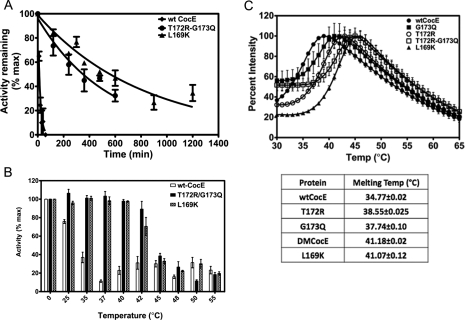
Computational studies (Gao et al., 2009) predicted that the following mutations could stabilize wt-CocE: N42V, D45R, F47K, F47R, W52L, V121D, T122A, Q123E, S159A, L163V, S167A, L169K, G171A, T172R, G173A, G173Q, L174R, S177Q, S179R, R182K, F189A, F189L, F189K, A193D, A194K, W220A and T254R. Positions of some of these mutations on the CocE structure are shown in Fig. 1. Single, double and triple site-directed mutations were incorporated into CocE and assessed for their stability at 37°C. Enzyme integrity was assessed by their capacity to convert cocaine to benzoic acid and egconine methyl ester. Only the T172R, G173Q, T172R/G173Q and the newly characterized L169K mutants significantly increased the stability of CocE over that of wt-CocE. Whereas the activity of the best previously characterized T172R/G173Q decays exponentially with τ1/2 of 370 min, 30-fold longer than wt-CocE (Gao et al., 2009), the L169K mutant improved the enzymatic stability even further (τ1/2=570 min) (Fig. 2A). To determine if these substitutions had a detrimental effect on catalytic efficiency, initial rates of CocE-mediated cocaine hydrolysis were used to determine Michaelis–Menten constants for wt-CocE and the thermostabilizing mutants (Table I). The kinetic constants, Vmax and Km, were both higher than the values reported previously for wt-CocE (Larsen et al., 2002) even though nearly identical methods were used to analyze the enzyme. We believe that the Vmax values were higher because our enzyme preparations had higher specific activity in comparison and the Km values were 10-fold higher because of the limitations of the spectrophotometric assay used. In order to maintain linearity in the initial phase of the cocaine metabolism, we needed to dilute the enzyme concentration further than previously published values to 12.5 ng/ml (Larsen et al., 2002). Even though our Km estimates appear larger than previously published, we deemed them sufficient for comparison purposes. The G173Q mutant did not have any deleterious effects on the catalytic efficiency, T172R and the T172R-G173Q double mutant showed ∼3-fold increase in Km compared to that of wt-CocE, whereas L169K mutant exhibited ∼8-fold increase in Km (Table I), perhaps consistent with the fact that it is positioned closer to the active site pocket (see below and Fig. 1). The triple mutant (T172R/G173Q/L169K) showed poor enzyme kinetics and did not display enhanced stabilization.
Fig. 1.
Structure of CocE. CocE is composed of three distinct structural domains (Domains I–III) colored gray, green and purple, respectively. Domain I contains the catalytic S117 and adopts a canonical α-/β-hydrolase fold. The locations of point mutations predicted by computational methods and tested for in vitro activity are shown as cyan spheres.
Fig. 2.
Enzymatic stability of wt-CocE and mutant CocE. (A) Decay in the capacity to convert cocaine to ecgonine methylester and benzoic acid was measured at 37°C. 50 ng/ml of wt-CocE (black), T172R-G173Q (blue) or L169K CocE (brown) were incubated at 37°C and the activity calculated (Xie et al., 1999) over time. τ1/2 values were measured from the resulting curves. L169K has a τ1/2 = 570 min. (B) Temperature-dependent decay in esterase activity. 50 ng/ml wt-CocE and mutant CocE were pre-incubated for 30 min at temperatures indicated (°C) and activity measured as described previously. The activities of each mutant remaining (as a percentage of the maximal activity, Vmax, without pre-incubation) following pre-incubation are shown. Wild-type-CocE (open bars) appears to inactivate between 30 and 35°C, whereas T172R/G173Q (solid bars) and L169K CocE (hatched bars) both display enhanced thermal stability (inactivation at 40–45°C). (C) Melting temperatures of wild-type and mutants forms of CocE. Protein unfolding was monitored on a ThermoFluor 384 reader using 1-anilinonaphthalene-8-sulfonic acid (1, 8-ANS) (100 μM final) at a protein concentration of 100 μg/ml. Melting curves of wt-CocE (closed circles), T172R (open circles), G173Q (closed squares), T172R-G173Q (open squares) and L169K (closed triangles) are shown. The melting temperatures observed from the melting curves are shown in the table.
Table I.
Kinetic behavior of wt-CocE and mutant CocE
| CocE variant | τ1/2 (min) | Kcat (s−1) | Km (mM) | Kcat/Km (s−1 M−1) |
|---|---|---|---|---|
| G173Q | 75.0 | 36.1 ± 1.11 | 0.0037 ± 0.0017 | 9.88 × 106 |
| L169K | 570 | 80.1 ± 10.8 | 0.044 ± 0.013 | 1.84 × 106 |
| T172R | 78.0 | 50.9 ± 15.9 | 0.017 ± 0.0047 | 3.06 × 106 |
| T172R/G13Q | 370 | 53.4 ± 0.65 | 0.017 ± 0.004 | 3.18 × 106 |
| T172R/G173Q/L169K | NA | No fit | No fit | NA |
| wt-CocE | 12.2 | 51.4 ± 16.8 | 0.0057 ± 0.0019 | 8.99 × 106 |
The metabolism of cocaine by purified preparations of wt-CocE, T172R, G173Q, T172R/G173Q or L169K CocE was measured as described in Materials and Methods section.
NA, not applicable; No fit, no Michaelis–Menton kinetics.
CocE mutants displaying enhanced stability at 37°C also display enhanced thermostability as confirmed by thermal inactivation assay and by monitoring protein unfolding (Fig. 2B and C). Treatment of wt-CocE at varying temperatures prior to measurement of activity exhibited a marked sensitivity to temperature where its activity plummets precipitously following treatment at 30 min at 30–35°C, whereas L169K and T172R/G173Q are inactivated at higher temperatures (40–45°C) (Fig. 2B).
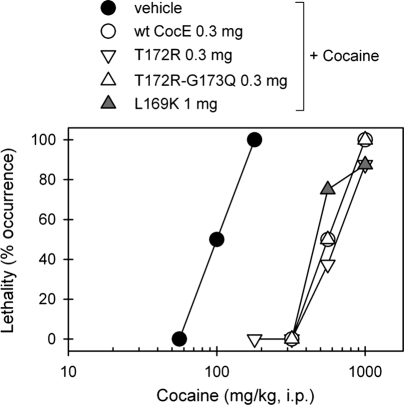
The trend in temperature-dependent inactivation of wt-CocE and mutants was consistent with the temperature-dependent degree of protein unfolding. Melting temperatures were predicted by following ANS binding (fluorescence) using a ThermoFluor reader (Fig. 2C and summary table). The T172R and G173Q each revealed increases in the melting temperature (Tm) by ∼3°C over wt-CocE, whereas in combination the mutations increased the Tm by 6°. L169K also improved the Tm by 6°. These increases were consistent with the earlier reported increase measured by circular dichroism assays (Gao et al., 2009). Thermostable mutants display prolonged capacity to protect against cocaine-induced lethality in mice. Pretreatment with wt-CocE, L169K, T172R or T172R/G173Q CocE (at 0.3 mg, or 9 mg/kg) administered i.v. 1 min prior to cocaine administration protected mice against lethality and shifted the LD50 value of cocaine from 100 mg/kg for the vehicle-treated group to 560 mg/kg (Fig. 3). L169K appeared less effective requiring larger doses (1 mg, or 30 mg/kg) to produce a similar ∼6-fold rightward shift in the cocaine dose-response curve, consistent with its decreased catalytic efficiency observed in vitro (Table I).
Fig. 3.
Protective effects of CocE against cocaine-induced toxicity. Wild-type or mutant CocE (mg) were administered to mice intravenously 1 min before cocaine administration (mg/kg, i.p.). Dose-response curves of cocaine-induced lethality in the absence or presence of wt-CocE or mutants were plotted. Each data point represents the percentage of mice (n = 6 for each dosing condition) exhibiting cocaine-induced lethality.
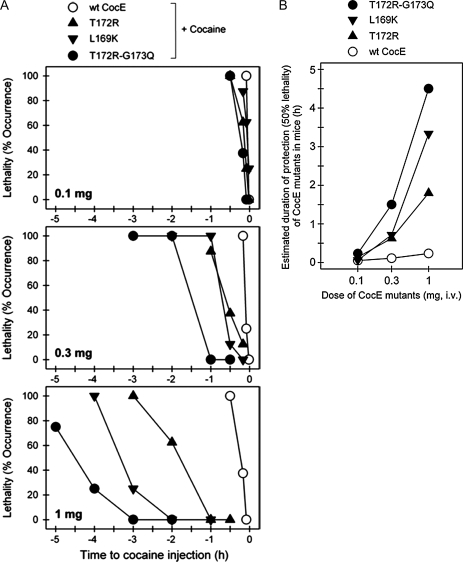
Pretreatment of mice with the thermostable mutants displayed enhanced protection against cocaine-induced lethality in a time-dependent manner that is consistent with enyzme stability measured in vitro at 37°C. Pretreatment (>30 min) with low doses of any CocE mutant (0.1 mg) was ineffective against a lethal dose of cocaine. Pretreatment with larger CocE doses (0.3 and 1 mg) appeared to be effective, the durations of which were dependent on the mutation (Fig. 4A). At the highest dose tested (1 mg), the enzyme pretreatment time necessary to result in 50% lethality (LT50) for wt-CocE, T172R and T172R/G173Q CocE were 14 min, 1.8 h and 4.5 h, respectively (Fig. 4B), again consistent with the in vitro data (Gao et al., 2009). The LT50 for L169K was approximately 3.3 h (Fig. 4B) correlating well with the extended in vitro τ1/2 of L169K.
Fig. 4.
Efficacy of wild-type and mutant CocE in vivo. (A) CocE protects against cocaine-induced lethality in a time-dependent manner. Wild-type (wt-CocE) or mutant CocE (as indicated) (0.1 mg, 0.3 mg, or 1 mg i.v.) was administered at different time points before cocaine administration (180 mg/kg, i.p.). Each data point represents the percentage of mice (n = 6 for each dosing condition) exhibiting cocaine-induced lethality. (B) Estimated duration of protection for 50% lethality. Time required to reach 50% lethality for CocE and each mutant was measured from the previous panel and was plotted against dosage.
Fig. 5.
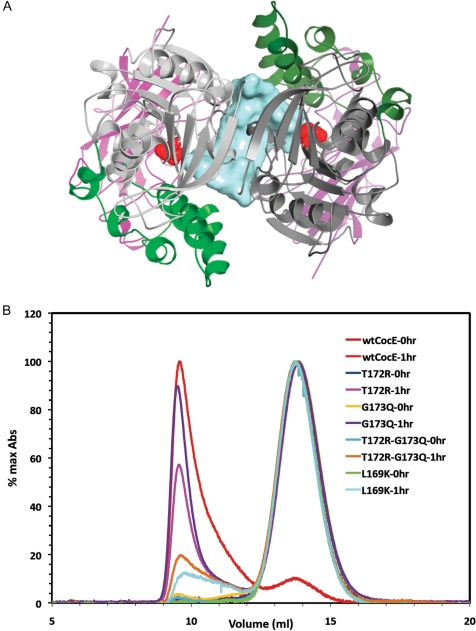
Quaternary structure of CocE. (A) X-ray crystal structure of the CocE homodimer, which is formed by a crystallographic 2-fold axis in crystals of CocE. The colors of one subunit are darker to differentiate the two monomers. The catalytic serine (S117) is shown as red spheres. A solvent-accessible channel formed at the dimer interface is colored blue. All three domains contribute to the dimer interface, which features a continuous β-sheet formed by symmetry-related strands in Domain I. (B) Size exclusion chromatography analysis of CocE and its mutants. Wild-type (wt-CocE) and mutant CocE (as indicated) were analyzed before and after incubation at 37°C for 1 h. Heat treatment leads to forms of CocE that elute in the void volume of the column, whereas non-treated CocE elutes as an apparent homodimer of molecular weight of ∼140 kDa.
Structural analysis of CocE variants
CocE contains three distinct domains. Domain I (residues 1–144 and 241–354) is composed of a canonical α/β-hydrolase fold. Domain II (residues 145–240) is a collection of seven α-helices inserted between strands β6 and β7 of Domain I. Domain III (residues 355–574) adopts a jelly roll-like topology (Fig. 1). All three domains contribute to the active site pocket. The L169K, T172R and G173Q point mutants exhibiting thermostable properties were all located within the same helix of Domain II (Fig. 6) (Larsen et al., 2002).
Fig. 6.
Overview of CocE and thermostabilizing mutations. Modeling of the L169K mutation onto the structure of CocE-T172R/G173Q shows how these stabilizing mutations cluster on H2 of Domain I (lighter space filled model), Domain II (helical region) and Domain III (darker space filled model) are shown. A portion of the DTT-carbonate adduct observed in the active site of our crystal structures is shown as ball and stick figures.
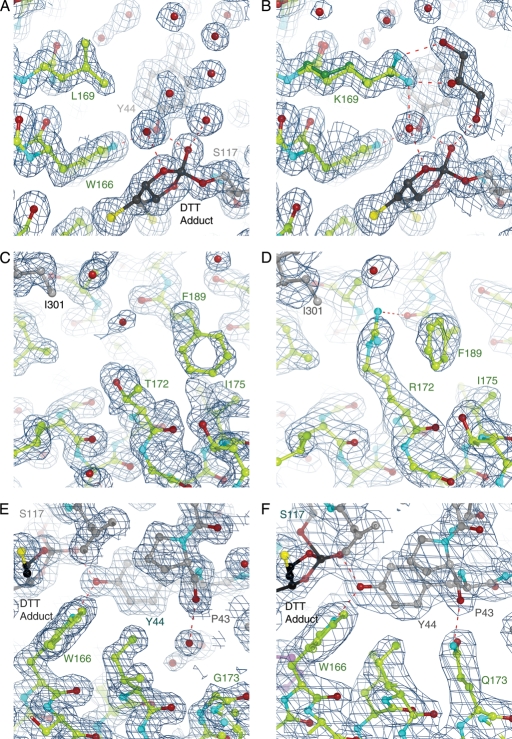
To understand how these mutations contribute to enzyme stability, we determined high-resolution X-ray crystal structures of wt-CocE (dmin = 1.5 Å), L169K (1.6 Å), T172R (2.0 Å), G173Q (2.5 Å) and T172R/G173Q (2.0 Å) CocE (Table II). The structure of ligand-free CocE (2.0 Å) was not previously reported and was determined for comparison in our study. All proteins crystallized in the same space group and retained essentially the same conformation as previously reported structures. The primary differences among the structures were in interactions formed by the mutant side chains, and in the conformation of the H2–H3 loop connecting the H2 (residues 159–175) and H3 (184–197) helices of Domain II, which exists in two to three distinct alternative conformations. Based on the great variability of this loop among the structures we determined, it was clear that the H2–H3 region of Domain II is the most dynamic region of the protein. The structures of the L169K, T172R, G173Q and T172R/G173Q mutants all showed well-ordered electron density for their mutated side chains (Fig. 7). In each case, the substitution appears to increase the number of inter- and intramolecular domain contacts and increase buried surface area, as described for each position below.
Table II.
Crystallographic data and refinement statistics
| T172R | G173Q | T172R/G173Q | L169K | wt-CocE (with DTT) | wt-CocE (no DTT) | |
|---|---|---|---|---|---|---|
| X-ray source | 23-ID D | 23-ID B | 23-ID D | 23-ID B | 21-ID D | 23-ID B |
| Wavelength (Å) | 0.9792 | 1.033 | 0.9793 | 1.033 | 1.000 | 1.033 |
| Dmin (Å) | 2.1 | 2.5 | 2.5 | 1.65 | 1.5 | 2.0 |
| Space group | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 |
| Cell constants (Å) | a = b = 106.3, c = 219.0 | a = b = 105.7, c = 221.6 | a = b = 106.2, c = 220.0 | a = b = 105.4, c = 222.2 | a = b = 105.5, c = 222.4 | A = b = 105.8, c = 222.2 |
| Unique reflections | 43 269 | 26 247 | 26 165 | 88 204 | 114 702 | 47 307 |
| Redundancy | 3.1 (2.8)a | 11.0 (11.0) | 6.0 (6.1) | 21.9 (21.9) | 4.3 (3.0) | 10.8 (11.0) |
| Rsym (%)b | 8.1 (53.6) | 16.4 (58.3) | 10.7 (55.9) | 8.9 (64.3) | 7.6 (54.4) | 12.5 (58.6) |
| Completeness (%) | 95.5 (97.3) | 90.8 (95.4) | 99.5 (100) | 99.9 (99.6) | 99.6 (100) | 99.83 (100) |
| I/σI | 21.3 (2.7) | 19.1 (4.3) | 19.5 (3.3) | 45.0 (5.4) | 35.1 (3.1) | 31.6 (4.8) |
| Refinement statistics | ||||||
| Resolution (Å) | 19.9–2.14 | 19.9–2.50 | 20.0–2.50 | 20.0–1.65 | 20.0–1.51 | 20–2.01 |
| Total reflections | 38 098 (2529)c | 24 975 (1800) | 25 949 (1833) | 87 941 (6321) | 114 339 (8299) | 49 838 (3533) |
| Protein atoms | 4700 | 4660 | 4655 | 4884 | 4883 | 4834 |
| Non-protein atoms | 544 | 361 | 296 | 837 | 907 | 651 |
| RMSD bond length (Å) | 0.012 | 0.012 | 0.010 | 0.007 | 0.008 | 0.009 |
| RMSD bond angles (°) | 1.279 | 1.247 | 1.209 | 1.117 | 1.210 | 1.146 |
| Est. coordinate error (Å)d | 0.078 | 0.107 | 0.119 | 0.039 | 0.031 | 0.071 |
| Ramachandran plot | ||||||
| Most favored (%) | 88.2 | 88.7 | 87.3 | 89.3 | 89.3 | 89.0 |
| Disallowed (%) | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 |
| Rworke | 13.8 (17.8) | 14.8 (20.7) | 14.5 (20.1) | 13.1 (14.3) | 13.0 (18.1) | 14.0 (16.0) |
| Rfreef | 18.0 (26.2) | 19.8 (28.1) | 20.8 (26.9) | 14.6 (17.1) | 15.3 (22.1) | 17.8 (23.1) |
| Rfinalg | 15.9 (19.4) | 16.3 (19.3) | 17.3 (22.5) | 15.5 (17.8) | 14.6 (21.9) | 16.7 (19.9) |
| RMSDh | 0.159 | 0.177 | 0.203 | 0.164 | 0.165 | 0.214 |
| Mean B values for all protein atoms | 31.5 | 41.6 | 43.6 | 19.2 | 14.4 | 25.0 |
aNumbers in parentheses correspond to the highest resolution shell of data; T172R: 2.18–2.10; G173Q: 2.59–2.5 Å; DM: 2.59–2.50 Å; L169K: 1.71–1.65 Å; wt-CocE: 1.56–1.51 Å.
bA cutoff of –1.0 I/σI was applied to the T172R data set.
cNumbers in parentheses correspond to the highest resolution shell of data; T172R: 2.19–2.14 Å; G173Q: 2.56–2.50 Å; T172/G173Q: 2.57–2.50 Å; L169K: 1.69–1.65 Å; wt-CocE: 1.55–1.51 Å.
dError determined using maximum likelihood in Refmac 5.2.
eRwork = ∑hkl‖Fobs(hkl)|−|Fcalc(hkl)‖/ ∑hkl |Fobs(hkl)|; no I/σ cutoff was used during refinement.
f5% of the truncated data set was excluded from refinement to calculate Rfree.
gFollowing model building and convergence of Rwork and Rfree, the final round of refinement was performed using all reflections.
hRMSD values for Cα atoms 5–173 and 183–574 against wild-type structure 1JU3. The previously reported structures 1JU3 and 1JU4 have an RMSD of 0.156 over these atoms.
Fig. 7.
Structural analysis of stabilizing CocE mutants. The crystal structures of wt-CocE (A, C and E, respectively) compared with T172R (B), G173Q (D) and L169K (F). The stabilizing effect of the mutants appears to result from enhanced interactions between H2 and H3 of Domain II or between subunits (T172R substitution) or from additional interdomain contacts (G173Q and L169K). The L169K side chain exhibits two conformations. Note that L169 is poorly ordered in the wt-CocE structure and its side chain is solvent exposed, which is expected to be destabilizing. A DTT-carbonate adduct (DBC) is evident in all structures that contain DTT (see Supplementary data). The 2|Fo|-|Fc| electron density maps were contoured at 1 σ.
T172R
A more extended alkyl chain at position 172 and the addition of a guanidinium moiety creates van der Waals contacts with the aromatic ring of F189 in H3 and a hydrogen bond between the guanidinium moiety to the backbone oxygen of F189 (Fig. 7A and B). Substitution of F189 to alanine did not alter the stabilizing properties of T172R suggesting that the enhanced stability is likely only to occur through the backbone oxygen of F189.
Interestingly, the guanidinium moiety of T172R also packs against the side chain of I301 of a 2-fold crystallographic symmetry-related chain of CocE in the crystal lattice (Fig. 7B). Subsequent analysis of the lattice packing revealed that many residues from all three domains of CocE are involved in close contacts between these two protein chains, suggesting that CocE is in fact a homodimer (Fig. 5A). Consistent with this idea, the presumptive CocE dimer interface buries ∼1400 Å2 of solvent-accessible surface area, which is comparable to the buried solvent-accessible surfaces areas found in larger antigen-antibody complexes (Wilson and Stanfield, 1994). The most prominent interaction at the 2-fold interface is formed between the symmetry-related β1 strands of Domain I, which generates a continuous β-sheet across the two subunits.
Size exclusion chromatography (SEC) studies reveal that CocE resolves as a single peak with an apparent molecular weight of ∼140 kDa (around 14 ml) most consistent with that of dimer (Fig. 5B). Heat treatment of wt-CocE or mutant CocE results in the appearance of a large molecular weight aggregate that elutes in the column void volume (around 9 ml). The time taken by each of these proteins to aggregate, however, correlates well with their respective in vitro decay time course. Wild-type CocE fully aggregates within 1 h of incubation at 37°C, while the thermostable mutant forms take 8-fold longer to aggregate (Fig. 5B). Although it was not clear whether the aggregate corresponds to denaturation of CocE or the formation of high order oligomers of folded proteins, the void volume fractions were inactive. The loss of CocE activity therefore appeared to correlate with a transition from a dimeric form to a much higher order aggregated state. It is likely that the stabilizing contacts formed at the dimer interface, such as those mediated or induced by T172R, could contribute to the enhanced thermostability. Considering both the intra- and intermolecular contacts, the T172R substitution leads to ∼33 Å2 additional buried accessible surface area for residues contacting the T172R side chain compared with wt-CocE (not considering F189, whose conformation is not fixed in the absence of the T172R substitution). The majority of this additional buried surface area is due to the contact with I301 across the dimer interface (Fig. 7B).
G173Q
The longer side chain of G173Q occupies what used to be a small water-filled pocket between Domains I and II in wt-CocE (Fig. 7C and D). The amide nitrogen of G173Q forms an interdomain hydrogen bond with the backbone carbonyl of P43 in Domain I, in good agreement with predictions from MD simulations (Gao et al., 2009). Interestingly, the analogous proline in Acetobacter α-amino acid hydrolase (Barends et al., 2006), the most similar structure in the PDB, also forms an interdomain hydrogen bond with a glutamine residue in Domain II. Additional van der Waals contacts are formed by the glutamine side chain both with the main and side chain of Y44, and the side chains of L77, F78 and I170, interactions that were not predicted by computational approaches (Gao et al., 2009). Compared to wt-CocE, the G173Q mutation leads to ∼45 Å2 of additional buried accessible surface area across the Domain I–Domain II interface suggesting that the enhanced stability is due to the enhanced domain–domain stability.
L169K
As did the native leucine side chain, the alkyl moiety of the L169K side chain contacts the phenyl rings of Y44 in Domain I and F408 in Domain III, leading to an additional 36 Å2 of buried surface area for these residues. Unlike L169 of wt-CocE, the K169 residue in the L169K mutant is well ordered, perhaps because and the Nζ atom of L169K is available to hydrogen bond with water molecules or the cryoprotectant, glycerol, in the active site pocket (Fig. 7E and F). The more favorable interactions of lysine relative to leucine with water may also contribute to the added stability of the L169K mutant. Modeling suggests that the longer side chain of lysine could sterically interfere with binding of the tropane ring of cocaine, which may account for the slightly higher Km exhibited by this mutant.
Discussion
To date, cocaine esterase is the most efficient enzyme for hydrolyzing cocaine, for decreasing cocaine levels in vivo, and for protecting against cocaine-induced lethality in rodents (Turner et al., 2002; Cooper et al., 2006; Ko et al., 2007). The effectiveness of this ‘antidote’ for cocaine toxicity in rodents suggests that CocE may be a potential therapeutic in humans. However, CocE displays considerable instability in vitro and in the blood stream with τ1/2 of ∼10–15 min. In comparison, tetrameric BchE remains in mouse plasma for 16 h and is active for up to 7 h post-injection (Duysen et al., 2002) and anti-cocaine antibodies (mAB2E2) remain in mouse circulation for 8.1 days (Norman et al., 2007). The clinical potential of CocE suggests that its duration of protection is, however, likely sufficient in acute overdose situations such as those due to snorting or injection (Landry et al., 1993). In cases involving massive overdoses, as is the case for ‘cocaine mules’ wherein large amounts of cocaine are released into the bloodstream over a long period of time, a longer acting CocE is desired. Furthermore, a longer lasting CocE would also likely be required to treat cocaine addiction. The short τ1/2 of wt-CocE in human plasma therefore represents a major obstacle in its development as a protein-based therapeutic for acute treatment of cocaine-induced lethality or abuse.
Here we provided in vitro and in vivo data demonstrating that the instability of wt-CocE observed in vivo may be a result of its thermal inactivation. The thermal sensitivity of CocE may reflect the fact that the microorganism from which CocE was isolated, Rhodococcus sp., thrives in the soil beneath coca plants under moderate temperatures around 10–18°C, much lower than the body temperature of rodents (37–38°C). We recently reported the use of computational approaches to identify two single amino residue substitutions that impart considerable improvements in the esterase's stability at 37°C (Gao et al., 2009). Computational modeling using molecular dynamics simulations predicted candidate, low energy conformations for these mutations at 37°C, however no crystallographic evidence was presented. Here we provide an expanded list containing 34-point mutations that were predicted to be stabilizing based on computational approaches and the identification of three mutations that displayed markedly enhanced thermal stability. While most candidates displayed properties not significantly better than wt-CocE, T172R and G173Q (previously identified) (Gao et al., 2009), and a new mutation, L169K, display superior thermal stable properties. Combining T172R and G173Q yielded a synergistic effect over the individual mutants thus creating an enzyme with an extended in vitro τ1/2 of 370 min, representing a 30-fold improvement over wt-CocE (τ1/2 ∼ 13 min). X-ray crystallographic analysis of the T172R/G173Q mutant presented here suggest that the substitutions increased the number of contacts between subunits (T172R with I301), between domains (G173Q of Domain II and P43 of Domain I), or within a domain (T172R with F189 within Domain II) leads to additional buried surface area. Correlation of enhanced stability with the increase in buried surface area in CocE is consistent with thermostabilizing mutations in other enzymes (Korkegian et al., 2005).
The location of thermostable substitutions in the H2 helix (within Domain II) and the structural heterogeneity of the H2–H3 loop itself suggested that the H2–H3 helical region is highly dynamic and may contribute strongly to the inactivation of CocE and aggregation. This mobility, however, may also be strongly linked to function, as truncation or complete removal of the loop between the H2 and H3 helices in CocE inactivated the enzyme (data not shown). Indeed, computational approaches identified Domain II as the most thermally unstable region of wt-CocE (Gao et al., 2009) and this provided the rationale for targeting potentially stable substitutions in this region. The introduction of residues that bridge the H2 and H3 helix or that increase contacts within Domain II, between domains or with the other subunit of the CocE homodimer, would therefore be expected to provide some degree of overall stabilization.
The modest contributions of T172R based on in vivo and in vitro measurements (τ1/2 = ∼78 min) represent a 6-fold improvement over wt-CocE, most likely via increased buried surface between H2 and H3 and across the dimer interface. By increasing interactions in the interface between Domain I and Domain II with the G173Q substitution, a similar protein stabilizing effect is achieved. Although the G173Q mutation only weakly stabilizes the in vitro activity of the enzyme, its combination with T172R yielded a substantially more stable form of CocE, taking advantage of the cooperativity between the enhanced inter- and intra-domain interactions at each of these positions.
Analysis of the crystal structure of CocE-L169K revealed that K169 forms additional van der Waals interaction with Y44 (Domain I) and F408 (Domain III). In addition, the Nζ atom of K169, which is observed to interact with glycerol in the crystal structure, contributes to the hydrogen bonding network of solvent in the active site cavity, whereas the side chain of the native residue (L169) is unfavorably solvent exposed. The interaction with glycerol likely represents an interaction with water in the active site. Glycerol was added to the crystals as a cryoprotectant, whose function is to displace water and prevent ice crystal formation. The combination of all these factors may account for the observed stability at 37°C of this mutant and the proximity to the active sites may account for the effect on the Km for cocaine.
The DTT-carbonate adduct observed in the active site of crystalline CocE was rather a surprise as it was not reported in the original CocE structures reported previously (Larsen et al., 2002; Turner et al., 2002). The adduct occurs through catalysis by the enzyme where it appears to be bound covalently to the catalytic S117 much in the overall manner as phenyl boronic acid (Larsen et al., 2002; Turner et al., 2002) (see supplementary data available at PEDS online). Superimposition of the active sites reveals that the plane of the dioxolane ring of the adduct overlaps the plane of the phenyl ring of both phenyl boronic acid- and benzoic acid-bound structures of CocE (Larsen et al., 2002; Turner et al., 2002). We surmise that the adduct was not observed in prior crystal structures of CocE since it is mutually exclusive with binding of benzoic acid or phenyl boronate, and not formed in catalytically challenged mutants. The appearance of DTT adduct in the active site appears to bear little relevance to our interpretation of the thermal stabilizing effects of the mutations because no effects of DTT on enzyme activity are observed below 10 mM concentrations (data not shown), which is in excess of the concentrations used for crystallization. Moreover, no effect of DTT was observed in thermostability assays that compared incubations in the presence or absence of DTT (data not shown).
Finally, the crystal structure of CocE and our SEC studies suggest that the enzyme exists as a dimer in solution. Incubation at 37°C induces protein aggregation and elution from a size exclusion column in the void volume of the column. Interestingly, the kinetics of inactivation of both T172R/G173Q and L169K reveal a two-phase inactivation. During the first phase, activity diminishes exponentially down to a plateau of approximately 35% of the maximal activity. There is residual activity in both T172R/G173Q (28%) and L169K CocE (19%) even after 8 h at 37°C, whereas wt-CocE, G173Q and T172R CocE lose all activity within 90 min (Gao et al., 2009). Further studies and the characterization of additional mutants, perhaps within the dimer interface, will help establish the importance and contribution of inter-subunit contacts to the stability of CocE.
In summary, we have shown that a multi-pronged approach combining computational, biochemical and structural analysis can be used to rationally develop and analyze variants of CocE that are significantly more stable than the native enzyme. These studies strongly suggest that these, or future thermostabilized variants of CocE, may serve as a novel therapeutic approach for the treatment of cocaine overdose and addiction.
Supplementary data
Funding
This work was supported by National Institute of Health National Institute of Drug Abuse grants DA021416, DA 025100, DA013930.
Supplementary Material
Footnotes
Edited by Albert Berghuis
References
- Baird T.J., Deng S.X., Landry D.W., Winger G., Woods J.H. J. Pharmacol. Exp. Ther. 2000;295:1127–1134. [PubMed] [Google Scholar]
- Barends T.R., Polderman-Tijmes J.J., Jekel P.A., Williams C., Wybenga G., Janssen D.B., Dijkstra B.W. J. Biol. Chem. 2006;281:5804–5810. doi: 10.1074/jbc.M511187200. [DOI] [PubMed] [Google Scholar]
- Benowitz N.L. Pharmacol. toxicol. 1993;72:3–12. doi: 10.1111/j.1600-0773.1993.tb01331.x. doi:10.1128/AEM.66.3.904-908.2000. [DOI] [PubMed] [Google Scholar]
- Bresler M.M., Rosser S.J., Basran A., Bruce N.C. Appl. Environ. Microbiol. 2000;66:904–908. doi: 10.1128/aem.66.3.904-908.2000. doi:10.1128/AEM.66.3.904-908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne S.P., Slaughter E.A., Couch R.A., Rudnic E.M., McLean A.M. Biopharm. Drug Dispos. 1998;19:309–314. doi: 10.1002/(sici)1099-081x(199807)19:5<309::aid-bdd108>3.0.co;2-9. doi:10.1002/(SICI)1099-081X(199807)19:5<309::AID-BDD108>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Carmona G.N., Baum I., Schindler C.W., Goldberg S.R., Jufer R., Cone E., Slaughter E., Belendiuk G.W., Gorelick D.A. Life Sci. 1996;59:939–943. doi: 10.1016/0024-3205(96)00392-x. doi:10.1016/0024-3205(96)00392-X. [DOI] [PubMed] [Google Scholar]
- Carrera M.R., Ashley J.A., Wirsching P., Koob G.F., Janda K.D. Proc. Natl Acad. Sci. USA. 2001;98:1988–1992. doi: 10.1073/pnas.041610998. doi:10.1073/pnas.041610998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera M.R., Kaufmann G.F., Mee J.M., Meijler M.M., Koob G.F., Janda K.D. Proc. Natl Acad. Sci. USA. 2004;101:10416–10421. doi: 10.1073/pnas.0403795101. doi:10.1073/pnas.0403795101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper Z.D., Narasimhan D., Sunahara R.K., Mierzejewski P., Jutkiewicz E.M., Larsen N.A., Wilson I.A., Landry D.W., Woods J.H. Mol. Pharmacol. 2006;70:1885–1891. doi: 10.1124/mol.106.025999. doi:10.1124/mol.106.025999. [DOI] [PubMed] [Google Scholar]
- Crumb W.J., Jr, Clarkson C.W. Biophys. J. 1990;57:589–599. doi: 10.1016/S0006-3495(90)82574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S.X., de Prada P., Landry D.W. J. Immunol. Methods. 2002;269:299–310. doi: 10.1016/s0022-1759(02)00237-5. doi:10.1016/S0022-1759(02)00237-5. [DOI] [PubMed] [Google Scholar]
- Duysen E.G., Bartels C.F., Lockridge O. J. Pharmacol. Exp. Ther. 2002;302:751–758. doi: 10.1124/jpet.102.033746. doi:10.1124/jpet.102.033746. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. doi:10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gao Y., Atanasova E., Sui N., Pancook J.D., Watkins J.D., Brimijoin S. Mol. Pharmacol. 2005;67:204–211. doi: 10.1124/mol.104.006924. doi:10.1124/mol.104.006924. [DOI] [PubMed] [Google Scholar]
- Gao D., Narasimhan D.L., Macdonald J., Brim R., Ko M.C., Landry D.W., Woods J.H., Sunahara R.K., Zhan C.G. Mol. Pharmacol. 2009;75:318–323. doi: 10.1124/mol.108.049486. doi:10.1124/mol.108.049486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick D.A. Drug Alcohol Depend. 1997;48:159–165. doi: 10.1016/s0376-8716(97)00119-1. doi:10.1016/S0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- Gorelick D.A., Gardner E.L., Xi Z.X. Drugs. 2004;64:1547–1573. doi: 10.2165/00003495-200464140-00004. doi:10.2165/00003495-200464140-00004. [DOI] [PubMed] [Google Scholar]
- Johanson C.E., Fischman M.W. Pharmacol. Rev. 1989;41:3–52. [PubMed] [Google Scholar]
- Jones T.A., Zou J-Y., Cowan S.W., Kjeldgaard M. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kantak K.M. Expert Opin. Pharmacother. 2003;4:213–218. doi: 10.1517/14656566.4.2.213. doi:10.1517/14656566.4.2.213. [DOI] [PubMed] [Google Scholar]
- Ko M.C., Bowen L.D., Narasimhan D., Berlin A.A., Lukacs N.W., Sunahara R.K., Cooper Z.D., Woods J.H. J. Pharmacol. Exp. Ther. 2007;320:926–933. doi: 10.1124/jpet.106.114223. doi:10.1124/jpet.106.114223. [DOI] [PubMed] [Google Scholar]
- Korkegian A., Black M.E., Baker D., Stoddard B.L. Science. 2005;308:857–860. doi: 10.1126/science.1107387. doi:10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B., Baker D. Proc. Natl Acad. Sci. USA. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. doi:10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry D.W., Zhao K., Yang G.X., Glickman M., Georgiadis T.M. Science. 1993;259:1899–1901. doi: 10.1126/science.8456315. doi:10.1126/science.8456315. [DOI] [PubMed] [Google Scholar]
- Larsen N.A., Turner J.M., Stevens J., Rosser S.J., Basran A., Lerner R.A., Bruce N.C., Wilson I.A. Nat. Struct. Biol. 2002;9:17–21. doi: 10.1038/nsb742. doi:10.1038/nsb742. [DOI] [PubMed] [Google Scholar]
- Lynch T.J., Mattes C.E., Singh A., Bradley R.M., Brady R.O., Dretchen K.L. Toxicol. Appl. Pharmacol. 1997;145:363–371. doi: 10.1006/taap.1997.8187. doi:10.1006/taap.1997.8187. [DOI] [PubMed] [Google Scholar]
- Mattes C.E., Lynch T.J., Singh A., Bradley R.M., Kellaris P.A., Brady R.O., Dretchen K.L. Toxicol. Appl. Pharmacol. 1997;145:372–380. doi: 10.1006/taap.1997.8188. doi:10.1006/taap.1997.8188. [DOI] [PubMed] [Google Scholar]
- Meijler M.M., Kaufmann G.F., Qi L., Mee J.M., Coyle A.R., Moss J.A., Wirsching P., Matsushita M., Janda K.D. J. Am. Chem. Soc. 2005;127:2477–2484. doi: 10.1021/ja043935e. doi:10.1021/ja043935e. [DOI] [PubMed] [Google Scholar]
- Mets B., et al. Proc. Natl Acad. Sci. USA. 1998;95:10176–10181. doi: 10.1073/pnas.95.17.10176. doi:10.1073/pnas.95.17.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzasalma T.M., Kranz J.K., Chan W., Struble G.T., Schalk-Hihi C., Deckman I.C., Springer B.A., Todd M.J. J. Biomol. Screen. 2007;12:418–428. doi: 10.1177/1087057106297984. doi:10.1177/1087057106297984. [DOI] [PubMed] [Google Scholar]
- Norman A.B., Tabet M.R., Norman M.K., Buesing W.R., Pesce A.J., Ball W.J. J. Pharmacol. Exp. Ther. 2007;320:145–153. doi: 10.1124/jpet.106.111781. doi:10.1124/jpet.106.111781. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. doi:10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Rogers C.J., Mee J.M., Kaufmann G.F., Dickerson T.J., Janda K.D. J. Am. Chem. Soc. 2005;127:10016–10017. doi: 10.1021/ja053086a. doi:10.1021/ja053086a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration, Drug Abuse Warning Network. DAWN Series D-30DHHS. 2006. Publication No. (SMA) 08-4339. [PubMed] [Google Scholar]
- Turner J.M., Larsen N.A., Basran A., Barbas C.F., 3rd, Bruce N.C., Wilson I.A., Lerner R.A. Biochemistry. 2002;41:12297–12307. doi: 10.1021/bi026131p. doi:10.1021/bi026131p. [DOI] [PubMed] [Google Scholar]
- Uhl G.R., Hall F.S., Sora I. Mol. Psychiatry. 2002;7:21–26. doi: 10.1038/sj.mp.4000964. doi:10.1038/sj/mp/4000964. [DOI] [PubMed] [Google Scholar]
- Vagin A.A., Steiner R.A., Lebedev A.A., Potterton L., McNicholas S., Long F., Murshudov G.N. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. doi:10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Wilson I.A., Stanfield R.L. Curr. Opin. Struct. Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. doi:10.1016/0959-440X(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Xie W., Altamirano C.V., Bartels C.F., Speirs R.J., Cashman J.R., Lockridge O. Mol. Pharmacol. 1999;55:83–91. doi: 10.1124/mol.55.1.83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.