Abstract
Weedy forms of crop species infest agricultural fields worldwide and are a leading cause of crop losses, yet little is known about how these weeds evolve. Red rice (Oryza sativa), a major weed of cultivated rice fields in the US, is recognized by the dark-pigmented grain that gives it its common name. Studies using neutral molecular markers have indicated a close relationship between US red rice and domesticated rice, suggesting that the weed may have originated through reversion of domesticated rice to a feral form. We have tested this reversion hypothesis by examining molecular variation at Rc, the regulatory gene responsible for grain pigmentation differences between domesticated and wild rice. Loss-of-function mutations at Rc account for the absence of proanthocyanidin pigments in cultivated rice grains, and the major rc domestication allele has been shown to be capable of spontaneous reversion to a functional form through additional mutations at the Rc locus. Using a diverse sample of 156 weedy, domesticated, and wild Oryzas, we analyzed DNA sequence variation at Rc and its surrounding 4 Mb genomic region. We find that reversion of domestication alleles does not account for the pigmented grains of weed accessions; moreover, we find that haplotypes characterizing the weed are either absent or very rare in cultivated rice. Sequences from genomic regions flanking Rc are consistent with a genomic footprint of the rc selective sweep in cultivated rice, and are compatible with a close relationship of red rice to Asian Oryzas that have never been cultivated in the US.
Keywords: crop weed evolution, de-domestication, grain color, red rice, Oryza sativa, proanthocyanidin
Introduction
Red rice is a conspecific weedy relative of cultivated Asian rice (Oryza sativa L.) that occurs in rice fields worldwide. In the southern US, red rice infestations can reduce crop harvests by up to 80% (Estorninos et al. 2005) and are estimated to create crop losses exceeding $45 million annually (Gealy et al. 2002). Control of red rice has proved difficult because of the weed’s morphological and agroecological similarity to cultivated rice. Moreover, the potential for gene flow from domesticated rice into red rice populations threatens the long-term effectiveness of weed control strategies based on herbicide-resistant crop varieties (Rajguru et al. 2005; Shivrain et al. 2007).
A defining feature of red rice is the dark-pigmented pericarp (bran) that gives the weed its common name. Like some other weed-associated traits, such as seed shattering and seed dormancy, pericarp pigmentation is a characteristic of wild Oryza species that was selected against during rice domestication (Sweeney et al. 2006; Sweeney et al. 2007). The vast majority of modern domesticated rice varieties lack proanthocyanidin pigments in the pericarp; this domestication trait arose through human selection for loss-of-function mutations at the Rc locus, which encodes a bHLH regulatory protein in the proanthocyanidin synthesis pathway (Sweeney et al. 2006). Selection for non-pigmented grains in the crop may potentially reflect human aesthetic preferences, selection against seed dormancy, selection for improved taste and cooking qualities, and/or selection for other domestication traits linked to Rc (Gu et al. 2004; Sweeney et al. 2007).
The major rc domestication allele, present in >97% of non-pigmented cultivars (Sweeney et al. 2007), is characterized by a 14-bp frameshift deletion in exon 7 (initially annotated as exon 6; see Sweeney et al. 2006; Furukawa et al. 2007). This mutation generates a truncated, nonfunctional gene product and the nonpigmented (‘white’) pericarp of domesticated rice (Sweeney et al. 2006; Furukawa et al. 2007). An independently evolved domestication allele, Rc-s, is characterized by a C→A substitution in exon 7 that creates a premature stop codon (Sweeney et al. 2006); this allele occurs at a frequency of <3% in rice varieties globally (Sweeney et al. 2007). Analyses of DNA sequence variation in the Rc genomic region by Sweeney et al. (2007) have revealed that the major rc domestication allele arose initially in japonica rice and was subsequently introgressed into other varieties through selective breeding during domestication. The size of this introgressed genomic sequence is <1 Mb in most varieties surveyed to date (Sweeney et al. 2007). In contrast, Rc-s most likely originated in aus rice, a variety-group cultivated in a limited area of the northern Indian subcontinent; dissemination of the Rc-s allele beyond this geographical region has been minimal (Sweeney et al. 2007).
Recently, two studies have documented instances of mutational reversion of the major rc domestication allele to a functional form. Brooks et al. (2008) demonstrated that a spontaneous red-pericarp variant of the US cultivar ‘Wells’ discovered in 2005 arose through a 1-bp deletion located 20-bp upstream of the original 14-bp deletion; this new mutation restores reading frame, protein function, and the proanthocyanidin-pigmented pericarp. Similarly, Lee et al. (2009) have documented a 1-bp deletion located 44 bp upstream of the loss-of-function deletion in a red-pericarp revertant of the Italian cultivar ‘Perla’. Given that two independent instances of rc spontaneous reversion have been confirmed in the four years since Rc was first molecularly characterized (Sweeney et al. 2006; Furukawa et al. 2007; Sweeney et al. 2007), it seems plausible that rc reversions could occur repeatedly during the history of rice cultivation and that this process could account for the emergence of a red-pericarp phenotype in cultivated rice fields.
To date, studies examining the evolutionary origin of red rice have focused on neutral genetic markers, and there is growing evidence from these studies that US weed strains are closely related to Asian domesticated rice. In particular, the two major phenotypic variants within US red rice, ‘blackhull awned’ (BHA) strains and ‘strawhull awnless’ (SH) strains, appear to be more closely related to domesticated rice of the aus and indica variety groups, respectively, than to any other groups within Oryza (Londo & Schaal 2007; Gealy et al. 2010; M. Reagon et al., University of Massachusetts, Amherst, unpublished) (see Figure 1A). Interestingly, neither indica nor aus varieties have ever been grown commercially in the US (Mackill & McKenzie 2003; Moldenhauer et al. 2004; Lu et al. 2005), and as there are no wild Oryza species native to North America, these patterns suggest that US red rice most likely arose through accidental introductions of indica-like and aus-like germplasm from Asia. Cultivated rice in the southern US is exclusively of the tropical japonica variety group (Mackill & McKenzie 2003), which is genetically distinct from indica and aus rice (Garris et al. 2005; Caicedo et al. 2007).
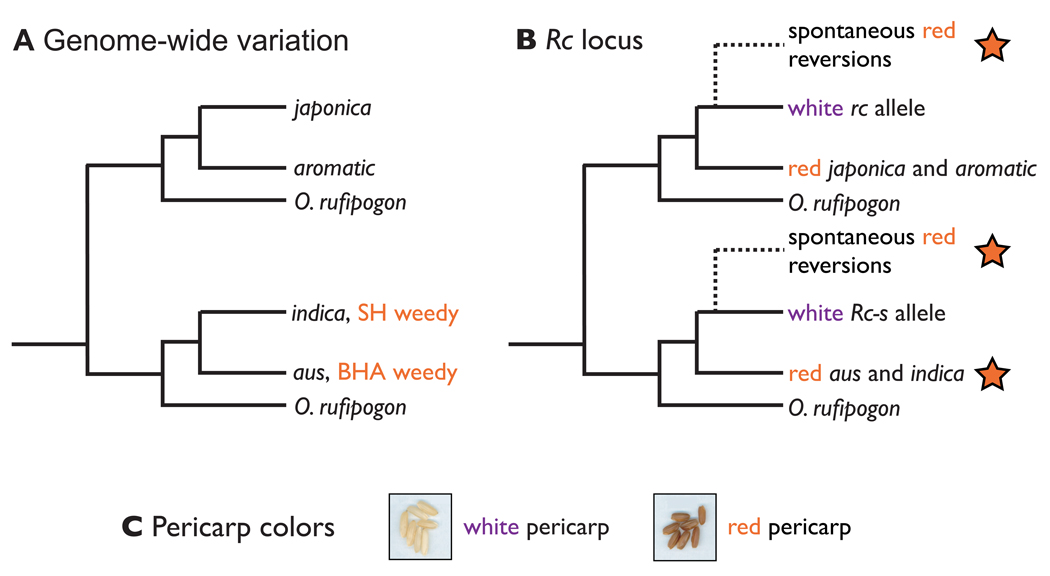
Figure 1. Relationships among Oryza species and varieties based on genome-wide variation and the Rc locus.
Schematic representations of the relationships between different Oryza species and variety groups based on A) genome-wide surveys of genetic variation (Garris et al. 2005; Caicedo et al. 2007) and B) genetic variation at the Rc locus. In B, potential relationships of the US weed haplotypes to cultivated varieties are indicated by red stars. Dotted lines indicate potential origins of functional Rc alleles through reversion from domestication alleles.
While neutral markers indicate a close relationship between US red rice and Asian cultivated varieties, these data do not provide information on whether the US weed strains are derived from Asian domesticates directly, or whether they are instead descended from undomesticated (wild and/or weedy) Oryza populations that are simply related to aus and indica rice. DNA sequences from the pericarp pigmentation gene, Rc, can provide a novel source of information for addressing this question. This gene underlies a phenotype that is nearly fixed in domesticated rice and absent in both wild and weedy Oryzas. Thus, if red rice were descended from white-pericarp crop varieties, the Rc alleles present in the weed would be reversions of domestication alleles back to a functional form, as has been documented in US and European cultivars (Brooks et al. 2008; Lee et al. 2009). Alternatively, if red rice were instead descended from undomesticated material or from the rare landraces that still have pigmented pericarps, the weed’s Rc alleles would be most closely related to alleles that never underwent loss-of-function mutations. These two possibilities need not be mutually exclusive, as there could be more than one evolutionary origin for the red pericarp phenotype present in weed populations (Figure 1B).
In this study, we evaluate patterns of DNA sequence variation at the Rc locus and surrounding genomic regions to determine the origin of the red pericarp characterizing US red rice. We have compared Rc haplotypes in the US weed to variation present in wild and domesticated rice. Our sampling of cultivated varieties includes all major variety groups that form genetically distinct subpopulations within the crop: indica and aus varieties (together composing the indica subspecies); and tropical japonica, temperate japonica, and aromatic varieties (together composing the japonica subspecies) (Garris et al. 2005; Londo et al. 2006; Caicedo et al. 2007). The cultivated O. sativa sampling includes landraces with pigmented pericarps as well as the more typical white-pericarp cultivars. In addition to the Rc locus, we have examined DNA sequence variation at five loci that flank this gene, spanning nearly 2 Mb upstream and 2 Mb downstream of Rc on chromosome 7. This genomic region is expected to extend beyond the region of introgressed japonica sequence carrying the common rc domestication allele in most crop varieties (Sweeney et al. 2007). Taken together, this combination of sampling and loci allows us to evaluate relationships at the Rc locus, where most crop varieties carry the common rc allele, as well as relationships outside of this region of introgression, where patterns of sequence diversity reflect the phylogenetic relationships among weeds and crop varieties. Our data reveal that reversion of domestication alleles has played a minimal role, if any, in the evolution of the US red rice phenotype; that there is minimal sharing of Rc haplotypes between weedy rice and red-pericarp crop landraces; and that Rc haplotypes in the weed are genealogically distinct from those that gave rise to the domestication alleles. Together these findings strongly suggest that US red rice has not originated through reversions of white-pericarp cultivars to a feral form.
Materials and Methods
Samples
Samples included both newly generated DNA sequence data and previously sequenced accessions that are publicly available. For the newly generated DNA sequences, the initial sample set consisted of 138 Oryza accessions from around the world (Table S1); seven accessions were eliminated from the dataset due to poor sequence quality, yielding a final set of 131 accessions. Analyses of the Rc locus also included 25 previously published sequences from cultivated O. sativa that represented accessions not already present in our sample set (Table S1). US red rice was represented by 58 samples from throughout the range of the rice cultivation in the Southern US, including Missouri, Arkansas, Mississippi, Louisiana, and Texas. These accessions were classified into four categories based on morphology (D. Gealy, USDA, Stuttgart, AR, pers. comm.) and a genome-wide survey of sequence variation (M. Reagon et al., University of Massachusetts, Amherst, unpublished). Classifications are as follows: 24 blackhull awned (BHA), 24 strawhull (SH), five accessions that were a rare intermediate brownhull awned morphology (BR), and five accessions identified as crop-weed admixed individuals (MIX) (Table S1). Weed phenotypic descriptions are based on the morphology of the majority of accessions present in each genetically distinct group, as identified from genome-wide sequence data (M. Reagon et al., University of Massachusetts, Amherst, unpublished). All but one of the weedy rice strains have pigmented (red) pericarps; the exception (1D10; Table S1) has been identified as a probable crop-weed hybrid (MIX) based on morphological characterization and genome-wide sequence data (M. Reagon et al., University of Massachusetts, Amherst, unpublished). All weedy accessions were generously provided by Dr. David Gealy from collections maintained by the USDA-ARS Dale Bumpers National Rice Research Center in Stuttgart, Arkansas; these accessions were all propagated through self-fertilization for two or more generations prior to use in the study (D. Gealy, USDA, Stuttgart, AR, pers. comm.).
Domesticated rice was represented by 13 US cultivars and 55 Asian landraces representing the diversity of varieties cultivated outside the southern US (Table 1). The US cultivars were obtained from the USDA-ARS and included eight modern cultivars released after 1930 as well as five historical crop varieties released before 1930, from which most modern US cultivars are derived. US cultivars, both modern and historical, are of the tropical japonica variety group, with extremely limited genetic contributions from temperate japonica and indica varieties at loci controlling traits such as semi-dwarfism and rice blast resistance (Mackill & McKenzie 2003). Sampled crop accessions from outside the US included 18 indica, nine aus, 16 tropical japonica, seven temperate japonica, and five aromatic landraces (Table 1). Oryza rufipogon (including samples labeled as the annual form O. nivara) was represented by 32 accessions from throughout the native range of the species (Table S1). In addition, samples of two other closely related Oryza species were included as potential progenitors of the weedy form and for use as outgroups. These included the Latin American species O. glumaepatula (2 accessions), and the Australasian species O. meridionalis (2 accessions). A portion of the crop accession from outside the US and O. rufipogon samples were kindly provided by Dr. Susan McCouch of Cornell University; others were obtained from collections maintained by the International Rice Research Institute in the Philippines. Samples of all the congeners used as outgroups were obtained from the International Rice Research Institute.
Table 1.
Oryza sativa samples used in Rc analyses. Sample sizes are indicated in parentheses. For white-pericarp accessions, samples are listed by domestication allele type. Details on accessions are provided in Table S1.
| O. sativa subpopulation | Pericarp color | |
|---|---|---|
| Asian domesticated variety groups (68) | Red | White (rc, Rc-s) |
| indica (18) | 3 | 15, 0 |
| aus (9) | 4 | 2, 3 |
| tropical japonica | ||
| — Asian cultivars (16) | 3 | 13, 0 |
| — US cultivars (13) | 0 | 13, 0 |
| temperate japonica (7) | 1 | 6, 0 |
| aromatic (5) | 0 | 4, 1 |
| US weedy rice (57) | ||
| Blackhull awned (BHA) | 24 | — |
| Strawhull awnless (SH) | 24 | — |
| Brownhull (BR) | 5 | — |
| Putative crop-weed hybrids (MIX) | 3 | 1, 0 |
DNA extraction, primer design, and sequencing
Plants were grown from seed in greenhouses at Washington University in St. Louis and the University of Massachusetts, Amherst. Fresh tissue was harvested and frozen in liquid nitrogen; DNA was extracted via a modified CTAB procedure (Gross et al. 2009). Primers were designed to PCR-amplify and sequence the coding region of Rc (6.4 kb) as well as the regions approximately 1.6 kb upstream and 0.8 kb downstream of the start and stop codons. Nineteen overlapping PCR fragments (∼400–600 bp) were used to sequence the region. Primers were designed using Primer3 (Rozen & Skaletsky 2000) and compared against the indica (93–11) and temperate japonica (Nipponbare) reference genome sequences in GenBank to ensure conservation and single-copy status. Primers were also designed to amplify and sequence ∼500 bp portions of five genes in genomic regions upstream and downstream of Rc; targeted loci were approximately 2.1 Mb, 1.1 Mb, and 0.3 Mb upstream, and 0.8 Mb and 2.0 Mb downstream of Rc. Previous research had shown that the length of the selective sweep surrounding the Rc locus was generally around 1 Mb (Sweeney et al. 2007), so these flanking fragments included locations that were likely to be both within and outside of the swept region. Flanking fragment primers were located in exons, with the sequenced regions containing intronic regions of the targeted loci. Targeted loci, their genomic locations, and primers are listed in Table S2 and Table S3.
PCR fragment amplification and Big Dye Terminator sequencing were performed according to conventional methods and separated on ABI capillary sequences by Cogenics™ in Connecticut and Texas. Missing fragments or poor quality data were finished using an ABI 3130 capillary sequencer at the Washington University Biology Departmental core facility. Sequencing techniques were standard and are available on request. One of the flanking fragments (rc3_005) was missing sequences for one aus and two US cultivars due to poor sequence quality; these three accessions were excluded from calculations for this locus and for calculations using concatenated flanking loci (Table S1). GenBank accession numbers for sequenced regions are XXXXXX-XXXXXX.
Data analysis
Sequence editing and alignment were performed using the PHRED, PHRAP, and Polyphred programs (Deborah Nickerson, University of Washington) and BioLign Version 4.0.6 (Tom Hall, North Carolina State University, Raleigh, North Carolina, United States). As O. sativa is predominately self-fertilizing, the majority of crop and weed samples were homozygous at sequenced loci. However, O. rufipogon outcrosses more frequently, and there were some heterozygous O. rufipogon individuals at each locus. Because sequences were not cloned, allelic phase was not known unambiguously, and analytical phasing was not successful for the Rc locus due to recombination within the sequenced region. Thus, all sequences were treated as genotypic, and whenever possible, unphased ambiguity codes were used for data analysis. It was necessary to generate arbitrarily phased ‘pseudohaplotypes’ for input into programs that could not accept ambiguity codes, but in these cases analyses were only conducted if they were not affected by SNP phase. The four weed accessions identified as putative crop-weed hybrids (MIX) (M. Reagon et al., University of Massachusetts, Amherst, unpublished) were excluded from calculations of diversity and divergence to avoid artificially inflating values for the weed, but they were included in the phylogenetic analysis.
Nei’s (1982) γST genetic distance between groups and net sequence divergence (Da; Nei 1987) were calculated as measures of genetic differentiation for the Rc locus and for the flanking regions, both concatenated and separately. Net sequence divergence controls for the amount of within-group variation by subtracting the average within-group diversity from the total divergence between populations. Sequence diversity within groups and values of Tajima’s (1989) D were also calculated for each sequenced region separately. Nucleotide diversity and divergence calculations were completed in DnaSP 5.0 (Librado & Rozas 2009).
Maximum likelihood (ML) analyses were performed for the Rc locus, with the best-fit model of nucleotide substitution selected in jModelTest 0.1.1 (Guindon & Gascuel 2003; Posada 2008) based on likelihood scores for 88 different models and the Akaike information criterion. ML trees were generated in RAxML (Stamatakis 2006b; Stamatakis et al. 2008) via the CIPRES web portal using the GTR model of molecular evolution with rate variation among sites. Because the lengths of Rc sequences from published GenBank accessions were shorter than the 9,576 bp region sequenced for the present study, the aligned Rc dataset for tree construction was 6,676 bp. Rate heterogeneity among sites was estimated using the GTR+CAT approximation, which is a fast computational substitution for the GTR+Γ model that can be applied to large datasets (>50 taxa) (Stamatakis 2006a). Bootstrap values were calculated via 1000 replicates of the dataset, and a consensus tree was generated using Consense from Phylip 3.68 (Felsenstein 2005). Neighbor joining (NJ) analyses based on the same dataset was performed using Phylip 3.68 (Felsenstein 2005). Distances for the neighbor joining analysis was calculated using the F84 model, and bootstrap values were calculated via 1000 replicates of the data.
Results
Distribution of Rc alleles and pericarp pigmentation
Analyses of the Rc locus were based on 156 Oryza accessions, including 125 O. sativa samples (68 cultivated rice accessions and 57 weed strains), 27 accessions of the wild progenitor O. rufipogon, and four accessions from related wild Oryza species used as outgroups (two accessions apiece of O. meridionalis and O. glumaepatula). A summary of O. sativa samples is shown in Table 1 (see also Table S1). Cultivated rice accessions include all five genetically distinct variety groups that are recognized in the crop (Garris et al. 2005; Caicedo et al. 2007), and the tropical japonica variety group is represented by both US and Asian cultivars (Table 1). Red rice accessions include 24 BHA strains, 24 SH strains, 5 samples that show a rare intermediate ‘brownhull awned’ (BR) phenotype, and 4 samples derived from individuals that have been identified as probable crop-weed hybrids (MIX) in morphological characterizations (D. Gealy, USDA, Stuttgart, AR, pers. comm.) and in an analysis of DNA sequences at 48 sequence tagged site (STS) loci (M. Reagon et al., University of Massachusetts, Amherst, unpublished). Together these Oryza samples comprise 131 newly sequenced accessions and 25 previously sequenced crop varieties (Sweeney et al. 2007; Table S1). The aligned length of the newly generated Rc sequences is 9,576 bp, which includes ∼1.6 kb of sequence immediately upstream of the Rc start codon, the transcribed portion of the gene, and ∼0.8 kb downstream of the stop codon.
For the cultivated rice accessions, all white-pericarp varieties carry one of the two previously reported domestication alleles, with the rare Rc-s allele restricted to one aromatic and three aus accessions (Table 1, Table S1). Among the weed strains, a single MIX individual has a white pericarp (accession 1D10, Table S1); this individual carries the common rc domestication allele, consistent with an origin of the phenotype via crop-weed hybridization (followed by one or more generations of selfing; see Methods). The other MIX accessions carry haplotypes that are either identical or nearly identical to the other red rice strains (described below). In contrast to red-pericarp phenotypes that have been reported in US and European crop cultivars (Brooks et al. 2008; Lee et al. 2009), no red-pericarp weed accessions carry haplotypes with the 14-bp deletion characterizing the rc domestication allele, nor are there any other nucleotide substitutions indicating mutational reversion from a non-functional gene copy. Thus, the pericarp pigmentation that characterizes US red rice is not derived through a process of reversion from a domestication allele.
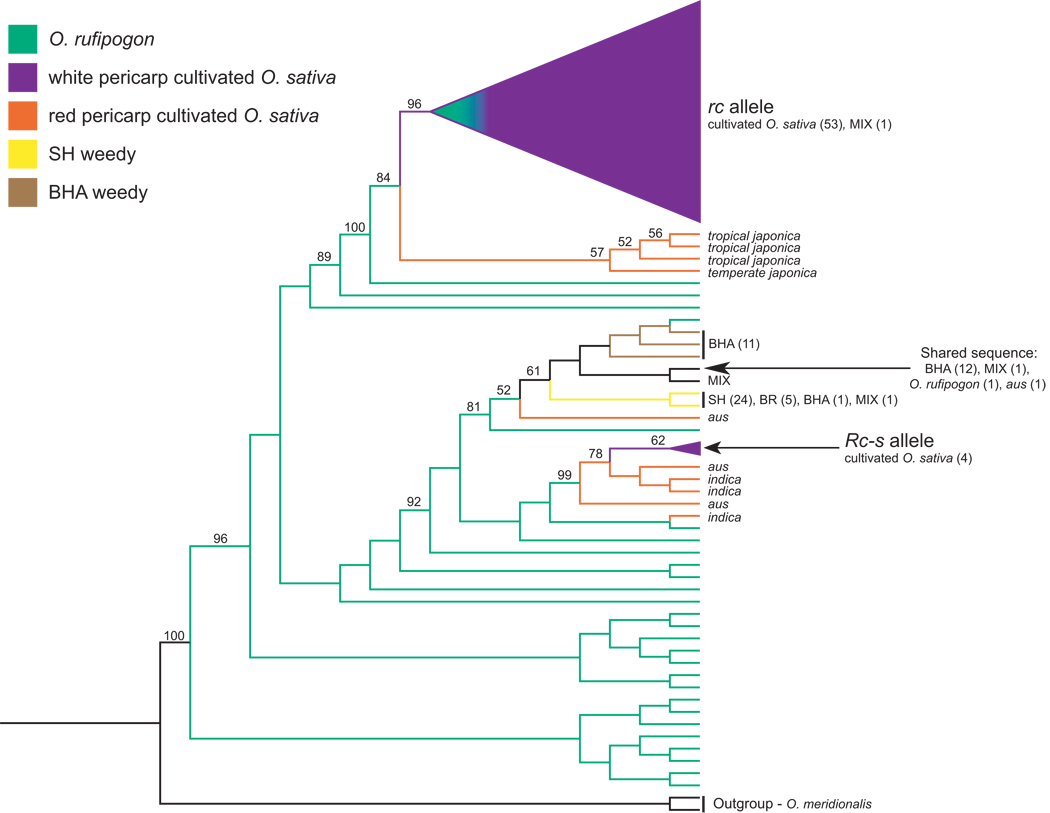
A ML tree of Rc haplotypes is shown in Figure 2. Haplotype relationships are consistent between this tree and a tree generated by NJ analysis (NJ results not shown). For crop accessions, inferred haplotype relationships are concordant with previous analyses (Sweeney et al. 2007); the common rc domestication allele is grouped with haplotypes of red-pericarp japonica landraces (84% bootstrap support), and the Rc-s allele is grouped with red-pericarp aus and indica landraces (99% bootstrap support). Red-pericarp weed accessions are characterized by eight closely related Rc haplotypes, which are grouped into a cluster with O. rufipogon and red-pericarp aus accessions (81% bootstrap support) (Figure 2). Out of these eight weed haplotypes, seven are unique to red rice. A single haplotype, observed in 12 BHA strains and one MIX accession, is also present in one red-pericarp aus landrace and a single O. rufipogon accession (Figure 2). The overall lack of haplotype sharing between weed strains and red-pericarp landraces indicates that the weeds are unlikely to be directly derived from the landraces represented by our sampling. Moreover, the genealogical distance between weed haplotypes and haplotype clusters containing the rc and Rc-s domestication alleles (Figure 2) indicates that the weed alleles are not closely related to those characterizing common cultivated rice varieties. Similarly, the absence of extensive haplotype sharing with O. rufipogon accessions suggests that the weed strains are not directly descended from the populations of this wild species represented by our sampling.
Figure 2. Maximum likelihood (ML) tree of Rc haplotypes.
Haplotypes carrying domestication alleles (rc, Rc-s) are grouped as triangles, with the size of the triangle roughly proportional to the number accessions carrying the allele. Colors indicate the identities of accessions carrying a haplotype, as indicated in the key. The BR and putative crop-weed hybrid (MIX) red rice accessions are not represented by separate colors, but are labeled where they occur on the tree. Numbers following labels indicate the number of individuals sharing a haplotype. Haplotypes represented by only one individual are labeled but not numbered, except for haplotypes unique to O. rufipogon, which are neither labeled nor numbered. Numbers at nodes represent percent bootstrap support; bootstrap values over 50% are shown. For simplicity, only O. meridionalis is used as the outgroup.
Rc nucleotide diversity and population differentiation
Silent site (synonymous and noncoding) nucleotide diversity at Rc across all cultivated O. sativa accessions is π =1.55 per kb (Table 2). For the subset of crop varieties carrying the common rc domestication allele, this value drops to π = 0.03 per kb, with a Tajima’s D value indicating a statistically significant deviation from neutrality (D = −1.9473, p < 0.05; Table 2); these patterns are consistent with the selective sweep previously reported for the domestication allele (Sweeney et al. 2007). This signature of selection is also evident as negative Tajima’s (1989) D values for individual variety groups possessing the rc allele (Table 2), although the pattern is statistically significant only for tropical japonicas (D = −1.9778, p < 0.05), which have the largest sample size (N= 29). For red-pericarp crop landraces, which have not been subject to domestication selection at Rc, the silent site nucleotide diversity is more than 100 times greater than the rc haplotype class (π = 3.14 per kb; Table 2), and is comparable to the genome-wide average for cultivated O. sativa based on a survey of 111 STS loci (π = 3.20 per kb; Caicedo et al. 2007). Silent site nucleotide diversity for the wild progenitor, O. rufipogon, is π = 5.19 per kb (Table 2); this value exactly matches the average level of polymorphism found in a survey of O. rufipogon accessions across 111 STS loci (π = 5.19 per kb; Caicedo et al. 2007).
Table 2.
Values of π, θW, and Tajima's D for total sites and silent sites per 1000 bases at the Rc locus, and average values across flanking fragments. Statistically significant values at p < 0.05 are indicated in bold font.
| Cultivated Oryza sativa |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Oryza rufipogon |
All cultivated |
white- pericarp (rc) |
red- pericarp cultivated |
indica | aus |
temperate japonica |
tropical japonica (All) |
tropical Japonica (US only) |
aromatic | All US weedy ricea |
BHA | SH | ||
| (N=27) | (N=68) | (N=53) | (N=11) | (N=18) | (N=9) | (N=7) | (N=29) | (N=13) | (N=5) | (N=56) | (N=24) | (N=24) | ||
| Rclocus | ||||||||||||||
| π per kb | Total sites | 4.62 | 1.27 | 0.04 | 2.65 | 1.32 | 2.29 | 0.04 | 0.07 | 0.02 | 1.80 | 0.34 | 0.06 | 0.00 |
| π per kb | Silent sites | 5.19 | 1.55 | 0.03 | 3.14 | 1.60 | 2.71 | 0.05 | 0.05 | 0.02 | 2.18 | 0.41 | 0.07 | 0.00 |
| θW per kb | Total sites | 6.58 | 1.68 | 0.19 | 2.33 | 1.73 | 2.01 | 0.06 | 0.22 | 0.04 | 2.16 | 0.17 | 0.18 | 0.00 |
| θW per kb | Silent sites | 7.52 | 1.90 | 0.16 | 2.64 | 2.09 | 2.32 | 0.07 | 0.14 | 0.04 | 2.62 | 0.18 | 0.22 | 0.00 |
| Tajima's D | Total sites | −1.0653 | −0.8099 | −1.9473 | 0.6593 | −0.9714 | 0.7122 | −1.0062 | −1.9778 | −1.1492 | −1.2441 | +2.6015 | −2.0832 | N/Ab |
| Flanking fragment average (Flanking fragment average without rc3_011)c | ||||||||||||||
| π per kb | Total sites | 6.13 | 4.09 | 3.76 | 0.82 | 0.67 | 4.55 | 0.00 | 1.39 | 0.00 | 7.75 | 0.50 | 0.17 | 0.43 |
| (3.25) | (1.95) | (1.71) | (1.03) | (0.83) | (2.48) | (0.00) | (0.64) | (0.00) | (1.68) | (0.62) | (0.21) | (0.53) | ||
| π per kb | Silent sites | 9.31 | 6.21 | 5.76 | 0.97 | 1.09 | 6.73 | 0.00 | 2.09 | 0.00 | 12.32 | 0.50 | 0.12 | 0.76 |
| (4.31) | (2.53) | (2.25) | (1.22) | (1.36) | (3.11) | (0.00) | (0.81) | (0.00) | (2.13) | (0.63) | (0.15) | (0.95) | ||
| θW per kb | Total sites | 6.32 | 3.55 | 3.65 | 0.67 | 0.61 | 3.77 | 0.00 | 2.36 | 0.00 | 7.75 | 0.35 | 0.44 | 0.22 |
| (4.60) | (1.65) | (1.70 | (0.84) | (0.76) | (2.09) | (0.00) | (1.28) | (0.00) | (1.68) | (0.44) | (0.55) | (0.28) | ||
| θW per kb | Silent sites | 9.11 | 5.40 | 5.56 | 0.80 | 1.01 | 5.58 | 0.00 | 3.50 | 0.00 | 12.32 | 0.32 | 0.39 | 0.39 |
| (5.93) | (2.14) | (2.20) | (1.00) | (1.26) | (2.62) | (0.00) | (1.60) | (0.00) | (2.13) | (0.40) | (0.49) | (0.49) | ||
| Tajima's Dd | Total sites | −0.4729 | 0.2903 | −0.0958 | 2.0119 | 0.5058 | 0.7963 | N/A | −1.3118 | N/A | 0.00 | 0.2648 | −1.3268 | N/A |
| (−0.8667) | (0.2521) | (−0.1556) | (2.0119) | (0.5058) | (0.6257) | N/A | (−1.3475) | N/A | (0.00) | (0.2648) | (−1.3268) | (N/A) | ||
Excluding the white-pericarp MIX accession.
N/A, cannot be calculated due to lack of polymorphisms
Average values without locus rc3_011 are indicated in parentheses.
Average across all fragments for which a Tajima's D value could be calculated.
For red-pericarp weed accessions, the silent site nucleotide diversity at Rc is nearly an order of magnitude lower than that of red-pericarp crop landraces (π = 0.41 per kb; Table 2). However, this observed nucleotide diversity is comparable to levels observed for the same set of weed accessions across 48 randomly selected STS loci (π per kb = 1.85 + 3.31 [mean + SD]; M. Reagon et al., University of Massachusetts, Amherst, unpublished). This finding suggests that genetic variation at Rc is consistent with a genome-wide genetic bottleneck in weed populations, most likely associated with their introduction into North America (M. Reagon et al., University of Massachusetts, Amherst, unpublished).
Tajima’s D for red rice is significantly positive (D = 2.6015, p < 0.05; Table 2). This pattern is created by Rc haplotype structure within the weed that closely corresponds to the major phenotypic variants: for haplotypes defined by SNPs only (i.e., excluding indels), weed accessions fall into two haplogroups (differing at six of the seven polymorphic sites), with one group consisting of all BHA strains but one (accession 1E08; Table S1), and the other consisting of all SH and BR strains plus the remaining BHA strain. This pattern is similar to the genome-wide differentiation detected between BHA and SH strains at neutral loci (Londo & Schaal 2007; Gealy et al. 2010; M. Reagon et al., University of Massachusetts, Amherst, unpublished); it is therefore likely to be reflecting historical population structure between the two weed subpopulations, with rare admixture events. The Rc haplotype structure in red rice also accounts for an absence of SNP polymorphism observed within the SH strains (Table 2), as well as a significantly negative Tajima’s (1989) D value for BHA strains (caused by the inclusion of the single divergent 1E08 sequence; Table 2).
In quantifications of population differentiation at the Rc locus, weed strains show the greatest differentiation from japonicas and the other cultivated varieties where the japonica-derived rc allele predominates (aromatics, indicas), and the least differentiation from the aus variety group and O. rufipogon (Table 3). These patterns are consistent between the two differentiation measures employed (γST and Da), and they are similar for both BHA and SH strains. They are also consistent with inferred Rc haplotype relationships, which indicate that the weeds are more closely related to O. rufipogon and aus accessions at this locus than to any other samples in the dataset (Figure 2). These patterns of differentiation at Rc stand in contrast to genome-wide patterns, which have indicated that SH strains are least differentiated from indica varieties (with very low differentiation between them), and that BHA strains are least differentiated from aus varieties and O. rufipogon (with somewhat greater differentiation than is observed between SH weeds and indicas) (Londo & Schaal 2007; M. Reagon et al., University of Massachusetts, Amherst, unpublished). The Rc-specific pattern is directly attributable to selective introgression of the rc allele across cultivated variety groups, as all white-pericarp indicas carry the japonica-derived domestication allele at this locus (Table 1; Figure 2; see also Sweeney et al. 2007).
Table 3.
Percent net sequence divergence (Da) between groups (above diagonal) and γST genetic distance between groups (below diagonal), for A) the Rc locus, and B) the flanking fragments (concatenated sequences). The US weedy group is not compared to BHA or SH weeds, as it comprises those groups.
| A) Rc gene | O. rufipogon | aus | indica | japonica | aromatic | US cultivars |
All US weedy |
BHA | SH | |
|---|---|---|---|---|---|---|---|---|---|---|
| O. rufipogon | 0.087 | 0.153 | 0.178 | 0.191 | 0.190 | 0.161 | 0.168 | 0.180 | ||
| aus | 0.070 | 0.220 | 0.261 | 0.280 | 0.280 | 0.074 | 0.074 | 0.108 | ||
| indica | 0.126 | 0.436 | 0.008 | 0.018 | 0.017 | 0.354 | 0.352 | 0.387 | ||
| japonica | 0.170 | 0.528 | 0.096 | 0.002 | 0.001 | 0.408 | 0.412 | 0.446 | ||
| aromatic | 0.068 | 0.419 | 0.113 | 0.035 | 0.001 | 0.431 | 0.437 | 0.470 | ||
| US cultivars | 0.191 | 0.594 | 0.219 | 0.049 | 0.152 | 0.431 | 0.437 | 0.470 | ||
| US weedy | 0.296 | 0.237 | 0.690 | 0.785 | 0.568 | 0.827 | N/A | N/A | ||
| BHA | 0.256 | 0.366 | 0.856 | 0.929 | 0.931 | 0.976 | N/A | 0.062 | ||
| SH | 0.279 | 0.441 | 0.891 | 0.952 | 0.994 | 0.997 | N/A | 0.907 | ||
|
B) Flanking Fragments |
O. rufipogon | aus | indica | japonica | aromatic | US cultivars |
All US weedy |
BHA | SH | |
| O. rufipogon | 0.179 | 0.128 | 0.474 | 0.269 | 0.654 | 0.155 | 0.195 | 0.143 | ||
| aus | 0.091 | 0.104 | 0.156 | 0.169 | 0.278 | 0.085 | 0.096 | 0.097 | ||
| indica | 0.102 | 0.251 | 0.459 | 0.392 | 0.661 | 0.030 | 0.078 | 0.007 | ||
| japonica | 0.313 | 0.235 | 0.628 | 0.074 | 0.013 | 0.521 | 0.575 | 0.490 | ||
| aromatic | 0.094 | 0.213 | 0.503 | 0.140 | 0.126 | 0.461 | 0.518 | 0.427 | ||
| US cultivars | 0.402 | 0.522 | 0.923 | 0.081 | 0.368 | 0.727 | 0.784 | 0.693 | ||
| US weedy | 0.241 | 0.171 | 0.127 | 0.669 | 0.406 | 0.825 | N/A | N/A | ||
| BHA | 0.245 | 0.299 | 0.527 | 0.777 | 0.627 | 0.968 | N/A | 0.052 | ||
| SH | 0.187 | 0.253 | 0.075 | 0.707 | 0.512 | 0.912 | N/A | 0.475 | ||
Flanking regions
For the set of 131 accessions newly sequenced at Rc, sequences were also generated for portions of five loci distributed up to 2 Mb upstream and 2 Mb downstream of the Rc locus (Figure 3, Table S2). Aligned lengths of DNA sequences range from 419 bp to 505 bp for the five regions. Silent site nucleotide diversity varies widely among these loci (Table S4 and Figure S1); one downstream fragment, rc3_011 (encoding LOC_OS07G14150), shows exceptionally high diversity due to the presence of two divergent haplogroups that transcend species and variety groups in the sample set. Because the diversity at rc3_011 is an order of magnitude higher for some groups compared to the other flanking fragments, average nucleotide diversity measures for flanking fragments were considered both with and without this locus for comparison purposes (Table 2; see also Table S4 and Figure S1).
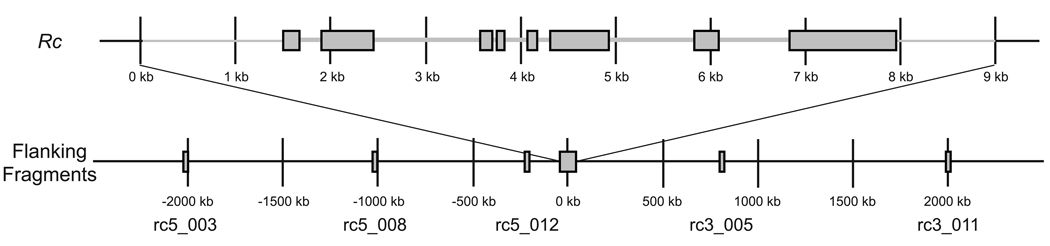
Figure 3. Schematic of sequenced regions.
Locations of the Rc locus with up- and downstream sequenced regions and flanking fragments on chromosome 7. At the Rc locus (top), exons are indicated as rectangles and the length of the sequenced region is shown in light grey. Distance of flanking fragments from the sequenced region of the Rc locus and names of the fragments are indicated below. Distances are approximately to scale, and are based on the Nipponbare reference sequence.
For cultivated O. sativa, average silent site nucleotide diversity across the flanking regions (π = 2.53 per kb; rc3_011 excluded) is similar to the previously reported genome-wide average of 3.20 (Caicedo et al. 2007) (Table 2). This pattern stands in contrast to the low diversity for cultivars at Rc (see above), an indication that the domestication-associated rc selective sweep does not dominate throughout the broader 4 Mb genomic region. Average values of silent site nucleotide diversity for O. rufipogon and US red rice (π = 4.31 and 0.63 per kb, respectively, with rc3_011 excluded; Table 2) are similar to values both at the Rc gene and from genome-wide assessments (see above) (Caicedo et al. 2007; M. Reagon et al., University of Massachusetts, Amherst, unpublished). When rc3_011 is included, values of π are elevated for populations containing both haplogroups (aromatic, aus, tropical japonica cultivated varieties; O. rufipogon), but are roughly unchanged for all other groups (Table 2).
For weed strains, genetic differentiation averaged across the flanking loci matches patterns revealed by genome-wide neutral markers (Londo & Schaal 2007; M. Reagon et al., University of Massachusetts, Amherst, unpublished), with very close genetic similarity between SH strains and indica cultivars, and moderately close similarity between BHA strains and aus varieties plus O. rufipogon. For a dataset consisting of concatenated sequences of the five flanking loci, γST measures (Nei 1982) indicate that BHA is least differentiated from O. rufipogon followed by aus, and that SH is least differentiated from indica followed by O. rufipogon (Table 3). For measures of Da, BHA and SH are collectively least differentiated from indica followed by aus varieties (Table 3). Oryza rufipogon shows low differentiation from both weedy and cultivated O. sativa in these measures; this is as expected, given that both groups are ultimately derived from O. rufipogon and share many polymorphisms with the wild species.
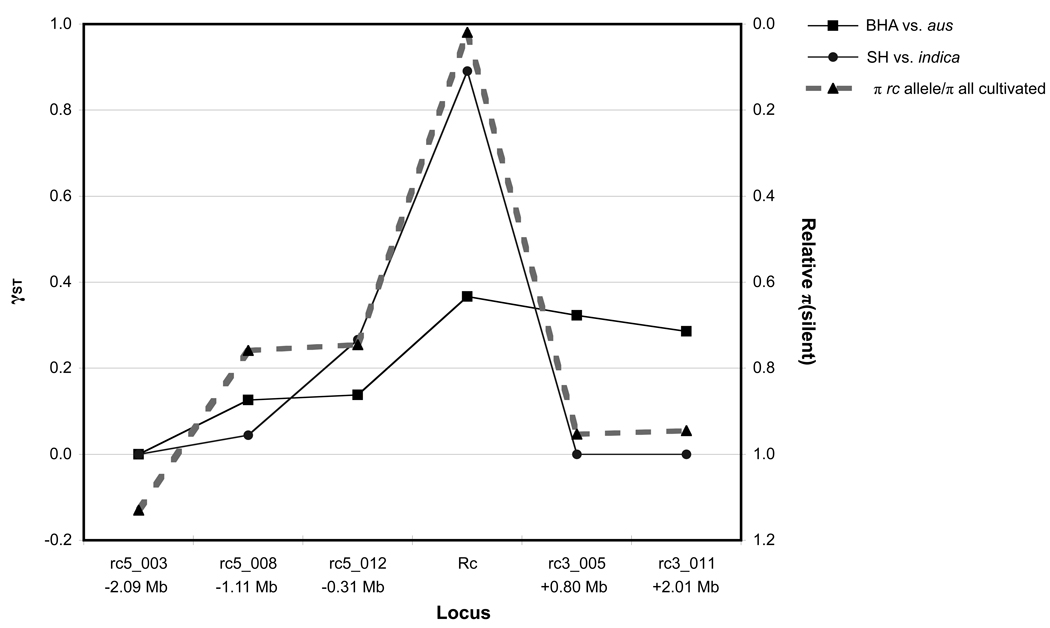
Of the five flanking loci, the nearest upstream locus, rc5_012 (encoding LOC_OS07G10600), is located ∼310 kb away from Rc (Figure 3; Table S2); it is therefore predicted to lie within the genomic region of rc allele introgression for some, but not all, introgressed rc haplotypes (Sweeney et al. 2007). The remaining four flanking loci are predicted to occur outside the ∼1 Mb boundaries of introgression for most rc haplotypes (Sweeney et al. 2007). To assess the extent to which the rc selective sweep affects patterns of weed-crop differentiation across the 4 Mb genomic region, we examined SH-indica and BHA-aus population differentiation at Rc and at each of the flanking loci, and we compared these patterns to the strength of the rc selective sweep at each locus (measured as the proportion of nucleotide diversity in the rc allele class compared to the diversity in all cultivated accessions at that locus) (Figure 4). For indica varieties, where virtually every white-pericarp cultivar carries the introgressed rc allele (Sweeney et al. 2007), the degree of population differentiation from SH weed strains closely matches the genomic footprint of the rc selective sweep: differentiation is highest at Rc, lower at the nearest flanking locus (rc5_012), and lowest at the loci falling outside the region of rc introgression. In contrast, for BHA-aus differentiation, correspondence to the footprint of the rc selective sweep is much less evident (Figure 4). This difference is consistent with the fact that, unlike indicas, most aus varieties in the sample set do not carry an introgressed rc allele (Table 1), so that the genomic impact of the rc selective sweep is lessened in this variety group.
Figure 4. Selection and weed-crop differentiation across the Rc genomic region.
Solid black lines indicate genetic differentiation (γST; left-hand Y axis) between red rice phenotypic variants (BHA and SH) and their closest crop relatives (aus and indica, respectively). The dashed gray line indicates an index of the rc selective sweep in cultivated rice (right-hand Y axis); the index is calculated as silent-site π for crop accessions carrying the rc domestication allele as a proportion of π for all crop accessions at that locus. Numbers below flanking fragment names indicate their approximate location in relation to the Rc locus (see Figure 3). Zero γST values for rc5_003 indicate that all red rice, aus, and indica accessions share an identical sequence at this locus.
Discussion
Rc and the evolutionary origin of red pericarps in US weedy rice
No visual trait so distinguishes US red rice from domesticated rice as the proanthocyanidin-pigmented pericarp that gives the weed its common name. Recent advances in our understanding of the genetic basis of pericarp color in wild and domesticated rice (Sweeney et al. 2006; Furukawa et al. 2007; Sweeney et al. 2007; Brooks et al. 2008; Lee et al. 2009) have now made it possible to examine the genetic basis and origin of this weed-associated trait. Pericarp color is largely controlled by the Rc locus: a functional Rc allele is required for the production of pericarp proanthocyanidins present in wild and weedy Oryzas, and the widespread nonfunctional rc allele results in the white pericarp that characterizes nearly all domesticated rice (Sweeney et al. 2006; Sweeney et al. 2007). If red rice were directly descended from de-domesticated (feral) white-pericarp varieties, the alleles present in the weed would likely be reversions of rc null alleles (Figure 1B). If red rice were instead derived from undomesticated material or from the rare landraces that still have red pericarps, the Rc alleles in the weed would be related to ancestral, functional alleles that did not undergo loss-of-function mutations (Figure 1B). A priori, at least two lines of evidence would seem to favor a de-domestication scenario. First, studies of genome-wide variation in US weeds have consistently shown that both BHA and SH weed forms are closely related to domesticated varieties of O. sativa (aus and indica varieties, respectively; Figure 1A) (Londo & Schaal 2007; Gealy et al. 2010; M. Reagon et al., University of Massachusetts, Amherst, unpublished); this would suggest that the weeds might well be directly descended from the crops with which they co-occur — the vast majority of which have white pericarps. Second, two independent instances of spontaneous rc allele reversion have already been documented in the four years since Rc was characterized at the molecular level (Brooks et al. 2008; Lee et al. 2009), and one of these reversion mutations likely occurred no earlier than 2005 (Brooks et al. 2008). Thus, mutational reversions from the common domestication allele have apparently occurred recurrently in cultivated rice fields.
Nonetheless, our examination of Rc alleles in US red rice and its possible progenitors does not support a crop reversion scenario, for either BHA or SH weeds. With the exception of a single white-pericarp weed accession that is a probable crop-weed hybrid (M. Reagon et al., University of Massachusetts, Amherst, unpublished), no US weeds carry the widespread 14-bp deletion in exon 7, nor do any accessions show polymorphisms consistent with reversion from nonfunctional alleles. Thus, mutational reversion from rc domestication alleles does not account for the weed-associated phenotype in US red rice. It is important to note that this conclusion is for North American weed populations and may not necessarily hold true for all weedy rice populations worldwide. Red rice infests crop fields in virtually all world regions where rice is grown, and, while currently limited, available genetic diversity data suggest that the genetic composition of the weed may vary widely by geographical region (e.g., Cao et al. (2006); reviewed by Gross and Olsen (2009)). Given the apparent ease with which the rc domestication allele can revert to a functional form (Brooks et al. 2008; Lee et al. 2009), it seems likely that this mechanism could account for the red pericarp in at least some weed strains on a global scale. Expanded sampling of weed populations from diverse geographical regions will be useful for testing this hypothesis.
Given that the functional Rc alleles in the US weeds are not derived from white-pericarp crop varieties, what can be determined about their origin? One possibility is that they are derived from crop landraces that still possess red-pigmented pericarps. We included red-pericarp indica, aus, temperate japonica, and tropical japonica landraces in our sampling to evaluate this possibility. With the exception of one weed haplotype that is shared with a single aus accession, most of the Rc haplotype diversity in the weeds is distinct from that characterizing red-pericarp landraces (Figure 2). In the one instance of crop-weed haplotype sharing, the haplotype is also present in the wild species O. rufipogon (Figure 2), suggesting that this may be a case of shared ancestral polymorphism (or potentially hybridization and gene flow from the crop into the O. rufipogon, although this would be of relatively little importance, because the allele is of the ancestral, functional class already found in the wild species). Thus, to the extent that our sampling of crop accessions can be considered representative of cultivated rice, our data suggest a limited role, if any, for red-pericarp landraces in the origin of the US weed alleles.
It should be noted that this finding does not rule out altogether a potential role for domesticated varieties in the origin of the weed Rc alleles. It is possible that Rc haplotype variation in the weeds represents variation that was once present in the crop (or in incipiently domesticated red-pericarp populations) but which was lost as the white-pericarp domestication alleles were selectively favored. It is also possible that more intensive sampling of extant red-pericarp landraces — specifically, indica and aus varieties — might reveal Rc haplotype similarity to weed strains that is not detected in the present analysis. In either of these cases, the weed populations would be descended primarily from domesticated germplasm, which is consistent with the close SH-indica and BHA-aus relationships observed in analyses of genome-wide neutral loci (Londo & Schaal 2007; Gealy et al. 2010; M. Reagon et al., University of Massachusetts, Amherst, unpublished).
Other than domesticated rice, the other potential origin for the red rice Rc alleles is undomesticated Oryza populations. In our dataset, just as most Rc haplotype variation in US red rice is not shared with domesticated rice, it is also largely absent from the O. rufipogon samples (Figure 2). However, whereas our sampling of domesticated rice is arguably representative, the same cannot necessarily be said for O. rufipogon. As a geographically widespread, outcrossing wild species, O. rufipogon shows much higher genetic diversity than the crop (Table 2; Caicedo et al. 2007), and the full genetic diversity of this species remains poorly characterized. Expanded collecting of O. rufipogon populations might be useful not only for further characterizing Rc haplotype diversity in comparison to red rice, but also for neutral marker assessments to clarify the roles of wild vs. domesticated populations in the weed’s evolutionary origins.
Why is red rice red?
The red-pigmented pericarp is nearly ubiquitous in US weedy rice, and aside from occasional reports of white-pericarp weed strains in Asia (Suh et al. 1992; Ushiki et al. 2005), proanthocyanidin pigmentation characterizes the great majority of red rice populations worldwide. This raises the question of whether proanthocyanidin pigmentation can be considered a weed-adaptive trait. Several lines of evidence suggest that the red pericarp is selectively favored, either directly or indirectly, in the US weed populations. First, we find no evidence for long-term persistence of nonfunctional rc alleles in US red rice populations. This is despite evidence of outcrossing rates from cultivated to weedy rice of between 0.1% and 1% (Messeguer et al. 2001; Zhang et al. 2003; Chen et al. 2004; Shivrain et al. 2007), which can potentially result in a large number of crop-weed hybrids when the density of weedy rice plants is high (documented at up to 40 plants/m2; Gealy 2005), and genetic evidence of crop-weed hybrids in population-level collections (Londo & Schaal 2007; M. Reagon et al., University of Massachusetts, Amherst, unpublished). We observed the rc domestication allele in only one MIX weed accession (1D10; Table S1), and this accession shows genome-wide evidence of descent from a very recent crop-weed hybridization event (M. Reagon et al., University of Massachusetts, Amherst, unpublished). Like cultivated rice, red rice is predominantly self-fertilizing, and so the recessive white-pericarp phenotype would be expressed in weed populations in generations following a crop-to-weed introgression event. The apparent absence of this domestication phenotype suggests that it is selectively disadvantageous in weed populations. Instead we observe haplotype structure consistent with selective maintenance of functional Rc alleles in the divergent SH and BHA weed populations (Table 2).
Additional evidence for the potential adaptive role of the red pericarp is provided by studies indicating that pericarp pigmentation may be important for seed dormancy. Strong seed dormancy is considered a classic weed-adaptive trait, allowing for the persistence of viable seeds over successive seasons (Baker 1965), and the dormancy of weedy rice in particular makes it difficult to prevent re-infestation of a field once the weed has been introduced (Cohn & Hughes 1981). Work involving weedy rice, Arabidopsis and wheat has all indicated that testa/pericarp pigmentation contributes to seed dormancy (Gfeller & Svejda 1960; Debeaujon et al. 2000; Groos et al. 2002). Moreover, genetic mapping in Asian weedy rice has localized a QTL for seed dormancy to the genomic region containing the Rc locus (Gu et al. 2004; Gu et al. 2005; Gu et al. 2006). Thus, a loss of pericarp pigmentation may be selectively deleterious in weedy rice populations. Of course, the possibility remains any patterns seen at the Rc locus in weedy rice are actually driven by selection at a closely linked locus; this can be evaluated through future genetic and functional studies.
Patterns of variation in the Rc genomic region
Examination of Rc in the context of its surrounding 4 Mb genomic region reveals contrasting evolutionary histories, not only for Rc in relation to flanking loci, but also for the weed populations in relation to cultivated O. sativa. At Rc, human selection for the white pericarp in domesticated rice has dramatically reshaped nucleotide diversity and haplotype distributions among variety groups. Consistent with a previous analysis by Sweeney et al. (2007) we observe patterns corresponding to a strong selective sweep associated with the japonica-derived rc domestication allele (Table 2), and a redistribution of haplotypes among variety groups resulting from selective introgression of this allele into all white-pericarp indicas as well as some white-pericarp aromatic and aus varieties (Table 1, Figure 2). The footprint of this rc selective sweep can be traced across the 4 Mb genomic region, with a declining impact on weed-crop genetic differentiation at increasing distances from Rc (Figure 4). This genomic signature closely matches expectations based on the work of Sweeney et al. (2007), who found that most selectively introgressed rc haplotypes extend < 1 Mb around the Rc locus.
Between the indica and aus variety groups, human selection for the rc domestication allele has disproportionately affected indica varieties: all white-pericarp indicas examined to date carry the japonica-derived rc allele, whereas white-pericarp aus varieties may carry either the rc allele or the independently-evolved Rc-s allele (Table 1; Sweeney et al. 2007). This difference between variety groups in the impact of rc selection has shaped the profile of weed-crop differentiation across the broader Rc genomic region. For the heavily-impacted indica varieties, the degree of differentiation from SH weeds closely matches the genomic footprint of the rc selective sweep: differentiation is highest at the Rc locus (where all white-pericarp indicas carry the introgressed japonica haplotype), and it declines at flanking loci in a pattern closely correlated with the declining impact of the rc sweep (Figure 4). In contrast, for aus varieties, a much weaker corresponding pattern is observed with respect to differentiation from BHA strains (Figure 4), consistent with the smaller impact of the rc sweep on this variety group.
The origin of weed characteristics
With the exception of herbicide resistance (Rajguru et al. 2005; Tan et al. 2007; Whaley et al. 2007), very little is known about the molecular basis and evolution of the traits that characterize conspecific crop weeds. This study represents a step in the direction of understanding the mechanisms by which these traits can arise in weed populations. For the case of US red rice pericarp color, our analyses of the Rc genomic region have revealed that this weed-associated trait reflects the presence of ancestral functional alleles, rather than novel mutations that arose from rc domestication alleles. Interestingly, this suggests a previously unrecognized constraint on the emergence of weed strains in the US: in marked contrast to North American crop weeds such as feral rye (Burger et al. 2006) and weedy Brassica (Gulden et al. 2008), red rice appears extremely unlikely to evolve in situ from US cultivated varieties — all of which carry the rc domestication allele. It will be interesting to see what future research reveals about the mechanisms of weedy trait evolution for other traits and other weed species.
Supplementary Material
Acknowledgements
The authors thank Jared L. Strasburg, Joshua S. Reece, and Seonghee Lee for assistance with data collection and advice regarding data analysis. We also thank the members of the Olsen Lab and Stuart F. McDaniel for comments on earlier drafts of the manuscript. Special thanks are given to staff of the Washington University Greenhouses for their expertise and assistance in the growing of rice accessions.
References
- Baker HG. Characteristics and modes of origin of weeds. In: Baker HG, Stebbins GL, editors. The Genetics of Colonizing Species. New York: Academic; 1965. pp. 147–172. [Google Scholar]
- Brooks SA, Yan W, Jackson AK, Deren CW. A natural mutation in rc reverts white-rice-pericarp to red and results in a new, dominant, wild-type allele: Rc-g. Theoretical and Applied Genetics. 2008;117:575–580. doi: 10.1007/s00122-008-0801-8. [DOI] [PubMed] [Google Scholar]
- Burger JC, Lee SKY, Ellstrand NC. Origin and genetic structure of feral rye in the western United States. Molecular Ecology. 2006;15:2527–2539. doi: 10.1111/j.1365-294X.2006.02938.x. [DOI] [PubMed] [Google Scholar]
- Caicedo A, Williamson S, Hernandez RD, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genetics. 2007;3:e163. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Lu B-R, Xia HUI, et al. Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in north-eastern China revealed by simple sequence repeat (SSR) markers. Annals of Botany. 2006;98:1241–1252. doi: 10.1093/aob/mcl210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LJ, Lee DS, Song ZP, Suh HS, Lu B-R. Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Annals of Botany. 2004;93:67–73. doi: 10.1093/aob/mch006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MA, Hughes JA. Seed dormancy in red rice (Oryza sativa) I. Effect of temperature on dry-afterripening. Weed Science. 1981;29:402–404. [Google Scholar]
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M. Influence of the Testa on Seed Dormancy, Germination, and Longevity in Arabidopsis. Plant Physiology. 2000;122:403–414. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estorninos LE, Jr, Gealy DR, Gbur EE, Talbert RE, McClelland MR. Rice and red rice interference. II. Rice response to population densities of three red rice (Oryza sativa) ecotypes. Weed Science. 2005;53:683–689. [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Seattle: Distributed by the author, Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- Furukawa T, Maekawa M, Oki T, et al. The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. The Plant Journal. 2007;49:91–102. doi: 10.1111/j.1365-313X.2006.02958.x. [DOI] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gealy DR. Gene movement between rice (Oryza sativa) and weedy rice (Oryza sativa) - a U.S. temperate rice perspective. In: Gressel J, editor. Crop Ferality and Volunteerism. Boca Raton: CRC Press; 2005. pp. 323–354. [Google Scholar]
- Gealy DR, Agrama HA, Eizenga GC. Exploring genetic and spatial structure of U.S. weedy red rice (Oryza sativa L.) in relation to rice relatives worldwide. Weed Science. 2010 In press. [Google Scholar]
- Gealy DR, Tai TH, Sneller CH. Identification of red rice, rice, and hybrid populations using microsatellite markers. Weed Science. 2002;50:333–339. [Google Scholar]
- Gfeller F, Svejda F. Inheritance of post-harvest seed dormancy and kernel colour in spring wheat lines. Canadian Journal of Plant Science. 1960;40:1–6. [Google Scholar]
- Groos C, Gay G, Perretant MR, et al. Study of the relationship between pre-harvest sprouting and grain color by quantitative trait loci analysis in a white×red grain bread-wheat cross. Theoretical and Applied Genetics. 2002;104:39–47. doi: 10.1007/s001220200004. [DOI] [PubMed] [Google Scholar]
- Gross BL, Olsen KM. Evolutionary genomics of weedy rice. In: Stewart CN, editor. Weedy and Invasive Plant Genomics. Ames: Wiley-Blackwell; 2009. pp. 83–98. [Google Scholar]
- Gross BL, Skare KJ, Olsen KM. Novel Phr1 mutations and the evolution of phenol reaction variation in US weedy rice (Oryza sativa L.) New Phytologist. 2009;184:842–850. doi: 10.1111/j.1469-8137.2009.02957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X-Y, Kianian SF, Foley ME. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa) Genetics. 2004;166:1503–1516. doi: 10.1534/genetics.166.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X-Y, Kianian SF, Hareland GA, Hoffer BL, Foley ME. Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa) Theoretical and Applied Genetics. 2005;110:1108–1118. doi: 10.1007/s00122-005-1939-2. [DOI] [PubMed] [Google Scholar]
- Gu XY, Kianian SF, Foley ME. Isolation of three dormancy QTLs as Mendelian factors in rice. Heredity. 2006;96:93–99. doi: 10.1038/sj.hdy.6800757. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Gulden RH, Warwick SI, Thomas AG. The biology of Canadian weeds. 137.Brassica napus L. and B. rapa L. Canadian Journal of Plant Science. 2008;88:951–996. [Google Scholar]
- Lee D, Lupotto E, Powell W. G-string slippage turns white rice red. Genome. 2009;52:490–493. doi: 10.1139/g09-025. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp187. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Londo JP, Chiang Y-C, Hung K-H, Chiang T-Y, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestication of cultivated rice Oryza sativa. Proceedings of the National Academy of Sciences of the USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londo JP, Schaal BA. Origins and population genetics of weedy red rice in the USA. Molecular Ecology. 2007;16:4523–4535. doi: 10.1111/j.1365-294X.2007.03489.x. [DOI] [PubMed] [Google Scholar]
- Lu H, Redus MA, Coburn JR, et al. Population structure and breeding patterns of 145 U.S. rice cultivars based on SSR marker analysis. Crop Science. 2005;45:66–76. [Google Scholar]
- Mackill DJ, McKenzie KS. Origin and characteristics of U.S. rice cultivars. In: Smith CW, editor. Rice: Origin, History, Technology, and Production. Hoboken: John Wiley & Sons, Inc; 2003. pp. 87–100. [Google Scholar]
- Messeguer J, Fogher C, Guiderdoni E, et al. Field assessments of gene flow from transgenic to cultivated rice (Oryza sativa L.) using a herbicide resistance gene as tracer marker. Theoretical and Applied Genetics. 2001;103:1151–1159. [Google Scholar]
- Moldenhauer KAK, Gibbons JH, McKenzie KS. Rice Varieties. In: Champagne ET, editor. Rice: Chemistry and Technology. St. Paul: American Association of Cereal Chemists, Inc; 2004. pp. 49–75. [Google Scholar]
- Nei M. Evolution of human races at the gene level. In: Bonne-Tamir B, Goodman R, editors. Human Genetics, Part A: The Unfolding Genome. New York: Alan R. Liss; 1982. pp. 167–181. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York, USA: Columbia University Press, New York; 1987. [Google Scholar]
- Posada D. ModelTest: Phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Rajguru SN, Burgos NR, Shivrain VK, Stewart JM. Mutations in the red rice ALS gene associated with resistance to imazethapyr. Weed Science. 2005;53:567–577. [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Shivrain VK, Burgos NR, Anders MM, et al. Gene flow between Clearfield(tm) rice and red rice. Crop Protection. 2007;26:349–356. [Google Scholar]
- Stamatakis A. Proceedings of the IPDP2006. Greece: Rhodos; 2006a. Phylogenetic models of rate heterogeneity: A high performance computing perspective. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006b;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Suh HS, Park SZ, Heu MH. Collection and evaluation of Korean red rices I. Regional distribution and seed characteristics. Korean Journal of Crop Science. 1992;37:425–430. [Google Scholar]
- Sweeney MT, Thomson MJ, Cho YG, et al. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genetics. 2007;3:e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MK, Preston C, Wang GX. Molecular basis of multiple resistance to ACCase-inhibiting and ALS-inhibiting herbicides in Lolium rigidum. Weed Research. 2007;47:534–541. [Google Scholar]
- Ushiki J, Ishii T, Ishikawa R. Morpho-physiological characters and geographical distribution of japonica and indica weedy rice (Oryza sativa) in Okayama Prefecture, Japan. Breeding Research. 2005;7:179–187. [Google Scholar]
- Whaley CM, Wilson HP, Westwood JH. A new mutation in plant ALS confers resistance to five classes of ALS-inhibiting herbicides. Weed Science. 2007;55:83–90. [Google Scholar]
- Zhang N, Linscombe S, Oard J. Out-crossing frequency and genetic analysis of hybrids between transgenic glufosinate herbicide-resistant rice and the weed, red rice. Euphytica. 2003;130:35–45. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.