Homeodomain-interacting protein kinase 2 (HIPK2) is a new CREB kinase for phosphorylation at Ser-271 but not Ser-133 in genotoxic stress and activates CREB transactivation function including brain-derived neurotrophic factor (BDNF) mRNA expression.
Abstract
CREB (cyclic AMP response element-binding protein) is a stimulus-induced transcription factor that plays pivotal roles in cell survival and proliferation. The transactivation function of CREB is primarily regulated through Ser-133 phosphorylation by cAMP-dependent protein kinase A (PKA) and related kinases. Here we found that homeodomain-interacting protein kinase 2 (HIPK2), a DNA-damage responsive nuclear kinase, is a new CREB kinase for phosphorylation at Ser-271 but not Ser-133, and activates CREB transactivation function including brain-derived neurotrophic factor (BDNF) mRNA expression. Ser-271 to Glu-271 substitution potentiated the CREB transactivation function. ChIP assays in SH-SY5Y neuroblastoma cells demonstrated that CREB Ser-271 phosphorylation by HIPK2 increased recruitment of a transcriptional coactivator CBP (CREB binding protein) without modulation of CREB binding to the BDNF CRE sequence. HIPK2−/− MEF cells were more susceptible to apoptosis induced by etoposide, a DNA-damaging agent, than HIPK2+/+ cells. Etoposide activated CRE-dependent transcription in HIPK2+/+ MEF cells but not in HIPK2−/− cells. HIPK2 knockdown in SH-SY5Y cells decreased etoposide-induced BDNF mRNA expression. These results demonstrate that HIPK2 is a new CREB kinase that regulates CREB-dependent transcription in genotoxic stress.
INTRODUCTION
CREB (cAMP response element-binding protein) belongs to the b-zip transcription factor family that contains a basic region for DNA binding and a leucine zipper domain involving dimerization within the same family members (Shaywitz and Greenberg, 1999; Mayr and Montminy, 2001). ATF1 (activating transcription factor 1; Hai et al., 1989) and CREM (cAMP response element modulator; Foulkes et al., 1991) are closely related to CREB in their amino acid sequences and functional domains (Mayr and Montminy, 2001; Lonze and Ginty, 2002). Besides a constitutive transactivator function of CREB through interaction with TORCs (transducers of regulated CREB; Conkright et al., 2003), CREB is a stimulus-coupled transcription factor that plays a crucial role in a wide range of signaling pathways such as those triggered by G-protein–coupled receptors, neurotrophin receptors, growth factor receptors, and calcium channels (Lonze and Ginty, 2002). CREB is also involved in neuroprotection under hypoxia and oxidative stress conditions (Lonze and Ginty, 2002). Stimulus-coupled activation of CREB has been extensively studied, in which PKA (protein kinase A; cAMP-dependent protein kinase) and several other serine/threonine kinases activate transactivation function of CREB through phosphorylation at Serine 133 located in the kinase-inducible domain (KID domain; Mayr and Montminy, 2001). The activation of CREB-mediated gene transcription is facilitated by subsequent recruitment of CREB-associated coactivator proteins such as CBP (CREB-binding protein; Chrivia et al., 1993) via the KID interaction (KIX) domain, whereas RGS13 (regulator of G-protein signaling) serves as a nuclear inhibitor of CREB transactivation function by interacting with Serine 133–phosphorylated CREB and CBP/p300 and decreasing both CREB binding to DNA and its interaction with CBP/p300 (Xie et al., 2008).
Besides CREB phosphorylation at Serine 133, calcium influx into neuronal cells was shown to induce CREB phosphorylation at two additional sites at Serines 142 and 143, in which the involvement of calmodulin kinase or casein kinase II was suggested by the effects of pharmacological inhibitors (Kornhauser et al., 2002). Phosphorylation of Serines 142 and 143 along with Serine 133 phosphorylation was required for efficient CREB-dependent transcription induced by calcium influx despite a decreased interaction between the CREB KID domain and the CBP KIX domain (Kornhauser et al., 2002). BDNF (brain-derived neurotrophic factor) is one of the major CREB-target genes regulated by calcium and plays a vital role in cell survival and adaptive neuronal responses (Shieh et al., 1998; Tao et al., 1998). CREB is also involved in genotoxic stress-coupled gene transcription during oxidative stress and DNA damage, such as ionizing radiation, UV light, and hydrogen peroxide. These stressors were shown to induce multiple and sequential phosphorylation of CREB leading to Serine 121 phosphorylation by the DNA damage-response kinase ATM (ataxia-telangiectasia–mutated) and casein kinases (Shanware et al., 2007). However, these phosphorylation events on CREB induced by DNA damage inhibited CREB–CBP interaction (Shanware et al., 2007), resulting in inhibition of CREB transcription activity (Shi et al., 2004).
Genotoxic stress activates a variety of protein kinases that relay signals to either cell cycle arrest or apoptosis. HIPK2 (homeodomain-interacting protein kinase 2; Kim et al., 1998), within a family of nuclear serine-threonine kinases that share significant homologies with the Dyrk dual specificity kinase family (Hofmann et al., 2000) has recently been characterized as a DNA-damage responsive kinase that plays a key regulatory role in both cell survival and apoptosis (Rinaldo et al., 2007). For instance, HIPK2 was shown to be proapoptotic in response to UV light exposure through direct phosphorylation of p53 at Serine 46, preventing MDM2-mediated p53 degradation and ultimately leading to activation of p53-dependent apoptotic pathways (D'Orazi et al., 2002; Hofmann et al., 2002). Accumulating evidence has shown that broader genotoxic stress agents including ionizing radiation (Dauth et al., 2007), DNA-damaging chemotherapeutic agents (such as cisplatin and doxorubicin; Di Stefano et al., 2004; Calzado et al., 2007; Rinaldo et al., 2007) and a cyclin-dependent kinase 2 inhibitor roscovitine (Wesierska-Gadek et al., 2007) activate HIPK2, which in turn activates p53. In addition to p53-dependent proapoptotic role of HIPK2, the transcriptional corepressor and antiapoptotic protein CtBP was shown to be phosphorylated at Serine 422 by HIPK2, resulting in CtBP degradation by the proteasome and promoting UV-induced apoptosis (Zhang et al., 2003). HIPK2 also plays an antiapoptotic role; for instance, HIPK2 is crucial for TGF-β–mediated survival of midbrain dopamine neurons through its interaction with Smads for activation of TGF-β target genes (Zhang et al., 2007). Interestingly, it was demonstrated that loss of HIPK2 caused increased proliferative potential in association with suppression of β-catenin–mediated activation of cyclin D1, and in the same study the loss of HIPK2 increased susceptibility to the two-stage carcinogenesis protocol (Wei et al., 2007), suggesting that HIPK2 has a tumor suppressor function.
To date, only a few HIPK2 protein substrates, their phosphorylation sites, and the biofunctional impact of phosphorylation have been identified and characterized (D'Orazi et al., 2002; Hofmann et al., 2002; Zhang et al., 2003; Aikawa et al., 2006; Kim et al., 2006; Wee et al., 2008), even though dozens of cellular proteins have been reported to be regulated by HIPK2 (Rinaldo et al., 2007). In this study we found that HIPK2 is a new CREB kinase that phosphorylates Serine 271 but not Serine 133 and regulates CREB-dependent gene transcription including the BDNF gene and genotoxic response.
MATERIALS AND METHODS
Cell Culture and Reagents
SH-SY5Y human neuroblastoma, K562 human erythroleukemia, and 293 human embryonal kidney cells transformed with adenovirus 5 DNA were purchased from American Type Culture Collection (ATCC, Manassas, VA). SH-SY5Y cells were cultured in a 1:1 mixture of Eagle's minimum essential medium (MEM) and HAM's F12 medium supplemented with nonessential amino acids and 10% fetal bovine serum (FBS, Mediatech, Manassas, VA). K562 were cultured in RPMI 1640 medium supplemented with 25 mM HEPES, 0.3g/l l-glutamine, and 10% FBS. 293 cells were cultured in MEM with 2 mM l-glutamine, Eagle's BBS, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 10% FBS. HIPK2+/+ and −/− mouse embryonic fibroblasts (MEFs), kindly provided by Dr. Huang at University of California San Francisco (Wiggins et al., 2004; Wei et al., 2007), were cultured in DMEM with 2 mM l-glutamine and 10% FBS. These cells were cultured at 37°C and 5% CO2 in a humidified atmosphere. Etoposide was purchased from BioMol (Plymouth Meeting, PA) and dissolved in methanol. Cisplatin was purchased from Calbiochem (La Jolla, CA) and dissolved in water at 1 mg/ml.
Plasmids, DNA transfection, and Luciferase Reporter Assay
Human CREBα341 cDNA was purchased from Invitrogen and subcloned into pCMV or pQE vector. CREB point mutations (133A, 271A, 133E, 271E, and double mutations) were constructed with the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mouse PKA cDNA was purchased from Clontech (Palo Alto, CA) and subcloned into cytomegalovirus (pCMV) vector. cDNAs for mouse CBP and human p300, kind gifts from Drs. R. Goodman (Vollum Institute, Oregon Health Science University) (Chrivia et al., 1993) and R. Eckner (New Jersey Medical School) (Eckner et al., 1994), were described previously (Tsuji et al., 1999). pCMVFlagHIPK2 and kinase dead (kd) mutant (K221R) plasmids (Kanei-Ishii et al., 2004) were used throughout the experiments along with similar effects observed with pcDNA3FlagHIPK2wt and K221A kindly provided by Dr. Schmitz (Justus-Liebig University, Germany; Hofmann et al., 2000, 2002). pLucMCS-luciferase reporter plasmid was purchased from Stratagene. CRE4-luciferase was constructed by insertion of four copies of an annealed double-strand oligonucleotide (5′-AGCCTGACGTCAGAG-3′) into pLucMCS-luciferase plasmid. Rat BDNFpIII-luciferase plasmid was generated by insertion of a 41-base pair annealed double-strand oligonucleotides (5′-AGCTCACGTCAAGGCAGCGTGGAGCCCTCTCGTGGACTCCC-3′) into pLucMCS-luciferase. Transient DNA transfection into MEFs was carried out by electroporation (X-Cell, Bio-Rad, Hercules, CA) with an optimized preset condition by Bio-Rad for NIH3T3 cells (exponential decay, 1000 μF, 155 V). Cells were suspended in 100 μl of media with plasmid DNA in a cuvette with a 0.2-cm gap. Transient DNA transfection into K562 cells was carried out with jetPEI (Polyplus, New York, NY) according to manufacturer's instructions. As a transfection internal control, 10 ng of pRL-null (Promega, Madison, WI) was simultaneously cotransfected. Luciferase assays were performed using Dual Luciferase Assay Reagents (Promega). Firefly luciferase expression was normalized by Renilla luciferase activity.
Immunoprecipitation and Western Blotting
One hundred to 500 μg of 293 whole cell lysates were used for immunoprecipitation with 1 μg of anti-HIPK2 (sc-10294, Santa Cruz Biotechnology, Santa Cruz, CA) and protein G-plus agarose (Calbiochem, IP04). Proteins separated on 7.5, 10, or 12.5% SDS-PAGE were transferred to Immobilon-P polyvinylidene difluoride membrane (Millipore, Bedford, MA) and incubated at 4°C overnight with primary antibodies for HA (HA11, Covance, Princeton, NJ, or SC-805, Santa Cruz Biotechnology), Flag (F1804, Sigma, St. Louis, MO), CREB (sc-240x, sc-186x, Santa Cruz Biotechnology), HIPK2, or CBP (sc-369x, Santa Cruz Biotechnology). Protein phosphatase treatment was carried out by incubating 45 μg of whole cell lysate from CREB or CREB plus HIPK2 transfected K562 cells with 20 U of lamda phosphatase (New England Biolabs, Ipswich, MA) for 30 min at 37°C before Western blotting with anti-CREB antibody. Nuclear fractionation was carried out using a nuclear extraction kit (Active Motif, Carlsbad, CA). Apoptosis was monitored by Caspase 3 cleavage in MEF cells treated with 2, 10, or 50 μM etoposide for 12–48 h. Total cell lysates were subjected to Western blotting using anti-Caspase 3 antibody (9665, Cell Signaling, Danvers, MA). After incubation with secondary antibodies conjugated with horseradish peroxidase, proteins were visualized using an ECL detection kit (HyGLO, Denville Scientific, Metuchen, NJ) or advanced ECL kit (GE Healthcare, Piscataway, NJ).
In Vitro Kinase Assay
His-tagged wild-type (wt) or mutant CREB proteins were expressed and purified with pQE vector system (Qiagen, Valencia, CA). They were incubated with 9 ng of recombinant HIPK2 (aa 165-564, Millipore) or 0.1 ng of PKA (Millipore) for 30 min at 30°C in the kinase buffer (10 mM HEPES, pH 7.4, 5 mM MgCl2, 1 mM DTT) containing 5 μCi ]γ-32P]ATP and 100 μM ATP. Samples were then separated on 10% SDS-PAGE and subjected to silver staining (Sigma) and autoradiography. For measuring HIPK2 activity in cells, Flag-HIPK2–transfected K562 cells were treated with 50 μM etoposide or 0.5 μg/ml cisplatin for 6 h and harvested in the lysis buffer containing 20 mM HEPES, pH 7.4, 150 mM NaCl, 12.5 mM β-glycerophosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 2 mM DTT, 1 mM Na3VO4, 0.5% Triton X-100, and 20 μM aprotinin. Cell lysates, 100 μg, were immunoprecipitated with anti-Flag antibody or control rabbit IgG in 1× kinase buffer containing 10 mM HEPES, pH 7.4, 5 mM MgCl2, 1 mM DTT, and 100 μM ATP. The immunoprecipitates were used for in vitro kinase assay using 1 μg of myelin basic protein (MBP, Sigma) as a substrate. After incubation at 30°C for 30 min or 22°C for 4 min, samples were separated on 12.5% SDS-PAGE, stained with Coomassie Brilliant Blue (CBB), and subjected to autoradiography.
Small Interfering RNA Transfection and real-time PCR
SH-SY5Y cells, 1 × 107, were electroporated with 100 pmol of nontargeting small interfering RNA (siRNA; 5′-UAGCGACUAAACACAUCAAUU-3′, D-001210-01; Dharmacon, Lafayette, CO) or siHIPK2 (target sequence: 5′-GAGAAUCACUCCAAUCGAA-3′, J-003266-10; Dharmacon) using Gene Pulser X-Cell (square wave, 25 ms, 110 V) in 100 μl FBS- and antibiotic-free media. After incubation of electroporated SH-SY5Y cells in the electroporation cuvette for 10 min at room temperature, the cells were suspended in 10 ml of growth media and plated with 2 ml cell suspension per well in six-well plates and incubated for 24 h. Cells were then treated with 2 or 10 μM etoposide, or methanol as a control for 48 h, and RNA was isolated with TRIzol reagent (Invitrogen). Real-time PCR was carried out to measure HIPK2, BDNF, or GAPDH mRNA with SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) in the presence of primer set for HIPK2 (QT00051485, Qiagen), BDNF (5′-CGCCATGCAATTTCCACTATCAATAATTTAAC-3′ and 5′-ACTTTTCAGTCACTACTTGTCAAAGTAACC-3′), or GAPDH (QT00079247, Qiagen).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assays were carried out as described previously (Sakamoto et al., 2009) using a ChIP assay kit (Millipore) with minor modifications. Briefly, 1 × 107 SH-SY5Y cells were transfected with 20 μg of plasmid DNA as indicated in figure legends and plated into two dishes of 35-mm plate. After incubation for 40 h, cells were subjected to chromatin cross-linking and preparation of cell lysates. Approximately one-tenth aliquots of cell lysate containing sheared DNA by sonication (Iwasaki et al., 2007) were immunoprecipitated with 2–4 μg each of antibodies for HA, Flag, CREB, CBP, or control goat IgG. PCR was performed in 50 μl reactions containing [32P]dCTP, Advantage 2 PCR polymerase mix (Clontech), and a pair of primers (5′-GCGCTGAATTTTGATTCTGGTAAT-3′, 5′-AATGGGAAAGTGGGTGGGAG-3′) to amplify 0.1-kb region of the human BDNF exon III promoter containing the CREB-binding site. The PCR samples were loaded and separated on an 8% acrylamide gel and subjected to autoradiography.
RESULTS
HIPK2 Phosphorylates CREB at Serine 271 But Not Serine 133
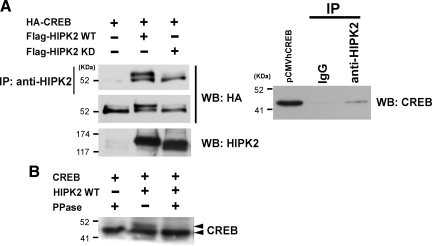
Our yeast-two hybrid screening for ATF1-interacting proteins cloned HIPK2 (unpublished observation). On the basis of the fact of high homology between CREB and ATF1 (Mayr and Montminy, 2001; Lonze and Ginty, 2002), we hypothesized that HIPK2 may also interact with CREB and regulate CREB function. To test this hypothesis, HIPK2 and HA-CREB expression plasmids were cotransfected, and their protein–protein interaction was examined by immunoprecipitation/Western blot. Immunoprecipitation of wtHIPK2 or kdHIPK2 with an anti-HIPK2 antibody coprecipitated HA-CREB (Figure 1A), suggesting that HIPK2, regardless of its kinase status, interacts with CREB. Immunoprecipitation of nontransfected 293 cells with the anti-HIPK2 antibody also coprecipitated endogenous CREB protein (Figure 1A). Of note is that cotransfection of HA-CREB along with wtHIPK2, but not kdHIPK2, induced a retarded form of HA-CREB on SDS-PAGE (Figure 1A). To test whether this retarded migration of CREB is due to phosphorylation of CREB, CREB- and wtHIPK2-transfected cell lysate was treated with protein phosphatase before loading on SDS-PAGE. Indeed, the protein phosphatase treatment abolished the retarded CREB band (Figure 1B), suggesting that HIPK2 expression caused CREB phosphorylation.
Figure 1.
HIPK2 interacts with CREB and induces CREB retardation on SDS-PAGE. (A) Left, 293 cells were transfected with HA-CREB alone, HA-CREB plus Flag-HIPK2 wild type (WT), or HA-CREB plus Flag-HIPK2 kinase dead (KD). Whole cell lysates prepared after 48-h incubation were immunoprecipitated with anti-HIPK2 antibody, and the immunoprecipitates were subjected to Western blotting with anti-HA antibody (top). The whole cell lysates were also analyzed by Western blotting for expression of transfected HA-CREB (middle) and HIPK2 (bottom). Right, whole cell lysate of nontransfected 293 cells was immunoprecipitated with goat IgG or anti-HIPK2 antibody, followed by Western blotting with anti-CREB antibody. pCMVhumanCREB-transfected cell lysate was loaded as a CREB positive control. (B) K562 cells were transfected with nontagged CREB, or CREB plus Flag-HIPK2-WT, and whole cell lysates were treated with protein phosphatase (PPase +) before SDS-PAGE and Western blotting with anti-CREB antibody.
The major regulatory mechanism of CREB is phosphorylation of Serine 133 by PKA, or several other kinases such as MSK1 (mitogen- and stress-activated kinase 1) and calcium-calmodulin kinases, leading to activation of transcription function of CREB (Mayr and Montminy, 2001). HIPK2 preferentially phosphorylates Ser/Thr sites adjacent to Proline residues (see Discussion). Serine 133 in CREB is flanked by Proline 132; therefore, we anticipated that HIPK2 might be another Serine 133 kinase. To test this possibility, Serine 133 to Alanine mutant CREB (133A) was coexpressed with HIPK2 in K562 cells, and cell lysates were subjected to Western blotting for the retarded migration of CREB. wtCREB reproducibly showed the retarded CREB band when wtHIPK2 was coexpressed (Figure 2A). Under this condition, the CREB 133A mutant gave rise to the same retarded band induced by HIPK2 (Figure 2A), suggesting that the CREB retardation, which was sensitive to phosphatase treatment (Figure 1B), was not due to Serine 133 phosphorylation.
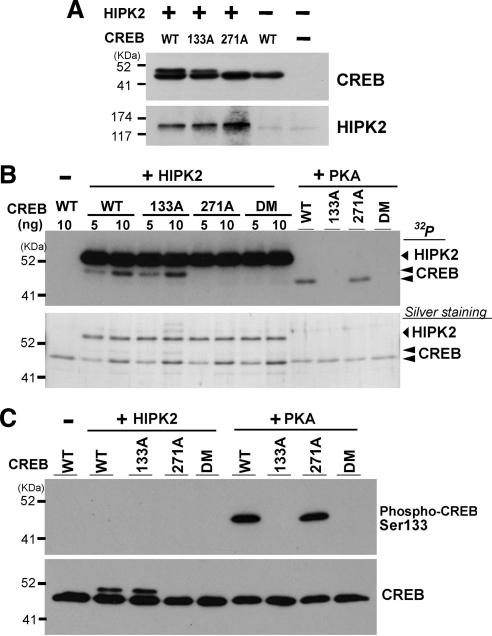
Figure 2.
HIPK2 phosphorylates CREB at Serine 271 but not Serine 133. (A) CREB-WT, CREB 133A, or CREB 271A mutant was cotransfected with FlagHIPK2-WT in K562 cells. Whole cell lysates were prepared after 48 h and subjected to SDS-PAGE and Western blotting with anti-CREB or anti-HIPK2 antibody. (B) Five or 10 ng of His-tagged recombinant CREB-WT, 133A, 271A, or 133A/271A double mutant (DM) was incubated with 9 ng recombinant HIPK2 (kinase domain aa. 165–564) in the presence of [γ-32P]ATP for 30 min at 30°C. Five nanograms of these recombinant CREB proteins was also incubated with recombinant PKA. Samples were loaded on SDS-PAGE, and phosphorylated bands were detected by autoradiograph. Comparable loading of the samples was verified by silver staining of the gel (bottom). (C) The same set of recombinant CREB proteins was incubated with recombinant HIPK2 or PKA in the presence of cold ATP, followed by SDS-PAGE and Western blotting with anti-phospho Serine 133 CREB (top) or CREB antibody (bottom).
There are eight potential phosphorylation sites in the human CREB protein by HIPK2, in which Serine or Threonine is adjacent to Proline (Ser-76, Ser-80, Ser-133, Thr-172, Ser-237, Thr-259, Ser-271, and Thr-276). Among these, amino acid sequences containing Ser-133, Thr-172, Ser-237, and Ser-271 of CREB are highly conserved in human ATF1 Ser-63, Thr-99, Ser-164, and Ser-198, respectively. Our preliminary experiments suggested that HIPK2 appears to phosphorylate ATF1 at Serine 198 (unpublished observation), suggesting that the corresponding CREB Serine 271 may be the HIPK2-mediated phosphorylation site. To test this possibility, we introduced Serine 271-to-Alanine mutation in CREB (271A) and performed coexpression with wtHIPK2 and CREB271A followed by Western blotting to detect the retarded migration. As shown in Figure 2A, the 271A mutation completely abolished the retarded migration of CREB, suggesting that HIPK2 phosphorylates CREB at Serine 271 and induces its slower migration in SDS-PAGE.
These results do not exclude the possibility of additional CREB phosphorylation sites by HIPK2, including Serine 133 that may not elicit the retarded migration. To test this possibility and to address whether HIPK2 directly phosphorylates CREB at Serine 271, we performed in vitro kinase assays by incubating recombinant wt or mutant CREB protein and HIPK2 in the presence of [γ-32P]ATP. As shown in Figure 2B, recombinant HIPK2 phosphorylated both wtCREB and 133A mutant CREB but failed to phosphorylate 271A or 133A/271A double mutated CREB (DM). These results indicate that HIPK2 directly phosphorylates CREB at Serine 271 and that Serine 271 is the sole phosphorylation site of CREB by HIPK2. As a control experiment, we incubated recombinant PKA with these recombinant CREB proteins to verify that our 133A CREB mutants are devoid of PKA-mediated phosphorylation. Indeed, PKA phosphorylated wt and 271A CREB proteins but failed to phosphorylate 133A and 133A/271A mutant CREB proteins (Figure 2B). Furthermore, we observed in Figure 2B that the phosphorylated recombinant CREB by HIPK2 (phospho-Serine 271 CREB) showed slower migration than the phosphorylated CREB by PKA (phospho-Serine 133 CREB), which is consistent with the CREB retardation seen in the cells (Figures 1 and 2A). To further rule out the possibility of CREB Serine 133 phosphorylation by HIPK2, recombinant CREB proteins incubated with HIPK2 or PKA in vitro were subjected to Western blotting with a phospho-Serine 133 CREB antibody (Figure 2C, top) or CREB antibody (Figure 2C, bottom). PKA-phosphorylated CREB proteins (wt and 271A) were detected by the phospho-Serine 133 CREB antibody but HIPK2-phosphorylated CREB proteins (wt and 133A, giving rise to the retarded CREB, Figure 2C, bottom) were not detected by the phospho-Serine 133 antibody. Taken all together, we concluded that HIPK2 phosphorylates CREB only at Serine 271.
Serine 271 Phosphorylation by HIPK2 Enhances a Transactivation Function of CREB
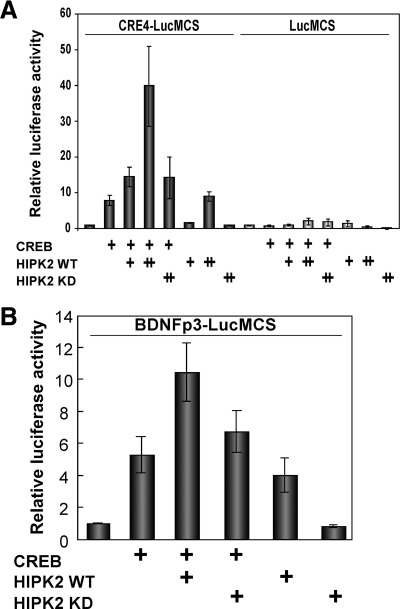
To explore the role of HIPK2 in CREB-dependent transcription, we performed CRE-luciferase reporter assays in cells transiently transfected with CREB and HIPK2. CREB alone activated tandem four copies of CRE-dependent luciferase (CRE4) transcription by approximately 10-fold in K562 cells (Figure 3A). Cotransfection of wt HIPK2 with CREB showed further activation, whereas the kdHIPK2 showed no effect (Figure 3A). Transfection of wtHIPK2 alone also slightly activated the CRE-dependent luciferase expression (Figure 3A), probably by the effect of HIPK2 on endogenous CREB-WT. We then asked whether a CRE enhancer derived from a CREB-target gene is also regulated by HIPK2. The exon III of BDNF gene has a functional CRE-enhancer, and we tested a BDNF-luciferase reporter. As shown in Figure 3B, the BDNF CRE enhancer was activated by CREB, and it was further activated by wtHIPK2 but not by kdHIPK2. These results suggest that HIPK2 activates CREB-dependent transcription in a kinase activity–dependent manner.
Figure 3.
HIPK2 activates CREB-dependent transcription in a kinase-dependent manner. (A) CREB, 0.1 μg, along with 0.1 μg (+) or 0.3 μg (++) HIPK2-WT or HIPK2-KD plasmids were cotransfected with pCRE 4 copies (CRE4)-luciferase or pTATA-luciferase reporter into K562 cells. (B) CREB, 0.1 μg, along with 0.3 μg HIPK2-WT or HIPK2kd plasmids were cotransfected with rat BDNF promoter III-luciferase into K562 cells. In both A and B, cells were harvested after 24 h to measure luciferase expression. Relative luciferase activity is shown by setting each reporter transfection alone as 1.0. Average of four experiments are shown with SEs.
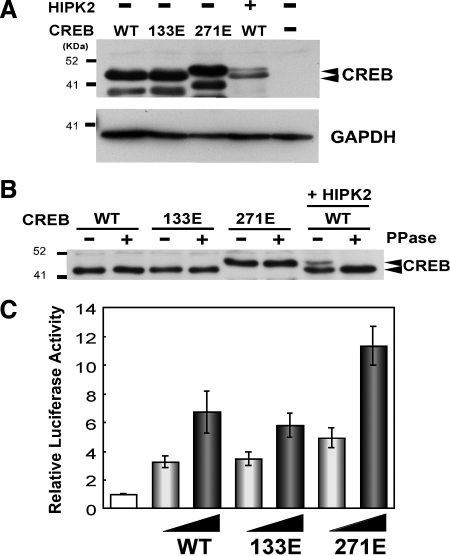
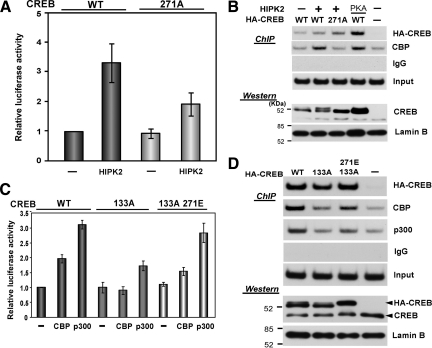
To further elucidate the role of HIPK2 in CREB phosphorylation at Serine 271, we generated the CREB Serine 271-to-glutamic acid mutant (271E) to mimic Serine 271 phosphorylated CREB. As a control, we generated the Serine 133 to glutamic acid mutant CREB (133E). First, we expressed these CREB mutants in K562 cells and tested whether the replacement of Serine with glutamic acid causes the retardation in SDS-PAGE. As shown in Figure 4A, 271E CREB showed the retarded migration that was similar to one induced by coexpression of wtCREB and HIPK2, whereas the 133E CREB mutant did not show the retarded migration. The retardation of 271E CREB was resistant to phosphatase treatment (Figure 4B), excluding the possibility of other CREB phosphorylation sites for the retardation. Then, we tested the effect of these glutamic acid CREB mutants on CRE-dependent transcription by a luciferase reporter assay. Compared with the CRE-dependent transcription activated by wtCREB or 133E CREB, 271E CREB activated it 1.8–2.0-fold higher than wt or 133E CREB (Figure 4C). No difference in the CREB activity between wt and 133E in this assay appears to be consistent with a previous report showing no significant impact of Serine 133 to glutamic acid replacement on CREB and coactivator interaction (Solt et al., 2006). Furthermore, cotransfection of wtCREB or Serine 271-to-Alanine mutant CREB (271A) along with HIPK2 in CRE4-luciferase assays demonstrated that the mutation of Serine 271 to Alanine diminished HIPK2-mediated activation (Figure 5A). Collectively, these results suggest that the phosphorylation of CREB at Serine 271 by HIPK2 enhanced the transactivation function of CREB.
Figure 4.
Replacement of CREB Serine 271 with glutamic acid caused the retarded migration in SDS-PAGE and enhanced CREB transactivation function. (A) CREB-WT, 133E, 271E mutant alone or CREB-WT plus Flag-HIPK2 was transfected into K562 cells. Whole cell lysates were prepared after 48 h and subjected to SDS-PAGE and Western blotting with anti-CREB antibody. Intact CREB proteins (retarded and nonretarded) are indicated with arrowheads. The smaller CREB proteins appear to be produced by unknown proteolytic cleavage. GAPDH Western blotting is shown as a loading control. (B) K562 cells were transfected with CREB-WT, CREB 133E, CREB 271E, or CREB-WT plus HIPK2. Whole cell lysates were treated with protein phosphatase (PPase+) before SDS-PAGE and Western blotting with anti-CREB antibody. (C) 0.1 or 0.3 μg of CREB-WT, Serine-to-glutamic acid mutant 133E or 271E expression plasmid was transfected with CRE4-luciferase reporter into K562 cells. Cells were harvested after 24 h incubation, and luciferase expression was measured. Luciferase expression from the reporter alone was set to 1.0. Average from eight independent experiments are shown with SEs.
Figure 5.
CREB phosphorylation at Serine 271 by HIPK2 increased CBP recruitment to the CRE site in the BDNF promoter. (A) CREB-WT or CREB 271A was transfected with HIPK2 and a CRE4-luciferase reporter into K562 cells. After 24 h, cells were harvested to measure luciferase expression. Relative luciferase activity (fold) is shown by setting CREB-WT alone as 1.0. Average of seven experiments with SEs are shown. (B) pCMVHA-CREBwt or 271A mutant was electroporated into 1 × 107 SH-SY5Y cells along with pCMVFlagHIPK2 or pCMVPKA and plated into two dishes. Forty-eight hours after electroporation, one group was used for ChIP assays for the CRE site in the BDNF promoter immunoprecipitated with control IgG, anti-CREB, or anti-CBP antibody (top). Another set of cells was used for isolation of nuclear proteins and 30 μg each was loaded on SDS-PAGE and analyzed by Western blotting with anti-CREB, or anti-lamin B (bottom). (C) pCMVCREBwt, 133A, or 133A271E was cotransfected with CRE4-luciferase reporter into 293 cells along with pCMV-CBP or pCMV-p300. Twenty-four hours after transfection, cells were harvested for luciferase assay. Luciferase expression from CRE4-luciferase with wtCREB was set to 1.0. Average from four independent experiments with SEs are shown. (D) 1 × 107 SH-SY5Y cells were electroporated with HA-CREBWT, 133A, 133A/271E, or empty vector and divided into two plates each. One set was subjected to ChIP assays for the CRE site in the BDNF promoter immunoprecipitated with anti-HA, anti-CBP, anti-p300, or control IgG (top). Another set of cells was used for isolation of nuclear proteins and 30 μg each was analyzed by Western blotting with anti-CREB or anti-lamin B (bottom).
Phosphorylation of CREB by HIPK2 Facilitates the Recruitment of the Coactivator CBP
To define the mechanism through which Serine 271 phosphorylation by HIPK2 enhances the transactivation function of CREB, we asked whether Serine 271 phosphorylation affects the DNA-binding ability of CREB. To this end, we expressed HA-tagged wt or 271A CREB together with HIPK2 in SH-SY5Y human neuroblastoma cells and performed CREB ChIP assays for the BDNF exon III CRE site. Under similar expression levels of wtCREB (with or without HIPK2 expression) and 271A CREB (Figure 5B, Western blot with nuclear extracts), binding of these CREB proteins to the BDNF promoter III was correlated with their CREB expression levels (Figure 5B). We also tested nuclear extracts isolated from these cells for their binding to a canonical CRE DNA sequence by gel retardation assays, in which no difference was observed in the ability of DNA binding between wtCREB and HIPK2-phosphorylated wtCREB or 271E CREB (Supplemental Figure S1). These results suggest that Serine 271 phosphorylation of CREB by HIPK2 did not alter the binding of CREB to the CRE site.
Because the CREB-dependent transcription is regulated by cooperation of CREB and CREB-associated proteins, we next examined whether the recruitment of CBP, a histone acetyltransferase serving as a coactivator of CREB, is affected by HIPK2-induced CREB Serine 271 phosphorylation. After expression of wtCREB alone or coexpression of wtCREB or 271A CREB mutant together with HIPK2 in SH-SY5Y cells, ChIP assays were performed to measure interaction of endogenous CBP with the CRE site in the BDNF promoter III. Coexpression of wtCREB along with PKA was used as a positive control of enhanced CBP recruitment to CREB via Serine 133 phosphorylation. Under equivalent expression levels of transfected CREB proteins, CBP recruitment to the BDNF promoter III was increased when wtCREB was coexpressed with HIPK2 or PKA (Figure 5B). Consistently, coexpression of 271A CREB mutant along with HIPK2 showed no increase in CBP recruitment (Figure 5B). Furthermore, in CRE4-luciferase reporter assays, cotransfection of wt or Serine 133A, or 133A271E double-mutated CREB with p300 or CBP demonstrated that CBP and p300 activated wtCREB but not 133A CREB; however, 133A271E regained the CBP/p300-mediated activation (Figure 5C). Consistently, ChIP assays in Figure 5D showed that decreased recruitment of CBP or p300 to 133A CREB compared with wtCREB was recovered by introduction of 271E phosphomimetic mutation in 133A CREB (133A/271E). Taken together, these results suggest that CREB phosphorylation at Serine 271 by HIPK2 increased the CBP/p300 recruitment or stabilized the recruited CBP/p300 in the CREB-binding complex.
Retarded Migration of Endogenous CREB by Etoposide via HIPK2
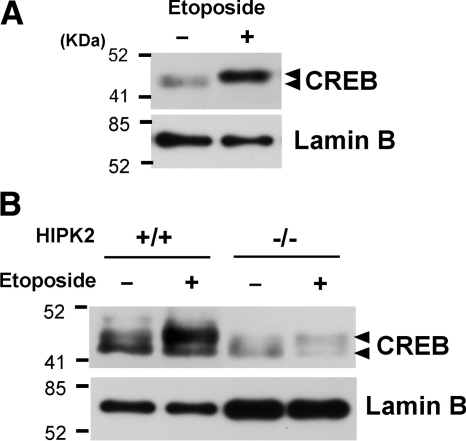
HIPK2 was shown to be activated by several genotoxic stress agents and plays a key regulatory role in both cell survival and apoptosis (Rinaldo et al., 2007). We next asked whether endogenous CREB protein is phosphorylated when HIPK2 is activated. To address this question, 293 cells were treated with etoposide for 6 h and examined the retardation of endogenous CREB protein on SDS-PAGE. As shown in Figure 6A, treatment with 50 μM etoposide showed increased expression of endogenous CREB along with retarded migration. To elucidate the involvement of HIPK2 in this retardation, wtHIPK2 (HIPK2+/+) MEFs or HIPK2 knockout MEF (HIPK2−/−) cells were treated with 10 μM etoposide, and nuclear extracts were separated on SDS-PAGE for retardation of endogenous CREB. As shown in Figure 6B, etoposide treatment significantly induced retarded migration of CREB in HIPK2+/+ MEF cells compared with HIPK2−/− cells. Collectively, these results suggest that endogenous CREB is phosphorylated by etoposide treatment via HIPK2.
Figure 6.
Retarded migration of endogenous CREB by etoposide via HIPK2. (A) 293 cells were treated with methanol (−) or 50 μM etoposide (+) for 6 h. Nuclear extracts were subjected to SDS-PAGE and Western blotting with anti-CREB antibody. Western blotting with anti-lamin B antibody is shown as a loading control. (B) HIPK2+/+ or −/− MEF cells were treated with 10 μM etoposide for 2 h. Nuclear extracts were subjected to SDS-PAGE and Western blotting with anti-CREB or anti-lamin B antibody.
Etoposide Induces CRE-dependent Transcription and BDNF mRNA via HIPK2
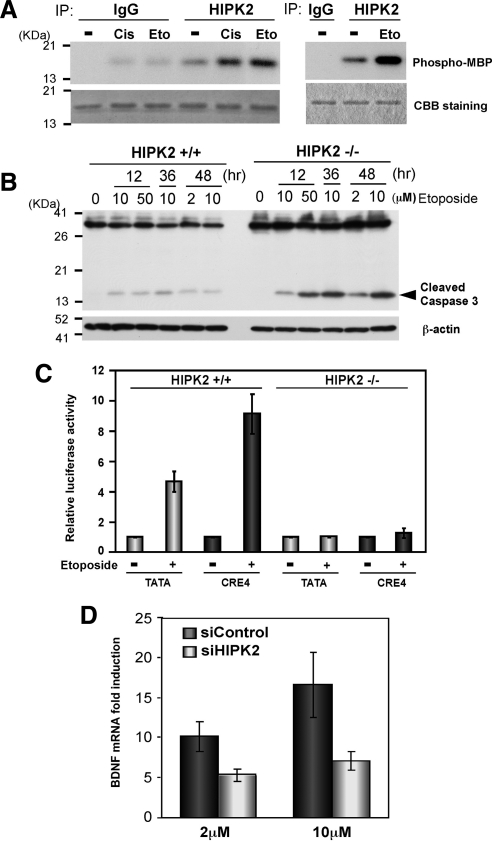
Our in vitro kinase assays with immunoprecipitates of HIPK2 also demonstrated that treatment of cells with etoposide as well as cisplatin activated HIPK2 (Figure 7A). We observed that HIPK2−/− MEFs are more susceptible to etoposide-induced Caspase 3 activation (Figure 7B) and cytotoxicity observed by increased number of cells detaching from culture plates. To understand a physiological role of HIPK2 in genotoxic stress and CREB-dependent gene regulation, we asked whether a DNA-damaging agent activates CRE-dependent transcription and whether it is regulated by HIPK2. To address these questions, HIPK2+/+ or HIPK2−/− cells (HIPK2 mRNA levels in Supplemental Figure S2) were transfected with CRE- or control- (TATA-) luciferase reporter, treated with 10 μM etoposide, and subjected to luciferase assay. In HIPK2+/+ MEF cells, etoposide activated CRE-dependent luciferase expression (9-fold), higher than the activation of the TATA-luciferase (4.5-fold). In contrast, HIPK2−/− MEF cells showed much lower luciferase expression and no difference in CRE- or TATA-luciferase expression even after etoposide treatment (Figure 7C). These results suggest that etoposide treatment activated the CRE-dependent transcription through HIPK2. CREB and CBP bind the promoter of BDNF exon III and it was regulated by HIPK2 in SH-SY5Y cells (Figure 5). We then tried to verify in SH-SY5Y cells whether etoposide induces BDNF mRNA in a HIPK2-dependent manner by knocking down HIPK2 with siRNA. SH-SY5Y cells transfected with nontargeting siRNA (siControl) or HIPK2 siRNA (siHIPK2) were treated with 2 or 10 μM etoposide and BDNF mRNA levels were measured. Given ∼70% knockdown of HIPK2 mRNA measured by RT-PCR (Supplemental Figure S2), etoposide-induced BDNF mRNA expression in siControl SH-SY5Y cells, but it was diminished in HIPK2-knockdown cells (Figure 7D). Collectively, these results suggest that HIPK2 regulates etoposide-induced CRE-dependent transcription, such as BDNF, that may be involved in cell survival against DNA damaging agents.
Figure 7.
Etoposide induces CRE-dependent transcription and BDNF mRNA via HIPK2. (A) FlagHIPK2-transfected K562 cells were treated with 0.5 μg/ml cisplatin or 50 μM etoposide for 6 h and harvested for immunoprecipitation with anti-HIPK2 antibody or control IgG. The immunoprecipitates from two independent experiments were incubated with recombinant MBP at 30°C for 30 min (left) or 22°C for 4 min (right) in the presence of [γ-32P]ATP and loaded on SDS-PAGE. Phosphorylated MBP was detected by autoradiography (top). Comparable protein loading was verified by Coomassie Brilliant Blue (CBB) staining (bottom). (B) Twenty-four hours after plating 5 × 106 HIPK2+/+ or −/− MEF cells, they were treated with indicated concentrations of etoposide for 12, 36, or 48 h. Whole cell lysates were analyzed by Western blotting with anti-Caspase 3 antibody. β-actin Western blot is shown as a loading control. (C) HIPK2+/+ or −/− MEF cells were transfected with TATA-luciferase or CRE4-luciferase and incubated for 24 h. Then cells were treated with 10 μM etoposide for 48 h and harvested for luciferase assay. Luciferase expression with each reporter without etoposide treatment was set to 1.0, and mean values from five independent experiments are shown; error bars, SEs. (D) 1 × 107 SH-SY5Y cells were transfected with nontargeting (siControl) or HIPK2-targeting (siHIPK2) siRNA and incubated for 24 h. Cells were then treated with 2 or 10 μM etoposide for 48 h for RNA isolation. BDNF exon III mRNA expression was analyzed with quantitative real-time PCR. BDNF mRNA expression in nontreated cells was set to 1.0; mean values from five independent experiments are shown; error bars, SEs.
DISCUSSION
In this study we have demonstrated that HIPK2 is a CREB Serine 271 kinase and regulates CREB-dependent transcription in response to a DNA-damaging agent, etoposide. HIPK2 preferentially phosphorylates Serine/Threonine sites adjacent to Proline: Ser46-Pro47 in p53 (D'Orazi et al., 2002; Hofmann et al., 2002), Pro421-Ser422-Pro423 in CtBP (Zhang et al., 2003), Pro296-Ser297 in Groucho (Choi et al., 2005), Ser249-Pro250 and Ser276-Pro277 in AML1 (Aikawa et al., 2006), and Thr281-Pro282, Thr304-Pro305, and Thr373-Pro374 in Pax6 (Kim et al., 2006). The amino acid sequences containing CREB Serine 271 are Val-Val-Met-Ala-Ser-Ser271-Pro, which is highly conserved in ATF1 as Val-Val-Met-Thr-Ser198-Pro. Indeed, our preliminary results suggested that ATF1 at Serine 198 appears to be phosphorylated by HIPK2 (Hailemariam and Tsuji, unpublished observation). We observed retarded migration of phosphorylated CREB at Serine 271 by HIPK2 (Figures 1 and 2), similar to the retarded migration of Groucho, AML1, and Pax 6 phosphorylated by HIPK2 (Choi et al., 2005; Aikawa et al., 2006; Kim et al., 2006), but not CREB phosphorylation at Serine 133 by PKA (Figure 2).
Protein phosphorylation has been shown to alter the stability of various transcription factors and coregulators including enhanced degradation of CtBP via Serine 422 phosphorylation by HIPK2 (Zhang et al., 2003); however, CREB Serine 271 phosphorylation does not appear to affect the protein stability because CREB expression levels after HIPK2 overexpression looked unchanged, along with the results showing no difference in protein expression of wtCREB and Ser271Ala mutant CREB when HIPK2 was coexpressed for 24–48 h (Figure 2A). Phosphorylation affects protein localization such as the cases of mitogen-activated protein (MAP) kinases. We tested whether phosphorylation of CREB by HIPK2 facilitates the nuclear localization of endogenous and transfected CREB by Western blotting of cytoplasmic and nuclear fractions after HIPK2 overexpression. Our results showed that endogenous and transfected CREB proteins in K562, SH-SY5Y, and 293 cells almost exclusively localized in nucleus regardless of HIPK2 overexpression or the Serine 271 mutated CREB (Supplemental Figure S3). Phosphorylation of CREB at Serine 133 was reported to increase DNA binding to some CRE sites despite more contradictory results that indicate no impact on the DNA binding ability of CREB upon Serine 133 phosphorylation (reviewed in Shaywitz and Greenberg, 1999). We tested this possibility by ChIP assays for the CRE sequence in the exon III of the BDNF promoter or by gel retardation assays for binding to the consensus CREB sequence (TGACGTCA; Figure 5, and Supplemental Figure S1), both of which suggested that CREB Serine 271 phosphorylation by HIPK2 does not modulate its DNA binding.
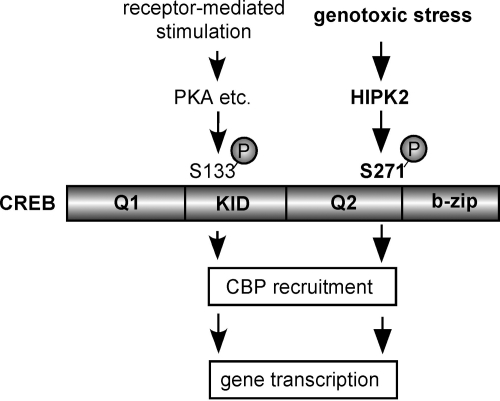
The transactivation function of CREB is regulated by interactions between CREB and its binding proteins. The interaction of CREB and CBP is primarily regulated by phosphorylation of CREB by PKA and other Serine 133 kinases, in which both the KID domain of CREB where Serine 133 is located and the KIX domain of CBP are involved in the interaction (Chrivia et al., 1993; Figure 8). Our ChIP assay showed that phosphorylated CREB by HIPK2 increased or stabilized CBP recruitment to the CRE site in the BDNF exon III promoter (Figure 5). The CREB 271A mutant showed decreased association with CBP compared with wtCREB despite the equivalent binding to the CRE site (Figure 5). Serine 271 in CREB is located adjacent to the second glutamine-rich region (Q2/CAD) and the basic region (Figure 8), involved in constitutive activation of CREB and DNA binding, respectively (Shaywitz and Greenberg, 1999). It was demonstrated that phosphorylation of Serine 142 and 143 in the CREB KID domain during calcium influx into neurons decreased the binding of CREB to the KIX domain of CBP, even though the CREB-dependent transcription was ultimately activated by the cooperative phosphorylation of CREB at Serine 133 by calcium signaling (Kornhauser et al., 2002). Similarly, ionizing radiation/ATM-induced phosphorylation of CREB at Serine 121 in the KID domain decreased interaction between CREB and CBP, resulting in decreased CREB transactivation function (Shi et al., 2004). These results suggest that additional phosphorylation events on CREB have the potential to alter interactions between CREB and CREB-binding proteins. In genotoxic stress and other yet uncharacterized conditions that activate HIPK2, cells may recruit the new mechanism of CREB activation and induction of CREB-target genes, that is through HIPK2-mediated phosphorylation of Serine 271 and facilitated interaction with CBP as demonstrated in this study (Figure 8). Furthermore, HIPK2 forms a complex with the coactivator p300 and AML1, phosphorylates p300 at multiple Serine/Threonine sites and activates p300 HAT activity and coactivator function (Aikawa et al., 2006). It is therefore likely that HIPK2 activates transcription of CREB target genes in genotoxic stress conditions not only through the increased interaction of CREB-CBP but also activation CBP/p300 via direct phosphorylation. Further investigation will be necessary for understanding additional roles of HIPK2 in CREB-dependent gene transcription.
Figure 8.
Regulation of CREB by HIPK2 in genotoxic stress. CREB is activated through phosphorylation of Serine 133 in the KID domain by PKA and related kinases in response to various receptor-mediated stimuli (Mayr and Montminy, 2001). This study demonstrated that genotoxic stress, such as etoposide, activates HIPK2, which in turn activates CREB through phosphorylation of Serine 271 adjacent to the Q2 domain and the b-zip domain of CREB. HIPK2 does not phosphorylate CREB at Serine 133. Phosphorylation of CREB at Serine 271 by HIPK2 increases CBP recruitment. CREB phosphorylation besides these two sites is discussed in the text and reviewed in Johannessen et al. (2004).
HIPK2 is expressed in various cell types, but its expression levels are relatively very low (Wang et al., 2001). Despite our trials of almost all commercially available anti-HIPK2 antibodies, we failed to detect expression of endogenous HIPK2 protein by Western or immunoprecipitation/Western approach. Our results using HIPK2+/+ and −/− MEF cells showed that etoposide activated CRE-dependent transcription only in HIPK2+/+ cells (Figure 7C). Furthermore, HIPK2-deficient MEF cells were highly susceptible to etoposide toxicity (Figure 7B). We speculated that the higher susceptibility to etoposide toxicity in HIPK2-deficient cells was, at least in part, due to the lack of expression of CRE-dependent genes serving as cell survival. Particularly in neuronal cells, CREB is a crucial transcription factor for cell survival and adaptive response (Lonze and Ginty, 2002), and BDNF is a CREB-regulated gene that plays a vital role in neuronal cell survival, differentiation, and adaptive responses (Shieh et al., 1998; Tao et al., 1998). We measured mRNA expression of several CREB-target genes in SH-SY5Y cells after etoposide treatment, in which BDNF mRNA expression showed highest induction with 10 μM etoposide within 24 h. The induction of BDNF mRNA by etoposide was 50% blocked by HIPK2 knockdown in SH-SY5Y cells (Figure 7D), suggesting that HIPK2 is involved in etoposide-induced BDNF mRNA expression. HIPK2 was demonstrated to be proapoptotic through p53 activation (D'Orazi et al., 2002; Hofmann et al., 2002) or CtBP degradation (Zhang et al., 2003) while also serving as a prosurvival factor through Smads-regulated TGF-β–dependent gene expression in midbrain dopamine neurons (Zhang et al., 2007). Cells may integrate these two opposing events or select either proapoptotic or prosurvival function of HIPK2, depending on the type of stress/environmental cues, magnitude of cell damage, and the type of cells and tissues.
In summary, this work has demonstrated that genotoxic stress activates another pathway of CREB regulation, in which HIPK2 phosphorylates CREB Ser-271 (but not Ser-133) and activates CREB-dependent transcription such as the BDNF gene through increased interaction with the coactivator CBP (Figure 8). These results shed light on a new regulatory mechanism of CREB in genotoxic stress.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Huang at University of California San Francisco for kindly providing us with the HIPK2 wt and knockout MEF cells. We thank Dr. Schmitz at Justus-Liebig University, Germany, for HIPK2 plasmids. We also thank Drs. Goodman and Eckner for CBP and p300 plasmids. This work was supported in part by National Institutes of Health (NIH) research grant DK-60007 to Y.T., GM068812 to J.N.-T., NIH Supplement Grant DK-60007S and National Institute of Environmental Health Sciences, National Institutes of Health Training Grant ES-007046 to K.H., and North Carolina State University Research Assistant Fellowship to B.-W.H.
Abbreviations used:
- ATF1
activating transcription factor-1
- BDNF
brain-derived neurotrophic factor
- CBP
CREB-binding protein
- ChIP
chromatin immunoprecipitation
- CRE
cyclic AMP-response element
- CREB
cyclic AMP response element–binding protein
- CREM
cyclic AMP response element modulator
- HIPK2
homeodomain-interacting protein kinase 2
- KID
kinase-inducible domain
- KIX
KID-interacting domain
- PKA
protein kinase A (cyclic AMP-dependent protein kinase).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0015) on June 23, 2010.
REFERENCES
- Aikawa Y., Nguyen L. A., Isono K., Takakura N., Tagata Y., Schmitz M. L., Koseki H., Kitabayashi I. Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 2006;25:3955–3965. doi: 10.1038/sj.emboj.7601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzado M. A., Renner F., Roscic A., Schmitz M. L. HIPK2, a versatile switchboard regulating the transcription machinery and cell death. Cell Cycle. 2007;6:139–143. doi: 10.4161/cc.6.2.3788. [DOI] [PubMed] [Google Scholar]
- Choi C. Y., Kim Y. H., Kim Y. O., Park S. J., Kim E. A., Riemenschneider W., Gajewski K., Schulz R. A., Kim Y. Phosphorylation by the DHIPK2 protein kinase modulates the corepressor activity of Groucho. J. Biol. Chem. 2005;280:21427–21436. doi: 10.1074/jbc.M500496200. [DOI] [PubMed] [Google Scholar]
- Chrivia J. C., Kwok R.P.S., Lamb N., Hagiwara M., Montominy M. R., Goodman R. H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Conkright M. D., Canettieri G., Screaton R., Guzman E., Miraglia L., Hogenesch J. B., Montminy M. TORCs: transducers of regulated CREB activity. Mol. Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- D'Orazi G., et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- Dauth I., Kruger J., Hofmann T. G. Homeodomain-interacting protein kinase 2 is the ionizing radiation-activated p53 serine 46 kinase and is regulated by ATM. Cancer Res. 2007;67:2274–2279. doi: 10.1158/0008-5472.CAN-06-2884. [DOI] [PubMed] [Google Scholar]
- Di Stefano V., Rinaldo C., Sacchi A., Soddu S., D'Orazi G. Homeodomain-interacting protein kinase-2 activity and p53 phosphorylation are critical events for cisplatin-mediated apoptosis. Exp. Cell Res. 2004;293:311–320. doi: 10.1016/j.yexcr.2003.09.032. [DOI] [PubMed] [Google Scholar]
- Eckner R., Ewen M. E., Newsome D., Gerdes M., DeCaprio J. A., Lawrence J. B., Livingston D. M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Foulkes N. S., Borrelli E., Sassone-Corsi P. CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- Hai T. W., Liu F., Coukos W. J., Green M. R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- Hofmann T. G., Mincheva A., Lichter P., Droge W., Schmitz M. L. Human homeodomain-interacting protein kinase-2 (HIPK2) is a member of the DYRK family of protein kinases and maps to chromosome 7q32–q34. Biochimie. 2000;82:1123–1127. doi: 10.1016/s0300-9084(00)01196-2. [DOI] [PubMed] [Google Scholar]
- Hofmann T. G., Moller A., Sirma H., Zentgraf H., Taya Y., Droge W., Will H., Schmitz M. L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- Iwasaki K., Hailemariam K., Tsuji Y. PIAS3 interacts with ATF1 and regulates the human ferritin H gene through an antioxidant-responsive element. J. Biol. Chem. 2007;282:22335–22343. doi: 10.1074/jbc.M701477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M., Delghandi M. P., Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kanei-Ishii C., et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18:816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. A., Noh Y. T., Ryu M. J., Kim H. T., Lee S. E., Kim C. H., Lee C., Kim Y. H., Choi C. Y. Phosphorylation and transactivation of Pax6 by homeodomain-interacting protein kinase 2. J. Biol. Chem. 2006;281:7489–7497. doi: 10.1074/jbc.M507227200. [DOI] [PubMed] [Google Scholar]
- Kim Y. H., Choi C. Y., Lee S. J., Conti M. A., Kim Y. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J. Biol. Chem. 1998;273:25875–25879. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- Kornhauser J. M., Cowan C. W., Shaywitz A. J., Dolmetsch R. E., Griffith E. C., Hu L. S., Haddad C., Xia Z., Greenberg M. E. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Lonze B. E., Ginty D. D. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Mayr B., Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Rinaldo C., Prodosmo A., Siepi F., Soddu S. HIPK 2, a multitalented partner for transcription factors in DNA damage response and development. Biochem. Cell Biol. 2007;85:411–418. doi: 10.1139/O07-071. [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Iwasaki K., Sugiyama H., Tsuji Y. Role of the tumor suppressor PTEN in antioxidant responsive element-mediated transcription and associated histone modifications. Mol. Biol. Cell. 2009;20:1606–1617. doi: 10.1091/mbc.E08-07-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanware N. P., Trinh A. T., Williams L. M., Tibbetts R. S. Coregulated ataxia telangiectasia-mutated and casein kinase sites modulate cAMP-response element-binding protein-coactivator interactions in response to DNA damage. J. Biol. Chem. 2007;282:6283–6291. doi: 10.1074/jbc.M610674200. [DOI] [PubMed] [Google Scholar]
- Shaywitz A. J., Greenberg M. E. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Shi Y., Venkataraman S. L., Dodson G. E., Mabb A. M., LeBlanc S., Tibbetts R. S. Direct regulation of CREB transcriptional activity by ATM in response to genotoxic stress. Proc. Natl. Acad. Sci. USA. 2004;101:5898–5903. doi: 10.1073/pnas.0307718101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh P. B., Hu S. C., Bobb K., Timmusk T., Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Solt I., Magyar C., Simon I., Tompa P., Fuxreiter M. Phosphorylation-induced transient intrinsic structure in the kinase-inducible domain of CREB facilitates its recognition by the KIX domain of CBP. Proteins. 2006;64:749–757. doi: 10.1002/prot.21032. [DOI] [PubMed] [Google Scholar]
- Tao X., Finkbeiner S., Arnold D. B., Shaywitz A. J., Greenberg M. E. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Tsuji Y., Moran E., Torti S. V., Torti F. M. Transcriptional regulation of the mouse ferritin H gene: involvement of p300/CBP adaptor proteins in FER-1 enhancer activity. J. Biol. Chem. 1999;274:7501–7507. doi: 10.1074/jbc.274.11.7501. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hofmann T. G., Runkel L., Haaf T., Schaller H., Debatin K., Hug H. Isolation and characterization of cDNAs for the protein kinase HIPK2. Biochim. Biophys. Acta. 2001;1518:168–172. doi: 10.1016/s0167-4781(00)00308-0. [DOI] [PubMed] [Google Scholar]
- Wee H. J., Voon D. C., Bae S. C., Ito Y. PEBP2-beta/CBF-beta-dependent phosphorylation of RUNX1 and p300 by HIPK2, implications for leukemogenesis. Blood. 2008;112:3777–3787. doi: 10.1182/blood-2008-01-134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Ku S., Ma G. K., Saito S., Tang A. A., Zhang J., Mao J. H., Appella E., Balmain A., Huang E. J. HIPK2 represses beta-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc. Natl. Acad. Sci. USA. 2007;104:13040–13045. doi: 10.1073/pnas.0703213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesierska-Gadek J., Schmitz M. L., Ranftler C. Roscovitine-activated HIP2 kinase induces phosphorylation of wt p53 at Ser-46 in human MCF-7 breast cancer cells. J. Cell. Biochem. 2007;100:865–874. doi: 10.1002/jcb.21211. [DOI] [PubMed] [Google Scholar]
- Wiggins A. K., Wei G., Doxakis E., Wong C., Tang A. A., Zang K., Luo E. J., Neve R. L., Reichardt L. F., Huang E. J. Interaction of Brn3a and HIPK2 mediates transcriptional repression of sensory neuron survival. J. Cell Biol. 2004;167:257–267. doi: 10.1083/jcb.200406131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Geiger T. R., Johnson E. N., Nyborg J. K., Druey K. M. RGS13 acts as a nuclear repressor of CREB. Mol. Cell. 2008;31:660–670. doi: 10.1016/j.molcel.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Pho V., Bonasera S. J., Holtzman J., Tang A. T., Hellmuth J., Tang S., Janak P. H., Tecott L. H., Huang E. J. Essential function of HIPK2 in TGFbeta-dependent survival of midbrain dopamine neurons. Nat. Neurosci. 2007;10:77–86. doi: 10.1038/nn1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Yoshimatsu Y., Hildebrand J., Frisch S. M., Goodman R. H. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115:177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.