Abstract
Purpose
Colorectal cancer prognosis is currently predicted from pathological staging, providing limited discrimination for Dukes’ stage B and C disease. Additional markers for outcome are required to help guide therapy selection for individual patients.
Experimental Design
A multi-site single-platform microarray study was performed on 553 colorectal cancers. Gene expression changes were identified between stage A and D tumors (three training sets) and assessed as a prognosis signature in stage B and C tumors (independent test and external validation sets).
Results
128 genes showed reproducible expression changes between three sets of stage A and D cancers. Using consistent genes, stage B and C cancers clustered into two groups resembling early-stage and metastatic tumors. A Prediction Analysis of Microarray (PAM) algorithm was developed to classify individual intermediate-stage cancers into stage A-like/good prognosis or stage D-like/poor prognosis types. For stage B patients, the treatment adjusted hazard ratio for six-year recurrence in individuals with stage D-like cancers was 10.3 (95% CI 1.3 to 80.0, P=0.011). For stage C patients, the adjusted hazard ratio was 2.9 (95% CI 1.1 to 7.6, P=0.016). Similar results were obtained for an external set of stage B and C patients. The prognosis signature was enriched for down-regulated immune response genes and up-regulated cell signaling and extracellular matrix genes. Accordingly, sparse tumor infiltration with mononuclear chronic inflammatory cells was associated with poor outcome in independent patients.
Conclusions
Metastasis-associated gene expression changes can be used to refine traditional outcome prediction, providing a rational approach for tailoring treatments to subsets of patients.
Keywords: colorectal cancer, gene expression, outcome prediction
Statement of Translational Relevance
Molecular markers are required to refine prediction of recurrence risk for colorectal cancer (CRC) to help guide the selection of adjuvant therapies for individual patients. This international single-platform microarray study demonstrates that metastasis-associated gene expression changes, identified across multiple sets of stage A and D cancers, can be used to improve outcome prediction for patients with Dukes’ stage B or C disease. Microarray data for training and test cases were produced at multiple sites, indicating good inter-institutional reproducibility required for clinical application. Our results improve our understanding of CRC progression, identifying putative signatures of down-regulated immune response genes and up-regulated cell signaling and extracellular matrix genes. Accordingly, low density of mononuclear chronic inflammatory cells within tumors was shown to be associated with poor prognosis in independent patients. Our candidate genes provide a good starting point for future study and potential targets for therapy.
Introduction
Colorectal cancer (CRC) is often detected at a stage when complete resection of the primary cancer is possible, yet 40 to 50% of patients who undergo potentially curative surgery alone relapse and die of metastatic disease (1). Patient risk of recurrence is currently largely predicted from the extent of spread of the primary tumor, and this is the major determinant of further clinical management. While the majority of patients with Dukes’ stage C (lymph-node positive) cancer receive a combination of 5-fluorouracil and oxaliplatin, adjuvant treatment is offered to only a subset of Dukes’ stage B (localized disease) patients presenting with specific high-risk clinical features including tumor perforation or invasion of adjacent organs (2). This approach is clearly sub-optimal, resulting in under-treatment of ~20% of stage B patients who will recur. Similarly, current adjuvant treatment is clearly ineffective in many stage C patients, with a recurrence rate of ~40% (3, 4), highlighting the need for treatment with more aggressive or newly emerging targeted therapies. There is an urgent need for biomarkers to refine traditional prediction of recurrence risk to enable better use of existing treatment options and the optimal development of novel individualized therapies.
Several studies have used microarray analysis on primary tumor specimens to identify gene expression signatures predictive of CRC prognosis (5–9). The general approach for signature discovery has been analysis of patients selected for good and poor outcomes (training set), followed by assessment of the signature in additional cases (test set). However, the performance and general applicability of published classifiers has been challenging to determine. Division of patients into training and test sets has often resulted in small sample sizes (5, 6, 8), and several studies did not formally assess a defined classifier, but rather the validity of candidate prognostic genes using cross-validation procedures (6, 8, 9). Furthermore, signature discovery based on outcome is generally confounded in patients undergoing adjuvant treatment (the majority of stage C patients), as it is difficult to distinguish markers of prognosis from markers of therapy response (7, 9).
Gene expression patterns have been shown to broadly differ between metastatic and non-metastatic colorectal cancers, implying that the acquisition of metastatic potential by the primary tumor is accompanied by specific changes in endogenous transcription and/or changes in the tumor micro-environment (10–13). This suggests an alternative approach to prognosis signature discovery, whereby expression differences between the extremes of stages of cancer (early-stage/stage A versus metastatic/stage D) could be used to predict recurrence in patients with intermediate stages of disease. Advantages of this approach are that tumor stage-based discovery does not require follow-up data, and that the confounding effect of previous therapy can be avoided by selecting patients who have not undergone such treatment.
In this international multi-site study, we evaluated this discovery strategy using data on CRCs from 553 patients analysed using a common microarray platform. Reproducible gene expression differences were identified between three training sets of stage A and D cancers, with the latter being represented by both primary and distant lesions. The feasibility of using consistent expression changes for classification of intermediate-stage cancers into groups resembling early-stage and metastatic lesions was assessed using unsupervised clustering on two sets of stage B and two sets of stage C tumors. A prognostic algorithm was developed to permit classification of individual test cancers into early-stage “good prognosis” or metastatic “poor prognosis” types, a requirement for clinical application. The prognostic value of this single-sample classifier was determined for stage B and C patients with long-term follow-up data. An external dataset of 99 stage B and C patients produced on an earlier version of our microarray platform was used for additional validation. To improve our understanding of the changes associated with metastatic progression in CRC, classifier genes were analyzed for functional category enrichment; a putative immune response signature was validated by histological analysis of tumor infiltrating mononuclear chronic inflammatory cells on 155 stage B and 166 stage C patients enrolled in the VICTOR clinical trial, a Phase III randomised placebo controlled study of rofecoxib (14).
Materials and Methods
Patients and gene expression microarray analysis
Fresh-frozen tumor specimens from 293 consecutive CRC patients were retrieved from the tissue banks of the Royal Melbourne Hospital, Western Hospital and Peter MacCallum Cancer Center in Australia, and the H. Lee Moffitt Cancer Center in the United States; individuals who had received preoperative chemo- and/or radiotherapy or for whom tumor-derived total RNA was inadequate for microarray analysis (RIN < 6) were excluded. All patients gave informed consent, and this study was approved by the medical ethics committees of all sites. Patient median age at diagnosis was 67 years (range 26 to 92 years). All specimens were derived from primary carcinomas and were snap-frozen in liquid nitrogen immediately after surgery for storage at −80°C. Cases comprised 44 stage A, 95 stage B, 93 stage C and 61 stage D cancers; 252 were localized to the colon and 40 to the rectum, with one case missing this information. 22 of 94 patients who had stage B disease and 64 of 91 patients who had stage C disease had received standard adjuvant chemotherapy (either single agent 5-fluouracil/capecitabine or 5-fluouracil and oxaliplatin) or postoperative concurrent chemoradiotherapy (50.4 Gy in 28 fractions with concurrent 5-fluorouracil) according to hospital protocols. All patients were assessed annually. For stage B and C patients, follow-up and additional clinical data including patient gender and TNM staging were collected by Biogrid Australia 1 for Australian patients and the Moffitt Cancer Center Tumor Registry for US patients. The median duration of follow-up was 47.8 months (range 0.9 to 118.6 months) for the 140 patients without recurrence, and 19.1 months (range 1.6 to 93.7 months) for the 48 patients with local or distant recurrence. The median follow-up for all 188 patients was 37.2 months (range 0.9 to 118.6 months).
Total RNA was extracted using Trizol reagent (Invitrogen) from CRC samples containing >60% tumor cells. All samples included showed good integrity of 18S and 28S ribosomal bands (RIN > 6) using a 2100 Bioanalyzer (Agilent Technologies). Total RNA was labeled and hybridized to HG-U133Plus2.0 GeneChip arrays (Affymetrix) according to the manufacturer’s instructions. The microarray data on a subset of 174 tumors have been published previously (NCBI Gene Expression Omnibus, GSE5206 and GSE13067).
In addition, published gene expression data were retrieved for 42 stage A CRCs, 83 stage B, 73 stage C and 62 stage D CRCs analyzed as part of the Expression Project for Oncology (expO) 2 using HG-U133Plus2.0 GeneChip arrays (Affymetrix) (Supplementary Table S1). Of the 62 stage D CRCs, 32 were primary cancer and 30 were metastectomy specimens. None of the primary cancer patients had received preoperative therapy, but 17 metastectomy specimens were from patients who had received adjuvant chemotherapy treatment prior to resection. Data processing and analysis were performed using the statistical software package R (15) and appropriate Bioconductor packages (16).
Identification of metastasis-associated gene expression changes
Consistent gene expression changes were identified between 44 stage A and 61 stage D CRCs from this study and 42 stage A and 62 stage D CRCs from expO. For the expO dataset, separate comparisons were performed for primary stage D cancers and distant metastases to identify gene expression maintained during metastatic spread. For each cohort, MAS5.0-calculated signal intensities were normalized using the quantile normalization procedure implemented in robust multiarray analysis (RMA) (17, 18) and the normalized data were log transformed (base 2). Probe sets which were not expressed or probe sets which showed a low variability across samples were excluded. Expression values were required to be above the median of all expression measurements in at least 25% of samples, and the interquartile range across the samples on the log scale was required to be at least 0.5. Genes mapping to sex chromosomes were excluded as cases were not matched by gender. A total of 6716 gene probes passed these filtering steps in all three sample sets.
Differentially expressed genes were identified using Significance Analysis of Microarrays (SAM) with a Wilcoxon rank-sum test and a false discovery rate (FDR) of 10% (19). Separate lists were generated for genes significantly up- or down-regulated in stage A CRCs as compared to stage D CRCs for each of the three comparisons. For differentially expressed genes identified repeatedly between cohorts, consistency of up- or down-regulation was assessed using Pearson’s chi-squared test.
Unsupervised clustering
For the 95 stage B and 93 stage C CRCs from this study and the 83 stage B and 73 stage C CRCs from expO, expression values of the identified metastasis-associated genes were mean- and sample-centered, followed by divisive hierarchical clustering using pair distances calculated as one minus the Spearman correlation coefficient as distance metric. Differences in median gene expression values were calculated for the samples within the two main branches of the resulting dendrogram. Relative up- or down-regulation of gene expression between these two groups was assessed for consistency with up- or down-regulation observed between early-stage and metastatic cancers using Pearson’s chi-squared test.
PAM classifier development and application
Based on metastasis-associated genes, a Prediction Analysis of Microarrays (PAM) (20) nearest shrunken centroid classifier was developed for separation of all primary stage A (n=86) and stage D (n=93) cancers (reference set). Microarray data were quantile normalized, followed by ten-fold cross-validation for increasing values of centroid shrinkage, designed to progressively eliminate noisy genes. Misclassification errors were calculated from this cross-validation procedure. Using the optimized PAM classifier, 95 stage B and 93 stage C CRCs were classified into stage A-like “good-prognosis” and stage D-like “poor-prognosis” types. MAS5.0-calculated signal intensities of stage B or C cancers were normalized against the reference set on a single-sample basis.
Functional category enrichment analysis
Functional category enrichment analysis was performed using the Functional Annotation Clustering tool on the Database for Annotation, Visualization and Integrated Discovery. 3 Metastasis-associated genes were classified according to their annotated role in biological process, molecular function, and cellular component from Gene Ontology (GO). 4 Category enrichment was tested against all human genes. P-values were adjusted using the Benjamini-Hochberg False Discovery Rate multiple testing correction.
Analysis of tumor infiltration with mononuclear chronic inflammatory cells
Haematoxylin and eosin (H&E) stained tissue sections of formalin-fixed paraffin-embedded CRC specimens were retrieved for 155 stage B and 166 stage C patients enrolled in the VICTOR clinical trial (14). The average density of mononuclear chronic inflammatory cells (comprising lymphocytes, plasma cells, and macrophages) was scored within tumor areas comprising more than 60% of neoplastic cells by two anatomical pathologists (MC and SP); areas of adenoma, ulceration and necrosis were excluded from the analysis. Mononuclear chronic inflammatory cell density was assessed at ×40 magnification and classified into low and moderate/high by each observer.
Statistical analysis
Associations between predicted stage A- and D-like cancers and clinical characteristics were separately assessed for stage B and C patients using Fisher’s exact test for categorical variables and the Welch two-sample t-test for continuous variables. For the outcome analysis, six-year recurrence was the primary endpoint. Disease-free survival was defined as the time of surgery to the first confirmed relapse. Censoring was performed when a patient died or was alive without recurrence at last contact. Cox proportional-hazards models were used to estimate survival distributions and hazard ratios and included the gene expression classifier, age at diagnosis, number of lymph nodes examined, N stage and adjuvant treatment. All statistical analyses were two-sided and considered significant if P<0.05.
Results
Expression changes between early-stage and metastatic CRCs
Reproducible gene expression changes between early-stage and metastatic CRCs were identified using 44 stage A and 61 stage D tumors from our laboratories, and 42 stage A and 62 stage D tumors from expO. Separate comparisons were performed for specimens derived from primary stage D cancers and distant metastases to identify changes maintained during metastatic spread. For each cohort, separate lists were generated for genes significantly up-or down-regulated in metastatic cancers, and for repeatedly identified genes consistency of up- or down- regulation was assessed (Table 1). All pair-wise comparisons of metastasis-associated changes were significant (P<0.001, chi-squared test), with more than 96% of changes being consistent in all cases. The level of consistency was high irrespective of whether the comparisons involved only primary metastatic cancers or primary stage D cancers and distant metastases. A total of 128 genes (163 probe sets, Supplementary Table S2) showed reproducible up- (71 genes) or down-regulation (57 genes) in metastatic cancers as compared to early-stage cancers across all three cohorts. Notably, two of out of the three comparisons solely involved primary cancers from patients who had not received preoperative therapy, thus excluding a confounding influence of treatment on classifier selection.
Table 1. Comparison of gene expression changes between early-stage and metastatic colorectal cancers across independent cohorts.
Analysis was performed for 44 stage A and 61 primary stage D CRCs from this study, 42 stage A and 32 primary stage D CRCs from expO, and 42 stage A CRCs and 30 distant metastases (stage D) from expO. For each cohort, genes (probe sets) differentially expressed between early-stage and metastatic cancers were identified using SAM and a FDR of 10%. For genes repeatedly identified between cohorts, consistency of up- or down-regulation in metastatic cancers was assessed using Pearson’s chi-squared test.
| expO: Stage A vs. D (primary) | |||
|---|---|---|---|
|
This study: Stage A vs. D (primary) |
Up-regulated | Down-regulated | P value |
| Up-regulated | 134 (57.3%) | 0 (0%) | < 0.001 |
| Down-regulated | 0 (0%) | 100 (42.7%) | |
| expO: Stage A vs. D (metastatic deposits) | |||
|
This study: Stage A vs. D (primary) |
Up-regulated | Down-regulated | |
| Up-regulated | 154 (41.3%) | 10 (2.7%) | < 0.001 |
| Down-regulated | 5 (1.3%) | 204 (54.7%) | |
| expO: Stage A vs. D (metastatic deposits) | |||
|
expO: Stage A vs. D (primary) |
Up-regulated | Down-regulated | |
| Up-regulated | 414 (49.9%) | 7 (0.8%) | < 0.001 |
| Down-regulated | 3 (0.4%) | 405 (48.9%) | |
Clustering of intermediate-stage CRCs using metastasis-associated genes
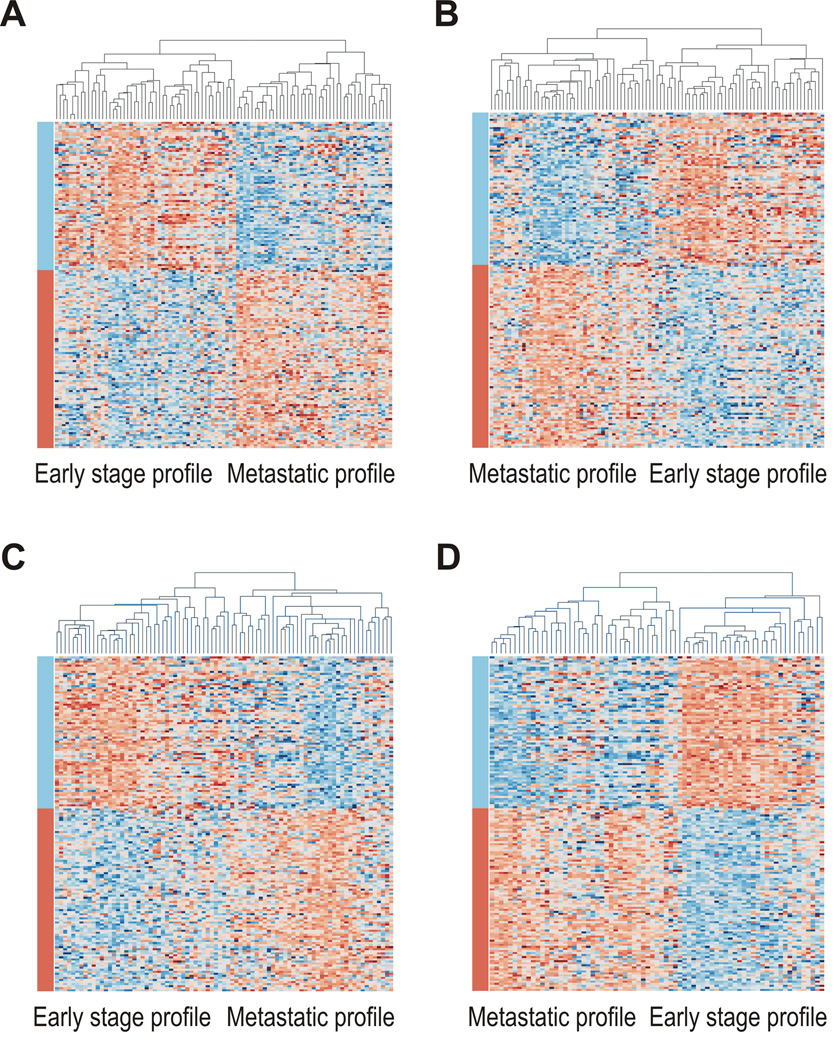
Feasibility of using our set of 128 metastasis-associated genes for classification of stage B and C CRCs into groups resembling early-stage and metastatic lesions was assessed using unsupervised clustering on four independent sample sets: 95 stage B and 93 stage C CRCs from this study, and 83 stage B and 73 stage C CRCs from expO (Fig. 1). For all four sets of tumors, the relative differences in median gene expression between the two main resulting clusters mirrored those identified between early-stage and metastatic lesions (Supplementary Table S3); more than 97% of changes were consistent for each comparison (P<0.001, chi-squared test).
Fig 1.
Unsupervised clustering for stage B and stage C colorectal cancers based on metastasis-associated genes. Clustering divides 95 stage B and 93 stage C cases from this study (A–B) and 83 stage B and 73 stage C cases from expO (C–D) into groups with early-stage and metastatic profiles. Samples are arranged along the x-axis and genes along the y-axis. Orange represents increased and blue decreased expression relative to the mean- and sample-centered scaled expression. Genes are grouped into those found to be down-regulated (blue) and up-regulated (orange) in metastatic cancers as indicated by the color bars.
Prognosis classification of intermediate-stage CRCs
To permit classification of individual test cancers into early-stage/good prognosis or metastatic/poor prognosis types - a requirement for clinical application - a PAM algorithm was developed using all 179 primary stage A and D cancers from this study and expO as a reference set (Supplementary Fig. S1). For each test cancer, microarray data were normalized against this reference set followed by sample classification into a stage A- or D-like type. Prior (expected) six-year recurrence probabilities were set as those presently observed for stage B and C patients (20% and 40%, respectively) (21).
The majority of test stage B (82 of 95, 86.3%) and stage C (77 of 93, 82.8%) CRCs were classified into stage A- and D-like types with a greater than 90% prediction probability (Supplementary Fig. S2). 45.1% (37 of 82) of stage B and 37.7% (29 of 77) of stage C cancers showed a stage A-like signature at this cut-off. For both groups of patients, class predictions were not associated with age at diagnosis, gender, tumor T stage, location, number of lymph nodes examined and adjuvant treatment (Table 2). However, stage C patients with stage D-like tumors tended to present with a higher node status (37.5% with N2 status, 18 of 48) than those with stage A-like tumors (13.8% with N2 status, 4 of 29; p=0.037, Fisher’s exact test), consistent with the anticipated classification by metastatic potential. The 13 stage B and 16 stage C patients who could not be confidently classified had clinical features similar to those patients who could be classified with confidence.
Table 2. Associations between clinical characteristics and PAM class predictions (stage A-like or D-like) based on 128 metastasis-associated genes for 95 stage B and 93 stage C colorectal cancer patients.
For single-sample PAM classification, prior (expected) six-year recurrence probabilities were set as 20% for stage B and 40% for stage C patients based on relapse rates observed in clinical practice (21). Class predictions with a >90% probability were scored. The Welch two-sample t-test was used for age and lymph nodes examined, the Fisher’s exact test for all other clinical variables.
| Stage B patients | Stage C patients | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stage A-like | Stage D-like | Not classified |
P value1 |
P value2 |
Stage A-like | Stage D-like | Not classified |
P value1 |
P value2 |
|
| (N=36) | (N=45) | (N=13) | (N=27) | (N=48) | (N=16) | |||||
| Age | ||||||||||
| Median, range | 66 (43–86) | 71 (30–92) | 71 (38–92) | 0.946 | 0.789 | 71 (26–84) | 63.5 (26–90) | 69 (30–81) | 0.147 | 0.719 |
| Gender | ||||||||||
| Female | 17 | 22 | 6 | 0.827 | 1 | 17 | 18 | 9 | 0.099 | 0.583 |
| Male | 20 | 23 | 7 | 12 | 30 | 7 | ||||
| T stage | ||||||||||
| T2 | - | - | - | 0.323 | 0.051 | 4 | 4 | 1 | 0.537 | 0.137 |
| T3 | 34 | 44 | 10 | 23 | 42 | 12 | ||||
| T4 | 3 | 1 | 3 | 2 | 2 | 3 | ||||
| N stage | ||||||||||
| N1 | - | - | - | - | 25 | 30 | 12 | 0.037 | 1 | |
| N2 | - | - | - | - | 4 | 18 | 4 | |||
|
Lymph nodes examined2 |
||||||||||
| Median, range | 15 (1–33) | 10 (1–31) | 14 (5–51) | 0.080 | 0.333 | 13 (3–37) | 14.5 (4–73) | 12 (6–43) | 0.391 | 0.506 |
| Tumor location | ||||||||||
| Colon | 31 | 38 | 13 | 1 | 0.203 | 25 | 44 | 14 | 0.466 | 0.680 |
| Rectum | 6 | 7 | 0 | 4 | 4 | 2 | ||||
|
Adjuvant therapy |
||||||||||
| No | 29 | 34 | 10 | 0.799 | 1 | 10 | 14 | 4 | 0.623 | 0.769 |
| Yes | 8 | 11 | 3 | 19 | 34 | 12 | ||||
The P values are for the comparison of stage A-like with stage D-like patients.
The P values are for the comparison of classified with not classified patients.
The number of lymph nodes examined was not available for five stage B and two stage C patients.
Metastasis-associated changes predict poor prognosis
Probabilities of disease-free survival were independently calculated for the 82 stage Band 77 stage C patients with “confident” class predictions (Supplementary Fig. S3). As anticipated, individuals with stage D-like cancers showed a poorer prognosis than individuals with stage A-like cancers in both cases. The estimated hazard ratio for recurrence was 10.6 (95% CI 1.3 to 82.0, P = 0.024, Wald test) for stage B, and 2.8 (95% CI 1.1 to 7.5, P = 0.035, Wald test) for stage C patients over a six-year follow-up period. Similar results were obtained when the analysis was adjusted for adjuvant treatment (stage B hazard ratio 10.3, 95% CI 1.3 to 80.0, P = 0.011; stage C hazard ratio 2.9, 95% CI 1.1 to 7.6, P = 0.016).
Comparison of the expression classifier and pathological staging
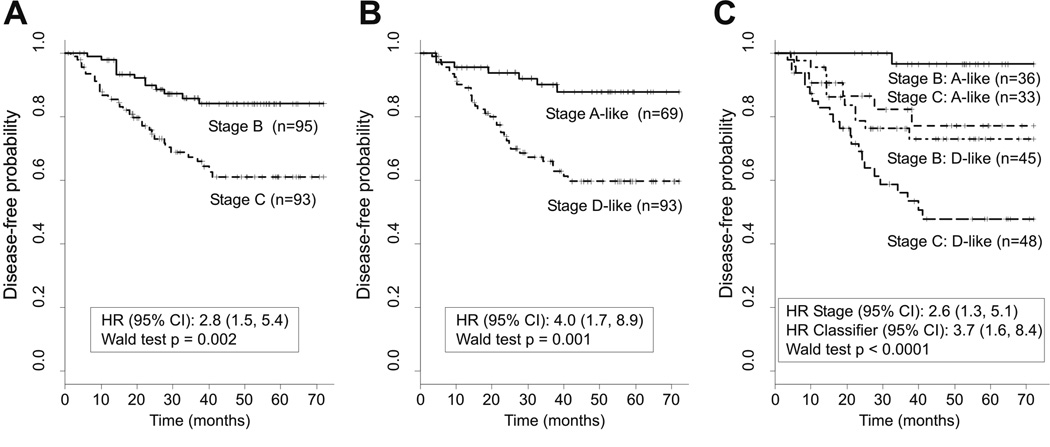
To assess the prognostic value of our 128-gene classifier, we compared it against pathological staging in stage B and C patients. For this comparison, expression-based classification was performed using the same prior recurrence probability of 30% for all patients. Individuals showed similar differences in outcomes when classified based on pathological staging or the expression classifier (Fig. 2 A–B). The estimated hazard ratio for recurrence was 2.8 for stage C patients as compared to stage B patients (95% CI 1.5 – 5.4; P = 0.002, Wald test), and 4.0 for patients with stage D-like cancers as compared to patients with stage A-like cancers (95%CI 1.7 – 8.9; P = 0.001, Wald test).
Fig 2.
Comparison of disease-free survival among stage B and C patients when grouped by (A) pathological staging, (B) the 128-gene PAM classifier, (C) and both pathological staging and the PAM classifier. For single-sample PAM classification, prior (expected) six-year recurrence probabilities were set as 30% for all cases. Class predictions with a >90% probability were scored.
Combining independent pathological staging and expression-based classification improved prediction of recurrence risk with broad separation into three groups of patients with different outcomes (Fig. 2C): (i) A good prognosis group consisting of stage B patients with stage A-like cancers showing a six-year disease-free survival probability of 96.5% (95% CI 90.1 – 100.0%); (ii) an intermediate prognosis group comprising stage B patients with stage D-like cancers and stage C patients with stage A-like cancers showing probabilities of 73.0% (95% CI 60.4 – 88.2%) and 77.1% (95% CI 62.2 – 95.7%), respectively; and (iii) a poor prognosis group of stage C patients with stage D-like cancers showing a probability of 47.9% (95% CI 34.7 – 66.1%).
Uni- and multivariate analyses
The prognostic value of our classifier was compared to clinical variables including patient age at diagnosis, the number of lymph nodes examined, N stage and adjuvant treatment using univariate Cox proportional-hazards regression analysis. T stage was not included as the majority of stage B (78 of 82) and stage C (65 of 77) cancers were of stage T3 (Table 2). For both stage B and C patients with “confident” class predictions (n=82 and n=77, respectively), our 128-gene classifier was the strongest predictor of outcome (Table 3). In stage B patients, adjuvant treatment was the only other clinical variable reaching statistical significance (P=0.042, Wald test). Stage B patients receiving adjuvant treatment showed a higher risk of six-year recurrence as compared to those who did not (HR=3.23, 95% CI=1.04–10.00), consistent with such therapy being offered specifically to selected high-risk individuals. In stage C patients, only N stage reached statistical significance besides the classifier (P=0.044, Wald-test), with N2 patients showing an increased risk of six-year recurrence as compared to N1 patients (HR=2.18, 95% CI=1.02–4.66).
Table 3.
Uni- and multivariate Cox proportional-hazard analysis of the risk of recurrence as a first event in 82 stage B and 77 stage C colorectal cancer patients as well as 55 stage C patients with N1 disease for whom confident PAM class predictions (stage A-like/good-prognosis or stage D-like/poor-prognosis) could be made using our 128-gene classifier.
For single-sample PAM classification, prior (expected) six-year recurrence probabilities were set as 20% for stage B and 40% for stage C patients based on relapse rates observed in clinical practice (21). Class predictions with a >90% probability were scored.
The P values were calculated using the Wald test.
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| Patients | Variable | Hazard ratio (95% CI) |
P value |
Hazard ratio (95% CI) |
P value |
|
Stage B (N=82) |
Classifier (Stage D- like vs stage A-like) |
10.60 (1.36–82.00) | 0.024 | 8.55 (1.07–68.61) | 0.043 |
| Age at diagnosis / 10 years |
0.83 (0.53–1.29) | 0.406 | 0.95 (0.54–1.65) | 0.850 | |
| LN examined / 5 | 0.74 (0.44–1.24) | 0.252 | 0.72 (0.40–1.29) | 0.280 | |
| Adjuvant treatment (Yes vs No) |
3.23 (1.04–10.00) | 0.042 | 5.77 (1.53–21.69) | 0.010 | |
|
Stage C (N=77) |
Classifier (Stage D- like vs stage A-like) |
2.85 (1.08–7.53) | 0.035 | 2.49 (0.90–6.94) | 0.080 |
| Age at diagnosis / 10 years |
0.98 (0.75–1.29) | 0.903 | 0.97 (0.71–1.34) | 0.870 | |
| N stage (2 vs 1) | 2.18 (1.02–4.66) | 0.044 | 2.18 (0.91–5.22) | 0.079 | |
| LN examined / 5 | 0.93 (0.76–1.14) | 0.495 | 0.85 (0.67–1.10) | 0.210 | |
| Adjuvant treatment (Yes vs No) |
0.47 (0.22–1.02) | 0.055 | 0.34 (0.14–0.80) | 0.014 | |
|
Stage C & N1 (N=55) |
Classifier (Stage D- like vs stage A-like) |
3.81 (1.07–13.5) | 0.038 | 3.67 (1.02–13.21) | 0.047 |
| Age at diagnosis / 10 years |
0.99 (0.67–1.46) | 0.960 | 0.94 (0.60–1.50) | 0.810 | |
| LN examined / 5 | 0.91 (0.66–1.27) | 0.590 | 0.93 (0.66–1.29) | 0.650 | |
| Adjuvant treatment (Yes vs No) |
0.72 (0.25–2.14) | 0.565 | 0.60 (0.17–2.05) | 0.410 | |
Assessment of whether the classifier was an independent factor predicting CRC prognosis was performed against all clinical variables (Table 3). The classifier was an independent predictor of six-year disease-free survival for stage B patients (P=0.043, Wald test) and showed a corresponding trend for stage C patients (P=0.080, Wald test). The decrease in the prognostic value of our classifier in the multivariate analysis for stage C patients was probably largely due to the observed positive association between class prediction and node status (Table 2). Accordingly, when analysis of stage C patients was limited to individuals with N1 disease, our classifier was an independent predictor of outcome (P=0.047, Wald test).
Classifier validation on an external dataset
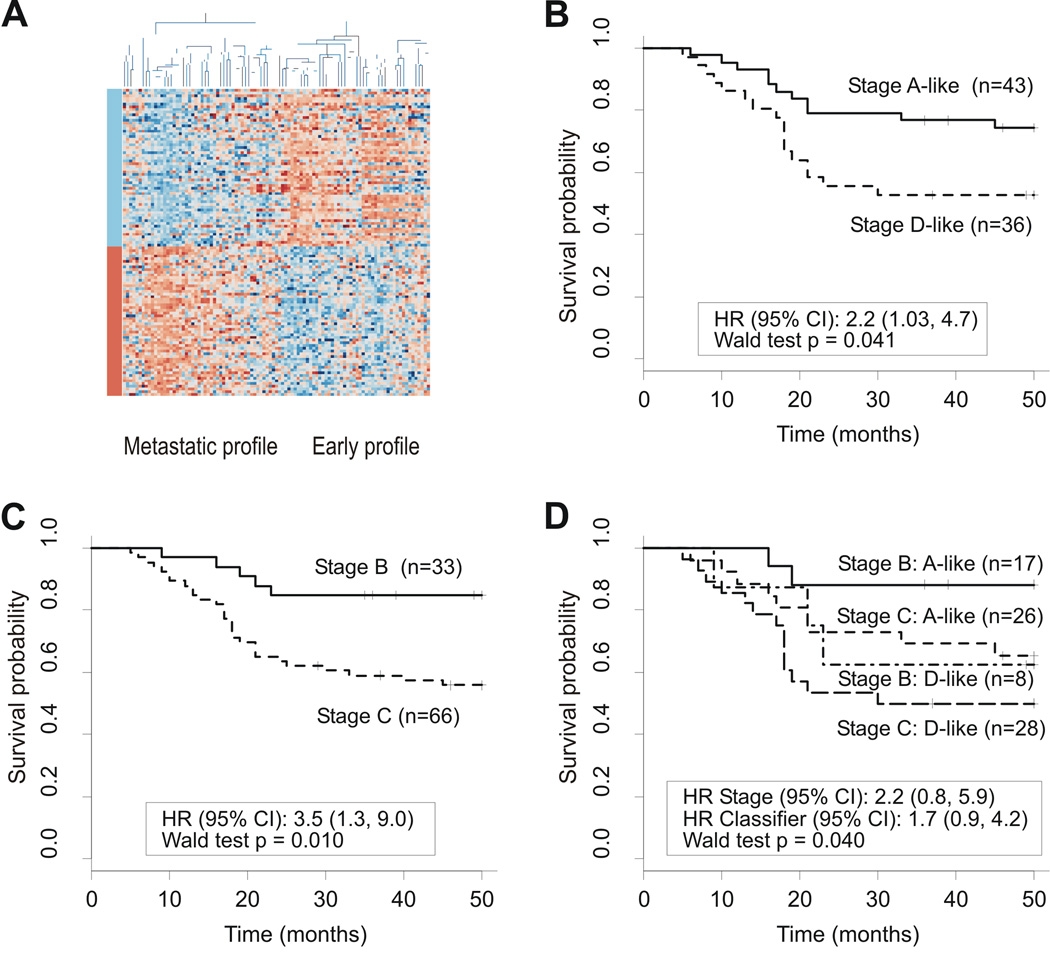
We identified an independent Danish colon cancer dataset comprising 33 Dukes’ stage B and 66 stage C patients. As these data were produced on HG-U133A rather than HG-U133plus2.0 GeneChip arrays (Affymetrix), our classifier was reduced from 163 to 113 available probe sets. Using this restricted gene signature, unsupervised clustering was found to divide these patients into the two expected groups showing median gene expression differences corresponding to those between early-stage and metastatic cancers (Fig. 3); again, more than 99% of changes were consistent (P<0.001, chi-squared test; details not shown). Single-sample PAM classification against our reference set of primary stage A and D cancers successfully divided patients into stage A-like/good prognosis and stage D-like/poor prognosis types based on overall survival (P=0.041, Wald test). When analysed by stage, the 113-gene classifier subdivided both Dukes stage B and C patients into good and poor prognosis groups.
Fig 3.
External prognosis classifier validation (Danish dataset). (A) Unsupervised clustering using metastasis-associated genes represented on HG-U133A GeneChip arrays. Samples are arranged along the x-axis and genes along the y-axis. Each square represents the expression level of a given gene in an individual sample. Orange represents increased expression and blue represents decreased expression relative to the mean- and sample-centered scaled expression of the gene across the samples. Genes are grouped into those found to be down-regulated (blue) and up-regulated (orange) in metastatic cancers as compared to early-stage cancers as represented by the color bars. The two main groups resulting from clustering show early-stage and metastatic profiles as indicated. (B) Survival curves generated using PAM classification show a significant difference in outcome. Class predictions with a >90% probability were scored. (C) Survival curves using Dukes’ staging criteria show a significant difference in outcome. (D) Survival curves grouped by both Dukes’ stage and molecular signature show that both stage B and C patients can be further subdivided into good and poor prognosis groups.
Assessment of prognostic value for individual classifier genes
To assess whether specific classifier genes were of particular prognostic value in our stage B and C patients, we performed Cox proportional-hazards regression analysis for individual probe sets adjusted for adjuvant treatment (Supplementary Table S4). As anticipated in both stage B and C patients, hazard ratios for probe sets up-regulated in metastatic cancers tended to be greater than one (81 of 89 (91.0%) and 82 of 89 (92.1%), respectively), whereas hazard ratios for probe sets down-regulated in metastatic cancers tended to be less than one (68 of 74 (91.9%) and 60 of 74 (81.1%), respectively). However, individual hazard ratios were statistically significant at an unadjusted P value of <0.05 for only a small proportion of probe sets in either stage B (28.2%, 46 of 163) or stage C (14.7%, 24 of 163) patients; only 10 probe sets, representing the VAT1, AKAP12, DCBLD2, WWTR1, ZNF532, IGJ, CTA-246H3.1, L06101, IGL@ and IGLJ3 genes, were significant for both stages. For consistent genes, hazard ratios ranged from 0.59 to 0.84 for down-regulated and 1.53 to 2.66 for up-regulated probe sets, lower than for the combined 128-gene classifier. When adjusting P values for multiple testing, expression of only one probe set, representing DCBLD2, remained significantly associated with outcome in stage B patients.
Functional clusters for classifier genes
For our 128-gene classifier, functional category enrichment analysis identified three significant GO annotation clusters, immune response, extracellular matrix (ECM) interaction and developmental process (Supplementary Table S5). When the signature was separated into genes showing up- or down-regulation in metastatic cancers as compared to early-stage cancers, the ECM interaction and developmental process clusters were found to specifically represent up-regulated genes. The ECM signature was further evident for a separate analysis of KEGG pathways (22), showing significant over-representation of genes for the ECM-receptor interaction (04512hsa) and focal adhesion (04510hsa) pathways. In contrast, the immune response cluster specifically represented down-regulated genes.
Validation of the immune response signature
To validate the observed association between downregulation of putative immune response genes and poor CRC prognosis, we assessed whether tumor infiltration with mononuclear chronic inflammatory cells predicted outcomes in 155 stage B and 166 stage C patients enrolled in the VICTOR clinical trial (14). Scores of average inflammatory cell density were concordant between two independent observers for 77% of cancers (kappa statistic 0.53; 95% CI=0.33–0.63) (23). Excluding samples with discordant scores, low density of mononuclear chronic inflammatory cells was significantly associated with poor recurrence-free survival (HR=2.00, 95% CI= 1.17–3.41; P=0.011, Wald test) over a six-year follow-up period when adjusted for patient age at diagnosis, tumor stage, adjuvant therapy and rofecoxib treatment.
Discussion
Molecular markers that predict CRC recurrence are required to improve the selection of therapies for individual patients. We hypothesized that gene expression differences between early-stage and metastatic cancers might predict recurrence for patients with intermediate stages of disease. Using three cohorts of early-stage and metastatic CRCs from multiple-sites, we identified 128 genes reproducibly associated with metastatic spread. The feasibility of using this signature for prediction of metastatic potential in stage B and C cancers was demonstrated using unsupervised clustering of five independent cohorts; all separated into two groups showing expression profiles corresponding to those observed for early-stage and metastatic lesions. An algorithm for single-sample classification was developed, which permitted scoring of individual test cases against a defined reference set of primary stage A and D cancers. As anticipated, intermediate-stage patients with stage D-like cancers showed a significantly worse prognosis than those with stage A-like cancers.
Controversy exists as to the benefit and use of adjuvant chemotherapy in stage B patients (24, 25). Our 128-gene classifier appeared to be a strong independent predictor of outcome in these patients. The difference in prognosis observed for expression-based classification in our patients was clinically significant, with an adjusted hazard ratio for recurrence in individuals with stage D-like cancers of 8.5 (95% CI, 1.1 – 68.6) for a six-year follow-up period. These results would justify a modification in the approach to adjuvant therapy. Low-risk patients could be reassured and not offered adjuvant treatment, whereas the most effective adjuvant therapy should be considered for high-risk patients.
Stage C patients are routinely offered adjuvant chemotherapy, but despite treatment approximately 40% of individuals relapse (3). Our classifier again identified subgroups with different outcomes: Firstly, it broadly distinguished between patients with different node status, with ~37% of stage D-like and ~14% of stage A-like tumours presenting with N2 disease. Secondly, for patients with N1 disease, our classifier was found to be an independent prognostic factor in multivariate analysis with an adjusted hazard ratio for recurrence in individuals with stage D-like cancers of 3.6 (95% CI, 1.02–13.2). Similar to N2 patients, N1 patients with stage D-like cancers showed particularly poor outcomes indicating a need for treatment with more aggressive regimes or with newly emerging targeted therapies.
Subsets of our 128 classifier genes appeared to represent three putative biological functions as indicated by functional category enrichment analysis; immune response, ECM interaction and cell signaling. Notably, genes suggested to belong to the same functional category showed consistent changes in gene expression between early-stage and metastatic lesions. Putative immune response genes, comprising multiple immunoglobulins (IGHA1, IGHG1, IGHM, IGH@, IGJ, IGKC, IGK@, IGL@, IGLJ3), chemokines (CCL20, CCL28, CXCL13) and proteasome genes (PSMB10, PSMB8, PSMB9), were down-regulated in metastatic/poor prognosis cancers, suggesting a role of the immune response in modulating CRC outcome. This potential association was supported by our systematic assessment of tumor infiltration with mononuclear chronic inflammatory cells in a large independent cohort of stage B and C patients enrolled in the VICTOR clinical trial. Consistent with our data, general enrichment of immune response genes has been reported for gene expression classifiers constructed by two previous microarray studies (5, 9), and poor survival from CRC has been associated with reduced numbers of tumor-infiltrating lymphocytes (26–30).
In contrast, genes up-regulated in metastatic cancers appeared to represent two broad functional categories, ECM interaction and cell signaling. Evidence for the former group was particularly strong, with multiple members identified from the ECM-receptor interaction KEGG pathways including integrins (ITGB1, ITGB5), collagen (COL5A1), fibronectin 1 (FN1), and secreted phosphoprotein 1 (SPP1). Notably, up-regulation of SPP1 has been noted and confirmed by previous microarray studies and shown to be associated with tumor progression, invasion and metastasis in multiple solid cancers including CRC (31–33). Up-regulated cell signaling genes appeared to represent a number of pathways believed to drive cancer progression and metastasis including the TGF-beta pathway through TGFB3 and latent TGF-beta binding protein 3 (LTBP3), the VEGF pathway through neuropilin 2 (NRP2) and fms-like tyrosine kinase 1 (FLT1), and the Wnt pathway through dapper homolog 1 (DACT1). Further validation and study of these metastasis-associate genes should inform our understanding of disease progression.
Previous studies have identified gene expression signatures for CRC prognosis by analyzing patients selected for good and poor outcomes, followed by signature validation in additional cases (5–9). Our approach was markedly different from this strategy, in that gene expression differences between early-stage and metastatic CRCs were evaluated as prognostic markers for patients with intermediate stages of disease. A number of previous studies had limited sample sizes (5, 6, 8) and solely focused on stage B or stage C patients (5, 6, 8). The analyses by Eschrich et al (7) and Lin et al (9) did comprise various stages of CRC, but did not adjust for adjuvant treatment, an important modifier of outcome. Importantly, several studies did not formally assess the performance of a single defined classifier in independent test samples, but rather assessed the validity of a set of candidate prognostic genes using cross-validation procedures (6, 8, 9). Our analysis of microarray data on 553 CRCs represents the largest multi-site study to date in which a single defined prognostic classifier was developed and subsequently evaluated in independent sets of both stage B and stage C patients. Furthermore, classifier validation was formally carried out using a prediction algorithm designed for single-sample classification.
Our classifier showed limited direct overlap with previously reported prognosis signatures (5–9). Overlapping genes included an ADAM metallopeptidase (ADAMTS12) (5), Kruppel-like factor 4 (KLF4) (6), SPP1 (7), discoidin (DCBLD2) (7), DACT1 (7), chloride intracellular channel 4 (CLIC4) (7), and PDZ binding kinase (PBK) (9). This may be due to multiple potential inter-study differences, including sample processing, microarray platforms, patient selection and the analytical tools used for signature discovery. Prospective classifier validation, and ultimately clinical application, will require adherence to standardized analysis protocols.
In summary, our results demonstrate that metastasis-associated gene expression changes can be used to refine traditional outcome prediction, providing a rational approach for tailoring treatments to subsets of patients. The gene expression changes accompanying the acquisition of metastatic potential by the primary tumor appear to reflect both changes in endogenous transcription and changes in the tumor microenvironment such as immune cells. Genes overexpressed in high-risk cancers are potential targets for the development of new anti-cancer drugs to prevent the development of metastatic disease.
Supplementary Material
Acknowledgments
The authors thank the Victorian Cancer BioBank and Biogrid Australia for the provision of specimens and clinical data.
Financial Support: Supported by National Cancer Institute grant R01-CA112215-01A2 (to T.J. Yeatman), the Jeannik M. Littlefield-AACR Grant in Metastatic Colon Cancer Research (to L. Lipton, P. Gibbs, O.M. Sieber), the CSIRO Preventative Health Flagship (to L. Lipton, P. Gibbs, O.M. Sieber) and the Hilton Ludwig Cancer Metastasis Initiative (to L. Lipton, P. Gibbs, O.M. Sieber). L. Lipton is supported by the Victorian Government through a Victorian Cancer Agency Clinical Researcher Fellowship.
Footnotes
References
- 1.Obrand DI, Gordon PH. Incidence and patterns of recurrence following curative resection for colorectal carcinoma. Diseases of the colon and rectum. 1997;40:15–24. doi: 10.1007/BF02055676. [DOI] [PubMed] [Google Scholar]
- 2.Benson AB, 3rd, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 3.Andre T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. The New England journal of medicine. 2004;350:2343–2351. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 4.Kuebler JP, Wieand HS, O'Connell MJ, et al. Oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: results from NSABP C-07. J Clin Oncol. 2007;25:2198–2204. doi: 10.1200/JCO.2006.08.2974. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Jatkoe T, Zhang Y, et al. Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol. 2004;22:1564–1571. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 6.Arango D, Laiho P, Kokko A, et al. Gene-expression profiling predicts recurrence in Dukes' C colorectal cancer. Gastroenterology. 2005;129:874–884. doi: 10.1053/j.gastro.2005.06.066. [DOI] [PubMed] [Google Scholar]
- 7.Eschrich S, Yang I, Bloom G, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23:3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 8.Barrier A, Boelle PY, Roser F, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol. 2006;24:4685–4691. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 9.Lin YH, Friederichs J, Black MA, et al. Multiple gene expression classifiers from different array platforms predict poor prognosis of colorectal cancer. Clin Cancer Res. 2007;13:498–507. doi: 10.1158/1078-0432.CCR-05-2734. [DOI] [PubMed] [Google Scholar]
- 10.Ki DH, Jeung HC, Park CH, et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. International journal of cancer. 2007;121:2005–2012. doi: 10.1002/ijc.22975. [DOI] [PubMed] [Google Scholar]
- 11.Grade M, Hormann P, Becker S, et al. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer research. 2007;67:41–56. doi: 10.1158/0008-5472.CAN-06-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamasaki M, Takemasa I, Komori T, et al. The gene expression profile represents the molecular nature of liver metastasis in colorectal cancer. International journal of oncology. 2007;30:129–138. [PubMed] [Google Scholar]
- 13.Fritzmann J, Morkel M, Besser D, et al. A Colorectal Cancer Expression Profile That Includes Transforming Growth Factor beta Inhibitor BAMBI Predicts Metastatic Potential. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 14.Pendlebury S, Duchesne F, Reed KA, Smith JL, Kerr DJ. A trial of adjuvant therapy in colorectal cancer: the VICTOR trial. Clin Colorectal Cancer. 2003;3:58–60. doi: 10.3816/CCC.2003.n.013. [DOI] [PubMed] [Google Scholar]
- 15.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Statist. 1996;5:299–314. [Google Scholar]
- 16.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics (Oxford, England) 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 18.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiser MR, Landmann RG, Kattan MW, et al. Individualized prediction of colon cancer recurrence using a nomogram. J Clin Oncol. 2008;26:380–385. doi: 10.1200/JCO.2007.14.1291. [DOI] [PubMed] [Google Scholar]
- 22.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic acids research. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Family medicine. 2005;37:360–363. [PubMed] [Google Scholar]
- 24.Quasar Collaborative G, Gray R, Barnwell J, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 25.Figueredo A, Coombes ME, Mukherjee S. Adjuvant therapy for completely resected stage II colon cancer. Cochrane database of systematic reviews (Online) 2008 doi: 10.1002/14651858.CD005390.pub2. CD005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zlobec I, Minoo P, Baumhoer D, et al. Multimarker phenotype predicts adverse survival in patients with lymph node-negative colorectal cancer. Cancer. 2008;112:495–502. doi: 10.1002/cncr.23208. [DOI] [PubMed] [Google Scholar]
- 27.House AK, Watt AG. Survival and the immune response in patients with carcinoma of the colorectum. Gut. 1979;20:868–874. doi: 10.1136/gut.20.10.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prall F, Duhrkop T, Weirich V, et al. Prognostic role of CD8+ tumor-infiltrating lymphocytes in stage III colorectal cancer with and without microsatellite instability. Human pathology. 2004;35:808–816. doi: 10.1016/j.humpath.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 29.Pages F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. The New England journal of medicine. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 30.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science (New York, NY. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 31.Thalmann GN, Sikes RA, Devoll RE, et al. Osteopontin: possible role in prostate cancer progression. Clin Cancer Res. 1999;5:2271–2277. [PubMed] [Google Scholar]
- 32.Agrawal D, Chen T, Irby R, et al. Osteopontin identified as lead marker of colon cancer progression, using pooled sample expression profiling. Journal of the National Cancer Institute. 2002;94:513–521. doi: 10.1093/jnci/94.7.513. [DOI] [PubMed] [Google Scholar]
- 33.Coppola D, Szabo M, Boulware D, et al. Correlation of osteopontin protein expression and pathological stage across a wide variety of tumor histologies. Clin Cancer Res. 2004;10:184–190. doi: 10.1158/1078-0432.ccr-1405-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.