Abstract
The Rieske nonheme iron aromatic ring-hydroxylating oxygenases (RHOs) NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1 have been implicated in the initial oxidation of high-molecular-weight (HMW) polycyclic aromatic hydrocarbons (PAHs), forming cis-dihydrodiols. To clarify how these two RHOs are functionally different with respect to the degradation of HMW PAHs, we investigated their substrate specificities to 13 representative aromatic substrates (toluene, m-xylene, phthalate, biphenyl, naphthalene, phenanthrene, anthracene, fluoranthene, pyrene, benz[a]anthracene, benzo[a]pyrene, carbazole, and dibenzothiophene) by enzyme reconstitution studies of Escherichia coli. Both Nid systems were identified to be compatible with type V electron transport chain (ETC) components, consisting of a [3Fe-4S]-type ferredoxin and a glutathione reductase (GR)-type reductase. Metabolite profiles indicated that the Nid systems oxidize a wide range of aromatic hydrocarbon compounds, producing various isomeric dihydrodiol and phenolic compounds. NidAB and NidA3B3 showed the highest conversion rates for pyrene and fluoranthene, respectively, with high product regiospecificity, whereas other aromatic substrates were converted at relatively low regiospecificity. Structural characteristics of the active sites of the Nid systems were investigated and compared to those of other RHOs. The NidAB and NidA3B3 systems showed the largest substrate-binding pockets in the active sites, which satisfies spatial requirements for accepting HMW PAHs. Spatially conserved aromatic amino acids, Phe-Phe-Phe, in the substrate-binding pockets of the Nid systems appeared to play an important role in keeping aromatic substrates within the reactive distance from the iron atom, which allows each oxygen to attack the neighboring carbons.
IMPORTANCE
Since the discovery of microbial ring-hydroxylating oxygenases (RHOs) in 1970, the sequences, structures, and enzyme biochemistry, including enantiospecific products, of RHOs have been studied and discussed extensively from the perspective of biodegradation, biotransformation, and biocatalysis processes. However, with all that effort to elucidate the enzymatic mechanisms of RHOs, little is known about the biochemistry and enzymology underlying high-molecular-weight (HMW) polycyclic aromatic hydrocarbon (PAH) degradation. We used Mycobacterium vanbaalenii PYR-1 Nid enzymes, the first type V RHO members to display an apparent substrate preference for HMW PAHs. Here, we examine the mechanism of the RHO reaction by integrating structural information of the NidAB and NidA3B3 enzymes with substrate and product data. This study gives us an understanding of how the model RHO systems of M. vanbaalenii PYR-1 metabolize HMW PAHs. The information obtained would also be helpful for successful application of RHO enzymes to the production of industrially and medically important chiral chemicals and the development of PAH bioremediation technologies.
High-molecular-weight (HMW) polycyclic aromatic hydrocarbons (PAHs), with four or more fused benzene ring compounds, are ubiquitous pollutants in nature. Because they are genotoxic, mutagenic, and carcinogenic and they bioaccumulate and persist in the environment, the fate of these compounds is of great public health concern (1). Mycobacterium vanbaalenii PYR-1 (2–4), a fast-growing scotochromogenic member of the genus Mycobacterium, is able to degrade a wide range of HMW PAHs, including fluoranthene, pyrene, 1-nitropyrene, benzo[a]pyrene, benz[a]anthracene, and 7,12-dimethylbenz[a]anthracene (5–8). The PYR-1 strain was isolated from petroleum-contaminated estuarine sediments and has been studied extensively with respect to the physiology, biochemistry, and genetics of bacterial metabolism of HMW PAHs (5, 9–14). Recently, the genome of M. vanbaalenii PYR-1 has been sequenced, allowing us to understand the molecular basis of PAH metabolism with the aim of improving the rate of degradation (14).
Among the 194 chromosomal genes that are likely involved in PAH degradation, we identified 21 diverse ring-hydroxylating oxygenase (RHO) genes in the genome of M. vanbaalenii PYR-1 (14). These enzymes, members of a Rieske nonheme iron oxygenase family, often initiate bacterial catabolism of aromatic compounds by incorporating molecular oxygen into aromatic substrates to form cis-dihydrodiols. Previously, two type V RHO enzymes, NidAB and NidA3B3 from M. vanbaalenii PYR-1, which exhibited moderate sequence identity (53% at the level of amino acid sequence between α subunits), have been shown to be involved in the initial oxidation of pyrene and fluoranthene (11, 15). Interestingly, proteomics methodologies revealed that expression of NidAB and NidA3B3 is highly dependent on the structural differences of the PAH substrates (12, 13, 16). NidAB enzyme was expressed when M. vanbaalenii PYR-1 was incubated with pyrene but was undetectable in fluoranthene-grown cells, whereas NidA3B3 enzyme was more abundant when the cells were incubated with fluoranthene (12, 13).
In recent years, oxygenase genes involved in the degradation of HMW PAHs have been reported for several environmental mycobacteria isolated from diverse geographic locations (17–21). They show high similarity in both sequence and gene organization with the nidAB and nidA3B3 genes of M. vanbaalenii PYR-1, suggesting that these enzyme systems play an important global role in the degradation of HMW PAH-contaminated sites.
Recently, molecular ecological approaches targeting RHO sequences to characterize microbial diversity in PAH-contaminated environments have been developed. In particular, studies using nidA and nidA3 genes for the functional assessment of the microbial community with respect to HMW PAH degradation have been recently reported (22–24). However, functional differences between these two genes have not been examined.
In this study, as the first step toward clarifying how NidAB and NidA3B3 are different in their substrate regiospecificities in the degradation of HMW PAHs by M. vanbaalenii PYR-1, we investigated their PAH substrate specificities and efficiencies, using 13 representative aromatic substrates. For the experiment, different electron transport chain (ETC) components were chosen on the basis of our recent RHO classification (Fig. 1) (25). We then coexpressed them with the NidAB or NidA3B3 oxygenase component to examine enzyme function. For structural understanding of the enzyme substrate specificity of Nid systems, in silico analysis was conducted. This study, building on the previous genetic, metabolic, and proteomics information, allows a better understanding of how M. vanbaalenii strain PYR-1 deals with HMW PAHs, including pyrene and fluoranthene.
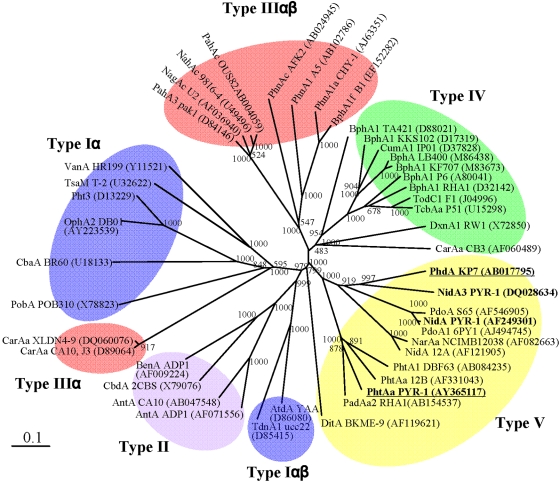
FIG 1.
Classification of 45 RHO enzymes. The phylogenetic tree, inferred by the neighbor-joining method, shows the relative positions of NidA and NidA3 among other RHOs based on the recent classification (25). Bootstrap values for 1,000 replicates are given. GeneID (GI) numbers were provided for RHO sequences whose detailed information can be found in the NCBI database. The bar indicates 10% sequence divergence. Underlining indicates the RHO systems from which the electron transport chain (ETC) components used in this study came.
RESULTS
Reconstitution of NidAB and NidA3B3.
Enzyme reconstitution assays in Escherichia coli BL21(DE3) were performed in the presence of different ETCs to determine the compatibility with the two oxygenase systems, NidAB and NidA3B3 (Table 1) (25). E. coli BL21(DE3)(pCEC35), lacking ETCs, did not result in expression of the functional enzymes and produced no metabolites, as previously shown with NidA3B3 (11). NidAB activity was reconstituted when combined with the two type V ETCs, consisting of a [3Fe-4S]-type ferredoxin and a glutathione reductase (GR)-type reductase (25). This enzyme system reconstituted with the ETC PhtAcAd from M. vanbaalenii PYR-1 (26) produced almost 7-fold-more pyrene dihydrodiol metabolites (Table 1) than with PhdCD (27). NidA3B3 was also functional with the two type V ETCs, transforming fluoranthene to cis-dihydrodiol metabolites with a higher transformation efficiency with PhdCD than PhtAcAc (Table 1).
TABLE 1.
RHO enzyme reconstitution study showing ETC compatibility with the NidAB and NidA3B3 systems
| Relative yield of dihydrodiol formation (%)b |
||
|---|---|---|
| Type V ETCa | NidABc | NidA3B3d |
| PhtAcAd | 905 (100) | 2,875 (15) |
| PhdCD | 129 (14) | 19,163 (100) |
Formation of dihydrodiol metabolites was measured based on the HPLC peak area (mAU × s). Representative results are shown for each of the enzyme reconstitution assays.
Formation of pyrene cis-4,5-dihydrodiol.
Formation of fluoranthene cis-2,3-dihydrodiol.
Functional analysis of the Nid enzyme systems.
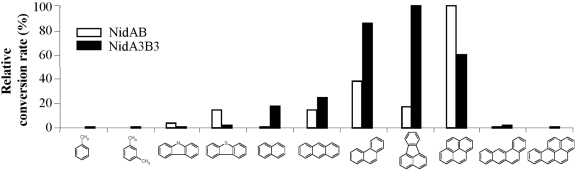
We investigated the substrate specificities of NidAB and NidA3B3 by testing 13 aromatic substrates, toluene, m-xylene, phthalate, biphenyl, naphthalene, phenanthrene, anthracene, fluoranthene, pyrene, benz[a]anthracene, benzo[a]pyrene, carbazole, and dibenzothiophene, which include eight aromatic compounds previously used as substrates (11). As shown in Fig. 2, the relative product yields of NidAB and NidA3B3 for the test compounds were compared on the basis of the metabolite profiles and high-performance liquid chromatography (HPLC) peak areas. The results indicated that the Nid systems oxidize a wide range of aromatic hydrocarbon compounds with overlapping substrate specificities. However, NidAB and NidA3B3 showed apparent substrate preferences for pyrene (100:57) and fluoranthene (15:100), respectively, with the highest conversion rates and product regiospecificity, whereas other aromatic substrates were converted at relatively low regiospecificity (Fig. 2 and 3).
FIG 2.
Diagram showing substrate preference by NidAB and NidA3B3, indicating that pyrene and fluoranthene were the most effective aromatic substrates, respectively, for these two enzymes. The relative conversion rate for each aromatic substrate is the sum of all metabolites produced, regardless of hydroxylation positions. The rates of pyrene and fluoranthene conversion were set at 100% for NidA and NidA3, respectively, and other substrates were calculated relative to the rates of pyrene and fluoranthene conversion. Biotransformation assays were conducted at least two times, and only representative results are shown as relative conversion rates. The chemical structures and compound names are shown in Fig. 3.
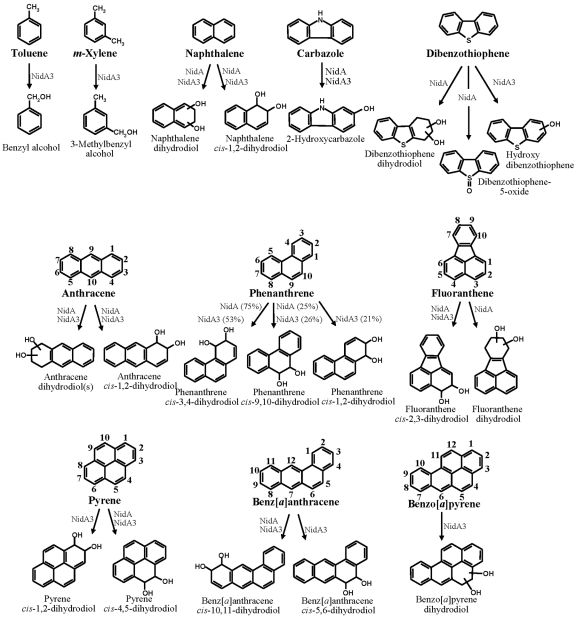
FIG 3.
Metabolite structures produced from aromatic substrates by NidAB and NidA3B3 enzymes. Biphenyl and phthalate were not included because no metabolite was produced for both Nid enzymes. The relative cis-dihydrodiol product ratios formed from phenanthrene by NidAB and NidA3B3 enzymes are shown in parentheses.
In the analysis of NidAB with toluene, m-xylene, phthalate, and biphenyl substrates, no hydroxy, dihydrodiol, or dihydroxy metabolites were formed. Incubation of NidAB with carbazole produced traces of a hydroxycarbazole (Fig. 3). In contrast, in the analysis of dibenzothiophene, three possible metabolites were found at 9.28 min (dibenzothiophene 5-oxide identified), 9.63 min (dibenzothiophenediol), and 10.29 min (dibenzothiophene dihydrodiol) (Fig. 3). Fragment ions of the metabolite dibenzothiophenediol (m/z 360 [M+˙]) included m/z 359 [M-1], 345 [M-15], 259 [M-101], 147, and 73. For the metabolite dibenzothiophene dihydrodiol (10.29 min), which had an apparent molecular ion at m/z 362, fragment ions included m/z 347 [M-1] and 239. In naphthalene analysis of NidAB and NidA3B3 incubation, naphthalene dihydrodiols that had a molecular ion at m/z 360 [M+˙] and major fragment ions at m/z 275 [M-31], 203, 191, 147, 128, 116, and 73 were observed. The major isomer formed was naphthalene cis-1,2-dihydrodiol. In the case of phenanthrene, two isomers with cis-dihydrodiol positioned at C-3 and C-4 (C-3,4) and C-9,10 were identified with different conversion rates, as they were previously with NidA3B3, but the isomer with cis-dihydrodiol at C-1,2 was not produced (Fig. 3 ). The metabolite profile of NidAB for the degradation of anthracene was identical to that of NidA3B3, with different conversion rates (Fig. 3). In the fluoranthene conversion by NidAB, two kinds of fluoranthene dihydrodiol metabolites were identified (Fig. 3), which is different from that by NidA3B3, which produces only fluoranthene cis-2,3-dihydrodiol (11). A different fluoranthene dihydrodiol with a retention time of 11.84 min, which had a molecular ion at m/z 380 [M+˙], was newly detected and produced at a concentration about 10 times less than cis-2,3-dihydrodiol based on the reconstructed ion chromatogram (RIC) peak area. In the case of pyrene, NidAB converted pyrene only to pyrene cis-4,5-dihydrodiol as previously shown (15) (Fig. 3), unlike NidA3B3, which produced both cis-4,5- and cis-1,2-dihydrodiol pyrene. In the benz[a]anthracene and benzo[a]pyrene experiments, NidAB produced traces of benz[a]anthracene cis-10,11-dihydrodiol, but not benzo[a]pyrene dihydrodiol.
The substrate specificity of NidA3B3 was determined with five aromatic hydrocarbons, and these results were then combined with the results of eight previously tested aromatic compounds (11). In the gas chromatography-electron ionization mass spectrometry (GC–EI-MS) analysis for toluene biotransformation, two possible metabolites were found at 3.80 (m/z 108 [M+˙]) and 5.30 (m/z 122 [M+˙]) min. The National Institute of Standards and Technology (NIST) library comparison of the trimethylsilyl (TMSi) esters of the two possible metabolites produced benzyl alcohol and benzoic acid (Fig. 3). In the case of m-xylene, one possible metabolite was found at 4.90 min, with an apparent molecular ion at m/z 122, identified as 3-methylbenzyl alcohol (Fig. 3). Dihydroxylated metabolites of toluene and m-xylene, however, were not detected in the HPLC and GC-MS analysis. The GC–EI-MS analysis for the biotransformation of carbazole provided two peaks at 11.4 and 11.8 min, which had identical mass spectra, with apparent molecular ions at m/z 183. These mass spectra matched moderately well to 2-hydroxycarbazole of the NIST Library, which is the only positional isomer of hydroxycarbazole in the library (Fig. 3). No angular oxidation product was detected from carbazole, indicating that NidA3B3 may not be able to perform the angular dioxygenation. In the case of dibenzothiophene, GC–EI-MS analysis after TMSi reaction provided one peak at 11.5 min, which had an apparent molecular ion at m/z 200. The mass spectra contained fragment ions at m/z 184, 172, and 171. This peak was identified tentatively as a hydroxy-dibenzothiophene (Fig. 3). For benzo[a]pyrene bioconversion studies, GC-MS analysis of the derivatized products detected one metabolite, eluting at 16.5 min, with a molecular ion at m/z 430, whose fragment ions are identical to a benzo[a]pyrene dihydrodiol (Fig. 3).
In silico analysis of NidA and NidA3.
Three-dimensional (3-D) protein model production of the Nid systems was performed in two steps, homology modeling and validation. The homology models of the α subunits NidA and NidA3 were constructed using the crystal structure of naphthalene dioxygenase (NDO) from Rhodococcus sp. strain NCIMB12038 (28) as a template. Amino acid sequence identities of NidA and NidA3 to the template are 48 and 53%, respectively. In addition to the basic structure check using PROCHECK, the models were further validated by the active-site topologies (see Fig. SA1 in the supplemental material) and ligand-docking analysis (see Table SA2 in the supplemental material) on the basis of their metabolite profiles. On the basis of the shape of the Nid active sites predicted from the metabolite profile (see Fig. SA1 in the supplemental material), the Nid models well reflected the dot-represented shape of substrate-binding pockets of both Nid systems. Docking test of the Nid models with phenanthrene also reflected the Nid enzyme biotransformation results (see Table SA2 in the supplemental material).
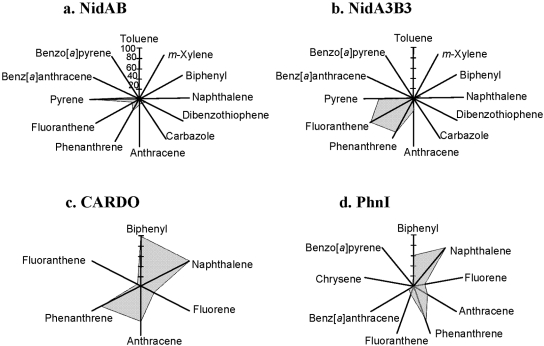
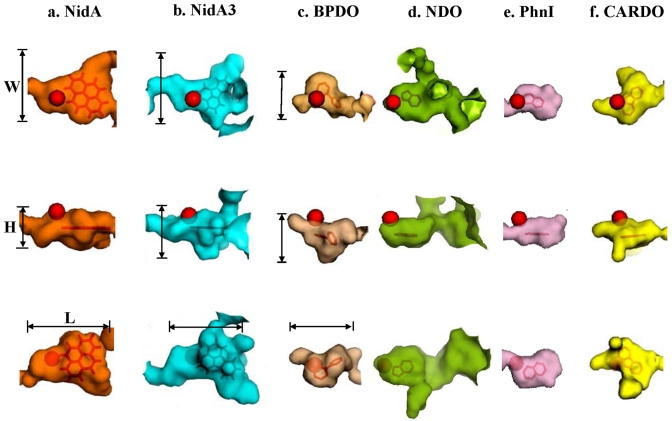
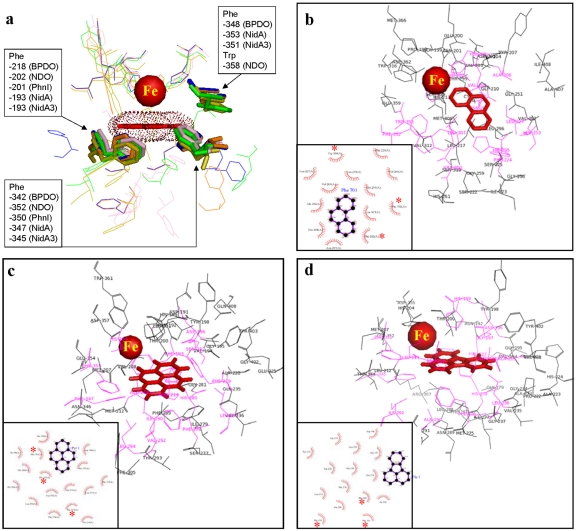
To address the relationship between substrate specificity and active site, we compared the Nid systems to other well-organized RHO information. As shown in Fig. 4, the substrate specificities of NidAB and NidA3B3 are different from those of other RHO enzymes. Pyrene and fluoranthene were the best substrates for NidAB and NidA3B3, respectively, whereas naphthalene and biphenyl were the most preferred substrates for PhnI from Sphingomonas sp. strain CHY-1 and carbazole 1,9a-dioxygenase (CARDO) from Pseudomonas sp. strain CA10 (29), respectively. As shown in Table 2, the amino acid sequence identities and root mean square deviation (RMSD) (Cα) of the Nid systems indicate their structural differences from other RHOs. In particular, the angular dioxygenase CARDO from Janthinobacterium sp. strain J3 showed the lowest structural similarity to the Nid systems. Table 3 and Fig. 5 show comparisons of the active sites, in which the volume of the active site corresponds to the area defined by the van der Waals surface of the residues that contribute to the overall topology of the active sites. The dimensions of the active sites were also measured to correlate them with the active-site volume. NidA and NidA3 showed the largest active sites in both volume (509 and 613 Å3, respectively) and size (14.5 Å by 7.4 Å by 14.8 Å and 15.8 Å by 10.0 Å by 17.8 Å [width, height, and length, respectively]). The presence of aromatic amino acids in the substrate-binding pockets is a common feature of many RHOs. The structural overlays of several RHO active sites on that of NDO derived from Pseudomonas putida NCIB 9816-4 and bound to phenanthrene revealed that the aromatic amino acids corresponding to Phe-202, Phe-352, and Trp-358 of the NDO are spatially conserved in the 3-D structure (Fig. 6a). In particular, two Phe residues, at positions 202 and 352 of the NDO, were perfectly conserved in these RHOs. The conserved aromatic amino acids in the active sites of NidA and NidA3 are Phe-193, Phe-347, and Phe-353 and Phe-193, Phe-345, and Phe-351, respectively. In the analysis of interaction between RHO active sites and their substrates, these conserved aromatic amino acids were revealed to be involved in the aromatic interactions (Fig. 6b, c, and d; see also Table SA3 in the supplemental material). Aromatic interactions in combination with hydrophobic interactions could play a role in substrate binding (see Fig. SA2 in the supplemental material). Residues of NidA and NidA3 in contact with the bound pyrene and fluoranthene, respectively, which are likely to be involved in aromatic and hydrophobic interactions, were proposed (Fig. SA2).
FIG 4.
Substrate ranges of NidAB, NidA3B3 from M. vanbaalenii PYR-1, CARDO from Pseudomonas sp. strain CA10, and PhnI from Sphingomonas sp. strain CHY-1. The relative conversion rate (as a percentage) for the test aromatic compounds with respect to the most favorable substrate (100%) was plotted.
TABLE 2.
Comparison of Nid proteins with other RHOs with known structures
| Sequence identity (%) with the following RHOc: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RHOa | Typeb | No. of amino acids | 1 | 2 | 3 | 4 | 5 | 6 | RMSD (Cα)d | Preferred substrate |
| 1. NidA | V | 455 | 100 | 0.0 | Pyrene | |||||
| 2. NidA3 | V | 444 | 53 | 100 | 0.2 | Fluoranthene | ||||
| 3. BPDO (1ULJ) | IV | 460 | 31 | 30 | 100 | 1.4 | Biphenyl | |||
| 4. NDO (1O7G) | IIIαβ | 449 | 29 | 31 | 31 | 100 | 1.3 | Naphthalene | ||
| 5. PhnI (2CKF) | IIIαβ | 454 | 31 | 28 | 36 | 41 | 100 | 1.4 | Naphthalene | |
| 6. CARDO (1WW9) | IIIα | 384 | 14 | 16 | 15 | 15 | 15 | 100 | 1.7 | Carbazole |
The RHO enzymes are from the following species: BPDO from Rhodococcus sp. strain RHA1; NDO from P. putida NCIB 9816-4; PhnI from Sphingomonas sp. strain CHY-1; and CARDO from Janthinobacterium sp. J3. The PDB accession numbers are provided in parentheses.
Based on the classification given in reference 25.
Sequence identity to the RHO. The RHO is referred to by the number shown in the RHO column.
RMSD of the models superimposed on NidA.
TABLE 3.
Comparison of the active sites of Nid systems with those of other RHOs with known structures
| Active-site measurementsc |
||||
|---|---|---|---|---|
| RHO | Active-site pocket vol (Å3)b | Width (Å) | Height (Å) | Length (Å) |
| NidA | 509 (607) | 14.49 | 7.44 | 14.84 |
| NidA3 | 613 (725) | 15.85 | 10.03 | 17.76 |
| BPDOa | 300 (367) | 8.30 | 9.61 | 13.49 |
| NDOa | 414 (550) | 10.40 | 6.54 | 12.26 |
| PhnI | 295 (404) | 9.05 | 6.15 | 12.41 |
| CARDO | 352 (473) | 11.25 | 6.75 | 17.44 |
The BPDO is from Rhodococcus sp. strain RHA1, while the NDO is from P. putida NCIB 9816-4.
The active-site volumes were measured using Pocketfinder and CASTp. The values in parentheses are the values found by using CASTp.
See Fig. 5.
FIG 5.
Surface plots of the substrate-binding pockets of RHO enzymes bound to their substrate. The mononuclear iron is represented as a red ball. The width (W), height (H), and length (L) are shown for NidA, NidA3, and BPDO. See Table 3 for detailed numerical data for the active sites compared and their sources (microorganisms).
FIG 6.
(a) Superposition of the five RHOs (NidA, NidA3, BPDO from Rhodococcus sp. strain RHA-1, NDO from P. putida NCIB 9816-4, and PhnI from Sphingomonas sp. strain CHY-1) substrate-binding pockets. The mononuclear Fe2+ and spatially conserved aromatic amino acids are represented as a red ball and a stick model, respectively. (b to d) Schematic diagrams show the interactions between the substrates and amino acid residues in the active sites for the following substrates: phenanthrene and NDO from P. putida NCIB 9816-4 (b), pyrene and NidA (c), and fluoranthene and NidA3 (d). The van der Waals interactions are indicated by pink shading. Asterisks indicate spatially conserved aromatic amino acids. Refer to Table SA3 in the supplemental material for the list of amino acid residues.
DISCUSSION
The comparative analysis of the substrate specificity and structural characteristics of NidAB and NidA3B3 from M. vanbaalenii PYR-1 can be summarized in the following features. First, there was a clear correlation between substrate preference and product regioselectivity in the metabolism of aromatic compounds. NidAB and NidA3B3 enzymes had a substrate preference for pyrene and fluoranthene, respectively. In addition, pyrene and fluoranthene were regiospecifically oxidized, resulting in only one type of metabolite, pyrene cis-4,5-dihydrodiol and fluoranthene cis-2,3-dihydrodiol, respectively. Other aromatic substrates were converted at relatively low regiospecificities, producing different isomeric dihydrodiol metabolites. Although several other RHOs have been shown to oxidize a wide range of PAHs with 2 to 5 benzene rings, the optimal substrates for these RHOs have never been HMW PAHs (18, 29, 30). The specific activity of these RHO enzymes gradually declines as substrate size increases. In this respect, NidAB and NidA3B3 are the first RHO enzyme systems that are able to oxidize a broad range of PAH substrates with an apparent substrate preference for the HMW PAHs. Second, Nid systems could also catalyze either mono-oxygenation or sulfoxidation. NidA3B3 oxidized toluene and m-xylene to benzyl alcohols by benzylic monohydroxylation (31), and NidAB catalyzed the sulfoxidation of dibenzothiophene to dibenzothiophene-5-oxide (32) (Fig. 3). Third, the molecular weight of aromatic substrates seems to have an effect on the conversion rate and type of oxygenation of the Nid enzyme systems. As shown in Fig. 3, there is a clear trend in the substrate preference of NidAB and NidA3B3 toward HMW PAHs. The preferred HMW substrates, pyrene and fluoranthene, whose conversion rates are high, were attacked mainly by dioxygenation reactions, whereas low-molecular-weight substrates, such as toluene and m-xylene, with low conversion rates, were attacked mainly by mono-oxygenation. It should also be noted that the molecular weights of substrates are also associated with structural features of the substrates. For example, the conversion rate of the four-aromatic-ring substrate pyrene, which is arranged in a planar pattern, is significantly higher than that of the other four-ring angularly arranged substrate, benz[a]anthracene (Fig. 3).
Structural characteristics of the active sites of Nid systems, revealed by a series of in silico analyses, appear to dictate the metabolic features of Nid enzymes. According to the regioselectivity and PAH conversion rate, pyrene and fluoranthene were the best substrates for NidAB and NidA3B3, respectively. When the sizes of the aromatic substrates were examined with respect to the active sites of the enzymes, the dimensions of active sites seemed least likely to be problematic from the standpoint of fit. In the case of fluoranthene, which has the van der Waals dimensions 11.4 Å (length) by 9.5 Å (width) by 3.8 Å (thickness) (33), the substrate-binding pocket of NidA3, with the approximate dimensions of 16 Å by 10 Å by 17 Å, satisfies the spatial requirement for the acceptance of fluoranthene (Fig. 5b and Table 3). Previously, PhnI from Sphingomonas sp. strain CHY-1 (34) and biphenyl 2,3-dioxygenase (BPDO) from Sphingobium yanoikuyae B1 (35) showed activity toward HMW PAH substrate. However, the pocket volumes of active sites of NidA and NidA3 are clearly larger than those two RHOs in both size and volume (Fig. 4 and Table 4), suggesting that the Nid systems have evolved to oxidize HMW PAHs more efficiently.
TABLE 4.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Invitrogen |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 galU galK Δ(ara-leu)7697 rpsL (Strr) endA1 nupG | Novagen |
| E. coli BL21(DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pGEM-T Easy | Apr; TA cloning vector | Promega |
| pCR2.1 topo | Apr; TA cloning vector | Invitrogen |
| pET-17b | Expression vector; Apr; T7 RNA polymerase promoter | Invitrogen |
| pALTER-EX2 | Expression vector; Apr Tcr; T7 RNA polymerase promoter, SP6 RNA polymerase promoter, Tac promoter | Promega |
| pBRCD | Gmr; pBBR1MCS-5 containing phdCD from Nocardioides sp. strain KP7 | 18 |
| pNCK33 | pET-17b containing nidA3B3 genes | 11 |
| pCEC35 | 1.93-kb nidAB genes cloned into the NheI-HindIII site of pET-17b | This study |
| pCEC36 | 1.43-kb phtAcAd genes cloned into the NcoI-BamHI site of pALTER-EX2 | This study |
Strr, streptomycin resistance; Apr, ampicillin resistance; Gmr, gentamicin resistance; Tcr, tetracycline resistance.
The structural information on active sites in the Nid systems also provided insight into substrate binding. Although there are no significant quantitative differences in the constituent amino acid residues in the active sites, the topologies of active sites were quite different (Fig. 5). As shown in Fig. 6 and in Fig. SA2 in the supplemental material, functionally important residues were selectively and spatially conserved in the active sites. The consensus residues for catalytic mononuclear iron in which the ferrous ion (Fe2+) is coordinated by a common structural motif, 2-His-1-carboxylate facial triad, were conserved (36). In addition, as revealed in the superimposition of several RHOs, the corresponding aromatic amino acids, Phe-202, Phe-352, and Trp-358 of NDO from P. putida NCIB 9816-4, are spatially conserved among RHOs, suggesting their structure-related functional roles (Fig. 6). Previous site-directed mutagenesis studies support the functional relevance of these residues (37–39). In the NDO from P. putida NCIB 9816-4, the substitutions Phe-202-Leu, Phe-352-Tyr, and Phe-352-Trp resulted in inactive enzymes, whereas the enzyme tolerated a wide range of substitutions near the active site (38, 39). Interestingly, the conserved amino acids contribute to substrate binding by aromatic interactions (Fig. 6b, c, and d; see also Table SA3 in the supplemental material). These aromatic side chains in the active site of RHOs, together with the 2-His-1-carboxylate motif, are considered to function in keeping their substrates within a reactive distance from the iron atom (Fig. 6; see also Fig. SA2 in the supplemental material). The structures of substrate-bound forms of RHOs experimentally showed that the substrate is positioned in almost the same place next to the iron, within about 5 Å (28, 40–43) (see Table SA1).
The Nid systems showed relatively low regiospecific oxidation of relatively small substrates, producing several isomeric dihydrodiol metabolites. These results clearly indicate the existence of multiple substrate binding modes in the active sites. The multiple substrate binding modes are most likely due to the substrate mobility in the active sites, as explained for other enzyme systems (44, 45). Considering the relatively huge space in the active sites of Nid systems, an aromatic substrate is likely to be more mobile as its size decreases. This phenomenon supports the hypothesis that the product selectivity is a result of substrate orientation, which is controlled by interactions with active-site residues (40). The low regiospecific oxidation of Nid systems toward small substrates also results in the relatively low conversion rate.
It is interesting to consider the roles of the Nid systems in the degradation of pyrene and fluoranthene by M. vanbaalenii PYR-1. The production of regiospecific metabolites of pyrene and fluoranthene with the best conversion rate indicates that the Nid RHO systems may have evolved to optimize both substrate/product specificity and enzyme activity. M. vanbaalenii PYR-1 oxidizes pyrene and fluoranthene by at least two and four different degradation routes, respectively (12, 13). For the degradation of pyrene, the route through C-4,5 dioxygenation is a productive pathway because it is channeled into the tricarboxylic acid (TCA) cycle, whereas the other route, via C-1,2 dioxygenation, forms O-methylated derivatives as dead-end products. The expression of NidAB, capable of regiospecifically catalyzing pyrene 4,5 dioxygenation, was tightly controlled to be upregulated in the presence of pyrene (12, 13), which is of great advantage to cells.
Degradation of fluoranthene by M. vanbaalenii PYR-1 is more complex with respect to the functional roles of RHOs. Fluoranthene is dioxygenated at the C-1,2, C-2,3, and C-7,8 positions. The C-1,2 dioxygenation route is merged into the C-2,3 dioxygenation route via 9-fluorenone-1-carboxylic acid, which can be further degraded directly or via 9-fluorenone by angular dioxygenation (13). The C-2,3 dioxygenation route is the preferential choice of M. vanbaalenii PYR-1 on the basis of the metabolic and proteomics data (13). The substrate specificity of NidA3B3 provided in this work supports the previous results. Since NidA3B3 catalyzes only C-2,3 dioxygenation of fluoranthene, another RHO(s) should be involved in the dioxygenation of fluoranthene at the C-1,2 and C-7,8 positions and angular dioxygenation of fluoranthene metabolites. However, considering the broad substrate selectivity, the possibility that the NidA3B3 enzyme is involved in the oxidation of metabolic intermediates produced from fluoranthene cannot be ruled out. Analysis of pyrene- and fluoranthene-treated M. vanbaalenii PYR-1 proteomes detected several other RHOs besides NidAB and NidA3B3 (12–14).
We studied the mechanism of RHO reaction by integrating in silico enzyme structural information with laboratory research that produced substrate specificity and metabolite data. The information, along with the previous polyomic data, gives us a more detailed understanding of how RHOs of M. vanbaalenii PYR-1 degrade HMW PAHs. Further structural biochemistry studies of the Nid systems will be launched to obtain direct evidence of the proposed hypothesis and help for successful application of RHO enzymes to the production of industrially and medically important chiral chemicals and the development of PAH bioremediation technologies.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains, vectors, and plasmids used in this study are listed in Table 4. E. coli strains were cultured at 37°C on Luria-Bertani (LB) medium or on phosphate-based mineral medium (46) with appropriate antibiotics (50 µg/ml) as described in a previous report (11). For gene expression in E. coli, the pET-17b (Novagen) and pALTER-EX2 (Promega) expression systems were used. Antibiotics, phthalate, naphthalene, biphenyl, carbazole, dibenzothiophene, anthracene, benz[a]anthracene, and benzo[a]pyrene were purchased from Sigma-Aldrich. Pyrene and phenanthrene were purchased from Chem Service. Fluoranthene was purchased from Fluka.
Construction of expression system and biotransformation experiment.
A DNA fragment of nidAB was PCR amplified from M. vanbaalenii PYR-1 genomic DNA, using the primer set EnidB-F-NheI (5′-AGCTAGCAACGCGGTTGCGGTCGATCGGGATAC-3′) and EnidB-R-HindIII (5′-TAAGCTTTCAAGCACGCCCGCCGAATGCGGG-3′). The underlined sequences are restriction enzyme recognition sites, which were inserted into the primers for convenient cloning. The two different ETC systems were constructed using their specific primer sets; for the phtAcAd genes from M. vanbaalenii PYR-1 (26), EphtAc-F-NcoI (5′-CCATGGGCGGAGTTATAAAGGCTAAGCGCG-3′) and EphtAd-R-BamHI (5′-GGATCCCTATGGTGATCGCGTTGCGGCGGT-3′) were used. The confirmed PCR amplicon was cloned into pALTER-EX2, resulting in plasmid pCEC36, containing phtAcAd. The previously used plasmid pBRCD (11, 18), containing PhdCD from Nocardioides strain KP7, was utilized again in this study. These plasmids were coexpressed in combination with pCEC35 and pNCK33, which contain the oxygenase genes nidAB and nidA3B3 of M. vanbaalenii PYR-1, respectively.
We investigated the substrate specificities of NidAB and NidA3B3 using the transformed E. coli whole-cell biotransformation method with the ETC components PhtAcAd and PhdCD, respectively, to compare the ranges of substrates accepted by the two Nid systems. Aromatic hydrocarbon substrates were added to a final concentration of 200 µM. We tested 13 aromatic substrates, toluene, m-xylene, phthalate, biphenyl, naphthalene, phenanthrene, anthracene, fluoranthene, pyrene, benz[a]anthracene, benzo[a]pyrene, carbazole, and dibenzothiophene, which include 8 previously used aromatic compounds (11). Carbazole was included to check the two Nid systems for angular dioxygenation, and dibenzothiophene was included to check for both sulfoxidation and angular dioxygenation, which possibly follows sulfoxidation (47, 48). The analysis and identification of metabolites using HPLC and GC-MS were performed as described in the previous studies with NidAB and NidA3B3 (11, 13).
In silico analysis.
The homology models of NidAB and NidA3B3 were generated using the Swiss-Model server (49) with the naphthalene dioxygenase (NDO) (Protein Data Base [PDB] accession no. 2B24A) from Rhodococcus sp. strain NCIMB12038 as a template (28). The structural model was checked using PROCHECK (European Bioinformatics Institute, Cambridge, United Kingdom). Superpose (version 1.0) was used for structural superposition and RMSD calculation of the models superimposed on the template. To predict the minimum size requirement of the substrate-binding pockets of NidA and NidA3, the regiospecificity of accepted aromatic substrates was used. To do so, the shape of the active site was roughly generated by gradual superimposition of a set of accepted aromatic compounds, according to their substrate preference, in which the double bonds of the attack site were matched in the same plane (see Fig. SA1 in the supplemental material). The results were then used to see how much it reflects its homology model generated from the Swiss-Model server. For docking analysis, enzyme-substrate complexes were generated on the basis of information on the substrate binding mode in the active site of previously determined RHO crystal structures, using PyMOL (http://www.pymol.org). The reliability of the docking process was verified on the basis of the substrate binding modes of NDO from P. putida NCIB 9816-4 (28, 40–43) and PhnI from Sphingomonas sp. strain CHY-1 (30, 34) (see Table SA1 in the supplemental material). The scoring function DrugScoreCSD (http://pc1664.pharmazie.uni-marburg.de/drugscore/) was adopted to score substrate binding affinity in the protein-ligand complexes. LPC and LIGPLOT were used to analyze and visualize protein-ligand interactions (50, 51). The volumes of active sites were measured using Pocketfinder (http://www.modelling.leeds.ac.uk/qsitefinder/) and CASTp (52). PyMOL (0.99RC6) was used to visualize the 3-D structures.
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
We thank J. B. Sutherland and H. Chen for critical review of the manuscript. We also thank Hong Wei Zhou, Department of Environmental Health, School of Public Health and Tropical Medicine, Southern Medical University, Guangzhou, China, for assistance with analytical experiments.
This work was supported in part by an appointment to the Postgraduate Research Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
The views presented in this article do not necessarily reflect those of the U.S. Food and Drug Administration.
Footnotes
Citation Kweon, O., S.-J. Kim, J. P. Freeman, J. Song, S. Baek, et al. 2010. Substrate specificity and structural characteristics of the novel Rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1. mBio 1(2):e00135-10. doi:10.1128/mBio.00135-10.
REFERENCES
- 1. Cerniglia C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351–368 [Google Scholar]
- 2. Heitkamp M. A., Cerniglia C. E. 1988. Mineralization of polycyclic aromatic hydrocarbons by a bacterium isolated from sediment below an oil field. Appl. Environ. Microbiol. 54:1612–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khan A. A., Kim S. J., Paine D. D., Cerniglia C. E. 2002. Classification of a polycyclic aromatic hydrocarbon-metabolizing bacterium, Mycobacterium sp. strain PYR-1, as Mycobacterium vanbaalenii sp. nov. Int. J. Syst. Evol. Microbiol. 52:1997–2002 [DOI] [PubMed] [Google Scholar]
- 4. Kim Y. H., Engesser K. H., Cerniglia C. E. 2005. Numerical and genetic analysis of polycyclic aromatic hydrocarbon-degrading mycobacteria. Microb. Ecol. 50:110–119 [DOI] [PubMed] [Google Scholar]
- 5. Cerniglia C. E. 2003. Recent advances in the biodegradation of polycyclic aromatic hydrocarbons by Mycobacterium species, p. 51–73 In Šašek V., Glaser J. A., Baveye P., The utilization of bioremediation to reduce soil contamination: problems and solutions. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 6. Moody J. D., Fu P. P., Freeman J. P., Cerniglia C. E. 2003. Regio- and stereoselective metabolism of 7,12-dimethylbenz[a]anthracene by Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 69:3924–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moody J. D., Freeman J. P., Fu P. P., Cerniglia C. E. 2004. Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 70:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moody J. D., Freeman J. P., Cerniglia C. E. 2005. Degradation of benz[a]anthracene by Mycobacterium vanbaalenii strain PYR-1. Biodegradation 16:513–526 [DOI] [PubMed] [Google Scholar]
- 9. Kim S. J., Kweon O., Cerniglia C. E. 2010. Degradation of polycyclic aromatic hydrocarbons by Mycobacterium strains, p. 1865–1880 In Timmis, Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany [Google Scholar]
- 10. Kweon O., Kim S. J., Cerniglia C. E. 2010. Genomic view of mycobacterial high molecular weight polycyclic aromatic hydrocarbon degradation, p. 1165–1178 In Timmis, Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany [Google Scholar]
- 11. Kim S. J., Kweon O., Freeman J. P., Jones R. C., Adjei M. D., Jhoo J. W., Edmondson R. D., Cerniglia C. E. 2006. Molecular cloning and expression of genes encoding a novel dioxygenase involved in low- and high-molecular-weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR1. Appl. Environ. Microbiol. 72:1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim S. J., Kweon O., Jones R. C., Freeman J. P., Edmondson R. D., Cerniglia C. E. 2007. Complete and integrated pyrene degradation pathway in Mycobacterium vanbaalenii PYR-1 based on systems biology. J. Bacteriol. 189:464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kweon O., Kim S. J., Jones R. C., Freeman J. P., Adjei M. D., Edmondson R. D., Cerniglia C. E. 2007. A polyomic approach to elucidate the fluoranthene degradative pathway in Mycobacterium vanbaalenii PYR-1. J. Bacteriol. 189:4635–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim S. J., Kweon O., Jones R. C., Edmondson R. D., Cerniglia C. E. 2008. Genomic analysis of polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Biodegradation 19:859–881 [DOI] [PubMed] [Google Scholar]
- 15. Khan A. A., Wang R. F., Cao W. W., Doerge D. R., Wennerstrom D., Cerniglia C. E. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:3577–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S. J., Kweon O., Cerniglia C. E. 2009. Proteomic applications to elucidate bacterial aromatic hydrocarbon metabolic pathways. Curr. Opin. Microbiol. 12:301–309 [DOI] [PubMed] [Google Scholar]
- 17. Pagnout C., Frache G., Poupin P., Maunit B., Muller J. F., Ferard J. F. 2007. Isolation and characterization of a gene cluster involved in PAH degradation in Mycobacterium sp. strain SNP11: expression in Mycobacterium smegmatis mc2155. Res. Microbiol. 158:175–186 [DOI] [PubMed] [Google Scholar]
- 18. Krivobok S., Kuony S., Meyer C., Louwagie M., Willison J. C., Jouanneau Y. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 185:3828–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Demanèche S., Meyer C., Micoud J., Louwagie M., Willison J. C., Jouanneau Y. 2004. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a Sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:6714–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller C. D., Hall K., Liang Y. N., Nieman K., Sorensen D., Issa B., Anderson A. J., Sims R. C. 2004. Isolation and characterization of polycyclic aromatic hydrocarbon-degrading Mycobacterium isolates from soil. Microb. Ecol. 48:230–238 [DOI] [PubMed] [Google Scholar]
- 21. Sho M., Hamel C., Greer C. W. 2004. Two distinct gene clusters encode pyrene degradation in Mycobacterium sp. strain S65. FEMS Microbiol. Ecol. 48:209–220 [DOI] [PubMed] [Google Scholar]
- 22. Debruyn J. M., Chewning C. S., Sayler G. S. 2007. Comparative quantitative prevalence of mycobacteria and functionally abundant nidA, nahAc, and nagAc dioxygenase genes in coal tar contaminated sediments. Environ. Sci. Technol. 41:5426–5432 [DOI] [PubMed] [Google Scholar]
- 23. Cébron A., Norini M. P., Beguiristain T., Leyval C. 2008. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDalpha) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 73:148–159 [DOI] [PubMed] [Google Scholar]
- 24. Margesin R., Labbe D., Schinner F., Greer C. W., Whyte L. G. 2003. Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine alpine soils. Appl. Environ. Microbiol. 69:3085–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kweon O., Kim S. J., Baek S., Chae J. C., Adjei M. D., Baek D. H., Kim Y. C., Cerniglia C. E. 2008. A new classification system for bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. BMC Biochem. 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stingley R. L., Khan A. A., Cerniglia C. E. 2004. Molecular characterization of a phenanthrene degradation pathway in Mycobacterium vanbaalenii PYR1. Biochem. Biophys. Res. Commun. 322:133–146 [DOI] [PubMed] [Google Scholar]
- 27. Saito A., Iwabuchi T., Harayama S. 2000. A novel phenanthrene dioxygenase from Nocardioides sp. strain KP7: expression in Escherichia coli. J. Bacteriol. 182:2134–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gakhar L., Malik Z. A., Allen C. C., Lipscomb D. A., Larkin M. J., Ramaswamy S. 2005. Structure and increased thermostability of Rhodococcus sp. naphthalene 1,2-dioxygenase. J. Bacteriol. 187:7222–7231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nojiri H., Nam J. W., Kosaka M., Morii K. I., Takemura T., Furihata K., Yamane H., Omori T. 1999. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J. Bacteriol. 181:3105–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jouanneau Y., Meyer C., Jakoncic J., Stojanoff V., Gaillard J. 2006. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry 45:12380–12391 [DOI] [PubMed] [Google Scholar]
- 31. Wackett L. P., Kwart L. D., Gibson D. T. 1988. Benzylic monooxygenation catalyzed by toluene dioxygenase from Pseudomonas putida. Biochemistry 27:1360–1367 [DOI] [PubMed] [Google Scholar]
- 32. Lee K., Brand J. M., Gibson D. T. 1995. Stereospecific sulfoxidation by toluene and naphthalene dioxygenases. Biochem. Biophys. Res. Commun. 212:9–15 [DOI] [PubMed] [Google Scholar]
- 33. Wammer K. H., Peters C. A. 2006. A molecular modeling analysis of polycyclic aromatic hydrocarbon biodegradation by naphthalene dioxygenase. Environ. Toxicol. Chem. 25:912–920 [DOI] [PubMed] [Google Scholar]
- 34. Jakoncic J., Jouanneau Y., Meyer C., Stojanoff V. 2007. The catalytic pocket of the ring-hydroxylating dioxygenase from Sphingomonas CHY-1. Biochem. Biophys. Res. Commun. 352:861–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferraro D. J., Brown E. N., Yu C. L., Parales R. E., Gibson D. T., Ramaswamy S. 2007. Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1. BMC Struct. Biol. 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferraro D. J., Gakhar L., Ramaswamy S. 2005. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338:175–190 [DOI] [PubMed] [Google Scholar]
- 37. Parales R. E. 2003. The role of active-site residues in naphthalene dioxygenase. J. Ind. Microbiol. Biotechnol. 30:271–278 [DOI] [PubMed] [Google Scholar]
- 38. Parales R. E., Lee K., Resnick S. M., Jiang H., Lessner D. J., Gibson D. T. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parales R. E., Resnick S. M., Yu C. L., Boyd D. R., Sharma N. D., Gibson D. T. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the alpha subunit. J. Bacteriol. 182:5495–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferraro D. J., Okerlund A. L., Mowers J. C., Ramaswamy S. 2006. Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase. J. Bacteriol. 188:6986–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Furusawa Y., Nagarajan V., Tanokura M., Masai E., Fukuda M., Senda T. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 342:1041–1052 [DOI] [PubMed] [Google Scholar]
- 42. Karlsson A., Parales J. V., Parales R. E., Gibson D. T., Eklund H., Ramaswamy S. 2003. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299:1039–1042 [DOI] [PubMed] [Google Scholar]
- 43. Colbert C. L., Couture M. M., Eltis L. D., Bolin J. T. 2000. A cluster exposed: structure of the Rieske ferredoxin from biphenyl dioxygenase and the redox properties of Rieske Fe-S proteins. Structure 8:1267–1278 [DOI] [PubMed] [Google Scholar]
- 44. Keizers P. H., de Graaf C., de Kanter F. J., Oostenbrink C., Feenstra K. A., Commandeur J. N., Vermeulen N. P. 2005. Metabolic regio- and stereoselectivity of cytochrome P450 2D6 towards 3,4-methylenedioxy-N-alkylamphetamines: in silico predictions and experimental validation. J. Med. Chem. 48:6117–6127 [DOI] [PubMed] [Google Scholar]
- 45. Prasad S., Mitra S. 2002. Role of protein and substrate dynamics in catalysis by Pseudomonas putida cytochrome P450cam. Biochemistry 41:14499–14508 [DOI] [PubMed] [Google Scholar]
- 46. Kim Y. H., Engesser K. H., Cerniglia C. E. 2003. Two polycyclic aromatic hydrocarbon o-quinone reductases from a pyrene-degrading Mycobacterium. Arch. Biochem. Biophys. 416:209–217 [DOI] [PubMed] [Google Scholar]
- 47. Nojiri H., Habe H., Omori T. 2001. Bacterial degradation of aromatic compounds via angular dioxygenation. J. Gen. Appl. Microbiol. 47:279–305 [DOI] [PubMed] [Google Scholar]
- 48. Bressler D. C., Fedorak P. M. 2000. Bacterial metabolism of fluorene, dibenzofuran, dibenzothiophene, and carbazole. Can. J. Microbiol. 46:397–409 [PubMed] [Google Scholar]
- 49. Schwede T., Kopp J., Guex N., Peitsch M. C. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sobolev V., Sorokine A., Prilusky J., Abola E. E., Edelman M. 1999. Automated analysis of interatomic contacts in proteins. Bioinformatics 15:327–332 [DOI] [PubMed] [Google Scholar]
- 51. Wallace A. C., Laskowski R. A., Thornton J. M. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134 [DOI] [PubMed] [Google Scholar]
- 52. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. 2006. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34:W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pei J., Kim B. H., Grishin N. V. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36:2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.