Abstract
Individuals of the fish Lithognathus mormyrus were exposed to a series of pollutants including: benzo[a]pyrene, pp-DDE, Aroclor 1254, perfluorooctanoic acid, tributyl-tin chloride, lindane, estradiol, 4-nonylphenol, methyl mercury chloride, and cadmium chloride. Five mixtures of the pollutants were injected. Each mixture included one to three compounds. A microarray was constructed using 4608 L. mormyrus hepatic cDNAs cloned from the pollutant-exposed fish. Most clones (4456) were sequenced and assembled into 1494 annotated unique clones. The constructed microarray was used to identify changes in hepatic gene expression profile on exposure to cadmium administered to the fish by feeding or injections. Thirty-one unique clones showed altered expression levels on exposure to cadmium. Prominently differentially expressed genes included elastase 4, carboxypeptidase B, trypsinogen, perforin, complement C31, cytochrome P450 2K5, ceruloplasmin, carboxyl ester lipase, and metallothionein. Twelve sequences have no available annotation. Most genes (23) were downregulated and hypothesized to be affected by general toxicity due to the intensive cadmium exposure regime. The concept of an operational multigene cDNA microarray, aimed at routine and fast biomonitoring of multiple environmental threats, is outlined and the cadmium exposure experiment has been used to demonstrate functional and methodological aspects of the biomonitoring tool. The components of the outlined system include: (1) spotted array, composed of both pollution-affected and constitutively expressed genes, the latter are used for normalization; (2) standard, repeatable labeling procedure of a reference transcript population; and (3) biomarker indices derived from the profile of expression ratio across the pollution-affected genes, between the field-sampled transcript populations and the reference.
Keywords: Biomarkers, Biomonitoring, cDNA microarray, Environment, Fish, Lithognathus mormyrus
Introduction
Anthropogenic impact on natural biota is a major environmental threat and its monitoring is an essential tool for designing and adjusting environmental policy. Sentinel organisms inhabiting an examined habitat and interrelating with almost all habitat components could serve as probes of changing environmental impacts.
Coordinated expression of genes is a basic biological phenomenon underlying almost all biological processes. Hence, the levels of expressed transcripts are widely used as experimental and diagnostic biomarkers in a variety of biological fields, including the evaluation of the biological effects of environmental pollutants. Assemblages of transcript biomarkers are adjustable, enabling the detection of a variety of present or future environmental threats via uniform instrumentation and methods. Such biomarkers may provide early warning of deleterious effects before substantial changes in species richness or diversity and may quickly prove or disprove the efficiency of a preventive environmental policy.
Despite their relatively high mobility, fish are generally considered important sentinels for pollution monitoring in aquatic systems. They are widespread in aquatic environments and play a major ecological role in the aquatic food web. Many studies have examined the environment-affected gene expression levels in sentinel fish species suggested as indicators of the biological effects of pollution. Many of these studies were cited in the reviews of Van der Oost et al. (2003) and Tom and Auslander (2005).
We chose Lithognathus mormyrus (striped sea bream) as a sentinel candidate. It is a bottom-dwelling fish widely distributed throughout the entire Mediterranean, including the Levantine basin and also the southeastern Atlantic (Bauchot and Hureau 1986). It inhabits polluted and pristine coastal sandy habitats and interacts with the water column but also with the bottom, through its bottom-feeding habits and burial behavior (Suau 1970). Its gut usually contains sediment engulfed with the ingested food (Froglia 1977; Buxton et al. 1984; Lasiak 1984). Similar to other members of its family, the Sparidae (Buxton and Garratt 1990; Micale and Perdichizzi 1994), it is a protandrous hermaphrodite (Besseau 1990; Kraljevic et al. 1995). The gonad is an ovotestis, sexually indistinguishable outside the summer reproductive season. Moreover, there are no external sex-distinguishing characters (Bauchot and Hureau 1986; Kraljevic et al. 1995).
As in other vertebrates, fish liver is commonly involved in xenobiotic detoxification (Van der Oost et al. 2003). The hepatic levels of environmentally affected transcripts and proteins in striped sea bream have been previously investigated for use as environmental biomarkers (Tom et al. 2002, 2003, 2004; Funkenstein et al. 2004). The cDNA microarray platform has been used with numerous organisms for a variety of applications including environmental sciences (Gracey et al. 2001; Williams et al. 2003, 2006, 2007; Koskinen et al. 2004; Snape et al. 2004; Cossins and Crawford 2005; Sheader et al. 2004, 2006; Reynders et al. 2006). However, the potential advantages of multigene response assays in the striped sea bream have not yet been examined via a cDNA microarray.
Cadmium is a major transition metal pollutant in Israeli waters and elsewhere (Herut and Kress 1997). Metallothionein is a well established and intensively studied biomarker of exposure to transition metals, including intensive studies of its transition metal-related induction mechanism (Coyle et al. 2002; Tom and Auslander 2005). Its induction by cadmium was quantitatively evaluated in L. mormyrus (Tom et al. 2004) and it is used here to indicate the effect of the cadmium administration and as a reference biomarker compared to the multigene expression profiles.
The goals of this study were to construct an equalized striped sea bream hepatic transcriptome, biased toward xenobiotic-affected genes and to use it to identify alterations in hepatic transcript expression profile on exposure to fed and injected cadmium. Implementation of multigene expression profiles as comprehensive environmental biomarkers is discussed, using the cadmium exposure experiment as a functional and methodological example.
Materials and Methods
Fish Maintenance and Experimental Manipulations
Fish used for the microarray construction or for the cadmium exposure experiment were similarly maintained and treated. L. mormyrus specimens (25 to 50 g/fish) were caught in their natural habitats along the Mediterranean coast of Israel, via a 20- to 22-mm mesh size nylon monofilament gillnet, and brought alive to the laboratory under aerated conditions. The fish used for the library construction or for the cadmium exposure experiment were taken from a single batch caught at the same date and site. The fish were kept in the maintenance facility a month before exposures and during the exposures in 700-liter clean seawater flow-through tanks under an ambient temperature regimen of 16°C to 21°C. They were fed commercial fish pellets (cat. no. 1369, Matmor central feed mill, Ashquelon, Israel), containing cadmium chloride when experimentally required.
Fish were exposed to toxic compounds as appropriate for the cDNA library construction or for the cadmium exposure experiment. They were killed by decapitation at the termination of the required exposure durations. Each killed individual was weighed, and its liver and gonads were immediately removed. Livers were divided into aliquots, snap frozen in liquid nitrogen, and kept at −80°C for later RNA purification. Gonads were fixed in 4% formaldehyde solution and used for examining vitellogenic activity according to the criteria determined by Funkenstein et al. (2004).
Total RNA and mRNA were extracted from frozen livers via commercial kits. Trireagent (Molecular Research Center, Cincinnati, OH) was used for total RNA extraction from livers and PolyATtract (Promega, Madison, WI) was used for mRNA extraction from total RNA. RNA and mRNA quality was tested by their spectrophotometric 260nm/280nm ratio and their separation on 1% formaldehyde-agarose gel (Sambrook and Russell 2001).
Microarray Construction
The cDNAs for constructing the microarray were produced from hepatic mRNA populations of individuals preexposed to a series of toxic compounds. Five xenobiotics mixtures, prepared from 10 separate compounds, were dissolved in suitable vehicles and injected into 5 groups of fish, on the first and third day of the experiment, as follows:
Two injections of 100 μl each, containing lindane (Supelco, Bellefonte, PA; 1 mg/ml), β-estradiol (Sigma-Aldrich, St. Louis, MO; 1 mg/ml), and Aroclor 1254 (polychlorinated biphenyl mixture, Supelco; 2 mg/ml).
Two injections of 50 μl each, containing benzo[a]pyrene (B[a]P, Fluka, St. Louis, MO; 4 mg/ml) and perfluorooctanoic acid (PFOA, Fluka; 8 mg/ml).
Two injections of 50 μl each, containing methyl mercury chloride (Sterm Chemical, Newburyport, MA; 1 μg/ml) and 4-nonylphenol (Fluka: 8 mg/ml).
Two injections of 50 μl each, containing 1,1 dichloro-2.2-bis [p-chlorophenyl] ethylene (pp-DDE, Chem Service; 8 mg/ml) and tributyl tin chloride (TBTC, Fluka; 1 mg/ml).
Two injections of 50 μl each, containing cadmium chloride (Sigma-Aldrich; 1 mg/ml).
Xenobiotics mixtures 1 to 4 were dissolved in corn oil and mixture 5 in saline. Noninjected fish served as a reference group. Fish were killed at the third and the seventh day after the second injection. Fish in reproductive activity arrest were selected for further processing. Arrest of reproductive activity was judged by the measured oocyte diameter, ranging 40 to 131 μm, only at the beginning of vitellogenesis (Funkenstein et al. 2004). Hepatic RNAs of fish injected with the same pollutant mixture were pooled and five transcript libraries were separately prepared from each of the five pools.
The libraries were constructed in two stages: (1) switching mechanism at the 5′ end of the RNA transcript (SMART) aimed at polymerase chain reaction (PCR) amplification of reverse transcribed cDNAs while reducing ribosomal RNA amplification (Clontech, Mountain View, CA) and (2) subtractive suppression hybridization (SSH), aimed at cloning roughly equally represented cDNAs from livers of the fish exposed to the five xenobiotics mixtures, subtracted from the reference unexposed RNA population (Diatchenko et al. 1999; PCR-select, Clontech). The SSH was aimed at forming a cDNA library biased toward genes differentially expressed in the exposed livers in comparison to the reference ones.
The produced DNA fragments were cloned into P-GEM-Teasy plasmid vector (Promega) and transformed into the Escheroichia coli strain JM109. The final cDNA assemblage contained 4608 clones, and it is equally composed of clones randomly selected from each of the five libraries. All the clones were sequenced. The obtained sequences were cleaned from both plasmid and poorly sequenced segments and were assembled via the CAP3 software (Huang and Madan 1999). The term “unique clone” is used here to describe a consensus sequence constructed by CAP3 from several identical clone sequences, fully or partially overlapping. Each unique clone was deposited in GenBank, receiving an identifying accession number.
The unique clones were annotated using BLASTX and BLASTN screening algorithms (Altschul et al. 1997). Gene Ontology (GO) annotations, aimed at providing unified system for gene function definitions (Ashburner et al. 2000), were also assigned to unique clones. BLAST2GO software (Conesa et al. 2005; http://www.blast2go.de/) was used to identify both the sequence and the GO annotations.
The 4608 clones were amplified from bacterial templates via standard PCR (5 min at 95°C; 32 cycles of 0.5 min at 95°C, 0.5 min at 68°C, and 1.5 min at 72°C; a final incubation of 4 min at 72°C; PCR system 9600, Applied Biosystems). The PCR-select universal primers located at both ends of each cloned DNA fragment were used for the amplification (Diatchenko et al. 1999; PCR-select, Clontech). PCR products were precipitated from the solution by 1 volume of isopropanol, redissolved in distilled water, and mixed at 1:1 ratio with printing buffer ×2 (20× saline sodium citrate [SSC] diluted to 0.6× SSC in dimethyl sulfoxide [DMSO]). The 20× SSC solution is composed of 3 M NaCl and 0.3 M Na citrate (pH 7.4). The presence of PCR product, its length, and its concentration were examined by running the purified products on a 1% agarose gel, concurrent with a DNA mass ladder. The clones were spotted in adjacent duplicates on modified glass slides (GAPS II slides, Corning, Corning, NY; MGII robot-printer, BioRobotics, Woburn, MA).
Cadmium Exposure Experiment
Three groups of fish participated in the experiment. Two of them (n = 4 in each group) were exposed to fed and injected CdCl2, respectively. The third reference group (n = 8) was not exposed to cadmium. Cadmium-containing food was prepared by grinding commercial food pellets followed by their repelleting as a mixture of 63% food powder, 35% CdCl2 aqueous solution (200 μg of CdCl2/ml) and 2.5% gelatin as solidifier. The reconstructed pellets were dried overnight at 45°C to 50°C. Cadmium content of the food was 57.25 μg/g of food and 31 food portions, each of 1% body weight, were given to the fish four times a week. Fish were killed 3 days after the last feeding and received 17.75 mg of Cd/kg body weight during the 57 days of the experiment. A dose of 2.5 mg Cd/kg body weight was intraperitoneally injected to the other group and the fish were killed 8 days after injection. mRNAs were extracted from reference and cadmium-affected fish livers and the mRNA preparations of each pair of reference fish were combined. Hence, four RNA preparations were present for each group of cadmium-exposed or reference fish.
Cadmium Concentration Measurements
Cadmium concentrations were determined on approximately 0.1 g of dry liver tissue or 0.5 g of ground food pellets. The samples were digested for 4 h at 140°C with concentrated nitric acid (65 wt %) in Uniseal, Teflon-lined, high-pressure decomposition vessels according to Herut and Kress (1997). The digests were diluted to 15 ml in clean plastic tubes with double-distilled water. Measurements were performed via Varian SpectrAA220 flame atomic absorption and SpectrAA880 graphite furnace. The accuracy and precision of the methods were evaluated on the basis of analysis of several international standard reference materials (DOLT3, fish liver; DORM-2 and IAEA 407, fish muscle). All elements gave results within 5% of the certified values.
Analysis of Multigene Expression Patterns on Exposure to Cadmium
Microarray Hybridizations and Their Analysis
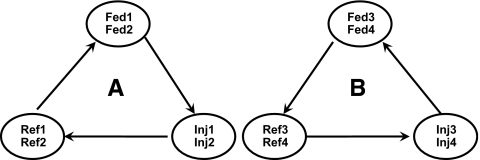
Differential hepatic gene expression between cadmium-fed or cadmium-injected fish and the reference ones was evaluated via hybridization of dual-labeled pairs of cDNA populations of cadmium-exposed or reference fish on microarray slides. Four replicate hybridization loops were designed; each contained three microarray slides hybridized to alternate pairs of individual labeled cDNAs from reference or exposed livers. The utilized fluorescent dyes were Cy3 and Cy5 and the labeling of each treatment or the reference was balanced between the two dyes to reduce dye effect (Fig. 1). The indirect fluorescent labeling of mRNA preparations through their conversion to cDNA, and the dual-label hybridization procedure on the microarray slides, were performed according to Yudkovski et al. (2007). An average of 4334 + 462 spots/slide out of 9216, did not meet the criteria of GenePix, the utilized imaging software.
Fig. 1.
Hybridization design. Each arrow connects two cDNA samples which are dual-hybridized on the same slide. Arrow tail, Cy3; arrowhead, Cy5. (A) and (B) are two alternative hybridization designs, aimed at balancing putative dye effect. Each design was performed twice with different individual fish
The analysis of differential gene expressions and its significance testing was carried out via the LIMMA software according to Yudkovski et al. (2007). In brief, binary comparisons of gene expression levels, which were done between each of the exposure routes and the reference, resulted with two descriptive parameters: (1) the M value, the clone-specific log2 expression ratio between each of the exposure routes and the reference, and (2) A = (log2(Cy3*Cy5))/2, the labeling intensity of each clone on the slide. MeanA is the average of A across the analyzed experiment. Two statistical, clone-specific parameters were obtained: the B statistic, the log-odds that a specific clone is differentially expressed (M ≠ 0), and the p-value that resulted from a moderated t-test.
In addition, the M-value profiles of 8 binary comparisons accomplished between each of the four cadmium-injected and the four fed individuals versus the averaged values of the reference fish, were clustered via the EXPANDER software. This software contains a battery of adjustable clustering methods (Sharan et al. 2003; http://www.cs.tau.ac.il/~rshamir/expander/expander.html). Only clones of meanA > 9, eliminating suspected faint fluorescence emissions, which were present in all analyzed binary comparisons, were included in the cluster analysis. They were arranged in a gene × M-value matrix that served as EXPANDER input. Clusters of clones characterized by similar expression patterns across the individual fish were determined using several clustering methods available in EXPANDER. Clusters revealing valuable expression patterns in the context of the experimental aims are presented and discussed below. M-values of redundant clones, belonging to the same unique clone, were averaged within each cluster.
Quantitative Real-Time PCR
Absolute measurements of metallothionein transcript level were performed according to Tom et al. (2004). Relative reverse transcriptase-real time PCR assays were performed to evaluate the relative expression level of target transcripts between two compared L. mormyrus RNA populations. The assays were performed according to Yudkovski et al. (2007). PCR reactions were accomplished for selected target genes and also for β-actin, which served as normalizing agent, using the primer pairs described in Table 3. Evaluation of relative expression was calculated from the ÄCP between two compared RNA populations, after their normalization to β-actin levels.
Table 3.
Real-time PCR results normalized to β-actin expression levels
| Accession no. | Amplified RNA | PCR primer pair | Real- time PCR ΔCP normalized to β-actin expression | M-value | Competence between PCR ΔCPs and M-value |
|---|---|---|---|---|---|
| DQ850666 | Metallothionein | F: AAGGGAAGACCTGCGACACTAG | 4.5 | 2.04 | + |
| R: TCACAAGGGCAGCAAGCA | |||||
| DQ850591 | Carboxypeptidase B | F: CTGCTCTGCGTAGTCTGTACGG | −2.1 | −2.09 | + |
| R: GGAATACTTCACACCCAGGTCG | |||||
| DQ850208 | Carboxyl ester lipase | F: CAGAAGATGAGGCTGCGTTACG | −2.2 | −1.51 | + |
| R: GAAGTACAGGTGATGGCACAAGC | |||||
| DQ849772 | Complement component C3 | F:TTACTTGGCCCTCTTGTGTGG | 0.9 | −1.14 | − |
| R: CAACAGGCAGTCAAATCAAGCC | |||||
| DQ849874 | Cytochrome P4502k5 | F: TGGTTGAAGCTCATCAGAATTCC | 0.6 | −1.3 | − |
| R: CCCTTGCATGGATTCAGATAGG | |||||
| DQ850737 | β-actin | F: CATCCTGCGTCTGGACTTGG | |||
| R: GATTTCCCTCTCGGCTGTGG |
M-values were obtained from the microarray analysis.
Results
Microarray Library Characterization
Most cDNAs comprising the cloned library, 4456 out of 4608, were successfully sequenced (97%). One thousand four hundred and ninety-four unique clones meeting GenBank length threshold (>50 bp) were identified (66% redundancy) and deposited in GenBank (accession numbers DQ849631 to DQ851090, DQ887346 to DQ887373, and DQ890524 to DQ890537). Elimination of ribosomal RNA cloning, one of the aims of the SMART procedure, was fully achieved in the randomly selected clones. The SSH procedure was aimed at roughly equalizing the representation of cDNAs in the constructed library, favoring differentially expressed genes in the target tissue in comparison to a reference one. Equal representation was partially achieved. The distribution of clone copy number revealed a major contribution of few genes to the redundancy, probably due to the less sufficient suppression of these abundant genes by the SSH. Most unique clones (1464) were roughly equally represented in 1 to 10 copies, whereas 33 unique sequences were present in 11 to 100 copies and 5 were present in 101 to 346 copies. Biasing the cloned transcriptome toward pollutant-affected genes was indicated by some of the revealed unique clones, previously established as environmental biomarkers affected by several of the pollutants applied here. These genes included cytochrome P4501A, vitellogenin, choriogenin, and metallothionein (Van der Oost et al. 2003; Tom and Auslander 2005).
BLAST resemblances to publicly available sequences having e-values >10−5 were filtered out, ending with 786 unique clones that showed similarity to a variety of genes from different organisms, mainly fish species. A detailed description of this microarray platform was deposited in the the NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/; Barrett et al. 2006) and are accessible through GEO platform accession number GPL5351.
Cohen et al. (2007) compared the sequences of this microarray platform with homolog cDNAs of several fish species, concluding that intrafamily hybridizations are beneficial, having tolerable lose of hybridized cDNAs. Slides of this microarray may be available to other researchers within the framework of scientific collaborations.
Exposure to Cadmium
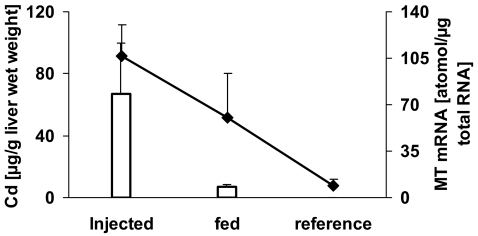
The average weight was not statistically different among the three experimental fish groups, averaging 40.8 ± 7.6 g across the experiment. All individuals experienced reproductive arrest, signified by the small oocyte diameter (<100 μM; Funkenstein et al. 2004). The bioavailability of the administered cadmium was indicated by the correspondingly increased hepatic cadmium and metallothionein transcript levels (Fig. 2). The injected fish revealed the highest cadmium level, after 8 days. The higher administered level of fed cadmium, in comparison to the injected dose, resulted in 10-fold lower hepatic level than that caused by injection. The lowest cadmium levels were measured in the reference fish (0.15 ± 0.03 μg Cd/g of liver wet weight). A similar trend was elucidated in the metallothionein transcript levels in the injected, fed, and reference fish, respectively (Fig. 2).
Fig. 2.
Hepatic levels of metallothionein transcript and cadmium in relation to the routes of cadmium applications. The line graph presents metallothionein transcript levels and the bar graph, cadmium levels. MT, Metallothionein; Cd, cadmium
The raw data from the hybridizations as well as LIMMA-analyzed data have been deposited in the NCBI’s GEO database and are accessible through GEO Series accession number GSE8005.
Clones that responded to the meanA > 9 threshold and were differentially expressed compared to the reference group, in at least one of the treatments, were stringently selected via two statistical criteria, p < 0.05 and B > 1.5. M values of significantly expressed clones that belong to one unique clone were averaged. High copy number unique clones in which few clones were significantly differentially expressed and unique clones that revealed no correspondence between M-values of fed and injected fish were also considered nondifferentially expressed. Consequently, 31 unique clones were considered genuinely affected by the exposure to cadmium. The characteristics of each selected unique clone include its accession number, the most resembling annotation, number of visualized clones (meanA > 9), the average M-value of significantly differentially expressed clones in each of the treatments, and their corresponding numbers (Table 1). Twelve out of the 31 sequences revealed no assigned function. Ten unique clones were differentially expressed by both cadmium application routes. Twenty-three of the 31 differentially expressed genes were downregulated; only 8 were upregulated. Only 12 significantly differentially expressed unique clones were revealed in the fed fish in comparison to 29 genes in the injected fish. In addition, M-values in 7 of the 10 unique clones that revealed differential expression on exposure to both routes of administration were closer to zero in the fed fish in comparison to the injected ones.
Table 1.
Differentially expressed striped sea bream hepatic unique clones after exposure to cadmium
| Accession no. | Gene annotation | Resembled species | Number of clones | Averaged M-values ± SD of differentially expressed unique clones | ||||
|---|---|---|---|---|---|---|---|---|
| Visualized clones of (meanA > 9) | Differentially expressed clones | |||||||
| FI | F | I | F | I | ||||
| Mitochondrial respiratory chain | ||||||||
| DQ850610 | Mitochondrial cytochrome b | Lithognathus mormyrus | 1 | 0 | 1 | 0 | −0.97* | |
| DQ849967 | Mitochondrial cytochrome c oxidase subunit III | Satyrichthys amiscus | 4 | 2 | 2 | 2 | −0.65 ± 0.11* | −0.88 ± 0.1* |
| Proteases | ||||||||
| DQ850573 | Elastase 4-like protein | Sparus aurata | 21 | 0 | 0 | 16 | −1.52 ± 0.31* | |
| DQ850591 | Carboxypeptidase B | Paralichthys olivaceus | 3 | 2 | 2 | 3 | −1.47 ± 0.34* | −2.09 ± 0.4* |
| DQ849726 | Trypsinogen-like protein | Sparus aurata | 19 | 0 | 0 | 9 | −1.76 ± 0.25* | |
| Protease inhibitors | ||||||||
| DQ851090 | Kininogen | Anarhichas minor | 1 | 0 | 1 | 0 | −0.86 | |
| DQ850843 | Heparin cofactor II | Brachidanio rerio | 4 | 1 | 1 | 2 | 1.05 | 0.71 ± 0.07 |
| Immune system | ||||||||
| DQ849772 | X- CO3_ONCMY and complement C31 | Paralichthys olivaceus | 1 | 0 | 0 | 1 | −1.14* | |
| DQ850497 | Perforin | Fundulus heteroclitus | 1 | 0 | 0 | 1 | −1.11 | |
| DQ849947 | OMY519930 C1R/C1S subunit of Ca2+-dependent complex | Oncorhynchus mykiss | 1 | 0 | 0 | 1 | 0.59 | |
| Oxidases | ||||||||
| DQ849874 | Cytochrome P450 (CYP2K5) | Oncorhynchus mykiss | 1 | 0 | 0 | 1 | −1.3 | |
| DQ849703 | Ceruloplasmin | Pseudopleuronectes americanus | 2 | 0 | 1 | 1 | 0.84* | 0.81* |
| DQ850163 | Glutathione peroxidase | Oplegnathus fasciatus | 1 | 0 | 0 | 1 | 0.66 | |
| Other enzymes | ||||||||
| DQ850208 | Carboxyl ester lipase | Brachidanio rerio | 2 | 0 | 0 | 1 | −1.51 | |
| Ribosomal proteins | ||||||||
| DQ849880 | Sop-prov protein | Xenopus laevis | 1 | 0 | 0 | 1 | −1.16 | |
| Metal-related proteins | ||||||||
| DQ850666 | Metallothionein | Lithognathus mormyrus | 3 | 1 | 1 | 3 | 1.26* | 2.04 ± 0.48* |
| DQ850735 | Putative transferrin | Acanthopagrus schlegelii | 9 | 2 | 2 | 4 | −0.64 ± 0.06 | −0.92 ± 0.18 |
| Other proteins | ||||||||
| DQ850849 | Vitellogenin | Pagrus major | 1 | 0 | 0 | 1 | 0.71 | |
| DQ850712 | Gastrula specific embryonic protein 1 | Epinephelus coioides | 48 | 3 | 3 | 8 | −0.51 ± 0.06 | −0.64 ± 0.08 |
| Unknowns | ||||||||
| DQ849656 | (Q5BXC0) Hypothetical protein | Schistosoma japonicum | 8 | 1 | 1 | 4 | −0.85 | −0.86 ± 0.31 |
| DQ849739 | CNS0FK72 full-length cDNA | Tetraodon nigroviridis | 8 | 0 | 1 | 1 | −0.64 | −0.58 |
| DQ850009 | 6 | 0 | 0 | 3 | 0.83 ± 0.3 | |||
| DQ850829 | 3 | 0 | 0 | 1 | −0.77 | |||
| DQ849818 | CNS0G66Q full-length cDNA | Tetraodon nigroviridis | 1 | 1 | 1 | 1 | −0.48 | −0.81 |
| DQ850314 | 1 | 0 | 0 | 1 | −1.42 | |||
| DQ850888 | 1 | 0 | 0 | 1 | 0.95 | |||
| DQ850970 | 1 | 0 | 0 | 1 | −0.48 | |||
| DQ851001 | 1 | 0 | 0 | 1 | −1.29 | |||
| DQ850538 | 1 | 0 | 0 | 1 | −1.72 | |||
| DQ849829 | 1 | 0 | 0 | 1 | −1.05 | |||
| DQ850030 | 1 | 0 | 0 | 1 | −1.39 | |||
Each unique clone is characterized by its accession number, short description of the most resembled sequence and GO annotations, number of clones of meanA > 9, significantly differentially expressed clones, and their average M-values. F, Cadmium fed; I, cadmium injected; FI, both treatments.
*Denotes correspondence with the results of Reynders et al. (2006).
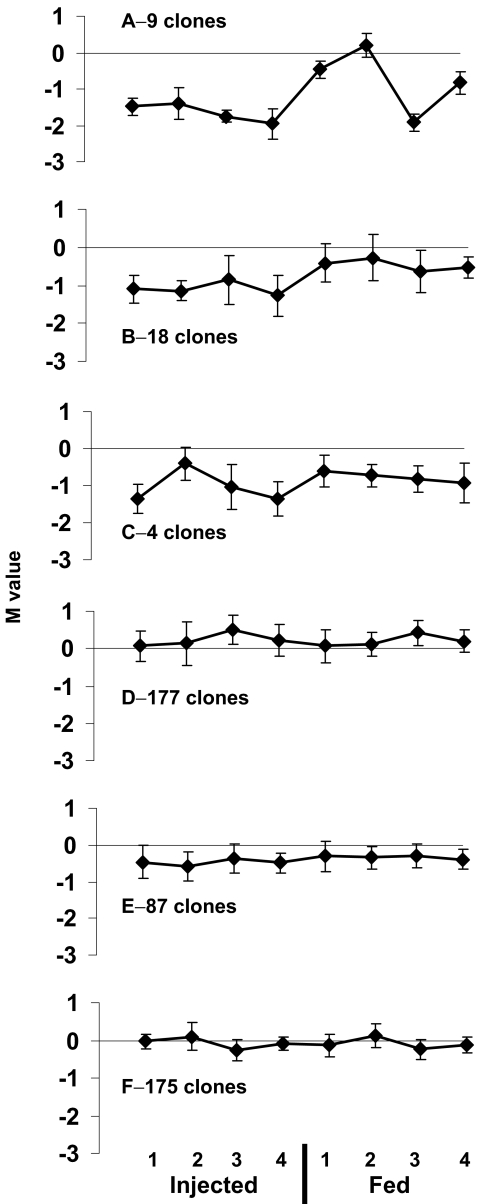
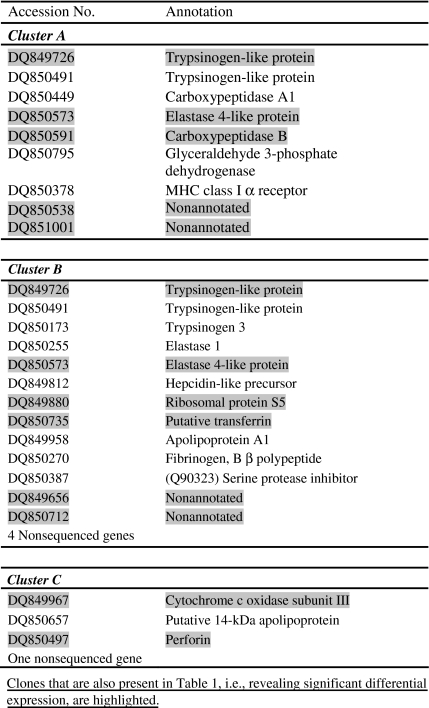
Eight M-value profiles were resulted from the binary comparisons of hepatic transcripts of single treated fish versus the averaged reference. These profiles were filtered only by the meanA > 9 criterion and were clustered via a variety of clustering procedures. The K-means clustering with seven predetermined groups was selected as suitable to demonstrate the characteristics of the operational microarray biomonitoring tool discussed below. However, two similar clusters were manually combined, ending with six clusters presented in Fig. 3. Clusters A to E demonstrated groups of differentially expressed genes contrasting with the more stably expressed genes of cluster F. The expression pattern of injected fish differed from that of the fed fish in clusters A to C and most of the unique clones belonging to these clusters were downregulated in comparison to the reference group. Injected fish revealed more uniform expression across individuals in comparison to the fed fish in clusters A and B and the opposite was true in cluster C. Cluster D elucidated a low general upregulation trend. Unique sequences of clusters A to C are listed in Table 2.
Fig. 3.
Clustering of M-value profiles obtained from binary comparisons of individual treated fish to the averaged reference. Injected 1–4 are cadmium-injected fish and Fed 1–4 are cadmium-fed fish
Table 2.
List of unique clones that were elucidated in clusters A to C (Fig. 3)
Quantitative PCR Validation of Results
The results of relative real-time PCR, normalized to β-actin are presented in Table 3, compared to the respective microarray M-values. Both M-values and ΔCPs represent the log2 expression ratio between compared RNA populations, and although evaluated by different methods, they can be qualitatively compared, demonstrating similar expression trends in three out of five accomplished PCR comparisons.
Discussion
It was well demonstrated, via both hepatic cadmium levels and gene expression data, that feeding is a less efficient route for exerting cadmium biological impact. Injection ended with higher hepatic cadmium levels. Moreover, injected fish revealed higher number of unique clones elucidating significant differential expression with higher absolute M-values, in comparison to cadmium-fed ones.
A relatively small number of unique clones were identified as differentially expressed on exposure to cadmium (Table 1), in comparison to the number of clones revealing M ≠ 0 (Table 2 and supplementary data, GEO accession number GSE8005), a consequence of the stringent selection criteria for genuine differential expression.
Putative inaccuracy of microarray results were intensively studied and discussed in the literature and mainly assigned to cross-gene hybridizations of related sequences (Wren et al. 2002; Flikka et al. 2004; Chen et al. 2006). Hence, real-time PCR is widely used as a validation tool of microarray results, being accurate, sensitive, and quantifiable. However, its results may also be biased. Unintentional choice of inadequate, i.e., differentially expressed normalizing agent, may cause a general shift of expression ratios of all examined genes to one direction in comparison to the M-values obtained via the array. Cross-gene nonspecific similarity of primer sequences, mostly within a gene family, may cause gene-specific cross-gene amplification. The obvious solution to these incompatibilities is using several normalizing agents on one hand and accomplishing a broad PCR validation effort containing many examined genes on the other. Both approaches would increase the statistical significance of the overall validation. Unfortunately, shortage of extracted RNA prevented more comprehensive validation effort here.
Individual pollution-affected transcripts can be used as stand-alone biomarkers (Tom and Auslander 2005). Two genes from the selected list of Table 1 can be considered stand-alone biomarker genes. One of them is the well established upregulated metallothionein. The clearly downregulated carboxypeptidase B can be also considered a candidate stand-alone biomarker owing to its prominent downregulation in both fed and injected fish, demonstrated in all its redundant clones. In contrast to metallothionein, specifically induced by transition metals, carboxypeptidase B belongs to the 23 downregulated unique clones elucidated via this experiment. This downregulation is interpreted by us to be a consequence of general toxicity caused by the high levels of toxic cadmium that were applied. Consequently, it is hypothesized that carboxypeptidase B may respond to a wider range of compounds, which cause general toxic effects. This interpretation is supported by a recent microarray-related study (Reynders et al. 2006) in which applying similar levels of cadmium in carps caused a similar downregulation of genes. The mutually affected genes in both studies are depicted in Table 1. In contrast, Williams et al. (2006) exposed flounders to assumedly environmentally realistic low levels of cadmium and examined its time-course impact on multigene expression, using cDNA microarray. They elucidated prominent changes in gene expression patterns. However, the affected genes were not similar to the ones mutually observed in the present study and in Reynders et al. (2006). Moreover, the increase in metallothionein transcript level in Williams et al. (2006) was mild and transient and lasted only 4 days. It was higher and permanent in both Reynders et al. (2006) and the present study. Hence, it can be concluded that metallothionein is not the most sensitive indicator of cadmium exposure, responding only to intensive exposure, whereas multigene expression pattern was changed even on exposure to low levels of cadmium. Future comprehensive examinations of fish hepatic multigene expression patterns on exposure to a wide range of cadmium concentrations and time scales may reveal the entire spectrum of the fish multigene response to cadmium.
Chaperon genes related to unfolded protein response were transiently revealed a short time after exposure, in the two studies cited above but not by us, probably owing to the absence of short- period expression examination.
Further description of functions of differentially expressed genes revealed by our microarray analysis (Table 1) are found in the following articles listed according to the subject genes: kininogen (Ylonen et al. 2002), heparin cofactor II (Hanumanthaiah et al. 2002; Law et al. 2006), C1R/C1S subunit of Ca2+-dependent complex (Wang and Secombes 2003), cytochrome P4502K5 (Arukwe and Goksoyr 1997), glutathione peroxidase 1 (Halliwell and Gutteridge 1999), carboxyl ester lipase (Whitcomb and Lowe 2007), perforin (Voskoboinik and Trapani 2006), elastase 4 (Suzuki et al. 2002), carboxypeptidase B (Srivastava et al. 2003), vitellogenin (Larkin et al. 2003), transferrin (Gomme et al. 2005; Lambert et al. 2005), ceruloplasmin (Bielli and Calabrese 2002; Hellman and Gitlin 2002).
A comprehensive response to multiple environmental perturbations is assumed by us to be better detected via a multigene-based biomarker. Therefore, a conceptual multigene based approach is outlined and discussed below using the present cadmium exposure experiment to demonstrate functional and methodological aspects. The suggested tool is an operational microarray composed of selected pollutant-affected genes. An additional group of constitutively expressed genes should be included in the array for normalization of the dual label. Defining normalization agents, rather than using the entire clone assemblage on the array, is required, as the biomarker genes that are differentially expressed are not suitable for normalization.
The results of exposure experiments such as the present one, performed in our or other fish species, may be used for the selection of differentially expressed genes. Cluster analysis may be used to identify constitutively expressed genes (Fig. 2, cluster F).
The suggested biomarker to be used by the operational microarray is the M-value profile obtained from hybridizing the hepatic cDNA populations of field-sampled sentinel fish against a common labeled control transcript mixture. A basic requirement for an environmental biomarker is long-term comparability over a wide geographical range. Therefore, the reference transcript mixture should be standardized, being easily and repeatedly produced. This aim can be accomplished via in vitro reverse transcribing an equally represented, linearized plasmid mixture composed of all microarray clones, concurrently incorporating fluorescent labeling, using any commercial in vitro transcription kit.
Intra- and intersample evaluation of alterations of the M-value profile should not be done using one index, as it may oversimplify the complexity of the profile. The number of differentially expressed genes, their average M-value, and their annotation may serve as biomarker parameters. Hence, a higher number of differentially expressed genes and higher average M-values may indicate biological effects of environmental factors in analogy to the analysis of our exposure experiment. The annotations of the expressed genes may indicate the nature of the perturbation, deduced from the functions of the annotated genes. It has to be emphasized that average M-values have to be calculated using absolute values, as both down- and upregulation should be equally considered as affected expression.
A complementary index may be obtained by clustering M-value profiles across individual fish. The clustering performed here (Fig. 2) may serve as a methodological example, using each treatment as an imaginary field sample. The clusters of differentially expressed genes (mainly clusters A to C, but also D and E) may be used to compare intra- and intersample differences of average M-values. Intrasample variability may indicate natural variability and/or different exposure history to pollutants, and intersample differences may point out different spatial and temporal sets of affecting environmental factors.
To summarize, a microarray platform was constructed from fish hepatic cDNAs and utilized to reveal cadmium effect on hepatic multigene expression patterns. A conceptual environmental biomonitoring approach using an operational cDNA microarray is outlined.
Acknowledgments
The study was supported by the EU projects GENIPOL (EVK3-CT-2001-00057) and MARINEGENOMICS EUROPE (Contract 505403). M.A. was a recipient of the Yohai Ben-Nun and Rieger scholarships. Efrat Shoham-Frider from the IOLR is thanked for the provision of the methyl mercury. The excellent animal care of Maria Sigal is appreciated. Dr. Miriam Kott Gutkowski from the Hebrew University of Jerusalem and Drs. Ofer Shenker and Vered Friedman from the Technion-Israel Institute of Technology in Haifa are thanked for provision of slide-related services.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST, a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arukwe A, Goksoyr A. Changes in three hepatic cytochrome P450 subfamilies during a reproductive cycle in turbot (Scophthalmus maximus L) J Exp Zool. 1997;277:313–325. doi: 10.1002/(SICI)1097-010X(19970301)277:4<313::AID-JEZ5>3.0.CO;2-S. [DOI] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock GC, The gene ontology consortium Gene ontology, tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R (2006) NCBI GEO: mining tens of millions of expression profiles, database and tools update. Nucleic Acids Res (NCBI, online) [DOI] [PMC free article] [PubMed]
- Bauchot ML, Hureau JC. Sparidae. In: Whitehead PJP, Bauchot ML, Hureau JC, Nielsen J, Tortonese E, editors. Fishes of the North- eastern Atlantic and the Mediterranean, Vol. II. London, UK: UNESCO; 1986. pp. 883–897. [Google Scholar]
- Besseau L. Etude Histo-Cytologique de la structure sexuelle d`une population de Lithognathus mormyrus (L) (Teleosteen, Sparide) Rapp Comm Int Medit. 1990;32:262–266. [Google Scholar]
- Bielli P, Calabrese L. Structure to function relationships in ceruloplasmin: a minmoonlightingmin protein. Cell Mol Life Sci. 2002;59:1413–1427. doi: 10.1007/s00018-002-8519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton CD, Garratt PA. Alternative reproduction styles in seabreams (Pisces, Sparidae) Environ Biol Fishes. 1990;28:113–124. doi: 10.1007/BF00751031. [DOI] [Google Scholar]
- Buxton CD, Smale MJ, Wallace JH, Cockcroft VG. Inshore small-mesh trawling survey of the Cape south coast Part 4 Contributions to the biology of some Teleostei and Chondrichthyes. S Afr J Zool. 1984;19:182–189. [Google Scholar]
- Chen Y, Chou C-C, Lu X, Slate E, Peck K, Xu W, Voit E, Almeida J. A multivariate prediction model for microarray cross-hybridization. BMC Bioinformat. 2006;7:101. doi: 10.1186/1471-2105-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R, Chalifa-Caspi V, Williams TD, Auslander M, George SG, Chipman JK, Tom M (2007) Estimating the efficiency of fish cross-species cDNA microarray hybridization. Mar Biotechnol (Available online) [DOI] [PubMed]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO, a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cossins AR, Crawford DL. Fish as models for environmental genomics. Nat Rev Genet. 2005;6:324–333. doi: 10.1038/nrg1590. [DOI] [PubMed] [Google Scholar]
- Coyle P, Philcox JC, Carey LC, Rofe AM. Metallothionein: the multipurpose protein. Cell Mol Life Sci. 2002;59:627–647. doi: 10.1007/s00018-002-8454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lukyanov S, Lau YFC, Siebert PD. Suppression subtractive hybridization, a versatile method for identifying differentially expressed genes. Methods Enzymol. 1999;303P:349–380. doi: 10.1016/S0076-6879(99)03022-0. [DOI] [PubMed] [Google Scholar]
- Flikka K, Yadetie F, Laegreid A, Jonassen I. XHM: a system for detection of potential cross hybridizations in DNA microarrays. BMC Bioinformat. 2004;5:117. doi: 10.1186/1471-2105-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froglia C. Feeding of Lithognathus mormyrus (L) in the Central Adriatic Sea (Pisces, Sparidae) Rapp Commun Intern Mer Medit. 1977;24:95–97. [Google Scholar]
- Funkenstein B, Dyman A, Levavi-Sivan B, Tom M. Application of real time PCR for quantitative determination of hepatic vitellogenin transcript levels in the striped seabream, Lithognathus mormyrus. Mar Environ Res. 2004;58:659–663. doi: 10.1016/j.marenvres.2004.03.058. [DOI] [PubMed] [Google Scholar]
- Gomme PT, McCann KB, Bertolini J. Transferrin: structure, function and potential therapeutic actions. Drug Discov Today. 2005;10:267–273. doi: 10.1016/S1359-6446(04)03333-1. [DOI] [PubMed] [Google Scholar]
- Gracey AY, Troll JV, Somero GN. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. PNAS. 2001;98:1993–1998. doi: 10.1073/pnas.98.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. 3. Oxford: Clarendon Press; 1999. p. 936. [Google Scholar]
- Hanumanthaiah R, Day K, Jagadeeswaran P. Comprehensive analysis of blood coagulation pathways in Teleostei: evolution of coagulation factor genes and identification of zebrafish factor VIIi. Blood Cells Mol Dis. 2002;29:57–68. doi: 10.1006/bcmd.2002.0534. [DOI] [PubMed] [Google Scholar]
- Hellman NE, Gitlin JD. Ceruloplasmin metabolism and function. Annu Rev Nutr. 2002;22:439–458. doi: 10.1146/annurev.nutr.22.012502.114457. [DOI] [PubMed] [Google Scholar]
- Herut B, Kress N. Particulate metals contamination in the Kishon River Estuary, Israel. Mar Pollut Bull. 1997;34:706–711. doi: 10.1016/S0025-326X(97)00018-0. [DOI] [Google Scholar]
- Huang X, Madan A. CAP3, A DNA sequence assembly program. Gen Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen H, Pehkonen P, Vehniainen E, Krasnov A, Rexroad C, Afanasyev S, Molsa H, Oikari A. Response of rainbow trout transcriptome to model chemical contaminants. Biochem Biophys Res Commun. 2004;320:745–753. doi: 10.1016/j.bbrc.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Kraljevic M, Dulcic J, Pallaoro A, Cetinic P, Jug Dujakovic J. Sexual maturation, age and growth of striped sea bream, Lithognathus mormyrus L, on the eastern coast of the Adriatic Sea. J Appl Ichthyol. 1995;11:1–8. doi: 10.1111/j.1439-0426.1995.tb00001.x. [DOI] [Google Scholar]
- Lambert LA, Perri H, Halbrooks PJ, Mason AB. Evolution of the transferrin family: conservation of residues associated with iron and anion binding. Comp Biochem Physiol B Biochem Mol Biol. 2005;142:129–141. doi: 10.1016/j.cbpb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Larkin P, Knoebl I, Denslow ND. Differential gene expression analysis in fish exposed to endocrine disrupting compounds. Comp Biochem Physiol B Biochem Mol Biol. 2003;136:149–161. doi: 10.1016/S1096-4959(03)00228-8. [DOI] [PubMed] [Google Scholar]
- Lasiak TA (1984) Aspects of the biology of three benthic-feeding teleosts from Kingmins Beach, Algoa Bay. S Afr J Zool 1951–1956
- Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micale V, Perdichizzi F. Further studies on the sexuality of the hermaphroditic teleost. J Fish Biol. 1994;45:661–670. doi: 10.1111/j.1095-8649.1994.tb00932.x. [DOI] [Google Scholar]
- Reynders H, Ven K, Moensa LN, Remortel P, Coen WM, Blust R. Patterns of gene expression in carp liver after exposure to a mixture of waterborne and dietary cadmium using a custom-made microarray. Aquat Toxicol. 2006;80:180–193. doi: 10.1016/j.aquatox.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sharan R, Maron-Katz A, Shamir R. CLICK and EXPANDER, a system for clustering and visualizing gene expression data. Bioinformatics. 2003;19:1787–1799. doi: 10.1093/bioinformatics/btg232. [DOI] [PubMed] [Google Scholar]
- Sheader DL, Gensberg K, Lyons BP, Chipman K. Isolation of differentially expressed genes from contaminant exposed European flounder by suppressive, subtractive hybridisation. Mar Environ Res. 2004;58:553–557. doi: 10.1016/j.marenvres.2004.03.044. [DOI] [PubMed] [Google Scholar]
- Sheader DL, Gensberg K, Lyons BP, Chipman JK. Oxidative stress response of European flounder (Platichthys flesus) to cadmium determined by a custom cDNA microarray. Mar Environ Res. 2006;62:33–44. doi: 10.1016/j.marenvres.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Snape JR, Maund SJ, Pickford DB, Hutchinson TH. Ecotoxicogenomics, the challenge of integrating genomics into aquatic and terrestrial ecotoxicology. Aquat Toxicol. 2004;67:143–154. doi: 10.1016/j.aquatox.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Srivastava AS, Kurokawa T, Suzuki T. Molecular cloning and cDNA sequence analysis of carboxypeptidases A1, A2 and B from the Japanese flounder Paralichthys olivaceus. Comp Biochem Physiol B Biochem Mol Biol. 2003;135:593–599. doi: 10.1016/S1096-4959(03)00123-4. [DOI] [PubMed] [Google Scholar]
- Suau P. Contribucion al estudio de la biologia de Lithognathus (Pagellus) Mormyrus L (Peces esparidos) Inv Pesq. 1970;34:237–265. [Google Scholar]
- Suzuki T, Srivastava AS, Kurokawa T. cDNA cloning and phylogenetic analysis of pancreatic serine proteases from Japanese flounder, Paralichthys olivaceus. Comp Biochem Physiol B Biochem Mol Biol. 2002;131:63–70. doi: 10.1016/S1096-4959(01)00487-0. [DOI] [PubMed] [Google Scholar]
- Tom M, Auslander M. Transcript and protein environmental biomarkers in fish-a review. Chemosphere. 2005;59:155–162. doi: 10.1016/j.chemosphere.2004.10.063. [DOI] [PubMed] [Google Scholar]
- Tom M, Myers CR, Waterman MR. Evaluating molar Cyp1a level in fish hepatic microsomes by competitive Elisa using recombinant membrane-free Cyp1a standard protein. Aquat Toxicol. 2002;59:101–114. doi: 10.1016/S0166-445X(01)00244-2. [DOI] [PubMed] [Google Scholar]
- Tom M, Shmul M, Shefer E, Chen N, Slor H, Rinkevich B, Herut B. Quantitative evaluation of hepatic cytochrome P4501A transcript, protein and catalytic activity in the striped sea bream, Lithognathus mormyrus. Environ Toxicol Chem. 2003;22:2088–2092. doi: 10.1897/02-275. [DOI] [PubMed] [Google Scholar]
- Tom M, Chen N, Segev M, Herut B, Rinkevich B. Quantifying fish metallothionein transcript by real time PCR for its utilisation as an environmental biomarker. Mar Pollut Bull. 2004;48:705–710. doi: 10.1016/j.marpolbul.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Oost R, Beyer J, Vermeulen NPE. Fish bioaccumulation and biomarkers in environmental risk assessment: a review. Environ Toxicol Pharmacol. 2003;13:57–149. doi: 10.1016/S1382-6689(02)00126-6. [DOI] [PubMed] [Google Scholar]
- Voskoboinik I, Trapani JA. Addressing the mysteries of perforin function. Immunol Cell Biol. 2006;84:66–71. doi: 10.1111/j.1440-1711.2005.01409.x. [DOI] [PubMed] [Google Scholar]
- Wang T, Secombes CJ. Complete sequencing and expression of three complement components, C1r, C4 and C1 inhibitor, of the classical activation pathway of the complement system in rainbow trout Oncorhynchus mykiss. Immunogenetics. 2003;55:615–628. doi: 10.1007/s00251-003-0622-5. [DOI] [PubMed] [Google Scholar]
- Whitcomb DC, Lowe ME. Human pancreatic digestive enzymes. Digest Dis Sci. 2007;52:1–17. doi: 10.1007/s10620-006-9589-z. [DOI] [PubMed] [Google Scholar]
- Williams TD, Gensberg K, Minchin SD, Chipman JK. A DNA expression array to detect toxic stress in European flounder (Platichthys flesus) Aquat Toxicol. 2003;65:141–157. doi: 10.1016/S0166-445X(03)00119-X. [DOI] [PubMed] [Google Scholar]
- Williams TD, Diab AM, George SG, Godfrey RE, Sabine V, Conesa A, Minchin SD, Watts PC, Chipman JK. Development of the GENIPOL European flounder (Platichthys flesus) microarray and determination of temporal transcriptional responses to cadmium at low dose. Environ Sci Technol. 2006;40:6479–6488. doi: 10.1021/es061142h. [DOI] [PubMed] [Google Scholar]
- Williams TD, Diab AM, George SG, Sabine V, Chipman JK. Gene expression responses of European flounder (Platichthys flesus) to 17-estradiol. Toxicol Lett. 2007;168:236–248. doi: 10.1016/j.toxlet.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Wren J, Kulkarni A, Joslin J, Butow R, Garner H. Cross-hybridization on PCR-spotted microarrays IEEE. Eng Med Biol Mag. 2002;21:71–75. doi: 10.1109/MEMB.2002.1046118. [DOI] [PubMed] [Google Scholar]
- Ylonen A, Helin J, Bogwald J, Jaakola A, Rinne A, Kalkkinen N. Purification and characterization of novel kininogens from spotted wolffish and Atlantic cod. Eur J Biochem. 2002;269:2639–2646. doi: 10.1046/j.1432-1033.2002.02927.x. [DOI] [PubMed] [Google Scholar]
- Yudkovski Y, Shechter A, Chalifa-Caspi V, Auslander M, Ophir R, Dauphin-Villemant C, Waterman M, Sagi A, Tom M (2007) Hepatopancreatic multi-transcript expression patterns in the crayfish Cherax quadricarinatus during the molt cycle. Insect Mol Biol (In press) [DOI] [PubMed]