Abstract
The chloroplasts of Euglena gracilis bounded by three membranes arose via secondary endosymbiosis of a green alga in a heterotrophic euglenozoan host. Many genes were transferred from symbiont to the host nucleus. A subset of Euglena nuclear genes of predominately symbiont, but also host, or other origin have obtained complex presequences required for chloroplast targeting. This study has revealed the presence of short introns (41–93 bp) either in the second half of presequence-encoding regions or shortly downstream of them in nine nucleus-encoded E. gracilis genes for chloroplast proteins (Eno29, GapA, PetA, PetF, PetJ, PsaF, PsbM, PsbO, and PsbW). In addition, the E. gracilis Pbgd gene contains two introns in the second half of presequence-encoding region and one at the border of presequence-mature peptide-encoding region. Ten of 12 introns present within presequence-encoding regions or shortly downstream of them identified in this study have typical eukaryotic GT/AG borders, are T-rich, 45–50 bp long, and pairwise sequence identities range from 27 to 61%. Thus single recombination events might have been mediated via these cis-spliced introns. A double crossing over between these cis-spliced introns and trans-spliced introns present in 5′-UTRs of Euglena nuclear genes is also likely to have occurred. Thus introns and exon-shuffling could have had an important role in the acquisition of chloroplast targeting signals in E. gracilis. The results are consistent with a late origin of photosynthetic euglenids.
Keywords: exon-shuffling, chloroplast-targeting, presequence, secondary endosymbiosis
1. Introduction
Euglena gracilis belongs to the order Euglenida, the protist phylum Euglenozoa, and the eukaryotic supergroup Excavata. The phylum Euglenozoa includes also the orders Kinetoplastida (including suborders Trypanosomatina and Bodonina) and Diplonemida. The monophyly of Euglenozoa has been suggested based on various common morphological features, e.g. discoidal mitochondrial cristae and a characteristic feeding apparatus,1,2 and on molecular phylogenies.3 Moreover, Euglenozoa share the presence of the modified base ‘J’ in the nuclear DNA.4 There is little evidence for the presence of signalling pathways regulating nuclear gene expression at the transcriptional level.5,6 The addition of non-coding capped spliced-leaders to nuclear pre-mRNAs via trans-splicing is also common among Euglenozoa.7–12
Euglena gracilis and other phototrophic euglenids possess chloroplasts surrounded by three membranes.13 These arose by a secondary endosymbiotic event in which an euglenozoan host engulfed a green alga.14–16 Chlorarachniophytes (belonging to the supergroup Rhizaria) possess complex green plastids with four envelope membranes and nucleomorph, obtained via an independent secondary symbiosis.17 While plastids of euglenids descended from a prasinophyte, chlorarachniophyte plastids most likely descended from an ulvophyte green algal endosymbiont.18
Many Euglena nuclear genes, mostly of symbiont (i.e. resulting from endosymbiotic gene transfer from the nucleus of the primary host cell to the nucleus of the secondary host cell), but also of host or other origin have acquired presequences for chloroplast targeting. Most presequences required for chloroplast import in Euglena are tripartite, comprising in order: N-terminal signal peptide for targeting to ER, the S/T-rich region resembling transit peptides of organisms possessing primary plastids, and the stop-transfer sequence serving as a membrane anchor (class I proteins, comprising also thylakoid-lumen-targeted class IB proteins possessing an additional hydrophobic thylakoid transfer domain).19–22 Therefore, the major part of the protein precursor stays ‘outside’ while passing through ER, Golgi apparatus, and membrane vesicles prior to their fusion with the outermost chloroplast membrane.19–21 A recent in-depth analysis of E. gracilis presequences revealed another set, the class II of nucleus-encoded plastid protein precursors.22 These lack the putative stop-transfer sequence and possess only a signal sequence at the N-terminus, followed by a transit-peptide-like sequence.22
The complete sequence of the E. gracilis chloroplast genome disclosed an unusually high number of introns: groups II and III introns, and even twintrons (introns within introns).23 However, little is known about introns in nuclear genes of euglenids, as only few genomic sequences from euglenids are available. Introns in the E. gracilis Lhcbm1 gene (according to the nomenclature of Koziol and Durnford,24 encoding light-harvesting chlorophyll a/b binding protein of photosystem II), RbcS genes (encoding small subunit of RuBisCo), and GapC (encoding cytosolic glyceraldehyde-3-phosphate dehydrogenase) do not possess consensus splicing borders (5′-GT/AG-3′) and structural characteristics of group I and II introns, and many of them are flanked by short direct repeats.25–27 These introns can form secondary structure, which could potentially bring together 5′- and 3′-ends, probably without the involvement of spliceosomes.25–28 However, E. gracilis contains also canonical introns, e.g. the 16 introns of the TubC genes (two gene copies encoding gamma-tubulin)28 or the introns in the fibrillarin gene.29,30 The 5′-ends of these introns can potentially base pair with U1 snRNA, suggesting that they are excised in a spliceosome-dependent manner.29 Introns with GT/AG borders are present also in the beta-tubulin gene of the non-photosynthetic euglenoid flagellate Entosiphon sulcatum.9 Furthermore, introns in E. gracilis TubA and TubB genes (encoding alpha- and beta-tubulin, respectively) are of conventional as well as of non-conventional type.28
Recombination events and exon-shuffling have been discussed by various authors as possibly involved in the addition of sequences encoding transit peptides (mitochondrial targeting signals) to nuclear genes for mitochondrial proteins.31–34 In an analogous manner, sequences encoding stroma-targeting peptides might have been added to nucleus-encoded genes for chloroplast proteins in organisms (Archaeplastida) possessing primary chloroplasts of cyanobacterial origin. Such exon-shuffling could occur via recombination processes mediated by introns. However, the identification of introns originally involved in exon-shuffling is problematic for nuclear genes encoding mitochondrial proteins, and for nuclear genes for proteins targeted to primary chloroplasts. The mitochondria arose via an alpha-proteobacterial endosymbiosis, which perhaps dates back to the origin of eukaryotes,35,36 and the cyanobacterial ancestry of primary plastids dates back to the origin of the Archaeplastida.37,38 Since then many intron integration/excision events occurred in various lineages39,40 making it almost impossible to identify introns, which were ancestrally involved in the acquisition of transit peptides. However, the secondary chloroplasts are the results of relatively recent endosymbioses of red and green algae in eukaryotic hosts (for reviews see refs 41–45). It has been suggested that recombination processes might have led to addition of presequences (or at least their parts) to nuclear genes for chloroplast proteins in organisms possessing secondary plastids.46,47 Perhaps the best evidence so far for the involvement of recombination processes mediated by introns in the acquisition of presequences and/or their parts came from the study of Kilian and Kroth48 which revealed the presence of a single intron either within the presequence region or shortly downstream of it in seven nucleus-encoded genes for plastid proteins (AtpC, FbaC1, PetJ, PsbM, PsbO, PsbU and Tpt1) in the diatom Phaeodactylum tricornutum possessing four-membrane-bounded plastids of red algal origin. In this study, we decided to extend this hypothesis to the flagellate E. gracilis possessing secondary chloroplasts of green algal origin.
2. Materials and methods
Euglena gracilis (Pringsheim strain Z, SAG 1224–5/25 Collection of Algae, Göttingen, Germany) was cultivated in 100 ml Erlenmeyer flasks containing 50 ml of a modified Cramer and Myers medium49 supplemented with ethanol (0.8%) and adjusted to pH 6.9. Medium was inoculated with 5 × 104 cells per ml. Cells were grown at 27°C with continuous illumination (30 μmol photons m−2 s−1). Cultures in the exponential growth phase were used for DNA isolation.
The protocol for genomic DNA isolation was used as described in the chapter 2.3.1. (Preparation of Genomic DNA from Plant Tissue) of Current Protocols in Molecular Biology50 with following modification: cells were harvested by centrifugation at 1000 × g (3 min), then washed twice with ice-cold ddH2O, and resuspended with buffer (100 mM Tris–Cl, pH 8; 100 mM EDTA, pH 8; 250 mM NaCl) containing 8 μl of proteinase K (Merck, 20 mg/ml) per 1 ml of buffer. 20% N-lauroylsarcosine (Sigma) was added and the mixture was incubated in waterbath at 55°C for 1 h. After the steps of extractions, centrifugation (6000 × g, 30 min, 4°C), DNA precipitation (2-propanol), centrifugation (7500 × g, 15 min, 4°C) and solubilization (TE buffer, pH 8), RNA was removed (RNase A, 15 min). Thereafter, phenol:chloroform (1:1) and chloroform:isoamylalcohol (24:1) extractions were performed each followed by centrifugation (7500 × g, 7 min). One-tenth volume of 3 M sodium acetate (pH 5.2) was added to the top phase, and DNA was precipitated with 96% ethanol at −20°C, centrifuged (8000 × g, 15 min, 4°C) and washed (70% ethanol). DNA was resuspended in the TE buffer (pH 8).
Primers were derived from six E. gracilis nuclear mRNA sequences encoding chloroplast proteins. Table 1 contains the accession numbers of these mRNAs (see refs 19, 26, 51–54) and the corresponding positions of primer sequences. Another four pairs of primers were derived from four E. gracilis nuclear EST sequences (see ref. 22) encoding chloroplast proteins: PetF (ferredoxin), PsaF subunit of photosystem I, and the PsbM and PsbW subunits of photosystem II. All these four ESTs possessed SL-leader sequence (TTTTTTTCG) at the 5′-end, and were used in previous analysis of presequences of E. gracilis.22 Table 2 contains the e-values, accession numbers of these ESTs used for the design of primers, and the positions corresponding to primer sequences in these ESTs.
Table 1.
List of primers derived from E. gracilis mRNA sequences
| mRNAa | Referenceb | Accession numberc | Forward primersd | Reverse primerse |
|---|---|---|---|---|
| Eno29 | 51 | AJ272112 | 65–84 | 298–279 |
| GapA | 26 | L21904 | 23–42 | 468–449 |
| Pbgd | 52 | X15743 | 183–202 | 511–492 |
| PetA | 53 | AF443625 | 49–68 | 422–403 |
| PetJ | 19 | AJ130725 | 89–108 | 386–367 |
| PsbO | 54 | D14702 | 40–59 | 674–655 |
aPrimers were derived from mRNA sequences of nucleus-encoded genes (Eno29, GapA, Pbgd, PetA, PetJ and PsbO) for chloroplast proteins (enolase, glyceraldehyde-3-phosphate dehydrogenase, porphobilinogen deaminase, cytochrome f, cytochrome c6, and 30 kDa protein of the oxygen-evolving complex, respectively).
bNumber of reference in the reference list in which the corresponding mRNA was characterized.
cAccesion numbers of mRNAs.
d,eThe numbers of primers correspond to the positions in mRNA sequences that can be found under the accession numbers (accession number) listed in the third column. For example, forward primer 65–84 (first row, fourth column) is identical to positions 65–84 of Eno29 mRNA sequence, which can be found under accession number AJ272112 and reverse primers 298–279 (first row, fifth column) is complementary to the sequence 279–298 of Eno29 mRNA.
Table 2.
List of primers derived from E. gracilis EST sequences
| EST producta | Accession numberb | Organism with the best BLASTX hit | E-value | Forward primersc | Reverse primersd |
|---|---|---|---|---|---|
| PetF | EG565162 | Euglena viridis | 100E−40 | 57–76 | 627–608 |
| PsaF | EG565174 | Chlamydomonas reinhardtii | 900E−36 | 30–49 | 499–480 |
| PsbM | EG565161 | Ostreococcus tauri | 700E−11 | 85–104 | 575–556 |
| PsbW | EG565140 | Bigellowiella natans | 300E−14 | 68–86 | 560–543 |
aThe name of protein product of ESTs from which primers were derived. PetF is plastid-targeted ferredoxin, PsaF is subunit F of photosystem I, and PsbM and PsbW are subunits M and W of photosystem II.
bAccession numbers of ESTs.
c,dThe numbers of primers correspond to the positions in corresponding ESTs. E.g. forward primer 57–76 (first row, fifth column) is identical to positions 57–76 of EST with accession number EG56162, and reverse primer 627–608 (first row, sixth column) is complementary to nucleotides 627–608 of this EST.
Primers were designed using Primer-BLAST (primer 3 and BLAST) to obtain similar melting temperature (60°C) for all primers. The effort was made to design primers such as to be able to amplify the whole presequence-encoding region and short part downstream of it (or as long part of this region as possible following our stringent primer design criteria).
The PCRs were performed in 50-µl reaction volume with the final concentration of Mg2+, primers and dNTPs as 2 mM, 0.2 µM and 0.5 mM, respectively. 100 ng of total E. gracilis DNA and 2.5 Units of Taq DNA polymerase (Invitrogen) were used per reaction. Samples were denatured by heating for 5 min at 94°C, subjected to 34 cycles of 30 s denaturation at 94°C, 1 min annealing at 58°C, and 2 min extension at 72°C, and a final cycle of 8 min at 72°C. PCR products were visualized on 1.5% agarose gels (TAE), purified using QIAquick PCR Purification Kit (Qiagene), and sequenced twice (using forward as well as reverse primers) using ABI 3130xl Genetic Analyzer (Applied Biosystems) and the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) according to suppliers’ protocols. The services of the Department of Molecular Biology (Faculty of Natural Sciences, Comenius University, Bratislava, Slovakia) were used for sequencing of PCR products.
The sequence data were analyzed using Chromas, BLAST and CLUSTAL W. Sequence identity of the intron sequences was computed by the global alignment (Needle tool from the EMBOSS suite with the default settings).55 Since the introns have unusual nucleotide composition, which may have inflated the scores, the statistical significance of each alignment score was computed by a permutation test. For each pair of introns, 100 000 random permutations of their bases were aligned, and the empirical distribution of scores was computed. Sequences were permuted by Shuffleseq from the EMBOSS suite,55 and the consensus splice sites (GT/AG) were kept in their original position in each permutation.
3. Results and discussion
The PCR products amplified using all primers were listed in Tables 1 and 2 (except those for Pbgd, PsbO, and PsbW) and total E. gracilis DNA as a template were about 50 bp longer than those expected for cDNA templates. In the cases of Pbgd, PsbO, and PsbW, PCR products were about 150, 90, and 250 bp longer, respectively.
Sequencing of seven PCR products revealed that each contained one 41–50 bp intron. The Pbgd PCR product contained three introns (48, 46, and 50 bp), the PsbO PCR product contained one 93 bp intron, and the PsbW PCR product contained two introns (48 and 195 bp). It is noteworthy, that the 195 bp psbW-i2 intron is present downstream of the stop codon in the 3′-UTR of PsbW gene. Thus the total number of introns identified in this study was 13. Except for the PsbO, PetJ, and the intron present in PsbW 3′-UTR, all introns are 45–50 bp in size and contain typical eukaryotic GT/AG consensus splicing borders (see Table 3 which also includes the accession numbers of the partial gene sequences containing introns identified in this study).
Table 3.
Introns in E. gracilis nucleus-encoded genes for chloroplast proteins identified in this study
| Intron | Accession number | Length | Borders | Percent AT | Percent T | Nucleotide position | Phase |
|---|---|---|---|---|---|---|---|
| eno29-i1 | GQ925702 | 48 | GT/AG | 62.50 | 37.50 | 166 | 1 |
| gapA-i1 | GQ925704 | 50 | GT/AG | 54.00 | 44.00 | 243 | 0 |
| pbgd-i1 | GQ925705 | 48 | GT/AG | 60.42 | 33.33 | 276 | 1 |
| pbgd-i2 | GQ925705 | 46 | GT/AG | 67.39 | 45.65 | 377 | 0 |
| pbgd-i3 | GQ925705 | 50 | GT/AG | 68.00 | 46.00 | 462 | 1 |
| petA-i1 | GQ925706 | 45 | GT/AG | 64.44 | 37.78 | 261 | 0 |
| petF-i1 | GQ925703 | 46 | GT/AG | 63.04 | 47.83 | 423 | 1 |
| petJ-i1 | GQ925707 | 41 | GT/TC or TT/CG | 63.41 | 34.15 | 304 or 305 | 1 or 2 |
| psaF-i1 | GQ925708 | 47 | GT/AG | 72.34 | 44.68 | 307 | 0 |
| psbM-i1 | GQ925709 | 47 | GT/AG | 55.32 | 38.30 | 411 | 1 |
| psbO-i1 | GQ925710 | 93 | ? | 68.82 | 44.09 | 517, 518 or 519 | ? |
| psbW-i1 | GQ925711 | 48 | GT/AG | 64.58 | 39.58 | 198 | 0 |
| psbW-i2 | GQ925711 | 195 | GA/GT | 54.36 | 31.79 | 505 | 1 |
All introns, except for psbW-i2, are present either in the presequence-encoding regions or shortly upstream of them. The table includes accession numbers of partial gene sequences containing corresponding introns, intron length (in nucleotides), intron borders, AT- and T-content of introns, and intron phase. Nt position is the position downstream of which the intron is inserted into the corresponding mRNA or EST sequence (for the accession numbers of mRNAs and ESTs see Tables 1 and 2).
It was impossible to determine borders and phase of the 93 bp-long intron in the PsbO gene, because it does not contain consensus borders, and TG sequence is present in mRNA, but also on both intron–exon borders. The splicing borders of intron in PsbO may be TG/TT, GA/TT or AC/TG. Similar problems with the determination of intron borders have been described for the Lhcb gene-encoding LHCPII protein,25 and for GapC-encoding cytosolic glyceraldehyde-3-phosphate dehydrogenase,26 because the introns in these genes are flanked by short direct repeats (2–5 bp) and do not possess consensus splicing borders. The 41 bp intron in the PetJ gene also does not show consensus splicing borders. A guanine nucleotide is present on both its intron–exon borders, thus its splicing borders might be GT/TC or TT/CG, and it is either in phase 1 or 2.
With the exception of the 195 bp intron in PsbW (psbW-i2, with GA/GT splicing borders and no direct repeat on intron–exon borders), all introns identified in this study are present either within the second half of the presequence-encoding region or shortly downstream of it. The 48 bp intron in Eno29 gene (eno29-i1) is present between the amino acid positions 166 and 167 of Eno29 mRNA (accession number AJ272112), while presequence-encoding region ends with the position 161 of this mRNA sequence. The 50 bp gapA-i1 was localized between the codons for aa 90 and 91 of the 127 aa GapA presequence. Sharif et al.51 reported a 139 aa presequence for Pbgd, whereas Durnford and Gray22 predicted a length of 151 aa. The C-terminus of the presequence region accounts for the difference between these two studies: the 48 bp pbgd-i1 is present in the codon for aa 85, the 46 bp pbgd-i2 is inserted between the codons for aa 119 and 120, and the 50 bp pbgd-i3 localizes to the codon for aa 144 of Pbgd preprotein (i.e. either within the end of the presequence region or shortly downstream of it). The 45 bp petA-i1 is present downstream of the codon 87 of the 147 aa presequence region. The 46 bp petF-i1 was found to be inserted into the codon-specifying aa 131 of the 138 aa presequence region of the EST-encoding ferredoxin (accession number EG565162). The 41 bp petJ-i1 localizes downstream of either nt 304 or 305 of PetJ partial mRNA sequence (accession number AJ130725), with nt 267 representing the end of the presequence-encoding region. The predicted 144 aa PsaF presequence harbours the 47 bp psaF-i1 downstream of codon 94. The 47 bp psbM-i1 is inserted into codon 131 of the PsbM presequence region (predicted to 154 aa). The 93 bp psbO-i1 was identified about 60 nt downstream of the PsbO presequence-encoding region. Finally, the 48 bp intron psbW-i1 localizes between the codons 66 and 67 of the predicted 82 aa PsbW presequence.
Taken together, in this study, 13 new intron sequences present in E. gracilis nuclear genes encoding chloroplast proteins have been described. In genes encoding chloroplast-targeted proteins Eno29, GapA, PetA, PetF, PetJ, PsaF, PsbO, PsbM and PsbW, one intron has been identified within the second half of presequence-encoding region or shortly downstream of it, while in gene encoding Pbgd, two introns were identified within the presequence and one at the presequence-mature peptide border encoding region. Importantly, the BLAST search revealed no significant primary sequence similarity of the introns identified in this study to either introns present in the E. gracilis chloroplast genome, or to any introns from other organisms in public databases.
Ten of 13 introns identified in this study are conventional, and are 45–50 bp long. Introns of similar size (44–53 bp) have been already described in some other E. gracilis nuclear genes, while some of them are conventional.26–29 The only shorter introns in euglenoid species known so far are three introns (27, 29 and 31 bp-long) present in hsp90 gene of the phagotrophic euglenid Peranema trichoforum.56 Of these, only one is conventional. Nevertheless, it should be mentioned that E. gracilis introns can widely vary in size,25–30 and the largest one identified so far is the conventional intron i1 (9.2 kb) in one of the two copies of the gamma-tubulin gene.28
Interestingly, the E. gracilis nuclear gene encoding chloroplast protein RbcS also contains an intron within the second half of presequence region. The size of this intron is 53 bp, it is in phase 0, but does not possess GT/AG borders.27 In the nuclear gene Lhcb (Lhcbm1), a 86 bp intron roughly separates presequence and mature peptide coding regions.25 This intron is also non-conventional, and it is impossible to determine its phase due to TG dinucleotide present on both intron–exon borders.25 Likewise, the 93 bp intron in the PsbO presequence is also flanked by TG dinucleotide and shares 46% primary sequence identity with the 86 bp intron in Lhcb.
Importantly, 10 of 14 E. gracilis introns known to be present in the second half of presequence-encoding regions or shortly downstream of them share various characteristic features: the length (45–50 bp), consensus GT/AG splicing borders, they are AT- and especially T-rich, and possess characteristic pyrimidine tracks at the 3′-ends. Moreover, the primary sequence identity of each two of these 10 introns ranges from 27 to 61% (Table 4). Notably, the 44 and 46 bp introns of conventional type present in the E. gracilis fibrillarin gene29 share 58% primary sequence identity, and the primary sequence identity of these 2 introns and 10 45–50 bp introns found in this study ranges from 32 to 55% (Table 4). Although not all alignment scores are statistically significant (Table 4), the sequence similarity together with other characteristics of these 44–50 bp E. gracilis introns suggests that recombination events between these introns can potentionally occur. In comparison, conventional introns present within or shortly downstream of presequence regions of nuclear-encoded plastid proteins from the diatom Phaeodactylum tricornutum are 183–410 bp long and their pairwise sequence comparison did not reveal significant sequence similarity.48
Table 4.
Primary sequence identity (top-right half) and alignment P-value (bottom-left half) of the selected introns from E. gracilis (44–50 bp long, with consensus GT/AG borders)
| eno29-i1 | gapA-i1 | pbgd-i1 | pbgd-i2 | pbgd-i3 | petA-i1 | petF-i1 | psaF-i1 | psbM-i1 | psbW-i1 | nop1p-i1 | nop1p-i3 (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| eno29-i1 | 45% | 60% | 27% | 42% | 36% | 61% | 56% | 55% | 47% | 55% | 42% | |
| gapA-i1 | 0.07 | 46% | 37% | 52% | 49% | 39% | 54% | 55% | 45% | 44% | 37% | |
| pbgd-i1 | 0.01 | 0.39 | 55% | 40% | 43% | 51% | 53% | 49% | 42% | 46% | 32% | |
| pbgd-i2 | 0.32 | 0.35 | 0.20 | 55% | 33% | 53% | 46% | 29% | 47% | 41% | 41% | |
| pbgd-i3 | 0.22 | 0.03 | 0.93 | 0.23 | 42% | 47% | 50% | 37% | 53% | 37% | 47% | |
| petA-i1 | 0.20 | 0.02 | 0.25 | 0.27 | 0.25 | 38% | 56% | 34% | 46% | 49% | 38% | |
| petF-i1 | 0.01 | 0.21 | 0.15 | 0.02 | 0.90 | 0.63 | 44% | 55% | 46% | 48% | 47% | |
| psaF-i1 | 0.33 | 0.15 | 0.05 | 0.74 | 0.13 | 0.03 | 0.05 | 38% | 40% | 46% | 39% | |
| psbM-i1 | 0.03 | 0.09 | 0.16 | 0.83 | 0.37 | 0.28 | 0.11 | 0.91 | 47% | 50% | 39% | |
| psbW-i1 | 0.01 | 0.06 | 0.20 | 0.34 | 0.05 | 0.24 | 0.26 | 0.07 | 0.27 | 50% | 46% | |
| nop1p-i1 | 0.04 | 0.00 | 0.26 | 0.44 | 0.83 | 0.28 | 0.36 | 0.02 | 0.01 | 0.35 | 58% | |
| nop1p-i3 | 0.38 | 0.31 | 0.34 | 0.95 | 0.58 | 0.65 | 0.23 | 0.62 | 0.25 | 0.93 | 0.04 |
Except for nop1p-i1 and nop1p-i3 (introns present in the gene-encoding nucleolar protein fibrillarin), all these introns are present either in the presequence-encoding regions or shortly upstream of them. The primary sequence identity was calculated as the number of identical nucleotide oppositions of two introns in a pairwise alignment divided by the length of the alignment. Statistically significant alignments with P-value ≤0.05 are shown in bold (see section Methods).
Kilian and Kroth48 suggested ‘semi-exon shuffling’ as a possible mechanism for the acquisition of presequence parts (e.g. signal peptides) in diatoms. The intron present within the presequence-encoding region of the donor gene might have recombined either with 5′-UTR of acceptor gene or with its transit peptide (likely transferred from the red algal symbiont nucleus to the host nucleus with the acceptor gene), while new 3′-AG intron border in the acceptor gene might have been generated by utilizing random AG nucleotides.48 However, the primary sequence similarity of 10 45–50 bp introns present within or shortly downstream of E. gracilis presequences, and the similarity between the 86 bp intron in the Lhcb and the 93 bp intron in the PsbO presequences, suggest exon-shuffling rather than ‘semi-exon shuffling’ as a likely mechanism for the acquisition of presequences or their parts in E. gracilis.
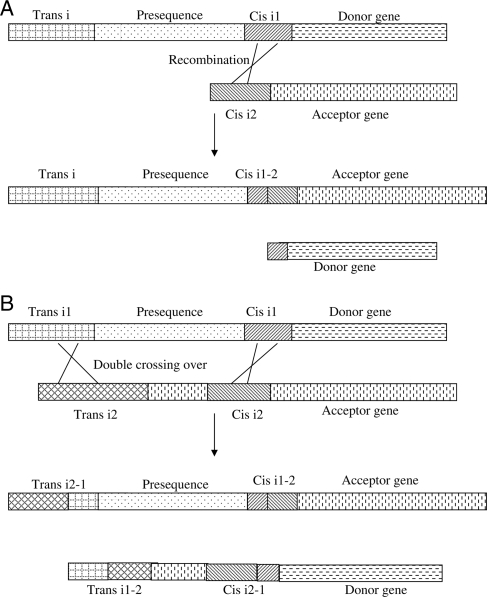
Two possible scenarios for presequence acquisition via exon-shuffling in euglenids are depicted in Fig. 1. The first one includes single recombination events between cis-spliced introns of donor gene (possessing the presequence region) and acceptor gene (Fig. 1A). Importantly, the acceptor may gain not only the presequence region, but also the trans-spliced intron necessary for the addition of capped spliced leader. Another mechanisms for presequence acquisition in E. gracilis involves double crossing over events, one occurring between trans-introns of donor and acceptor gene, and the second involving adjacent cis-spliced introns of donor and acceptor gene (Fig. 1B). The cis-intron in the donor gene in Fig. 1 is placed right at the border of the presequence-mature peptide-encoding region for illustration. However, it could also be present within the presequence-encoding region (most likely in the second half of it). It should be mentioned that the presequence regions of E. gracilis chloroplast precursor proteins have been predicted to vary from 61 to 233 aa,22 and the shortest one currently known (that of Eno29) possibly comprises only 47 aa.51 Thus the addition(s) of shorter parts of presequence region from donor genes to acceptor genes might have resulted in targeting to chloroplasts. In addition, three introns identified in Pbgd gene might represent an example of how the presequence-encoding regions were generated via recombination events mediated by introns.
Figure 1.
Possible mechanisms for the acquisition of presequences in the ancestor of phototrophic euglenids. A single recombination event mediated via cis-spliced introns (Cis i1 and Cis i2) can result in the addition of presequence (or its part) from donor gene to acceptor gene (A). The donor gene would also acquire trans-spliced intron (Trans i) necessary for the addition of capped SL-leader ensuring translation of acceptor gene mRNA. Note that intron Cis i1 is shown to be present exactly at the presequence-mature peptide border encoding region for illustration in (A), but it can be present also within second half of presequence-encoding region or shortly downstream of it. Intron Cis i2 is also shown to be present at 5′-end of the acceptor gene for illustration in (A), but it can be also present within 5′-end of the acceptor gene. Another possible mechanism for the acquisition of chloroplast-targeting signals (B) may involve double crossing over, i.e. the first recombination event occurring between trans-spliced introns Trans i1 and Trans i2, present at the 5′-ends of donor and acceptor gene, respectively, and the second recombination event occurring between cis-spliced intron (Cis i1) present at the presequence-mature peptide border-encoding region of the donor gene and cis-spliced intron (Cis i2) present somewhere within the 5′-end of the protein-coding region of the acceptor gene.
It has once been suggested that euglenids and trypanosomatids might have acquired their plastids prior to their divergence, followed by plastid loss in the trypanosomatid clade.57 However, the cladistic analysis of gene loss inferred from complete plastid genome sequences,58 and the morphological characters shared by eukaryotrophic and phototrophic euglenids but absent from osmotrophic and bacteriotrophic euglenids, and trypanosomes strongly suggest a more recent origin of photosynthetic euglenoids.59,60 The presence of short conventional introns (sharing 27–61% sequence identity) within the second half or shortly downstream of Euglena presequence-encoding regions is indicative of a relatively recent acquisition of chloroplast-targeting signals in Euglena. This is consistent with, and adds additional support for a relatively recent origin of euglenoid secondary plastids, later than the endosymbiosis of the evolutionarily ancient red algae leading to diatoms. Anyway, the repertoire for creating novel targeting sequences or for replacing the transit sequences from the primary host cell by bi- or tripartite presequences did already exist. This applies for the α-proteobacterial endosymbiosis leading to mitochondria and the above-mentioned secondary endosymbiosis leading to chromophytes, respectively: exon-shuffling at the DNA level via appropriately placed introns enabling recombination. Our data suggest that euglenids also made use of this mechanism, probably as the last in a row.
Although nuclear gene sequence data of euglenids are fragmentary, it seems that nuclear genes of euglenids possess many cis-spliced introns. In contrast, wide-scale genome data from parasitic kinetoplastids are available, but very few cis-spliced introns from trypanosomes were reported so far, including a 11 bp intron in the gene for tRNA(tyr) of Trypanosoma cruzi and Trypanosoma brucei,61 and 653 and 302 bp introns in the gene for poly(A) polymerase of T. brucei and T. cruzi, respectively.62 One might argue that almost complete loss of cis-spliced introns in trypanosomes arose through parasitic life style, as did the overall compaction of nuclear genomes of trypanosomes including fairly short intergenic spacers with polycistronic transcription63,64 and overlapping genes.65 However, cis-spliced introns seem to be rare in both parasitic and free-living kinetoplastids, and this general condition could pre-date the adoption of parasitism by the trypanosomatid lineage.66 The euglenid lineage with numerous cis-spliced introns—as opposed to the kinetoplastid lineage—likely was better pre-adapted for the acquisition of chloroplast-targeting presequences, and thus for the successful integration of an algal symbiont.
Funding
This work was supported by grants from the Ministry of Education of the Slovak Republic (VEGA 1/0416/09, to J. K.; and VEGA 1/0118/08, to R. V.), by grants from Comenius University, Bratislava, Slovakia (UK/144/2007, and UK/208/2009 to M. V.), and by grant P19683 from the Austrian ‘Fonds zur Förderung der wissenschaftlichen Forschung’ to W. L.
Footnotes
Edited by Satoshi Tabata
References
- 1.Triemer R.E., Farmer M.A. An ultrastructural comparison of the mitotic apparatus, feeding apparatus, flagellar apparatus and cytoskeleton in euglenoids and kinetoplastids. Protoplasma. 1991;164:91–104. doi:10.1007/BF01320817. [Google Scholar]
- 2.Simpson A.G.B. The identity and composition of the Euglenozoa. Arch. Protistenkd. 1997;148:318–328. [Google Scholar]
- 3.Simpson A.G.B., Roger A.J. Protein phylogenies robustly resolve the deep-level relationships within Euglenozoa. Mol. Phylogenet. Evol. 2004;30:201–212. doi: 10.1016/s1055-7903(03)00177-5. doi:10.1016/S1055-7903(03)00177-5. [DOI] [PubMed] [Google Scholar]
- 4.Dooijes D., Chaves I., Kieft R., Dirks-Mulder A., Martin W., Borst P. Base J originally found in Kinetoplastida is also a minor constituent of nuclear DNA of Euglena gracilis. Nucleic Acids Res. 2000;28:3017–3021. doi: 10.1093/nar/28.16.3017. doi:10.1093/nar/28.16.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koumandou V.L., Natesan S.K.A., Sergeenko T., Field M.C. The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics. 2008;9:298. doi: 10.1186/1471-2164-9-298. doi:10.1186/1471-2164-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vesteg M., Vacula R., Burey S., et al. Expression of nucleus-encoded genes for chloroplast proteins in the flagellate Euglena gracilis. J. Eukaryot. Microbiol. 2009;56:159–166. doi: 10.1111/j.1550-7408.2008.00383.x. doi:10.1111/j.1550-7408.2008.00383.x. [DOI] [PubMed] [Google Scholar]
- 7.Tessier L.H., Keller M., Chan R.L., Fournier R., Weil J.H., Imbault P. Short leader sequences may be transferred from small RNAs to pre-mature mRNAs by trans-splicing in Euglena. EMBO J. 1991;10:2621–2625. doi: 10.1002/j.1460-2075.1991.tb07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonen L. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- 9.Ebel C., Frantz C., Paulus F., Imbault P. Trans-splicing and cis-splicing in the colourless euglenoid, Entosiphon sulcatum. Curr. Genet. 1999;35:542–550. doi: 10.1007/s002940050451. doi:10.1007/s002940050451. [DOI] [PubMed] [Google Scholar]
- 10.Frantz C., Ebel C., Paulus F., Imbault P. Characterization of trans-splicing in Euglenoids. Curr. Genet. 2000;37:349–355. doi: 10.1007/s002940000116. doi:10.1007/s002940000116. [DOI] [PubMed] [Google Scholar]
- 11.Campbell D.A., Thomas S., Sturm N.R. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;4:1231–1240. doi: 10.1016/j.micinf.2003.09.005. doi:10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Liang X.H., Haritan A., Uliel S., Michaeli S. Trans and cis splicing in trypanosomatids: mechanism, factors, and regulation, Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefort-Tran M., Pouphile M., Freyssinet G., Pineau B. Structural and functional significance of the chloroplast envelope of Euglena, immunocytological and freeze fracture study. J. Ultrastruct. Res. 1980;73:44–63. doi: 10.1016/0022-5320(80)90115-x. doi:10.1016/0022-5320(80)90115-X. [DOI] [PubMed] [Google Scholar]
- 14.Gibbs S.P. The chloroplasts of Euglena may have evolved from symbiotic green algae. Can. J. Bot. 1978;56:2883–2889. doi:10.1139/b78-345. [Google Scholar]
- 15.Morden C.W., Delwiche C.F., Kuhsel M., Palmer J.D. Gene phylogenies and the endosymbiotic origin of plastids. Biosystems. 1992;28:75–90. doi: 10.1016/0303-2647(92)90010-v. doi:10.1016/0303-2647(92)90010-V. [DOI] [PubMed] [Google Scholar]
- 16.Ahmadinejad N., Dagan T., Martin W. Genome history in the symbiotic hybrid Euglena gracilis. Gene. 2007;402:35–39. doi: 10.1016/j.gene.2007.07.023. doi:10.1016/j.gene.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 17.Rogers M.B., Gilson P.R., Su V., McFadden G.I., Keeling P.J. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol. Biol. Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. doi:10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- 18.Turmel M., Gagnon M.-C., O'Kelly C.J., Otis C., Lemieux, C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol. Biol. Evol. 2009;26:631–648. doi: 10.1093/molbev/msn285. doi:10.1093/molbev/msn285. [DOI] [PubMed] [Google Scholar]
- 19.Vacula R., Steiner J.M., Krajčovič J., Ebringer L., Löffelhardt W. Nucleus-encoded precursors to thylakoid lumen proteins of Euglena gracilis possess tripartite presequences. DNA Res. 1999;6:45–49. doi: 10.1093/dnares/6.1.45. doi:10.1093/dnares/6.1.45. [DOI] [PubMed] [Google Scholar]
- 20.van Dooren G.G., Schwartzbach S.D., Osafune T., McFadden G.I. Translocation of proteins across multiple membranes of complex plastids. Biochim. Biophys. Acta. 2001;1541:34–53. doi: 10.1016/s0167-4889(01)00154-9. doi:10.1016/S0167-4889(01)00154-9. [DOI] [PubMed] [Google Scholar]
- 21.Sláviková S., Vacula R., Fang Z., Ehara T., Osafune, T., Schwartzbach, S.D. Homologous and heterologous reconstitution of Golgi to chloroplast transport and protein import into the complex chloroplasts of Euglena. J. Cell Sci. 2005;118:1651–1661. doi: 10.1242/jcs.02277. doi:10.1242/jcs.02277. [DOI] [PubMed] [Google Scholar]
- 22.Durnford D.G., Gray M.W. Analysis of Euglena gracilis plastid-targeted proteins reveals different classes of transit sequences. Eukaryot. Cell. 2006;5:2079–2091. doi: 10.1128/EC.00222-06. doi:10.1128/EC.00222-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallick R.B., Hong L., Drager R.G., et al. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 1993;21:3537–3544. doi: 10.1093/nar/21.15.3537. doi:10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koziol G.A., Durnford D.G. Euglena light-harvesting complexes are encoded by multifarious polyprotein mRNAs that evolve in concert. Mol. Biol. Evol. 2008;25:92–100. doi: 10.1093/molbev/msm232. doi:10.1093/molbev/msm232. [DOI] [PubMed] [Google Scholar]
- 25.Muchhal U.S., Schwartzbach S.D. Characterization of the unique intron-exon junctions of Euglena gene(s) encoding the polyprotein precursor to the light-harvesting chlorophyll a/b binding protein of photosystem II. Nucleic Acids Res. 1994;22:5737–5744. doi: 10.1093/nar/22.25.5737. doi:10.1093/nar/22.25.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henze K., Badr A., Wettern M., Cerff R., Martin W. A nuclear gene of eubacterial origin in Euglena gracilis reflects cryptic endosybioses during protist evolution. Proc. Natl. Acad. Sci. U.S.A. 1995;92:9122–9126. doi: 10.1073/pnas.92.20.9122. doi:10.1073/pnas.92.20.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tessier L.H., Paulus F., Keller M., Imbault P. Structure and expression of Euglena gracilis nuclear rbcS genes encoding the small subunits of the ribulose 1,5-bisphosphate carboxylase/oxygenase: A novel splicing process for unusual intervening sequences. J. Mol. Biol. 1995;245:22–33. doi: 10.1006/jmbi.1994.0003. [DOI] [PubMed] [Google Scholar]
- 28.Canaday J., Tessier L.H., Imbault P., Paulus F. Analysis of E. gracilis alpha-, beta- and gamma-tubulin genes: introns and pre-mRNA maturation. Mol. Genet. Genomics. 2001;265:153–160. doi: 10.1007/s004380000403. doi:10.1007/s004380000403. [DOI] [PubMed] [Google Scholar]
- 29.Breckenridge D.G., Watanabe Y.-I., Greenwood S.J., Gray M.W., Schnare M.N. U1 small nuclear RNA and spliceosomal introns in Euglena gracilis. Proc. Natl. Acad. Sci. USA. 1999;96:852–856. doi: 10.1073/pnas.96.3.852. doi:10.1073/pnas.96.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell A.G., Watanabe Y., Charette J.M., Gray M.W. Unusual structure of fibrillarin cDNA and gene structure in Euglena gracilis: evolutionary conservation of core proteins and structural predictions for methylation-guide box C/D snoRNPs throughout the domain Eukarya. Nucl. Acids Res. 2005;33:2731–2791. doi: 10.1093/nar/gki574. doi:10.1093/nar%2Fgki574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wischmann C., Schuster W. Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett. 1995;374:152–156. doi: 10.1016/0014-5793(95)01100-s. doi:10.1016/0014-5793(95)01100-S. [DOI] [PubMed] [Google Scholar]
- 32.Long M., de Souza S.J., Rosenberg C., Gilbert W. Exon shuffling and the origin of the mitochondrial targeting function in plant cytochrome c1 precursor. Proc. Natl. Acad. Sci. USA. 1996;93:7727–7731. doi: 10.1073/pnas.93.15.7727. doi:10.1073/pnas.93.15.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patthy L. Genome evolution and the evolution of exon-shuffling—a review. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. doi:10.1016/S0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- 34.Adams K.L., Daley D.O., Whelan J., Palmer J.D. Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell. 2002;14:931–943. doi: 10.1105/tpc.010483. doi:10.1105/tpc.010483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin W., Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. doi:10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 36.Vesteg M., Krajčovič J. Origin of eukaryotic cells as a symbiosis of parasitic α-proteobacteria in the periplasm of two-membrane-bounded sexual pre-karyotes. Comm. Integr. Biol. 2008;1:104–113. doi: 10.4161/cib.1.1.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McFadden G.I., van Dooren G.G. Evolution: red algal genome affirms a common origin of all plastids. Curr. Biol. 2004;14:R514–R516. doi: 10.1016/j.cub.2004.06.041. doi:10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez-Ezpeleta N., Brinkmann H., Burey S.C., et al. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. doi:10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 39.Jeffares D.C., Mourier T., Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. doi:10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Basu M.K., Rogozin I.B., Deusch O., Dagan T., Martin W., Koonin E.V. Evolutionary dynamics of introns in plastid-derived genes in plants: saturation nearly reached but slow intron gain continues. Mol. Biol. Evol. 2008;25:111–119. doi: 10.1093/molbev/msm234. doi:10.1093/molbev/msm234. [DOI] [PubMed] [Google Scholar]
- 41.Cavalier-Smith T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae) Phil. Trans. R. Soc. Lond. B. 2003;358:109–134. doi: 10.1098/rstb.2002.1194. doi:10.1098/rstb.2002.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer J.D. The symbiotic birth and spread of plastids: How many times and whodunit? J. Phycol. 2003;39:4–11. doi:10.1046/j.1529-8817.2003.02185.x. [Google Scholar]
- 43.Gould S.B., Waller R.F., McFadden G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. doi:10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 44.Keeling P.J. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Eukaryot. Microbiol. 2009;56:1–8. doi: 10.1111/j.1550-7408.2008.00371.x. doi:10.1111/j.1550-7408.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- 45.Vesteg M., Vacula R., Krajčovič J. On the origin of chloroplasts, import mechanisms of chloroplast-targeted proteins, and loss of photosynthetic ability. Folia Microbiol. 2009;54:303–321. doi: 10.1007/s12223-009-0048-z. doi:10.1007/s12223-009-0048-z. [DOI] [PubMed] [Google Scholar]
- 46.Waller R.F., Keeling P.J., Donald R.G.K., et al. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. doi:10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaap D., van Poppel N.F., Vermeulen A.N. Intron invasion in protozoal nuclear encoded plastid genes. Mol. Biochem. Parasitol. 2001;115:119–121. doi: 10.1016/s0166-6851(01)00269-9. doi:10.1016/S0166-6851(01)00269-9. [DOI] [PubMed] [Google Scholar]
- 48.Kilian O., Kroth P.G. Presequence acquisition during secondary endosymbiosis and the possible role of introns. J. Mol. Evol. 2004;58:712–721. doi: 10.1007/s00239-004-2593-z. doi:10.1007/s00239-004-2593-z. [DOI] [PubMed] [Google Scholar]
- 49.Cramer M., Myers J. Growth and photosynthetic characteristics of Euglena gracilis. Arch. Mikrobiol. 1952;17:384–403. doi:10.1007/BF00410835. [Google Scholar]
- 50.Ausubel F.M., Brent R., Kingston R.E., et al. Current Protocols in Molecular Biology. John Wiley & Sons, Inc., Boston, Massachusetts; 1996. [Google Scholar]
- 51.Hannaert V., Brinkmann H., Nowitzki U., et al. Enolase from Trypanosoma brucei, from amitochondriate protist Mastigamoeba batamuthi, and from the chloroplast and cytosol of Euglena gracilis: pieces in the evolutionary puzzle of the eukaryotic glycolytic pathway. Mol. Biol. Evol. 2000;17:989–1000. doi: 10.1093/oxfordjournals.molbev.a026395. [DOI] [PubMed] [Google Scholar]
- 52.Sharif A.L., Smith A.G., Abell C. Isolation and characterization of cDNA clone for a chlorophyll synthesis enzyme from Euglena gracilis. The chloroplast enzyme hydroxymethylbilane synthase (porhobilinogen deaminase) is synthetised with a very long transit peptide in Euglena. Eur. J. Biochem. 1989;184:353–359. doi: 10.1111/j.1432-1033.1989.tb15026.x. doi:10.1111/j.1432-1033.1989.tb15026.x. [DOI] [PubMed] [Google Scholar]
- 53.Santillán-Torres J.L., Ateia A., Claros M.G., Gonzáles-Haplhen D. Cytochrome f and subunit IV, two components of the photosynthetic bf complex typically encoded in the chloroplast genome, are nucleus-encoded in Euglena gracilis. Biochim. Biophys. Acta. 2003;1604:180–189. doi: 10.1016/s0005-2728(03)00058-6. doi:10.1016/S0005-2728(03)00058-6. [DOI] [PubMed] [Google Scholar]
- 54.Shigemori Y., Inagaki J., Mori H., Nishimura M., Takahashi S., Yamamoto Y. The presequence of the precursor to the nucleus-encoded 30 kDa protein of photosystem II in Euglena gracilis Z includes two hydrophobic domains. Plant Mol. Biol. 1994;24:209–215. doi: 10.1007/BF00040587. doi:10.1007/BF00040587. [DOI] [PubMed] [Google Scholar]
- 55.Rice P., Longden I., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. doi:10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 56.Breglia S.A., Slamovits C.H., Leander B.S. Phylogeny of phagotrophic euglenids (Euglenozoa) as inferred from Hsp90 gene sequences. J. Eukaryot. Microbiol. 2007;54:86–92. doi: 10.1111/j.1550-7408.2006.00233.x. doi:10.1111/j.1550-7408.2006.00233.x. [DOI] [PubMed] [Google Scholar]
- 57.Hannaert V., Saavedra E., Duffieux F., et al. Plant-like traits associated with metabolism of Trypanosoma parasites. Proc. Natl. Acad. Sci. USA. 2003;100:1067–1071. doi: 10.1073/pnas.0335769100. doi:10.1073/pnas.0335769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nozaki H., Ohta N., Matsuzaki M., Misumi O., Kuroiwa T. Phylogeny of plastids based on cladistic analysis of gene loss inferred from complete plastid genome sequences. J. Mol. Evol. 2003;57:377–382. doi: 10.1007/s00239-003-2486-6. doi:10.1007/s00239-003-2486-6. [DOI] [PubMed] [Google Scholar]
- 59.Leander B.S., Triemer R.E., Farmer M.A. Character evolution in heterotrophic euglenids. Eur. J. Protistol. 2001;37:337–356. doi:10.1078/0932-4739-00842. [Google Scholar]
- 60.Leander B.S. Did trypanosomatid parasites have photosynthetic ancestors? Trends Microbiol. 2004;12:251–258. doi: 10.1016/j.tim.2004.04.001. doi:10.1016/j.tim.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 61.Schneider A., Martin J., Agabian N. A nuclear encoded tRNA of Trypanosoma brucei is imported into mitochondria. Mol. Cell Biol. 1994;14:2317–2322. doi: 10.1128/mcb.14.4.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mair G., Shi H., Li H., et al. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA. 2000;6:163–169. doi: 10.1017/s135583820099229x. doi:10.1017/S135583820099229X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Myler P., Andleman L., deVos T., et al. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein coding genes. Proc. Natl. Acad. Sci. USA. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. doi:10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horn D. Nuclear gene transcription and chromatin in Trypanosoma brucei. Int. J. Parasit. 2001;31:1157–1165. doi: 10.1016/s0020-7519(01)00264-8. doi:10.1016/S0020-7519(01)00264-8. [DOI] [PubMed] [Google Scholar]
- 65.Liniger M., Bodenmüller K., Pays E., Gallati S., Roditi I. Overlapping sense and antisense transcription units in Trypanosoma brucei. Mol. Microbiol. 2001;40:869–878. doi: 10.1046/j.1365-2958.2001.02426.x. doi:10.1046/j.1365-2958.2001.02426.x. [DOI] [PubMed] [Google Scholar]
- 66.Simpson A.G.B., Lukeš J., Roger A.J. The evolutionary history of kinetoplastids and their kinetoplasts. Mol. Biol. Evol. 2002;19:2071–2083. doi: 10.1093/oxfordjournals.molbev.a004032. [DOI] [PubMed] [Google Scholar]